Significance
Currently, chemotherapy is still the only treatment option for ALT cancers. ALT cells rely heavily on homologous recombination to maintain their telomere length and are more susceptible to replication stress. A better understanding of how ALT cells respond to replication stress at their telomeres will help to uncover novel therapeutic strategies to treat ALT cancers. Here we found that depletion of FANCM or its obligatory binding partners induces pronounced replication stress at ALT telomeres. We thus propose that FANCM-depleted ALT cells can be used as an endogenous replication stress model. Most importantly, we propose that the new synthetic lethal interactions discovered in our study can be explored for targeting ALT cancers.
Keywords: FANCM, BRCA1, BLM, replication stress, ALT telomeres
Abstract
In the mammalian genome, certain genomic loci/regions pose greater challenges to the DNA replication machinery (i.e., the replisome) than others. Such known genomic loci/regions include centromeres, common fragile sites, subtelomeres, and telomeres. However, the detailed mechanism of how mammalian cells cope with the replication stress at these loci/regions is largely unknown. Here we show that depletion of FANCM, or of one of its obligatory binding partners, FAAP24, MHF1, and MHF2, induces replication stress primarily at the telomeres of cells that use the alternative lengthening of telomeres (ALT) pathway as their telomere maintenance mechanism. Using the telomere-specific single-molecule analysis of replicated DNA technique, we found that depletion of FANCM dramatically reduces the replication efficiency at ALT telomeres. We further show that FANCM, BRCA1, and BLM are actively recruited to the ALT telomeres that are experiencing replication stress and that the recruitment of BRCA1 and BLM to these damaged telomeres is interdependent and is regulated by both ATR and Chk1. Mechanistically, we demonstrated that, in FANCM-depleted ALT cells, BRCA1 and BLM help to resolve the telomeric replication stress by stimulating DNA end resection and homologous recombination (HR). Consistent with their roles in resolving the replication stress induced by FANCM deficiency, simultaneous depletion of BLM and FANCM, or of BRCA1 and FANCM, leads to increased micronuclei formation and synthetic lethality in ALT cells. We propose that these synthetic lethal interactions can be explored for targeting the ALT cancers.
Faithfully replicating its genome is vital for the fitness and health of a mammalian cell. The replisome frequently encounters a variety of impediments throughout the genome. The temporary/transient slowing or stalling of the replication fork is referred to as replication stress (1, 2). The list of endogenous sources of replication stress is still growing. Nonetheless, the well-recognized sources of replication stress include unrepaired DNA lesions, mis-incorporated ribonucleotides, unique DNA sequences that are prone to form secondary structures (e.g., G-quadruplex or G4), collision of the replication fork with the transcriptional machinery, an RNA–DNA hybrid (or R-loop) that is formed between a nascent RNA and the adjacent displaced single-stranded DNA (ssDNA), common fragile sites (CFS), and tightly packed genomic regions, such as heterochromatin (2). Because of these constant challenges faced by the replisome, mammalian cells have developed elaborate and complex strategies to resolve the replication stress and ensure the successful completion of DNA replication (1–4).
One of the endogenous loci/regions that frequently pose challenges to the replisome is the telomere. Mammalian telomeres are tandem repetitive DNA sequences [the large majority as (TTAGGG)n] located at the end of every linear chromosome. The addition of TTAGGG is catalyzed by an enzyme called telomerase, a large ribonucleoprotein complex with reverse transcriptase activity. The length of mammalian telomeres varies from 10 to 15 kb in humans and 25–50 kb in mice (5). Because most somatic cells do not express telomerase, they experience progressive telomere shortening with each cell cycle, which eventually leads to cellular senescence and aging of the organism (6). One of the hallmarks of cancers is their replicative immortality (7). To achieve that, cancer cells have to overcome the telomere dysfunction-induced cell cycle arrest or cell death by adopting one of the two telomere maintenance mechanism (TMMs): (i) 85–90% of cancers reactivate the expression of telomerase (8), and (ii) the remaining 10–15% of cancers use the HR-based ALT pathway (9, 10).
The unique sequence and structure of mammalian telomeres render them especially challenging to the replisome. First, one of the strands of telomere is rich in guanines (thus called the G-rich strand) and is prone to the formation of G4s (11, 12). Second, the subtelomeric/telomeric noncoding RNA, TERRA, was shown to form an R-loop (13, 14). Third, we showed previously that DNA replication can be initiated from the telomeres; however, the majority of the DNA replication near the end of a mammalian chromosome is initiated from the subtelomeres (15), suggesting that the subtelomeric/telomeric regions lack DNA replication origins. In addition, de Lange and colleagues observed that low dosage of aphidicolin, a DNA polymerase inhibitor, dramatically increases the incidence of fragile telomeres (16). Together, these two features indicate that telomeres may be a special type of CFS. It is also known that ALT cells have more heterogeneous telomeres and that some of them can be quite long (17). Therefore, ALT telomeres may be even more prone to express the fragility. Fourth, heterochromatins are enriched at the telomeric and subtelomeric regions of the genome (18). Finally, to protect the end of linear chromosomes, there are a variety of high-order DNA–DNA and DNA–protein structures at the telomeres. For example, the telomeric G-strand overhang folds back, invades the internal double-stranded regions of telomere, and forms the telomeric loop (T-loop) as well as the displacement-loop (D-loop) (19, 20). The T-loop and D-loop are further stabilized by the Shelterin complex and other proteins (21, 22). These various high-order structures may also slow down the progression of the replisome. Taken together, mammalian telomeres pose constant challenges to the replisome, and replication stress often takes place at the telomeres, especially in ALT cells (16, 23, 24). Therefore, a robust replication stress response is vital for the successful replication of telomeres.
FANCM is an evolutionarily conserved ATP-dependent DNA helicase/translocase (25, 26). It belongs to the Fanconi anemia (FA) family of genes, the most recognized function of which is to repair the interstrand cross-linking (ICL) lesions (27, 28). FANCM and its three obligatory binding partners, FAAP24, MHF1, and MHF2, are part of the FA core complex, which catalyzes the monoubiquitination of FANCD2 and FANCI (ID2) (29–33). In addition, FANCM can also promote checkpoint activation and replication traversal across the ICL lesion (34–37). Furthermore, FANCM also plays a critical role in replication stress response. For example, in cells treated with hydroxyurea (HU), FANCM binds the checkpoint protein HCLK2 and promotes the activation of ATR-mediated checkpoint response (34). FANCM also helps to stabilize the stalled replication fork and promote replication fork restart (38). Intriguingly, a study by Constantinou and colleagues suggests that, even in unperturbed human cells, FANCM can modulate the steady progression of replication elongation (39). Consistent with its important functions in replication stress response, FANCM-deficient cells are also hypersensitive to a variety of replication stress inducers, including HU, aphidicolin, ultraviolet (UV), and camptothecin (39–41).
Sporadic evidence suggests that there may be a connection between the FA pathway and telomere biology. Many FA patients manifest progressive telomere shortening (42–50). Surprisingly, most FA knockout mice did not show pronounced telomeric defects (51). This is likely due to the exceptional length of mouse telomeres, which are two to five times longer than those of humans. In support of this notion, under the high turnover condition, the hematopoietic cells from the FANCC-deficient mice experienced faster telomere attrition (52). Furthermore, in the absence of telomerase, the bone marrow cells of the FANCC-deficient mouse showed a higher incidence of telomere sister-chromatid exchange (tSCE), an indicator of dysfunctional telomeres. In human keratinocytes, Duensing and colleagues showed that FANCD2 localizes to the HPV-16 E6-induced ALT-associated promyelocytic leukemia (PML) bodies (APBs) (53). APBs are thought to be the sites where damaged ALT telomeres are clustered and repaired (10). Another study by Andreassen and colleagues showed, that in a small population of ALT cells, FANCD2 can be found at the telomeres and APBs (54). This APB localization is dependent on FANCA and FANCL, suggesting that the monoubiquitination of FANCD2 may be important for its telomere association. Furthermore, Andreassen and colleagues showed that depletion of FANCA or FANCD2 increases the frequency of telomere-free chromosome ends and decreases the tSCE, suggesting that the recruitment of monoubiquitinated FANCD2 to the ALT telomeres promotes HR. Although there is no report that human FANCM is involved in TMM, intriguingly its budding yeast homolog, Mph1, does localize to yeast telomeres and promotes telomere uncapping and premature senescence in the absence of telomerase (55, 56).
It has been observed that acute depletion of FANCM in unperturbed human cancer cells causes increased formation of γ-H2AX foci, a commonly used marker for DNA damages (34). Genetic deletion of FANCM in chicken DT40 cells also induces increased γ-H2AX foci (38). However, the cause and the nature of the DNA damages in these FANCM-deficient cells are unknown. In the process of characterizing the nature and locations of these DNA damages, we found that depletion of FANCM or of one of its obligatory binding partners in ALT cells induces replication stress, which primarily occurs at telomeres. In support of the notion that the FANCM complex suppresses replication stress at ALT telomeres, using the telomere-specific single-molecule analysis of replicated DNA (SMARD) technique, we found that depletion of FANCM dramatically reduced the replication efficiency at ALT telomeres. We therefore propose that depletion of FANCM in ALT cells can be used as an endogenous replication stress model. Mechanistically, we found that, in this endogenous replication stress model, BLM and BRCA1, two of the most important HR proteins, are actively recruited to the replication stress sites and help to resolve the replication stress by stimulating DNA end resection and promoting HR. Finally, we found that codepletion of FANCM and BLM or of FANCM and BRCA1 dramatically increases the micronuclei formation and induces synthetic lethality in ALT cells.
Results
Depletion of FANCM Induces Replication Stress at ALT Telomeres.
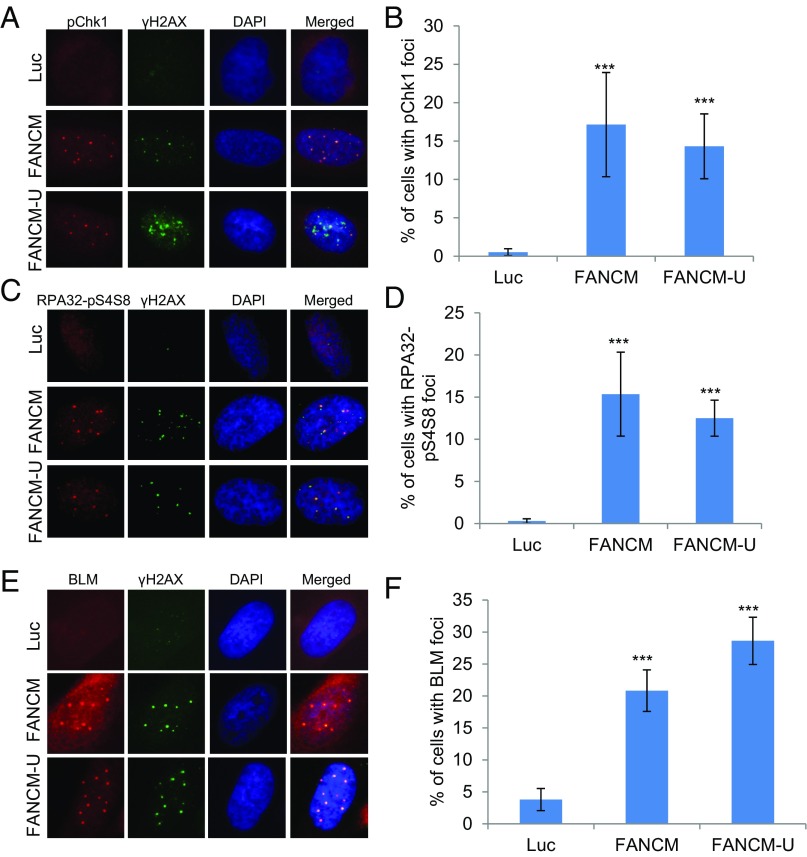
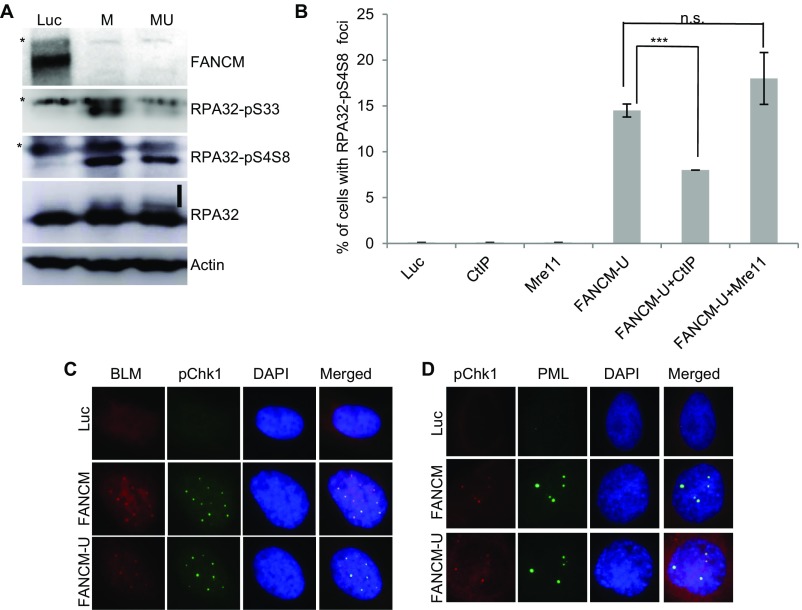
Based on the potential roles of FANCM in DNA replication, we hypothesized that the DNA damages seen in FANCM-deficient cells are due to replication stress (34, 38). Replication stress primarily activates the ATR-DNA-PKcs-Chk1 pathway whereas double-stranded DNA breaks (DSBs) mainly activate the ATM-Chk2 pathway (4, 57). In response to replication stress, Serine-345 of Chk1 (Chk1-pS345) and Serine-33 of RPA32 (RPA32-pS33) are primarily phosphorylated by ATR (58–60), whereas Serine-4 and Serine-8 of RPA32 (RAP32-pS4S8) are primarily phosphorylated by DNA-PK (61). All three phosphorylation events are widely used as the surrogate markers of replication stress. To test our hypothesis, FANCM-depleted U2-OS cells were probed with antibodies that specifically recognize Chk1-pS345, RPA32-pS33, and RPA32-pS4S8. In Luciferase (Luc) siRNA transfected cells, very few cells showed positive Chk1-pS345 and RPA32-pS4S8 foci (<1%) (Fig. 1 A–D). In stark contrast, depletion of FANCM with two different siRNAs induces bright and discreet Chk1-pS345 and RPA32-pS4S8 foci in ∼15% of the cells. Consistent with the increased RPA32-pS4S8 foci, depletion of FANCM also caused a dramatic increase of RPA32-pS33 and RPA32-pS4S8 by immunoblotting (Fig. S1A). Collectively, these data suggest that the DNA damages induced in FANCM-deficient cells are due to DNA replication stress.
Fig. 1.
Depletion of FANCM induces replication stress. U2-OS cells were transfected with siRNA targeting Luciferase (Luc) or two different siRNA targeting FANCM (FANCM and FANCM-U). Cells were then stained with antibodies recognizing γH2AX or Chk1-pS345 (A and B, labeled as pChk1), RPA32-pS4S8 (C and D), or BLM (E and F). All nuclei were also stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments. Standard two-sided t test: ***P < 0.001.
Fig. S1.
Chk1-pS345 foci colocalize with the BLM and PML foci. (A) U2-OS cells were transfected with siRNA targeting either Luciferase (Luc) or two different siRNA against FANCM (labeled as M and MU). Cell lysates were prepared and blotted with antibodies as indicated on the right. Asterisks indicate cross-reaction bands. The vertical line marks the phosphorylated RPA32. (B) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), CtIP, Mre11, FANCM (FANCM-U), or in combination. Cells were then costained with antibodies recognizing TRF1 or RPA32-pS4S8. More than 200 cells were counted for each sample. All error bars are SD obtained from two different experiments. (C and D) U2-OS cells were transfected with siRNA targeting either Luciferase (Luc) or two different siRNA against FANCM (labeled as FANCM and FANCM-U). Cells were then costained with antibodies recognizing Chk1-pS345 (labeled as pChk1) and BLM (C) or Chk1-pS345 and PML (D). All nuclei were also stained with DAPI. Standard two-sided t test: ***P < 0.001. n.s., not significant.
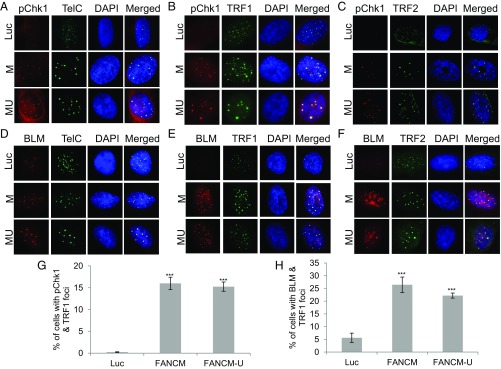
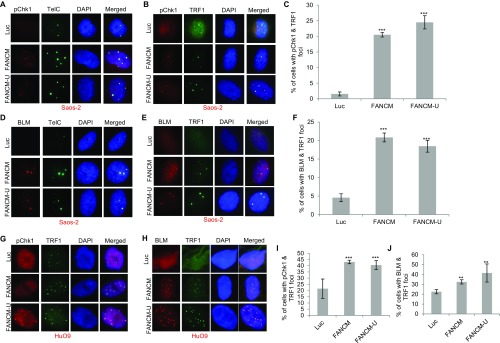
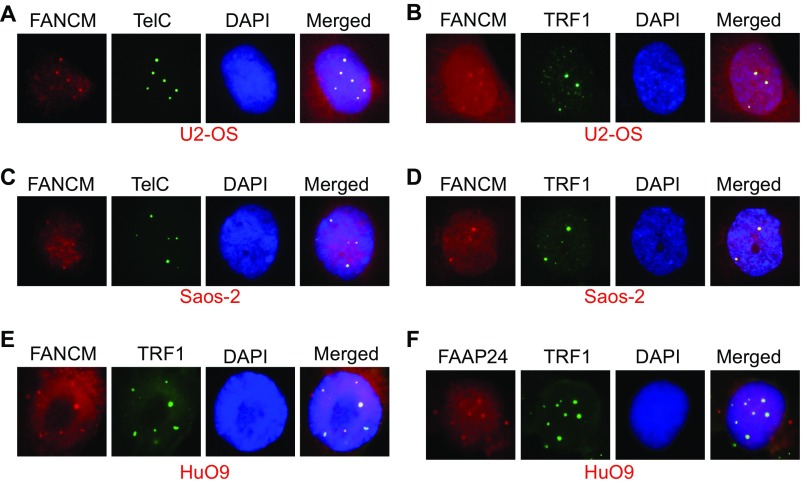
In addition to being the markers for checkpoint activation of the ATR-DNA-PKcs-Chk1 pathway, the increased level of RPA32-pS4S8 and RPA32-pS33 and their ability to form discrete foci by immunofluorescent staining also strongly correlates with the formation of ssDNA and the activation of DNA end resection (62). Robust DNA end resection promotes the repair and restart of the stalled replication fork through the high-fidelity HR pathway (1, 63). It is known that, in response to DSBs, CtIP and the MRN complex (Mre11-NBS1-Rad50) play a critical role in the DNA end resection step of HR (57). To test if CtIP and the MRN complex are also important for the DNA end resection in FANCM-deficient cells, we codepleted CtIP and FANCM or Mre11 and FANCM. Intriguingly, only the depletion of CtIP significantly reduced the RPA32-pS4S8 foci formation (Fig. S1B), suggesting that CtIP, but not the MRN complex, promotes the DNA end resection in FANCM deficiency-induced replication stress. One of the key factors of HR in response to the replication stress is BLM, a crucial DNA helicase functioning in multiple steps of HR (64, 65). Next, we tested whether BLM is also recruited to these replication stress sites. When stained with an antibody recognizing BLM, 20–30% of FANCM-depleted cells showed strong BLM foci (Fig. 1 E and F). Most importantly, the BLM foci colocalize well with both γ-H2AX and Chk1-pS345 foci, suggesting that BLM is also actively recruited to these replication stress sites (Fig. 1E and Fig. S1C). During the immunofluorescent foci (IF) analysis, we noted that the nuclear Chk1-pS345, RPA32-pS4S8, and BLM foci are quite large and intense, which reminded us of the pattern of PML bodies. Indeed, when costained with a PML antibody, the Chk1-pS345 foci seen in FANCM-depleted cells colocalize with the PML bodies (Fig. S1D). U2-OS is a commonly used ALT cell model (66). ALT cells are known to form a unique nuclear punctate structure, called the APBs (67). APBs are a subtype of the PML bodies, where the damaged ALT telomeres are clustered and repaired (10). The nice colocalization of Chk1-pS345, RPA32-pS4S8, and BLM foci with PML suggests that depletion of FANCM in ALT cells induces replication stress likely at the telomeres. To test this hypothesis, we probed the FANCM-depleted cells with three different telomere markers, TelC [a telomere-specific peptide nuclei acid (PNA) probe], and TRF1 and TRF2, two key components of the Shelterin complex (68). As seen in Fig. 2, the Chk1-pS345 and BLM foci colocalize with all three telomere markers, indicating that the replication stress indeed occurs primarily at the ALT telomeres. In addition, these data also suggest that the replication stress seen in FANCM-depleted cells is less likely due to the loss of TRF1 and TRF2 at the telomeres, which are known to cause DNA damages at telomeres (16). We also noted that the telomeric foci that are colocalized with the Chk1-pS345, RPA32-pS4pS8, and BLM foci tend to be bigger and brighter than normal telomere staining, suggesting that these foci are likely the clustered telomeres that are experiencing replication stress. Similar results are also observed in two other ALT cells, Soas-2 and HuO9, but not in two telomerase-positive cells (TEL+), HeLa and MG63 (Fig. S2). Taken together, our data strongly suggest that FANCM functions to suppress the replication stress at the ALT telomeres. Consistent with this notion, FANCM itself and one of its associated proteins, FAAP24, can be found at the telomeres by immunofluorescent staining in a small population of unperturbed U2-OS, Saos-2, and HuO9 cells (Fig. S3), the telomeres of which are presumably experiencing endogenous replication stress.
Fig. 2.
Depletion of FANCM induces replication stress at ALT telomeres. U2-OS cells were transfected with siRNA targeting Luciferase (Luc) or two different siRNA targeting FANCM (FANCM or M and FANCM-U or MU). (A–C and G) siRNA transfected cells were costained with an antibody recognizing Chk1-pS345 (labeled as pChk1) together with a PNA probe recognizing the G-rich strand of telomeres (A, TelC), an antibody recognizing TRF1 (B), or TRF2 (C). (D–F and H) siRNA transfected cells were costained with an antibody recognizing BLM, together with TelC (D), an antibody recognizing TRF1 (E), or TRF2 (F). All nuclei were also stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments. Standard two-sided t test: ***P < 0.001.
Fig. S2.
FANCM deficiency induces replication stress in Saos-2 and HuO9 cells. Saos-2 (A–F) or HuO9 (G–J) cells were transfected with siRNA either targeting Luciferase (Luc) or two different siRNA targeting FANCM (FANCM and FANCM-U). Cells were then costained with either a PNA probe recognizing the G-rich strand of telomeres (TelC) or different antibodies as indicated on the top in A, B, D, E, G, and H. All nuclei were also stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from two different experiments. Standard two-sided t test: **P < 0.01, ***P < 0.001.
Fig. S3.
FANCM and FAAP24 foci colocalize with large telomere foci in three ALT cells. U2-OS (A and B), Saos-2 (C and D), or HuO9 (E and F) cells were costained with an antibody recognizing FANCM, FAAP24, or TRF1 or a PNA probe recognizing the G-rich strand of telomeres (TelC).
Depletion of the Three Obligatory Binding Partners of FANCM also Induces Replication Stress at ALT Telomeres.
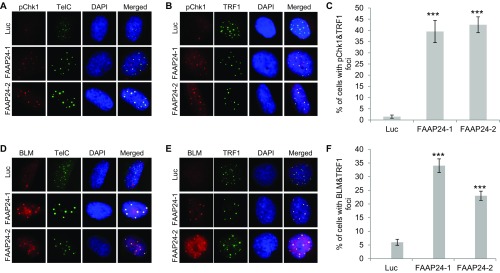
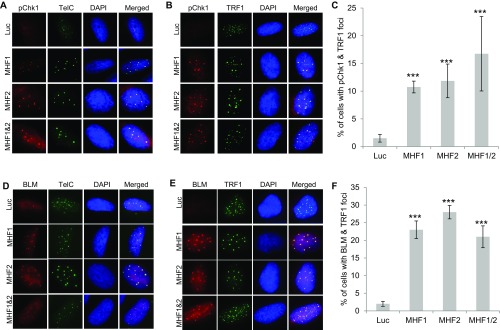
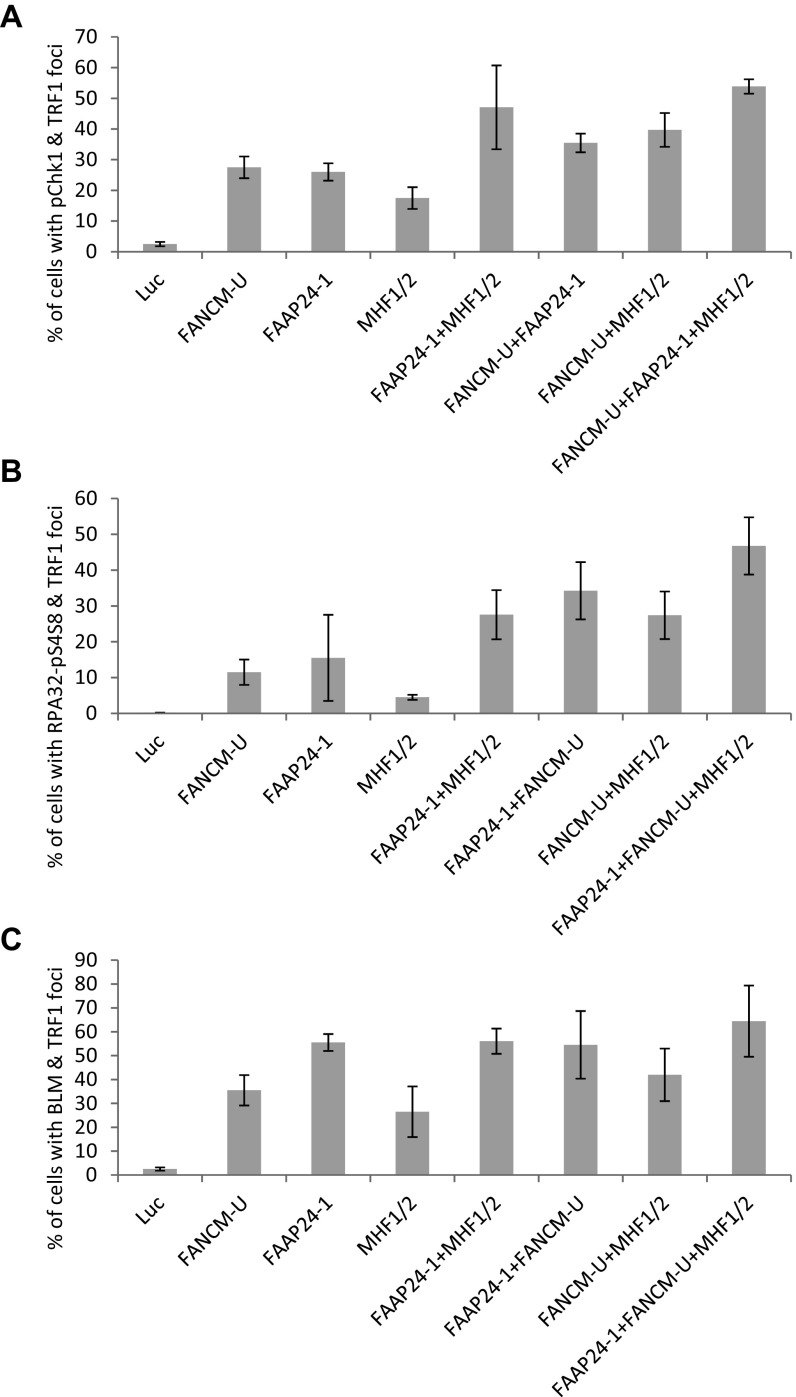
As mentioned earlier, various functions of FANCM also require its three obligatory binding partners, FAAP24, MHF1, and MHF2. FAAP24 binds the C terminus of FANCM and functions as part of the FA core complex to promote the monoubiquitination of ID2 (31). MHF1 and MHF2 are two histone-fold proteins that form a stable and stoichiometric heterodimer (MHF1/2). MHF1/2 binds the N terminus of FANCM and also functions as part of the FA core complex to promote the monoubiquitination of ID2 (32, 33). Next, we tested whether FAAP24 and MHF1/2 have similar functions as FANCM at ALT telomeres. Depletion of FAAP24 using two different siRNA induces a dramatic increase of Chk1-pS345 and BLM foci, both of which colocalize with ALT telomeres (Fig. S4). Similarly, depletion of MHF1 or MHF2 individually, or in combination also causes a dramatic increase of Chk1-pS345 and BLM foci, both of which colocalize nicely with ALT telomeres as well (Fig. S5). Intriguingly, in comparison with the depletion of FANCM or FAAP24, depletion of MHF1/2 induces fewer Chk1-pS345, RPA32-pS4S8, or BLM foci (Fig. S6), suggesting that FAAP24 and FANCM play a more important role in suppressing the replication stress at ALT telomeres than MHF1/2. Codepletion of FAAP24 and MHF1/2, or FAAP24 and FANCM, or FANCM and MHF1/2, or all four proteins induces a moderate increase of the Chk1-pS345 and RPA32-pS4S8 foci (Fig. S6), suggesting that there is a mild additive effect of the components of the FANCM-FAAP24-MHF1/2 complex in checkpoint activation and DNA end resection. Collectively, our data firmly established that depletion of one or more components of the FANCM-FAAP24-MHF1/2 complex induces pronounced replication stress primarily at ALT telomeres.
Fig. S4.
Depletion of FAAP24 induces replication stress at the ALT telomeres. U2-OS cells were transfected with siRNA targeting Luciferase (Luc) or two different siRNA targeting FAAP24 (FAAP24-1 and -2). Cells were then costained with an antibody recognizing Chk1-pS345 (labeled as pChk1, A–C) or BLM (D–F) together with a PNA probe recognizing the G-rich strand of telomeres (TelC) or an antibody recognizing TRF1. All nuclei were stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments. Standard two-sided t test: ***P < 0.001.
Fig. S5.
Depletion of MHF1, MHF2, or MHF1 and -2 induces replication stress at the ALT telomeres. U2-OS cells were transfected with siRNA targeting Luciferase (Luc), MHF1, MHF2, or MHF1 and -2. Cells were then costained with an antibody recognizing Chk1-pS345 (labeled as pChk1, A–C) or BLM (D–F), together with a PNA probe recognizing the G-rich strand of telomeres (TelC) or an antibody recognizing TRF1. All nuclei were stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments. Standard two-sided t test: ***P < 0.001.
Fig. S6.
Depletion of the component of the FANCM-FAAP24-MHF1/2 complex individually or in combination induces replication stress at the telomeres. U2-OS cells were transfected with siRNA as indicated. Cells were then costained with an antibody recognizing TRF1 together with an antibody recognizing Chk1-pS345 (A), RPA32-pS4S8 (B), or BLM (C). More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments.
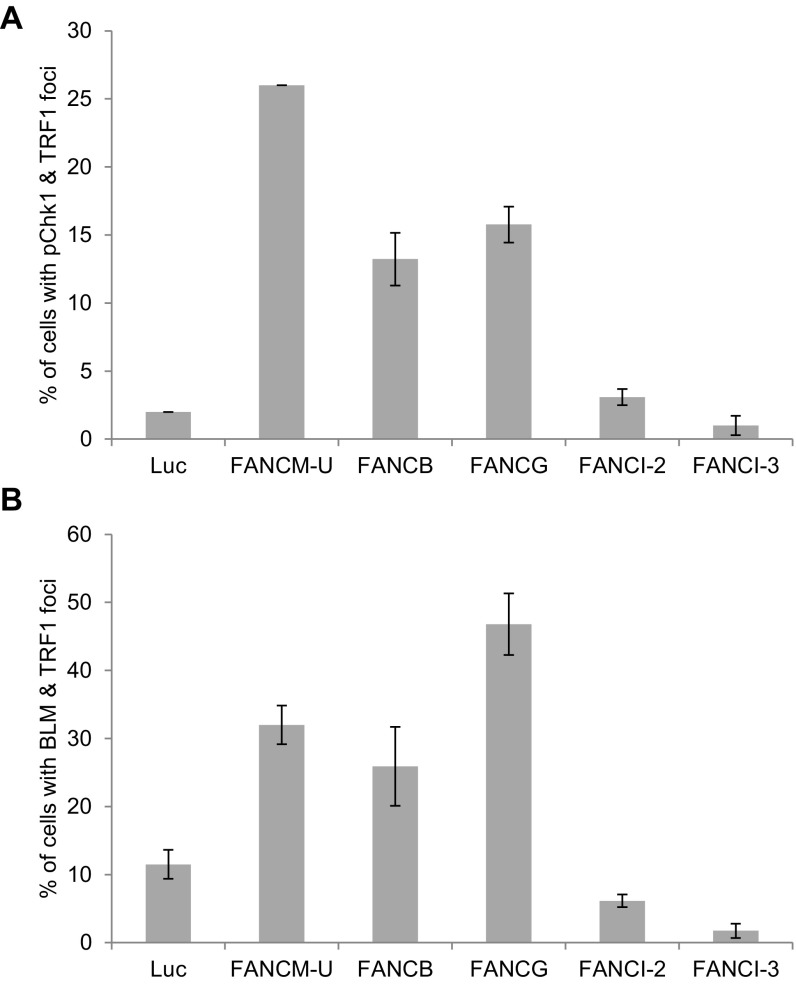
To test whether other FA proteins have a similar function at ALT telomeres as FANCM, we depleted two other FA core components, FANCB and FANCG, as well as FANCI and analyzed the foci formation of Chk1-pS345 and BLM (Fig. S7). Depletion of FANCI does not induce foci formation of Chk1-pS345 or BLM at ALT telomeres, suggesting that FANCI is likely not involved in suppressing the replication stress at ALT telomeres. Interestingly, depletion of FANCB or FANCG induced a moderate increase of Chk1-pS345 foci but much less than the depletion of FANCM, suggesting that FANCM plays a more important role in suppressing the checkpoint activation at ALT telomeres than other FA proteins. Surprisingly, depletion of FANCB or FANCG induced as much BLM foci as the depletion of FANCM, suggesting that the entire FA core complex may play an active role in regulating the recruitment of BLM to ALT telomeres.
Fig. S7.
The effect of different FA proteins on the Chk1-pS345 and BLM foci formation. U2-OS cells were transfected with siRNA as indicated. Cells were costained with an antibody recognizing TRF1 together with an antibody recognizing Chk1-pS345 (A) or BLM (B). All nuclei were stained with DAPI. More than 200 cells were counted for each sample. All error bars are SD obtained from three different experiments.
Depletion of FANCM Reduces the Replication Efficiency of Telomeric DNA.
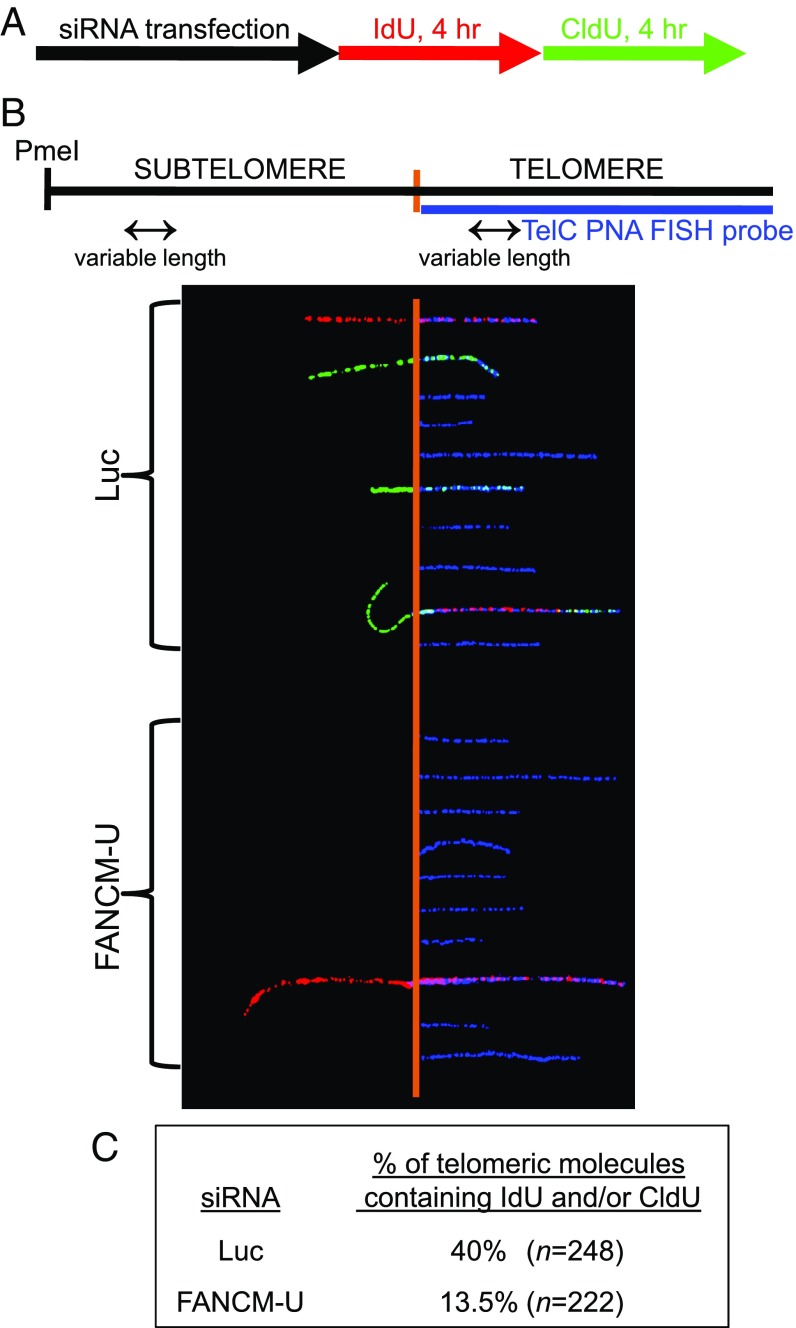
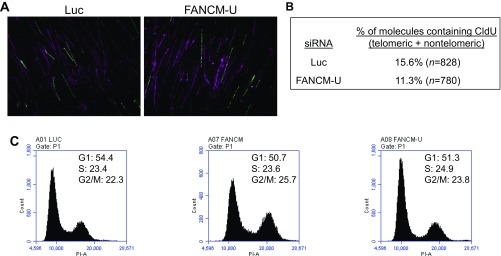
To directly examine the role of FANCM in telomere replication, we performed a SMARD assay in the subtelomeric and telomeric regions of FANCM-depleted cells (15). Briefly, siRNA transfected cells were sequentially pulse-labeled with 5-iodo-2′-deoxyuridine (IdU) for 4 h and then with 5-chloro-2′-deoxyuridine (CldU) for 4 h (Fig. 3A). Genomic DNA was digested and separated by pulsed-field gel electrophoresis (PFGE). Genomic DNA enriched with subtelomeres and telomeres was identified by Southern blot using a telomeric PNA probe. Isolated subtelomeric and telomeric DNA was then stretched on slides and stained with antibodies that recognize IdU and CldU. Telomeres were visualized by fluorescence in situ hybridization (FISH) using the telomeric PNA probe. Representative images of the subtelomeric and telomeric fibers are shown in Fig. 3B. Through counting the number of telomeric fibers containing halogenated nucleotides (IdU and/or CldU), we observed a threefold reduction of the replication efficiency of telomeric DNA in FANCM-depleted ALT cells (Fig. 3C). Additionally, we also examined the genome-wide DNA replication by probing the same DNA with antibodies that recognize the ssDNA (to visualize the total DNA fibers) and CldU (Fig. S8 A and B) (69). We observed only a slight reduction of genome-wide DNA replication (1.4-fold reduction genome-wide vs. threefold reduction at telomeres). The cell cycle profile of FANCM siRNA transfected cells is also comparable to that of Luc siRNA transfected cells (Fig. S8C). Therefore, the telomeric SMARD analysis strongly supports our conclusion that, in the absence of FANCM, replication forks are more prone to pause/stall at ALT telomeres, thus leading to increased replication stress.
Fig. 3.
Depletion of FANCM reduces the replication efficiency of telomeric DNA. (A and B) U2OS cells were transfected with siRNA targeting Luciferase (Luc) or FANCM (FANCM-U). Twenty-four hours after the second siRNA transfection, cells were sequentially pulse-labeled with IdU for 4 h and then with CldU for 4 h. SMARD was performed on telomeric DNA PmeI fragments isolated from these cells as detailed in Materials and Methods, with fragments ranging from 160 to 200 kb that contained subtelomeric DNA of variable lengths. The telomeric DNA molecules of variable lengths were identified by FISH using telomeric PNA probes (TelC, blue), and incorporated IdU and CldU were detected by indirect immunofluorescence with antibodies recognizing IdU (red) and CldU (green). The diagram of the PmeI fragment including the subtelomeric and telomeric DNA and the representative images of subtelomeric/telomeric fragments is shown. The vertical orange lines mark the boundary between the subtelomere and telomere. (C) The percentage of telomeric molecules containing IdU and/or CldU (i.e., the ratio of labeled versus unlabeled fragments) is presented. This percentage indicates the relative efficiency of DNA replication during the IdU/CldU labeling period.
Fig. S8.
Genome-wide DNA replication analysis and cell cycle analysis of FANCM-deficient cells. (A and B) U2OS cells were transfected with siRNA targeting Luciferase (Luc) or FANCM (FANCM-U). Twenty-four hours after the second siRNA transfection, cells were sequentially pulse-labeled with IdU for 4 h and then with CldU for 4 h. SMARD was performed on PmeI fragments isolated from these cells as detailed in Materials and Methods, with fragments ranging from 160 to 200 kb that are enriched with telomeric and subtelomeric DNA. The genomic DNA molecules of variable lengths were identified by an antibody recognizing the ssDNA Ab (purple), and the incorporated CldU was detected by an antibody recognizing the CldU (green). The percentage of DNA molecules that contained CldU vs. those recognized by the ssDNA Ab is presented (B). This percentage indicates the relative efficiency of genome-wide DNA replication during the CldU-labeling period. (C) U2-OS cells were transfected with siRNA targeting either Luciferase (Luc) or two different siRNA targeting FANCM (FANCM and FANCM-U). Cells were then fixed, stained with propidium iodide (PI), and analyzed using a C6 flow cytometer.
In Response to the Replication Stress at ALT Telomeres, BLM Promotes DNA End Resection and Is Synthetically Lethal with FANCM.
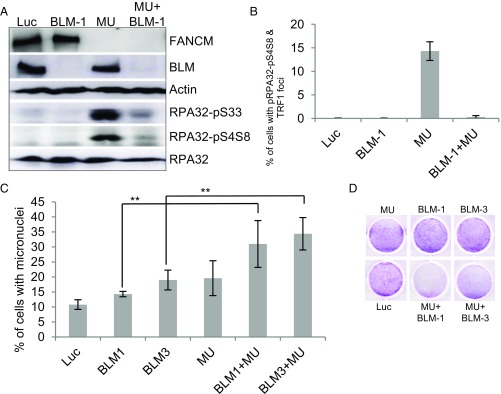
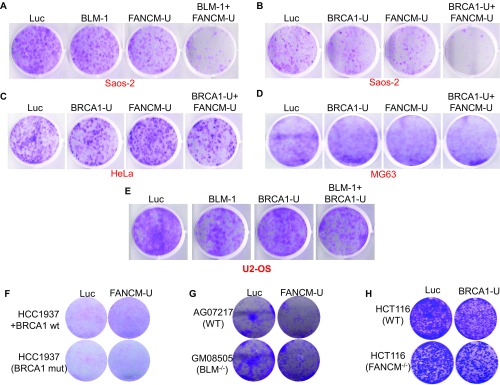
BLM has been implicated in a variety of DNA metabolic functions including DNA replication, DNA replication stress response, and telomere biology (64, 65). Mechanistically, BLM plays an important role in DNA end resection during HR and at telomeres (70, 71). Here, we found that depletion of FANCM induces pronounced DNA end resection and BLM foci formation (Fig. 1 C–F and Fig. S1A). To test whether BLM stimulates the DNA end resection in FANCM-depleted cells, we codepleted BLM and FANCM and monitored DNA end resection by Western blot as well as IF. As seen in Fig. 4 A and B, depletion of BLM dramatically attenuated DNA end resection in FANCM-deficient cells, indicating that BLM is indeed required for DNA end resection during the replication stress response at ALT telomeres. It is well known that ALT cells rely on HR to maintain the length of their telomeres (10). Reduced DNA end resection will impair HR and hinder the DNA repair processes. Consistent with this notion, we observed a dramatic increase of cells with micronuclei when FANCM and BLM were codepleted and also a strong synthetic lethal interaction between FANCM and BLM in both U2-OS and Saos-2 (Fig. 4 C and D and Fig. S9A). Collectively, our data suggest that when ALT telomeres encounter replication stress, BLM is recruited to these telomeres and stimulates DNA end resection.
Fig. 4.
BLM promotes DNA end resection and DNA repair at the ALT telomeres. (A and B) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), BLM (BLM-1), FANCM (MU), or both BLM and FANCM (BLM-1+MU). Cell lysates were blotted with antibodies as indicated on the right (A). The siRNA transfected cells were also costained with antibodies recognizing either RPA32-pS4S8 or TRF1 (B). More than 200 cells were counted for each sample, and the percentage of cells with both RPA32-pS4S8 and TRF1 foci was plotted. (C and D) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), two different siRNA targeting BLM (BLM-1 and BLM-3), FANCM (MU), or both BLM and FANCM. The nucleus of siRNA transfected U2-OS cells was stained with DAPI. More than 200 cell nuclei were analyzed, and the percentage of cells with micronuclei was plotted (C). The viability of the same siRNA transfected cells in C was analyzed by crystal violet assay as detailed in Materials and Methods (D). All error bars are SD and obtained from three independent experiments. Standard two-sided t test: **P < 0.01.
Fig. S9.
Cell viability analysis. The viability of siRNA transfected Saos-2 (A and B), HeLa (C), MG63 (D), U2-OS (E), HCC1937 (F), AG07217 and GM08505 (G), or HCT116 (H) were analyzed by crystal violet assay as detailed in Materials and Methods.
In Response to Replication Stress at ALT Telomeres, BRCA1 Is also Actively Recruited to the Telomeres, and the Recruitment of BLM and BRCA1 to the Damaged Telomeres Is Mutually Dependent.
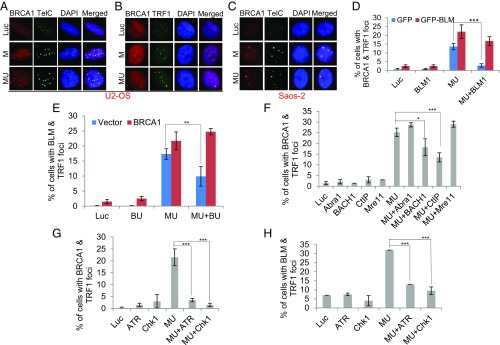
BRCA1 is another important player involved in a variety of DNA metabolic functions including checkpoint activation and HR (72). Intriguingly, BRCA1 was also found at ALT telomeres and shown to stimulate the helicase activity of BLM in an in vitro helicase assay using telomeric DNA as the substrate (73). In response to various types of DNA damages, BRCA1 is actively recruited to the damage sites. To test whether BRCA1 can also be recruited to the damaged ALT telomeres, we costained FANCM-depleted cells with an antibody recognizing BRCA1 and telomere markers in both U2-OS and Saos-2 cells (Fig. 5 A–C). In Luc siRNA transfected cells, a small percentage of cells manifested BRCA1 foci. However, these BRCA1 foci rarely colocalize with telomeres. In stark contrast, the large majority of the BRCA1 foci (>90%) seen in FANCM-depleted cells colocalize with ALT telomeres. Interestingly, depletion of BLM abolished the BRCA1 foci, which are fully rescued by the expression of the BLM-1 siRNA- resistant GFP-BLM (Fig. 5D). Conversely, depletion of BRCA1 also reduced the BLM foci, which are fully rescued by the expression of the BRCA1 ORF (Fig. 5E; BRCA1-U or BU siRNA targets the 3′ UTR of the endogenous BRCA1 mRNA, and therefore BRCA1 ORF is resistant to the degradation by the BU siRNA). In a previous study, we identified at least three different BRCA1-containing complexes: BRCA1 A-complex (Abra1/Abraxas-BRCA1-BARD1), B-complex (BACH1/BRIP1/FANCJ-BRCA1-BARD1), and C-complex (CtIP-BRCA1-BARD1) (74). To test which one of these complexes is important for BRCA1 foci formation, we codepleted Abra1 and FANCM, or BACH1 and FANCM, or CtIP and FANCM (Fig. 5F). Intriguingly, we found that depletion of CtIP has the most profound effect on the BRCA1 foci. Depletion of BACH1 mildly affects the BRCA1 foci whereas depletion of Abra1 has no effect. Finally, because ATR and Chk1 are the most important kinases that regulate the replication stress response, we tested whether they also regulate the foci formation of BRCA1 and BLM in FANCM-deficient cells. Consistent with our hypothesis that FANCM deficiency induces replication stress at ALT telomeres, depletion of either ATR or Chk1 abolishes the foci formation of BRCA1 and BLM (Fig. 5 G and H). Taken together, we have demonstrated that in FANCM-deficient ALT cells, both BRCA1 and BLM are actively recruited to the telomeres experiencing replication stress and that their recruitment is interdependent and also dependent on both ATR and Chk1. In addition, we show that CtIP and BACH1, but not Abra1, are also important for the recruitment of BRCA1.
Fig. 5.
The molecular mechanism behind the recruitment of BLM and BRCA1 to ALT telomeres. (A–C) BRCA1 localized to ALT telomeres. U2-OS cells (A and B) or Saos-2 cells (C) were transfected with siRNA targeting either Luciferase control (Luc) or two different siRNA targeting FANCM (M and MU). Cells were then costained with an antibody recognizing BRCA1 together with a PNA probe recognizing the telomere (A and C, TelC) or an antibody recognizing TRF1 (B). (D) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), BLM (BLM1), FANCM (MU), or both BLM and FANCM (BLM1+MU). Cells were then transfected with either GFP or GFP-BLM that is resistant to the BLM1 siRNA and costained with antibodies recognizing either BRCA1 or TRF1. (E) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), BRCA1 (BU), FANCM (MU), or both BRCA1 and FANCM (BU+MU). Cells were then transfected with either Vector or BRCA1 ORF and costained with antibodies recognizing either BLM or TRF1. (F–H) U2-OS cells were transfected with different siRNAs and then costained with antibodies recognizing TRF1, BRCA1 (F and G), or BLM (H). More than 200 cells were counted for each sample. All error bars are SD and obtained from two independent experiments. Standard two-sided t test: *P < 0.05, **P < 0.01, ***P < 0.001.
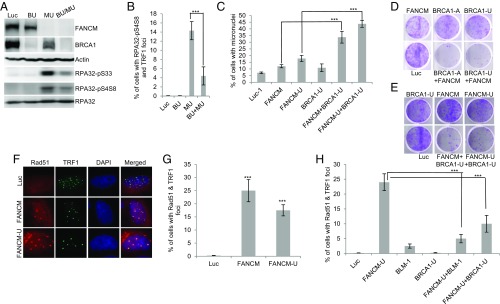
In Response to Replication Stress at ALT Telomeres, BRCA1 Promotes DNA End Resection and Is Synthetic Lethal with FANCM.
During the repair of DSBs, the most important function of BRCA1 is to counter 53BP1 and stimulate DNA end resection so that DSBs can be repaired preferentially via the high-fidelity repair pathway—HR (72). Interestingly, we recently showed that, in cells treated with UV, which is also considered a replication stress inducer, BRCA1 also promotes DNA end resection (75). Next, we tested whether BRCA1 also plays a role in DNA end resection at telomeres in FANCM-deficient ALT cells. As seen in Fig. 6 A and B, codepletion of BRCA1 and FANCM severely impedes the phosphorylation of RPA32 at both Serine-4/Serine-8 and Serine-33 by Western blot and IF, suggesting that BRCA1 is indeed required for DNA end resection at ALT telomeres. Consistent with this notion, we observed a dramatic increase of cells with micronuclei when FANCM and BRCA1 were codepleted (Fig. 6C) and a strong synthetic lethal interaction between FANCM and BRCA1 in both U2-OS and Saos-2 (Fig. 6 D and E and Fig. S9B). Intriguingly, we did not observe a synthetic lethal interaction between FANCM and BRCA1 in two TEL+ cells: HeLa and MG63 (Fig. S9 C and D). We did not observe a synthetic lethal interaction between BLM and BRCA1 in U2-OS either (Fig. S9E), suggesting that BLM and BRCA1 likely function in the same pathway. Taken together, our data suggest that the synthetic lethal interaction between BRCA1 and FANCM or BLM and FANCM occurs primarily in ALT cells. Consistent with this notion, we did not observe any synthetic lethal interaction between BRCA1 and FANCM or BLM and FANCM when using either disease cell lines (for BRCA1 and BLM) or a genetic knockout cell line (for FANCM) (Fig. S9 F–H) because none of them are known ALT cells.
Fig. 6.
BRCA1 and BLM cooperatively promotes DNA end resection and homologous recombination at ALT telomeres. (A) U2-OS cells were transfected with siRNA targeting Luciferase (Luc), BRCA1 (BU), FANCM (MU), or both BRCA1 and FANCM (BU+MU). Cell lysates were blotted with antibodies as indicated on the right. The siRNA transfected cells were also costained with antibodies recognizing either RPA32-pS4S8 or TRF1 (B). (C) The nuclei of siRNA transfected U2-OS cells were stained with DAPI. More than 200 nuclei were analyzed, and the percentage of cells with micronuclei was plotted. (D and E) The viability of the siRNA transfected cells was analyzed by crystal violet assay as detailed in Materials and Methods. (F and G) U2-OS cells were transfected with siRNA targeting either Luciferase (Luc) or two different siRNA targeting FANCM (FANCM and FANCM-U). Cells were then costained with antibodies recognizing Rad51 or TRF1. All nuclei were also stained with DAPI. (H) U2-OS cells were transfected with different siRNA and then costained with antibodies recognizing either Rad51 or TRF1. More than 200 cells were counted for each sample. All error bars are SD and obtained from three independent experiments. Standard two-sided t test: ***P < 0.001.
To directly measure HR activity at ALT telomeres, we examined the Rad51 foci formation in FANCM-depleted cells. Amazingly, more than 20% of FANCM-depleted cells also showed robust formation of Rad51 foci (Fig. 6 F and G). Most importantly, the formation of Rad51 foci in FANCM-deficient cells is dependent on both BLM and BRCA1 (Fig. 6H). Collectively, our data strongly suggest that, in FANCM-deficient cells, BLM and BRCA1 act in an epistatic pathway to promote DNA end resection and HR to repair and restart the stalled replication fork at ALT telomeres.
Discussion
In most studies on replication stress response, investigators use either chemicals or UV to induce replication stress. However, with these replication stress inducers (RSI), it is very difficult to pinpoint the genomic regions/loci where the replication stress occurs. In addition, these RSIs often induce replication stress through different mechanisms and produce a mixture of DNA damages. Therefore, a model where the genomic location of the replication stress is well-defined and the replication stress is induced under physiological condition is urgently needed. In our study, we found that depletion of FANCM or its obligatory binding proteins in ALT cells induces replication stress predominantly at telomeres. Therefore, we propose that depletion of the FANCM complex in ALT cells can be used as a genomic loci-specific (i.e., ALT telomeres) DNA replication perturbation system. We refer to this replication stress system as MR-SAT (FANCM deficiency induced replication stress at ALT telomere). Although we have firmly established that depletion of the FANCM complex induces replication stress at ALT telomeres, the detailed mechanism behind the exact causes of the replication stress certainly warrants further investigation. Intriguingly, a recent study showed that FANCM is capable of unwinding a generic R-loop structure and that its translocase activity is required for this unwinding activity (76). It is known that R-loops can form at telomeres and that the aberrant accumulation of this R-loop is capable of inducing replication stress (13, 14, 77). We are currently investigating whether FANCM can unwind the R-loop formed between TERRA and telomeric DNA and whether accumulation of this R-loop causes the replication stress in our MR-SAT system. FANCM belongs to the SF2 family of helicase; however, currently there is no published data demonstrating that FANCM is capable of unwinding a DNA–DNA duplex. Another possibility is that the FANCM complex may help to resolve the G4 structures at telomeres. The studies of testing this hypothesis are also under way.
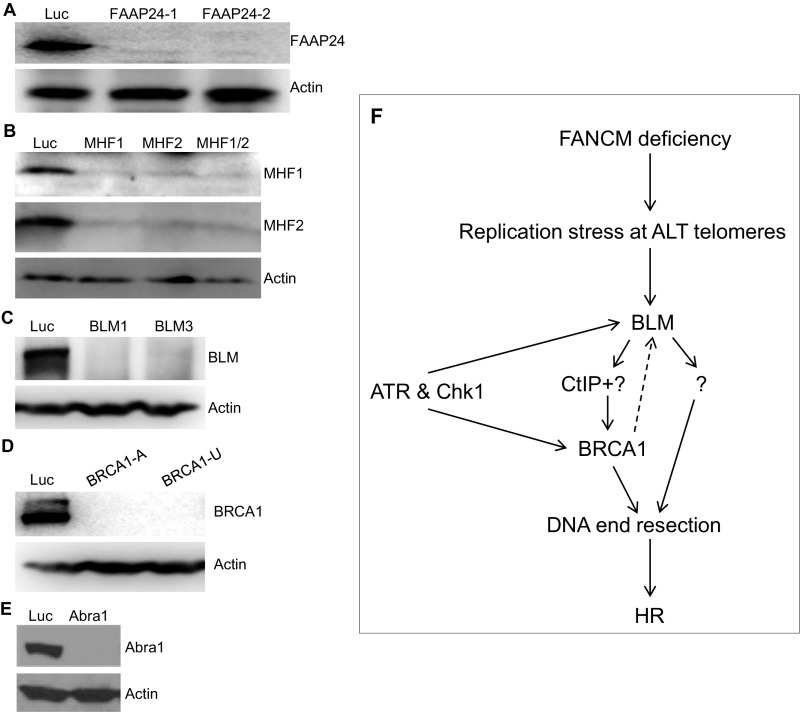
BLM has well-recognized roles in telomere biology. It associates with telomeres in both TEL+ cells and ALT cells (78, 79). Intriguingly, Stavropoulos and colleagues showed that, in ALT cells, BLM interacts with TRF2 and that overexpression of BLM in ALT cells increases telomeric DNA, suggesting that BLM may play an important role in ALT telomere synthesis (79). The proposed functions of BLM at the telomere include suppressing the fragile-telomere phenotype (16), processing the late-replicating intermediate structure (78), and aiding fork progression possibly through unfolding the G4s (80, 81). Here we show that, similar to its role during DSB repair (70), BLM is also required for DNA end resection and HR in our MR-SAT system. The interaction between BLM and BRCA1 was identified previously; however, the functional importance of this interaction was unknown (82). Multiple studies have found that BRCA1 localizes to telomeres; however, its function at telomeres was unclear (72, 73). Here we show that BRCA1 is also required for DNA end resection and HR in our MR-SAT system, possibly by recruiting BLM to the stressed telomeres. In support of the epistatic interaction between BLM and BRCA1 at ALT telomeres, codepletion of FANCM and BLM or of FANCM and BRCA1, but not BLM and BRCA1, dramatically induces the micronuclei formation and synthetic lethality. Intriguingly, in FANCM-depleted TEL+ cells (HeLa and MG63), we did not observe any pronounced increase of Chk1-pS345, RPA32-pS4S8, and BLM foci, nor did we observe any synthetic lethal interaction between FANCM and BLM or FANCM and BRCA1 (Fig. S9 C, D, and F–H). Although the number of cell lines tested here is limited (three ALT cells and five TEL+ cells), our data strongly suggest that FANCM, BLM, and BRCA1 play an important role in alleviating the replication stress at ALT telomeres. Based on the data presented in this study, we propose a model of how ALT cells respond to replication stress at their telomeres (Fig. S10): when a replication fork stalls at or near telomeres, both BLM and BRCA1 are actively recruited to the stalled replication fork. Similar to the replication stress response induced by exogenous RSIs, the recruitment of BLM and BRCA1 is highly regulated by both ATR and Chk1. In addition, BACH1 and CtIP also play a role in the recruitment of BRCA1. Together, BLM and BRCA1 then stimulate DNA end resection and help to repair and restart the stalled replication fork by the HR pathway.
Fig. S10.
Knockdown efficiency of the siRNAs targeting FAAP24, MHF1, MHF2, BLM, BRCA1, or Abra1. U2-OS cells were transfected with siRNA targeting Luciferase (Luc), two different siRNA targeting FAAP24 (FAAP24-1 and -2) (A), MHF1, MHF2, or both MHF1 and MHF2 (MHF1/2) (B), two different siRNA targeting BLM (BLM-1 and -3) (C), two different siRNA targeting BRCA1 (BRCA1-A and -U) (D), or Abra1 (E). Cell lysates were prepared and blotted with antibodies as indicated on the right. (F) A diagram of how ALT cells suppress FANCM deficiency-induced replication stress at telomeres.
In recent years, synthetic lethality has emerged as an attractive strategy for overcoming the cytotoxic effect of most conventional chemotherapy drugs (83, 84). Based on the synthetic lethal interactions identified in our study, simultaneously inhibiting the enzymatic activity of FANCM and BLM could be explored as a targeted therapy to treat ALT cancers, which currently can be treated only by conventional chemotherapy.
Materials and Methods
U2-OS, Saos-2, HeLa, and MG63 cells were purchased from ATCC. A SV40 transformed wild-type fibroblast cell line (AG07217) and a SV40 transformed Bloom syndrome fibroblast cell line (GM08505) were purchase from the Coriell Institute for Medical Research. HuO9, a human osteosarcoma cell line, and the FANCM knockout cell line (in HCT116) were generously provided by Lee Zou, Harvard Medical School, Boston, and Lei Li, MD Anderson Cancer Center, Houston, respectively. HuO9 was grown in RPMI-1640 supplemented with 10% FBS and penicillin and streptomycin. All other cells were grown in DMEM supplemented with 10% FBS and penicillin and streptomycin. All cells were cultivated at 37 °C in a humidified incubator with 5% CO2.
SI Materials and Methods
siRNA.
All siRNAs used in this paper were purchased from Dharmacon: Luciferase (UCGAAGUAUUCCGCGUACGUUdTdT); FANCM (AAGCUCATAAAGCUCUCGGAAdTdT) and FANCM-UTR (AAAGACCUCUCACAAUAUUdTdT) (85); FAAP24-1 (CCGGAUGAGUGAACAAUACUUdTdT) and FAAP24-2 (AUUUUCGAGGAUGGCUUGACAdTdT) (36); MHF1 (L-032895-01) and MHF2 (L-016829-01) (32); FANCB (M-016941-01); FANCG (M-011899-00); FANCI-2 (UUAACAAGGUGUCCACACAGCUGCCdTdT) and FANCI-3 (GCUGGUGAAGCUGUCUGGUUCUCAdTdT) (86); BLM-1 (CCCACUACUUUGCAAGUAAdTdT) and BLM-3 (GAUCAAUGCUGCACUGCUUdTdT); BRCA1-A (CAACAUGCCCACAGAUCAAdTdT) and BRCA1-U (GAAGGAGCUUUCAUCAUUCdTdT); ATR (CCUCCGUGAUGUUGCUUGAdTdT) (87); Chk1 (AAGCGUGCCGUAGACUGUCCAdTdT) (88); Abral (UAUUAGUGGUAACGUGAUAdTdT); BACH1 (AGUCAAGAGUCAUCGAAUAdTdT) (86); CtIP (GCUAAAACAGGAACGAAUCdTdT) (62); and Mre11 (GGAGGUACGUCGUUUCAGAdTdT) (89).
Human cells were transfected twice with 50 nM siRNA using RNAiMAX (Invitrogen).
Antibodies Used for Immunoblotting.
The following antibodies were used for immunoblotting: actin (Santa Cruz, sc-1616); BLM (Bethyl, A300-110A and A300-120A); RPA32-pS4/S8 (Bethyl, A300-245A); RPA32-pS33 (Bethyl, A300-246A); RPA32 (Bethyl, A300-244A); and BRCA1 (Millipore, OP93). Antibodies recognizing Abra1, FANCM, FAAP24, MHF1, and MHF2 are generous gifts from Bin Wang, MD Anderson Cancer Center, Houston, and Weidong Wang, National Institute of Aging, Bethesda.
Immunofluorescent Staining.
U2-OS cells were transfected twice with different siRNA and then replated on coverslips. Cells were then used for immunostaining on day 4 or day 5 (siRNA rescue experiments) after siRNA transfection. Briefly, cells were first fixed with 3% paraformaldehyde containing 2% sucrose for 10 min, then permeablized with Triton X-100 solution on ice for 5 min, and stained with different primary antibodies (as indicated later) and the appropriate Alexa-405 (Invitrogen), Alexa-488 (Invitrogen), and Alexa-546 (Invitrogen)-conjugated secondary antibodies. Images were taken with an Olympus upright fluorescent microscope. Antibodies used for immunofluorescent staining include Chk1-pS345 (Cell Signaling, 2348); RAP32-pS4S8 (Bethyl, A300-245A); BLM (Bethyl, A300-110A and A300-120A); BRCA1 (Bethyl, A301-377A); anti–γ-H2AX (Millipore, 05–636) and (Bethyl, A300-081A); Rad51 (Santa Cruz, sc-8349); TRF1 (abcam, ab10579); and TRF2 (Millipore, 05–521). Anti-BRCA1 antibody raised in rabbit is a generous gift from Jeff Parvin, Ohio State University, Columbus, OH. The antibodies recognizing FANCM and FAAP24, which were raised in rabbits, were generous gifts from Weidong Wang.
Immunofluorescent and Telomere Fluorescent in Situ Hybridization Staining.
Immunofluorescent and telomere fluorescent in situ hybridization was performed as previously described (23). The telomere PNA probe (TelC) was purchased from PNA Bio (F1004).
SMARD.
The SMARD assay was performed essentially as described previously (15). Briefly, 48 h after the siRNA transfection, U2-OS cells were sequentially labeled with 30 μM IdU (4 h) and 30 µM CldU (4 h). DNA isolation and processing for SMARD were as described previously (15). Following Pme I digestion, telomeric DNA fragments ranging from 160 to 200 kb were resolved by PFGE, identified by Southern blot, and then isolated. The Pme I-digested DNA was stretched on microscope slides coated with 3-aminopropyltriethoxysilane (Sigma). After stretching, the DNA was denatured in alkali-denaturing buffer (0.1 N NaOH in 70% ethanol and 0.1% β-mercaptoethanol) for 12 min and fixed by addition of 0.5% glutaraldehyde for 5 min. Telomeric DNA was identified by hybridization with a Biotin-OO-(CCCTAA)4 PNA probe (TelC, Biosynthesis) followed by incubation with Alexa Fluor 350-conjugated NeutrAvidin (Molecular Probes), followed by two rounds of incubation first with a biotinylated anti-avidin antibody (Vector) and then with the Alexa Fluor 350-conjugated NeutrAvidin. Incorporated halogenated nucleotides were detected with a mouse anti-IdU monoclonal antibody (BD) and a rat anti-CldU monoclonal antibody (Accurate) followed by Alexa Fluor 568-conjugated goat anti-mouse (Invitrogen) and Alexa Fluor 488-conjugated goat anti-rat secondary antibodies (Invitrogen). An antibody recognizing the single-stranded DNA was purchased from Millipore (anti-DNA antibody, clone 16–19, MAB3034).
Cell Viability Assay.
siRNA transfected U2-OS were plated on 12-well plates, 2,000–4,000 cells per well. Cells were allowed to grow 7–10 d, fixed, and then stained with 1% crystal violet (75).
siRNA Rescue Experiments.
U2-OS cells were first transfected twice with siRNA. Cells were then transfected with different plasmids using the GenJet In Vitro Transfection Reagent for U2-OS cells (SignaGen Laboratories, SL100489-OS). Twenty-four hours later, cells were plated on a coverslip for immunostaining.
Acknowledgments
We thank Drs. Lee Zou, Lei Li, Jeff Parvin, Bin Wang, and Weidong Wang for reagents and Charles Pavia and Larry Stepp for critical reading of the manuscript. This research was supported by the generous startup fund from the College of Osteopathic Medicine at New York Institute of Technology (D.Z.), and NIH/NIGMS Grant 5R01-GM045751, Empire State Stem Cell Fund through New York State contract C024348, and the Tri-Institutional Stem Cell Initiative funded by the Starr Foundation (C.L.S).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708065114/-/DCSupplemental.
References
- 1.Berti M, Vindigni A. Replication stress: Getting back on track. Nat Struct Mol Biol. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst) 2015;32:149–157. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco MA. Telomeres and human disease: Ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 6.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 9.Dilley RL, Greenberg RA. ALTernative telomere maintenance and cancer. Trends Cancer. 2015;1:145–156. doi: 10.1016/j.trecan.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 11.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam EY, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 14.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 15.Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol. 2012;197:253–266. doi: 10.1083/jcb.201112083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 19.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 21.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 22.Pfeiffer V, Lingner J. Replication of telomeres and the regulation of telomerase. Cold Spring Harb Perspect Biol. 2013;5:a010405. doi: 10.1101/cshperspect.a010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cesare AJ, et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 24.Suram A, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31:2839–2851. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29:1777–1788. doi: 10.1101/gad.266593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 28.Michl J, Zimmer J, Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016;35:909–923. doi: 10.15252/embj.201693860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosedale G, et al. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 31.Ciccia A, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh TR, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collis SJ, et al. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang M, et al. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, et al. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell. 2013;49:997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh TR, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackford AN, et al. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21:2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- 42.Ball SE, et al. Progressive telomere shortening in aplastic anemia. Blood. 1998;91:3582–3592. [PubMed] [Google Scholar]
- 43.Leteurtre F, et al. Accelerated telomere shortening and telomerase activation in Fanconi’s anaemia. Br J Haematol. 1999;105:883–893. doi: 10.1046/j.1365-2141.1999.01445.x. [DOI] [PubMed] [Google Scholar]
- 44.Adelfalk C, et al. Accelerated telomere shortening in Fanconi anemia fibroblasts: A longitudinal study. FEBS Lett. 2001;506:22–26. doi: 10.1016/s0014-5793(01)02869-1. [DOI] [PubMed] [Google Scholar]
- 45.Hanson H, Mathew CG, Docherty Z, Mackie Ogilvie C. Telomere shortening in Fanconi anaemia demonstrated by a direct FISH approach. Cytogenet Cell Genet. 2001;93:203–206. doi: 10.1159/000056985. [DOI] [PubMed] [Google Scholar]
- 46.Callén E, et al. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum Mol Genet. 2002;11:439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 47.Li X, et al. Abnormal telomere metabolism in Fanconi’s anaemia correlates with genomic instability and the probability of developing severe aplastic anaemia. Br J Haematol. 2003;120:836–845. doi: 10.1046/j.1365-2141.2003.04225.x. [DOI] [PubMed] [Google Scholar]
- 48.Uziel O, et al. Oxidative stress causes telomere damage in Fanconi anaemia cells: A possible predisposition for malignant transformation. Br J Haematol. 2008;142:82–93. doi: 10.1111/j.1365-2141.2008.07137.x. [DOI] [PubMed] [Google Scholar]
- 49.Joksic I, et al. Dysfunctional telomeres in primary cells from Fanconi anemia FANCD2 patients. Genome Integr. 2012;3:6. doi: 10.1186/2041-9414-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alter BP, Giri N, Savage SA, Rosenberg PS. Telomere length in inherited bone marrow failure syndromes. Haematologica. 2015;100:49–54. doi: 10.3324/haematol.2014.114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarkar J, Liu Y. Fanconi anemia proteins in telomere maintenance. DNA Repair (Amst) 2016;43:107–112. doi: 10.1016/j.dnarep.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee DB, et al. FANCC suppresses short telomere-initiated telomere sister chromatid exchange. Hum Mol Genet. 2010;19:879–887. doi: 10.1093/hmg/ddp556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spardy N, Duensing A, Hoskins EE, Wells SI, Duensing S. HPV-16 E7 reveals a link between DNA replication stress, Fanconi anemia D2 protein, and alternative lengthening of telomere-associated promyelocytic leukemia bodies. Cancer Res. 2008;68:9954–9963. doi: 10.1158/0008-5472.CAN-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009;37:1740–1754. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luke-Glaser S, Luke B. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PLoS One. 2012;7:e42028. doi: 10.1371/journal.pone.0042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva S, et al. Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance. Genes Dev. 2016;30:700–717. doi: 10.1101/gad.276204.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashley AK, et al. DNA-PK phosphorylation of RPA32 Ser4/Ser8 regulates replication stress checkpoint activation, fork restart, homologous recombination and mitotic catastrophe. DNA Repair (Amst) 2014;21:131–139. doi: 10.1016/j.dnarep.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6:a016477. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 67.Yeager TR, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 68.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 69.Calderano SG, et al. Single molecule analysis of Trypanosoma brucei DNA replication dynamics. Nucleic Acids Res. 2015;43:2655–2665. doi: 10.1093/nar/gku1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kibe T, Zimmermann M, de Lange T. TPP1 blocks an ATR-mediated resection mechanism at telomeres. Mol Cell. 2016;61:236–246. doi: 10.1016/j.molcel.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosen EM. BRCA1 in the DNA damage response and at telomeres. Front Genet. 2013;4:85. doi: 10.3389/fgene.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Acharya S, et al. Association of BLM and BRCA1 during telomere maintenance in ALT cells. PLoS One. 2014;9:e103819. doi: 10.1371/journal.pone.0103819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tian F, et al. BRCA1 promotes the ubiquitination of PCNA and recruitment of translesion polymerases in response to replication blockade. Proc Natl Acad Sci USA. 2013;110:13558–13563. doi: 10.1073/pnas.1306534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwab RA, et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arora R, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stavropoulos DJ, et al. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 80.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 83.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 84.Hartman JL, 4th, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 85.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zou J, et al. FancJ regulates interstrand crosslinker induced centrosome amplification through the activation of polo-like kinase 1. Biol Open. 2013;2:1022–1031. doi: 10.1242/bio.20135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem. 2010;285:1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]