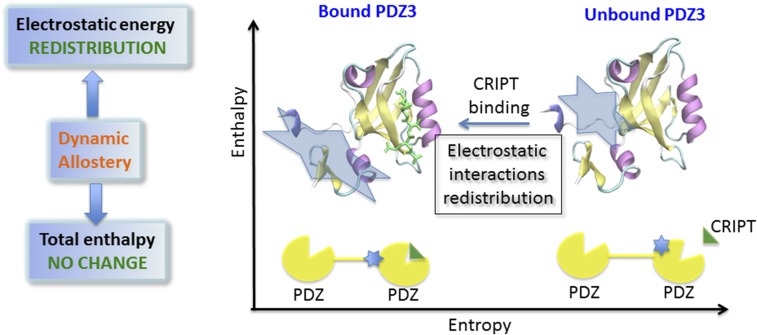
A perturbation at one site of the protein could cause an effect at a distant site. This important biological phenomenon, termed the “allosteric effect,” is essential for protein regulation and cell signaling, playing an important role in cellular function. Its fundamental functional significance has inspired numerous works aiming to understand how allostery works. Allostery can involve large, or unobserved, subtle (mainly side-chain) conformational changes (1). Conformational changes are driven by enthalpy. The term “dynamic allostery” was coined by Cooper and Dryden in the early 1980s to describe allostery “even in the absence of a macromolecular conformational change” (2). Cooper and Dryden argued that dynamic allostery is primarily an entropy effect. However, numerous works have been published over the last 20 y taking “dynamic allostery” to imply a complete absence of conformational change because the authors did not observe such changes (1). Importantly, “dynamic allostery” without observable conformational changes is still ruled by a population shift between two “distinct” states where a new energetic redistribution favorable for the allosteric (functional) state is either dominated by entropy, enthalpy, or both. Few studies questioned whether enthalpy plays a role in dynamic allostery as well (1). In PNAS, Kumawat and Chakrabarty (3) demonstrate that indeed even in dynamic allostery enthalpy plays a role by redistributing internal energies, especially electrostatic interaction energies, among residues upon perturbation (Fig. 1).
Fig. 1.
The scheme of the electrostatic basis of dynamic allostery in the PDZ3 domain protein. Dynamic allostery has no significant conformational change. Upon binding of the peptide (CRIPT), there is no significant enthalpy change, but Kumawat and Chakrabarty (3) reported a redistribution of the electrostatic energies. Such redistribution may propagate to other PDZ domains for the proteins to execute their function.
Electrostatics is an established player in function. Decades ago, Warshel (4) and Warshel et al. (5) demonstrated its critical role in protein catalysis and Perutz (6) in allostery. Recently, rearrangements of electrostatic networks were reported for conformationally driven allostery (7, 8). The report of hidden electrostatic interactions in dynamic allostery in PNAS (3) argues that redistribution of electrostatic interaction energy could be universal in allosteric proteins, with or without significant conformational change.
Proteins embrace redistribution of electrostatic interaction energies to execute their functions. For example, myosin, a motor protein, binds to the negatively charged ATP, which redistributes the electrostatic bond network, thus altering the charge of the actin-binding site, leading to myosin’s dissociation from actin (7). Thus, myosin redistributes electrostatic interaction energy in allostery to execute its function of dissociation from actin.
The PDZ domain protein is a well-known cell signaling protein. The major functions of PDZ domains include recognition of the disordered C-terminal peptide motifs of hundreds of receptors and ion-channel proteins, dimerization with other modular domains, and phospholipid binding. As a dynamic regulator of cell signaling, PDZ function can be modulated allosterically (9). In the PDZ3 domain protein, the peptide recognition site is located at the hydrophobic β2–α2 region. Mutations of distal residues on the PDZ3 domain or deletion of the distal α3 helix (10) allosterically affect the binding affinity at the recognition site without causing significant conformational change. By contrast, binding of the peptide at the recognition site changes the dynamics of the PDZ3 domain, albeit still without large conformational change. Using molecular dynamics simulations and analysis of protein interaction energy, Kumawat and Chakrabarty (3) show that the perturbation of the binding of a peptide in the β2–α2 region leads to the side-chain rearrangement of certain charged residues to form new favorable electrostatic interactions. As a result, some of the previously favorable electrostatic interactions in the unbound state become unfavorable. Like a domino effect, additional side-chain rearrangements occur to release this energetic stress, altering the electrostatic and hydrogen bond network, especially in the N- and C-terminal regions, consistent with the known allosteric sites. How does this energetic redistribution affect PDZ3 function? PDZ domains are usually sequential on the protein chain. The energetic redistribution in one PDZ domain can propagate through an allo-network (11) to a second PDZ domain, thus affecting cell signaling. It is difficult to detect this dramatic redistribution of energy in experiments, because the energetic redistribution does not change the total interactive energy due to the cancellation of the favorable and unfavorable interactions.
Kumawat and Chakrabarty (3) demonstrated the hidden electrostatic basis on PDZ3 domain protein, raising the question of whether the electrostatic interaction redistribution is a common feature among “dynamics-driven” allosteric proteins with minor conformational change, or whether it is a unique feature of the PDZ3 domain protein. PDZ3 recognizes the peptide through its hydrophobic pocket, but a positively charged residue, K5, initiates the rearrangement of the H-bonded network, leading to changes in electrostatic interaction energies (3). One question that arises is whether the charged residue at the binding interface is critical for the electrostatic interaction energy redistribution. For example, cAMP-catabolite activator protein (CAP) has been considered a classic dynamics-driven allosteric protein. Binding of one cAMP to CAP decreases the binding affinity for a second cAMP allosterically; however, even though NMR suggested absence of conformational change, subsequent cAMP-CAP crystal structures (12, 13) indicated that large conformational changes are necessary for its DNA binding function (14). Interestingly, mutations of CAP to reduce its contact with cAMP can enhance this negative allosteric cooperativity (15). Note that charged residues are involved in the interaction interface between CAP and cAMP. One may then speculate that these charged residues, which are perturbed by binding, trigger redistribution of the internal electrostatic interaction energy and thus play a role in CAP allosteric function. Protein kinases were also proposed to be dynamics-driven allosteric proteins (16), where allostery is orchestrated by a dynamic hydrophobic core (17). Like the butterfly effect, a single mutation in cancer can remodel the whole cell (18). Interestingly, many of the frequent cancer mutations involve charged residues, such as the BRAF V600E cancer mutation. This raises the question of whether these charged residue mutations trigger redistribution of internal energies within the protein and in between proteins through allo-network (11) shifts of side chains, which could partly explain the butterfly effect in cancer.
Electrostatic interactions are not the only source for energetic redistribution. Kumawat and Chakrabarty (3) report that Van de Waals interactions and H-bonded networks are both involved in energetic redistribution to some extent. The energy of a protein represents all potential local and long-range interactions within and between residues. In one protein, multiple amino acid interaction networks can be identified to describe different interactions (19). Upon perturbation, one or more amino acid interaction networks could be altered, to different extents, and the energy could be redistributed. The protein is not an isolated system but interacts with its environment including water molecules. Kumawat and Chakrabarty (3) noticed significant local solvation environment change for certain residues, indicating that the energetic redistribution is not only within or between proteins; protein–water interactions also contribute to the energetic redistribution.
How can we unify the electrostatic basis and dynamic basis in dynamic allostery? Previous studies advanced entropy as a driving force in dynamic allostery. Recently, a few computational and experimental reports suggested that La Châtelier’s principle may be behind dynamic allostery. Using a rigid residue scan molecular dynamics simulation-based method, Kalescky et al. (20, 21) reported that decreasing local entropy by increasing individual residue rigidity of PDZ domain proteins often leads to increasing global configurational entropy. Recently, biophysical experiments performed by Law et al. (22) revealed that the volume of PDZ3 domain protein is coupled with local internal motion, following La Châtelier’s principle. Interestingly, deletion of α3 helix at the distal site decreases binding affinity but increases the protein volume and entropy. From the standpoint of energetic redistribution, the loss of local entropy triggers the redistribution of global entropy to balance the local entropy loss. Thus, dynamic allostery can also be viewed as the redistribution of entropy. However, the arrangement of side chains to have the population shift of the amino acid networks (electrostatic, H-bonded network, etc.) is a result of side-chain dynamics. Therefore, we can unify the enthalpy and entropy basis of dynamic and conformational allostery through energy redistribution and propose that allosteric function can be executed through the internal energy redistribution upon the perturbation of external energy. The Kumawat and Chakrabarty (3) observations are thus important in refocusing current views. Rather than classifying allostery as entropic or dynamic, they contend that for the PDZ3 domain protein it should be viewed in terms of internal redistribution and population shift of specific electrostatic interactions. Future studies should explore the generality of these findings and their predictions in cell signaling and drug discovery.
Acknowledgments
This was supported in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health Contract HHSN261200800001E.
Footnotes
The authors declare no conflict of interest.
See companion article on page E5825.
References
- 1.Nussinov R, Tsai CJ. Allostery without a conformational change? Revisiting the paradigm. Curr Opin Struct Biol. 2015;30:17–24. doi: 10.1016/j.sbi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 3.Kumawat A, Chakrabarty S. Hidden electrostatic basis of dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. 2017;114:E5825–E5834. doi: 10.1073/pnas.1705311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warshel A, et al. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 6.Perutz MF. Electrostatic effects in proteins. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Ohnuki J, Takano M. Dielectric allostery of protein: Response of myosin to ATP binding. J Phys Chem B. 2016;120:13047–13055. doi: 10.1021/acs.jpcb.6b10003. [DOI] [PubMed] [Google Scholar]
- 8.Ohnuki J, Sato T, Takano M. Piezoelectric allostery of protein. Phys Rev E Stat Nonlin Soft Matter Phys. 2016;94:012406. doi: 10.1103/PhysRevE.94.012406. [DOI] [PubMed] [Google Scholar]
- 9.Ivarsson Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 2012;586:2638–2647. doi: 10.1016/j.febslet.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nussinov R, Tsai CJ, Csermely P. Allo-network drugs: Harnessing allostery in cellular networks. Trends Pharmacol Sci. 2011;32:686–693. doi: 10.1016/j.tips.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao RR, Lawson CL. Structure of catabolite activator protein with cobalt(II) and sulfate. Acta Crystallogr F Struct Biol Commun. 2014;70:560–563. doi: 10.1107/S2053230X14005366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- 14.Sharma H, Yu S, Kong J, Wang J, Steitz TA. Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding. Proc Natl Acad Sci USA. 2009;106:16604–16609. doi: 10.1073/pnas.0908380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsend PD, et al. The role of protein-ligand contacts in allosteric regulation of the Escherichia coli catabolite activator protein. J Biol Chem. 2015;290:22225–22235. doi: 10.1074/jbc.M115.669267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornev AP, Taylor SS. Dynamics-driven allostery in protein kinases. Trends Biochem Sci. 2015;40:628–647. doi: 10.1016/j.tibs.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, et al. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci Adv. 2017;3:e1600663. doi: 10.1126/sciadv.1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart JR, et al. The butterfly effect in cancer: A single base mutation can remodel the cell. Proc Natl Acad Sci USA. 2015;112:1131–1136. doi: 10.1073/pnas.1424012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Rourke KF, Gorman SD, Boehr DD. Biophysical and computational methods to analyze amino acid interaction networks in proteins. Comput Struct Biotechnol J. 2016;14:245–251. doi: 10.1016/j.csbj.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalescky R, Zhou H, Liu J, Tao P. Rigid residue scan simulations systematically reveal residue entropic roles in protein allostery. PLOS Comput Biol. 2016;12:e1004893. doi: 10.1371/journal.pcbi.1004893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalescky R, Liu J, Tao P. Identifying key residues for protein allostery through rigid residue scan. J Phys Chem A. 2015;119:1689–1700. doi: 10.1021/jp5083455. [DOI] [PubMed] [Google Scholar]
- 22.Law AB, Sapienza PJ, Zhang J, Zuo X, Petit CM. Native state volume fluctuations in proteins as a mechanism for dynamic allostery. J Am Chem Soc. 2017;139:3599–3602. doi: 10.1021/jacs.6b12058. [DOI] [PubMed] [Google Scholar]