Abstract
The goal of modern Clinical Decision Support (CDS) systems is to provide physicians with information relevant to their management of patient care. When faced with a medical case, a physician asks questions about the diagnosis, the tests, or treatments that should be administered. Recently, the TREC-CDS track has addressed this challenge by evaluating results of retrieving relevant scientific articles where the answers of medical questions in support of CDS can be found. Although retrieving relevant medical articles instead of identifying the answers was believed to be an easier task, state-of-the-art results are not yet sufficiently promising. In this paper, we present a novel framework for answering medical questions in the spirit of TREC-CDS by first discovering the answer and then selecting and ranking scientific articles that contain the answer. Answer discovery is the result of probabilistic inference which operates on a probabilistic knowledge graph, automatically generated by processing the medical language of large collections of electronic medical records (EMRs). The probabilistic inference of answers combines knowledge from medical practice (EMRs) with knowledge from medical research (scientific articles). It also takes into account the medical knowledge automatically discerned from the medical case description. We show that this novel form of medical question answering (Q/A) produces very promising results in (a) identifying accurately the answers and (b) it improves medical article ranking by 40%.
Keywords: Question Answering, Medical Information Retrieval, Clinical Decision Support
1. INTRODUCTION
In their everyday practice, physicians make a variety of clinical decisions regarding the care of their patients, e.g. deciding the diagnosis, the test(s) or the treatment that they prescribe. Clinical Decision Support (CDS) systems have been designed to help physicians address the myriad of complex clinical decisions that might arise during a patient’s care [11]. By leveraging the fact that patient care is documented in electronic medical records (EMRs), one of the goals of modern CDS systems is to anticipate the needs of physicians by linking EMRs with information relevant for patient care. Such relevant information can be retrieved from bio-medical literature. Recently, the special track on Clinical Decision Support in the Text REtrieval Conference (TREC-CDS) [26], has addressed the challenge of retrieving bio-medical articles relevant for to a medical case when answering one of three generic medical questions: (a) “What is the diagnosis?”; (b) “What test(s) should be ordered?”; and (c) “Which treatment(s) should be administered?”. The TREC-CDS track did not rely on a collection of EMRs, instead it used an idealized representation of medical records in the form of 30 short medical case reports, each describing a challenging medical case. Thus, systems developed for the TREC-CDS challenge were provided with a list of topics, consisting of (1) a narrative describing the fragments from the patient’s EMRs that were pertinent to the case; (2) a summary of the medical case and (3) a generic medical question. Systems were expected to use either the medical case description or the summary to answer the question by providing a ranked list of articles available from PubMed Central [31] containing the answers. As only one of the three generic questions was asked in each topic, the expected medical answer type (EMAT) of the question was diagnosis, test or treatment. Figure 1 illustrates three examples of topics evaluated in the 2015 TREC CDS, one example per EMAT. Figure 1 also illustrates the correct answer of each of the questions.
Figure 1.

Examples of topics evaluated in the 2015 TREC CDS track.
In the 2015 TREC-CDS track a new task was offered, in which for questions having the EMAT ∈ {test, treatment}, the patient’s diagnosis was provided (shown in Figure 1). The results for this new task, as reported in [26] were superior to the results for the same topics when no diagnoses were provided. This observation let us to believe that when knowing even a partial answer to the question, the ability to retrieve relevant bio-medical literature was significantly improved. Moreover, we asked ourselves if identifying the answers to the medical questions could be performed with acceptable accuracy. More importantly, we wondered if we should first try to find the answer and then rank the relevant scientific articles for a given question. It was clear to us from the beginning that answer identification would be a harder problem, unless we could tap into a new form of knowledge and consider answering the questions directly from a knowledge base (KB). Question answering (Q/A) from KBs has experienced a recent revival. In the 60’s and 70’s, domain-specific knowledge bases were used to support Q/A, e.g. the Lunar Q/A system [34]. With the recent growth of KBs such as DBPedia [3] and Freebase [7], new promising methods for Q/A from KBs have emerged [9, 35, 5]. These methods map questions into sophisticated meaning-representations which are used to retrieve the answers from the KB.
In this paper, we present a novel Q/A from KB method used for answering the medical questions evaluated in TREC-CDS. First, instead of relying on an existing large KB, we automatically generated a very large medical knowledge graph from a publicly available collection of EMRs. As reported in [23], the medical case descriptions from the TREC-CDS topics were generated by consulting the EMRs from MIMIC-II [17]. Consequently, we used all the publicly available EMRs provided by MIMIC-III (a more recent superset of the EMRs in MIMIC-II) to automatically generate a very large probabilistic knowledge graph designed to encode knowledge acquired from medical practice. Second, instead of retrieving answers directly from the KB, the answers were obtained through probabilistic inference methods. Third, instead of identifying the answers from relevant PubMed articles, we used the answers inferred from the knowledge graph to select and rank the PubMed articles which contain them. In this way, we replaced the architecture of the typical system that was evaluated in TREC-CDS, illustrated in Figure 2(a) with a new architecture, illustrated in Figure 2(b). The new architecture identifies the ranked list of answers to the questions as well as the ranked list of scientific articles that contain them. While current state-of-the-art systems that were evaluated in TREC-CDS first processed the topics and then used query expansion methods to enhance the relevance of the retrieved scientific articles from PubMed, as reported in [23] and illustrated in Figure 2(a), we relied on a knowledge graph encoding the clinical picture and therapy of a large population of patients documented in the MIMIC III EMR database. The automatic generation of the knowledge graph involved: (1) medical language processing to identify medical concepts representing signs/symptoms, diagnoses, tests and treatments, as well as their assertions; (2) the cohesive properties of the medical narratives from the EMRs; and (3) a factorized Markov network representation of the medical knowledge. As illustrated in Figure 2(b), this probabilistic knowledge graph was used for inferring the answer of the TREC-CDS topics, which were processed to discern the medical concepts and their assertions in the same format as the nodes from the knowledge graph. We experimented with three different probabilistic inference methods to identify the most likely answers for each of the TREC-CDS topics evaluated in 2015. By participating in the challenge, we had access to the correct answers1, thus we could evaluate the correctness of the answers identified by our novel Q/A from KB method. Moreover, the inferred answers allowed us to produce a ranking of the scientific articles that contained them and thus define a novel, answer-informed relevance model. Our main contributions in this paper are:
Answering medical questions related to complex medical cases from an automatically generated knowledge base derived from a vast, publicly available EMR collection;
Using probabilistic inference to identify answers from a vast medical knowledge graph;
Combining medical knowledge derived from an EMR collection with medical knowledge derived from relevant scientific articles to enhance the quality of probabilistic inference of medical answers – a combination that unifies medical knowledge characterizing medical practice (from EMRs) with medical knowledge characterizing medical research (from scientific articles); and
Using the likelihood of the automatically discovered answers to the question associated with a topic to produce a novel ranking of the relevant scientific articles containing the answers.
Figure 2.
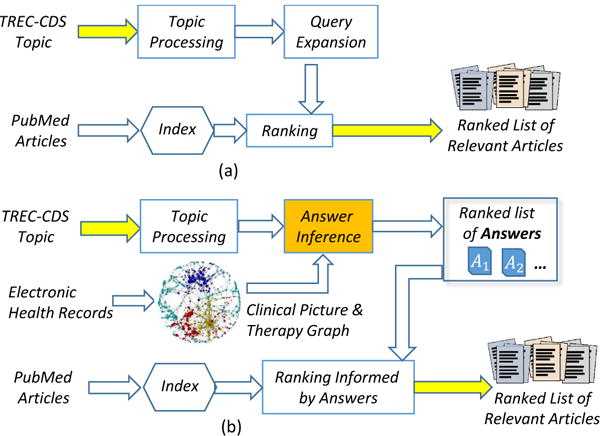
Architectures of medical question answering systems for clinical decision support
The remainder of the paper is organized as follows. Section 2 details the answer inference in the new architecture for Q/A-CDS. Section 3 details the automatic generation of the medical knowledge graph while Section 4 presents the three forms of probabilistic inference of answers we experimented with. Section 5 discusses the experimental results and Section 6 summarizes the conclusions.
2. AN ARCHITECTURE FOR INFERRING MEDICAL ANSWERS
The cornerstone of our medical Q/A method for clinical decision support (CDS) is the derivation of the answers to a topic’s question from a vast medical knowledge graph, generated automatically from a collection of EMRs. The medical knowledge base contained approximately 634 thousand nodes and 14 billion edges, in which each node represents a medical concepts and the belief value (or assertion, as described in Section 3.3) associated with it. We automatically identified four types of medical concepts: signs/symptoms, tests, diagnoses and treatments (as detailed in Section 3.2). However, identifying medical concepts is not sufficient to capture all the subtleties of medical language used by physicians when expressing medical knowledge. Medical science involves asking hypotheses, experimenting with treatments, and formulating beliefs about the diagnoses and tests. Therefore, when writing about medical concepts, physicians often use hedging as a linguistic means of expressing an opinion rather than a fact. Consequently, clinical writing reflects this modus operandi with a rich set of speculative statements. Hence, automatically discovering clinical knowledge from EMRs needs to take into account the physician’s degree of belief by qualifying the medical concepts with assertions indicating the physician’s belief value (e.g. HYPOTHETICAL, PRESENT, ABSENT) as detailed in Section 3.3. It should be noted that the same medical language processing techniques were used to process the EMRs, the medical topics, and the (relevant) scientific articles from PubMed.
To represent the relations spanning the medical concepts in our knowledge graph, we modeled the cohesive properties of the narratives from EMRs (details are provided in Section 3.1). In order to use this graph to infer answers to medical questions we cast the medical knowledge graph as a factorized Markov network, which is a type probabilistic graphical model. We call this graphical model the clinical picture and therapy graph (CPTG) because it enables us to compute the probability distribution over all the possible clinical pictures and therapies of patients. For a given topic t, the set of medical concepts and their assertions discerned from t is interpreted as a sketch of the clinical picture and therapy described in the topic, represented as Z(t). Thus, answering the medical question associated with t amounts to determining which medical concept in the CPTG (with the same type as the expected medical answer type or EMAT) has the highest likelihood given Z(t). In addition to the medical sketch Z(t), we believed that answering medical questions with the CPTG could benefit from combining the medical sketch of a topic with knowledge acquired from individual scientific articles deemed relevant to the topic. This belief is also motivated by our observation that the joint distribution represented by the CPTG favors more common concepts, whereas the topics evaluated in the TREC-CDS correspond to complex medical cases, rather than common cases. Thus, we believe that the combination of Z(t) with all the medical concepts (and their assertions) derived from a scientific article l relevant to the topic generates a more complete view of a possible clinical picture and therapy for a patient than the one discerned only from the topic. Therefore, we consider this extended set of medical concepts and assertions for a topic t as its extended sketch, denoted as EZ(t,l). Regardless of which sketch z ∈ {Z(t),EZ(t,l)} of a topic is used, we discovered the most likely answer â to the medical question associated with t by discovering the medical concept which, when combined with the sketch, produces the most likely clinical picture and therapy. Formally:
| (1) |
where the set A denotes all the concepts in the CPTG with the same type as the EMAT, and P(·) refers to the probability estimate provided by the CPTG.
The architecture of the medical QA system which operated on the medical graph that we derived automatically from a collection of EMRs (i.e. the CPTG) is represented in Figure 3. When the Z(t) is used in the inference of answers, we have Case 1, as illustrated in Figure 3, in which the topic is processed to derive Z(t) by automatically identifying medical concepts and assertions from either the description or the summary of the topic. When EZ(t,l) (the extended sketch of the topic) is used, in addition to the concepts (and their assertions) obtained from processing the topic, new knowledge from a relevant scientific article l needs to be derived. It is to be noted that from each relevant article for a topic t we generated a distinct extended sketch which combines Z(t) with the medical concepts (and their assertions) discovered in the article. In Figure 3, this is represented as Case 2, in which the first step is to derive (and expand) a query for the topic.
Figure 3.
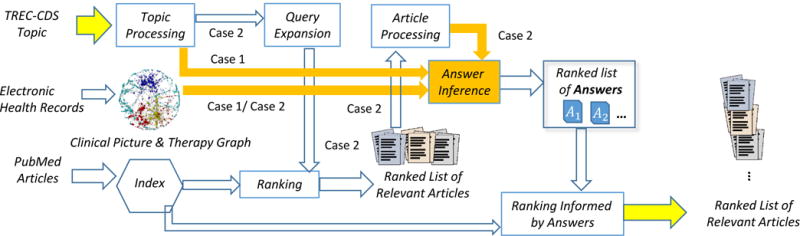
An architecture that implements two different cases for answering medical questions for clinical decision support
When processing the topic to generate a query, deciding whether to use individual words or concepts is important. In TREC-CDS, there were systems that used all the content words from the description to produce the query, while other systems considered only medical concepts. Both the Unified Medical Language System (UMLS) [6] and MeSH [18] were commonly used as ontological resources for medical concepts. In the architecture represented in Figure 3, we opted to use medical concepts rather than words, identifying the signs, symptoms, diagnoses, tests and treatments mentioned in the topic. We expanded each query with medical concepts from UMLS which share the same concept unique identifier (CUI) as any concept detected from the topic, obtaining synonyms, and, in some cases, hyponyms, and hypernyms. Some medical concepts (e.g. “crystalloid solution”) or their synonyms (e.g. “sodium chloride”) are phrases rather than single words. Consequently, the resulting expanded query consists of lists of key-phrases which are processed by a relevance model to retrieve and rank articles from an index of PubMed articles. The index was generated from the PubMed Central Open Access Subset. We used a snapshot of these articles from January 21, 2014 containing a total of 733,138 articles which were provided by the TREC-CDS organizers. In TREC-CDS, systems implemented a variety of relevance models, as reported in [23]; we experimented with several relevance models, discussed in the evaluation section. For each of the first 1,000 relevant articles2, which we denote as L, we automatically identified the medical concepts and their assertions. This enabled us to generate for each article, lr ∈ L its extended medical sketch of the topic t, denoted as EZ(t,lr).
A close inspection of the contents of the extended medical sketches obtained for many scientific articles indicated the inclusion of many medical concepts which presented no relevance to the topic. This reflects the fact that many of the scientific articles in PubMed Central document unexpected or unusual medical cases – often in non-human subjects. This created a serious problem in the usage of the extended medical sketches to infer answers from the CPTG. Specifically, because the likelihood estimate of an answer enabled by the CPTG is based on the observed clinical pictures and therapies of patients in the MIMIC clinical database, non-relevant scientific articles which contained common diagnoses, treatments, tests, signs, or symptoms had a disproportionately large impact on the ranking of answers. In order to solve this problem, we refined the ranking of answers provided in Equation 1 in order to incorporate the relevance of the scientific article used for creating each extended medical sketch. Thus, in Case 2 illustrated in Figure 3, we produced the answer ranking by using a novel probabilistic metric, namely the Reciprocal-Rank Conditional Score (RRCS). RRCS considers for each article in L, (1) the conditional probability of the answer given the extended sketch associated with that article, i.e. EZ(t,lr), as well as (2) the relevance rank of the article, represented by the rank r of lr in L. Formally, the new ranking of answers to a question associated with topic t generated by the RRCS metric is defined as:
| (2) |
The ranking of answers based on the RRCS reflects both the likelihood of the answer according to EZ(t,lr) as well as the relevance of each scientific article.
In addition to ranking medical answers, we also use the CPTG to rank scientific articles based on the answers they contain. In Case 1, when the medical sketch of a topic Z(t) was used for inferring the answer, the ranked list of answers was produced by relying entirely on the knowledge from the medical sketch and from the EMR collection. Therefore, the set of scientific articles that contain at least one of the answers of the medical question for a topic t in Case 1 needs to be retrieved. Hence, a query in disjunctive form of all the inferred answers is used. When the relevance model uses the query against the index, it provides a list of ranked relevant articles L. We denote by Yi all answers found in a relevant article ranked on position i of L. This allows us to define the relevance of scientific articles responding to the answer of a topic t as:
| (3) |
Equation 3 represents a ranking of each scientific article li ∈ L based on the likelihood of the answers in the article given the medical sketch derived for the topic.
In contrast, in Case 2 represented in Figure 3, when EZ(t,l) is used for inferring the answers, the set of ranked relevant scientific articles L is known, as it was already used to provide the new knowledge for each EZ(t,li). Moreover, each answer found in any scientific article from L is necessarily part of its extended medical sketch, thus Yi ⊆ EZ(t,li). Consequently, we define the relevance of scientific articles responding to the answer of a topic t as:
| (4) |
In this way the relevance of an article responding to the question of a topic is computed by comparing the likelihood of the extended medical sketch which includes the answers found in the article against the likelihood of the extended medical sketch which does not contain the answers found in the article. Clearly, the ability to infer the probability of a combination of medical concepts (and their assertions), i.e. the capability of determining P(·), enabled us to produce the new, answer-informed ranking models for answers as well as scientific articles given in Equations 1–4.
3. GENERATING THE CLINICAL PICTURE AND THERAPY GRAPH
As defined in [25] the clinical picture constitutes the clinical findings about a patient (e.g. medical problems, signs, symptoms, and tests) that might influence the diagnosis. In addition, therapy is defined as the set of all treatments, cures, and preventions included within the management plan for a patient. Moreover the clinical picture may vary significantly between patients with the same disease and may even vary between different points in time for the same patient during the course of his/her disease. Therefore, in order to capture the variation in the clinical picture and therapy of a patient population, we created a clinical picture and therapy graph (CPTG) in which each node corresponds to a medical concept qualified by its assertion. Inspired by the approach reported in [12], we represented the CPTG as a 4-partite graph in which partitions of nodes represent all the signs/symptoms , all the diagnoses , all the tests and all the treatments which were automatically recognized in the MIMIC-III3 collection of EMRs. By considering four partitions of medical concepts, we need to encode all six possible types of relations between each partition of nodes in the CPTG. The relations types are: (1) , between signs/symptoms and diagnoses; (2) , between signs/symptoms and tests; (3) , between signs/symptoms and treatments; (4) , between diagnoses and tests; (5) , between tests and treatments; and (6) , between diagnoses and treatments. We take advantage of the fact that any k-partite graph can be interpreted as a factorized Markov network [16], and encode the strength of these relations using mathematical factors. Figure 4 illustrates the factorized Markov network corresponding to the CPTG.
Figure 4.
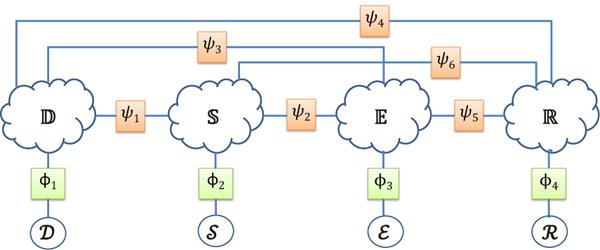
Factorized Markov network modeling the likelihood of any possible clinical picture and therapy.
A factorized Markov network is a type of Probabilistic Graphical Model which represents knowledge in terms of (1) statistical random variables, and (2) mathematical factors (or functions), which assign a real value to each potential assignment of a set of random variables (known as the factor’s scope) allowing us to represent the strength of the relationships between the random variables in the model. In this representation, each possible medical concept (i.e. each node) is interpreted as a binary random variable. This allows any possible clinical picture and therapy (CPT) to be encoded by assigning a value of 1 to the random variable associated with each concept in the CPT which is asserted to be PRESENT, CONDUCTED, ORDERED, or PRESCRIBED, and a value of 0 to the random variable associated with each medical concept which is asserted to be ABSENT. Every other medical concept (i.e. those mentioned with another assertion, or which were not mentioned in the CPT) is considered a latent variable whose value is later inferred. This random variable representation allows us to encode any possible combination of medical concepts (and their assertions), which we represent as , in which represents the random variables corresponding to diagnoses, represents the random variables corresponding to signs/symptoms, represents the random variables corresponding to tests, and represents the random variables corresponding to treatments. We can estimate the likelihood of as:
| (5) |
Using the maximum likelihood estimate provided by , we have defined four factors which represent the (prior) probability of a CPT containing combinations of medical concepts with the same type: (1) , the likelihood of a CPT containing diagnoses in ; (2) , the likelihood of a CPT containing the signs/symptoms given by ; (3) , the likelihood of a CPT containing the tests in ; and (4) , the likelihood of a CPT containing the treatments in .
The clinical picture and therapy of a patient also involves relationships between medical concepts of different types. To model these six types of relations, we considered six additional factors: (1) , the correlation between all the diagnoses in and all the signs/symptoms in ; (2) , the correlation between all the signs/symptoms in and all the tests in ; (3) , the correlation between all the diagnoses in and all the tests in ; (4) , the correlation between all the diagnoses in and all the treatments in ; (5) , the correlation between all the tests in and all the treatments in ; and (6) , the correlation between all the signs/symptoms in and all the treatments in .
All ten factors enable us to infer the probability of any possible clinical picture and therapy, such as the sketch z ∈ {Z(t),EZ(t,l)} of the clinical picture and therapy discussed in Section 2. By definition, an (extended) sketch is a set of medical concepts and their assertions. Thus, we can encode z in terms of the random variables corresponding to the diagnoses , signs/symptoms , tests , and treatments the (extended) sketch contains, such that As before, all the remaining random variables (i.e. ) are left as latent variables. Using the CPTG (illustrated in Figure 4), we can compute the probability of a medical sketch z:
| (6) |
Or, the more compact equivalent notation:
| (7) |
where, for each factor, we ignore all medical concepts which do not have the semantic types expected by that factor’s scope (e.g. ). As defined, the probability distribution given in Equation 7 is the product of the ten factors defined above. Each of these factors depends on the maximum likelihood estimate of how many CPTs in the EMR collection contain various combinations of medical concepts. Thus, computing P(z) relies on the ability to automatically recognize medical concepts and their assertions in EMRs, as described in Sections 3.1 and 3.2.
3.1 Identification of Medical Concepts
Our methodology for automatically recognizing medical concepts in clinical texts benefits from the general framework developed by the 2010 shared-task on Challenges in Natural Language Processing for Clinical Data [30] provided by the Informatics for Integrating Biology at he Bedside (i2b2) and the United States Department of Veteran’s Affairs (VA). In this challenge, identification of medical concepts in clinical narratives targeted three categories: medical problems, treatments, or tests. In this work, we have extended this framework to also distinguish two sub-types of medical problems: (1) signs (observations from a physical exam) and symptoms (observations by the patient); and (2) the diagnoses, including co-morbid diseases or disorders. Figure 6 illustrates our methodology.
Figure 6.
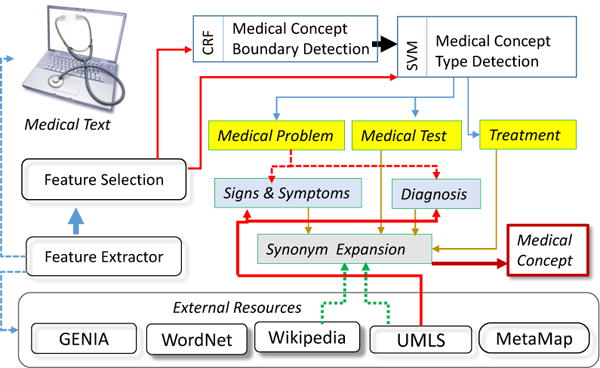
System for Medical Concept Recognition
We followed the framework reported by [22] in which medical concept identification was cast as a three-stage classification problem, using 72,846 annotations of medical concepts and their assertions provided by the i2b2 challenge. In the first stage, a conditional random field (CRF) is used to determine the boundaries (starting and ending tokens) of each medical concept. In the second stage, a support vector machine (SVM) was used to classify each type of medical concept into a medical problem, treatment, or test.
Classification relied primarily on lexical features, as well as concept type information from UMLS and Wikipedia and predicate-argument semantics resulting from automatic feature selection as described in [22]. The resources used for feature extraction are The Unified Medical Language System (UMLS) [6], MetaMap [2], the GENIA project [14], WordNet [10], PropBank [15], the SwiRL semantic role labeler [29], and Wikipedia.
Unlike the work of [22], we introduced a third and final stage, in which we project each concept onto the UMLS ontology and classify it as a sign or symptom if the UMLS semantic type is SYMPTOM OR SIGN or FINDING, and as a diagnosis otherwise. Finally, synonyms for each medical concept are provided by (1) identifying all UMLS atoms that share the same concept unique identifier (CUI) and (2) groups of article titles in Wikipedia which redirect to the same article. Thus, we can account for synonymous concepts in the CPTG by combining all the nodes corresponding to synonymous concepts. Figure 5 illustrates the results of medical concept recognition.
Figure 5.

Example of medical concepts and their assertions discerned from the description and summary of medical topic 32 (illustrated in Figure 1) as well as from the relevant PubMed article PMC3132335. Medical concepts mentioned in the text are typeset in boldface, while synonymous concepts are not.
3.2 Recognizing the Medical Assertions
We followed the framework reported in [22] in which the belief status (or assertion type) of a medical concept is determined by a single SVM classifier. Medical assertions were categorized as PRESENT, ABSENT, POSSIBLE, HYPOTHETICAL, CONDITIONAL, or ASSOCIATED-WITH-SOMEONE-ELSE which were defined in the 2010 i2b2 challenge only for medical problems. Note that we have extended these assertion values to qualify tests and treatments as well as previously reported in [13]. We considered and annotated five new assertion values to encompass the physicians’ beliefs about tests and treatments: PRESCRIBED, ONGOING, and SUGGESTED for treatments as well as ORDERED and CONDUCTED for tests. We have produced an additional set of 2,349 new annotations for the new assertion values as previously reported in [13].
Our assertion classification methodology relies on the same feature set and external resources reported by [22]: UMLS, MetaMap, NegEx [8] and the Harvard General Inquirer [28]. These external resources, along with lexical features and statistical information about the assertions assigned to previous mentions of the same medical concept in the same document were used to train a multi-class SVM by following the framework reported in [22].
4. ANSWER INFERENCE
Inferring the answers to the question associated with a topic, as defined in Equations 1–4, relies on the ability of the CPTG to model the distribution over all possible CPTs. However, evaluating this distribution(defined in Equation 7) can be prohibitively expensive (it requires storing counts). Another problem stems from the significant sparsity in clinical data: for a particular combination of medical concepts, we may not find a CPT which exactly matches the given combination (i.e. may be zero). For example, if the diagnoses in the CPT are , we may not find any patient documented in the EMR collection who is diagnosed with all the diagnoses with the same assertions as in . Consequently, we would infer the likelihood of this sketch to be zero. Instead, we would prefer to consider patients whose clinical picture and therapies are similar to those provided in the sketch, relaxing the maximum likelihood estimation requirements. For this purpose, we considered three alternative inference techniques: (1) approximate inference based on the notion of Bethe free-energy, (2) pair-wise variational inference, and (3) inference based on interpolated smoothing.
4.1 Bethe Free-energy Approximation
We first considered state-of-the-art methods for approximate inference. Unlike other approaches for estimating inference, approximate inference techniques guarantee certain upper bounds on the error between their approximate probability and the true probability of the distribution. In the graphical modeling community, the most commonly used approximate inference algorithm is that of Loopy Belief Propagation [20] wherein variables and factors repeatedly exchange messages until, at convergence, the full distribution is estimated. More recently, approximate inference approaches have considered interpreting the distribution of a set of random variables as the information energy present in a physical system. In this setting, the distribution of all possible clinical pictures and therapies given in Equation 7 is cast as the energy J:
This allows us to then define the “Free Energy” of the system as follows:
| (8) |
where U(z) is the energy and H(z) is the entropy. As shown in [32] and [36], the minimum fixed points of the free energy equation are equivalent to fixed points of the iterative Loopy Belief Propagation algorithm. This means that minimizing the free energy in Equation 8 obtains the same solution as running iterative loopy belief propagation on Equation 7 until convergence. Moreover, the Free Energy can be approximated using the Bethe approximation which transforms our original potentially infinite message passing problem into a simple, convex, linear programming problem based on pair-wise information:
where
and
Thus, we can approximate P(z) from Equation 7 by minimizing FB over τ:
| (9) |
where τ must satisfy the following conditions:
| (10a) |
| (10b) |
| (10c) |
By representing the constraints in Equations 10a–10c as Lagrangian multipliers, we approximated the joint probability of any clinical picture and therapy from Equation 7 by using straight-forward stochastic gradient descent4. In our implementation, we used the publicly available Hogwild software for parallel stochastic gradient descent [21].
4.2 Pair-wise Variational Inference
In addition to approximate inference by Bethe-free energy, we wanted to know whether simpler approximations would suffice. An obvious and much simpler strategy for relaxing the maximum likelihood estimates to better handle sparsity would be to define each factor using the association between all pairs of concepts in the factor. In this way, the four same-typed factors (ϕi) can be assigned to the product of all pair-wise MLE estimates in the CPTG:
Likewise, the factors ψ1…ψ6 can be similarly defined:
By using these alternative pair-wise definitions, we were able to estimate the joint distribution in Equation 7 by considering a a graph with a much simpler (i.e. pair-wise) structure. This approximation smooths the likelihood of a particular sketch by considering the likelihood of each pair of concepts in the sketch, rather than the likelihood of the entire sketch at once.
4.3 Inference Using Interpolated Smoothing
The pair-wise variational inference method defined in sub-section 4.2 still suffers from sparsity problems: if the likelihood for any pair of medical concepts is zero, then the joint probability will be zero. Moreover, the pairwise approach does not discriminate between the level of similarity between a given CPT (e.g. z) and each CPT used to generated the CPTG. We defined the level of similarity between two CPTs as the number of concepts contained in both CPTs. Thus, the levels of similarity range from perfectly similar (all |z| concepts in common) to perfectly dissimilar (0 concepts in common). In order to account for each of these levels of similarity, we interpolated the likelihood of a sketch z (or CPT), with the likelihoods of all CPTs formed by subsets of medical concepts in z. Although this would typically require enumerating all 2|z| subsets of z and, thus, would be computationally intractable, we reduced the complexity to be linear in the size of the EMR collection by casting the inference problem as an information retrieval problem.
By first indexing the medical concepts present in each patient’s EMRs, we were able to compute the smoothed likelihood of a particular sketch through a series of constant-time Boolean retrieval operations. Specifically, by indexing the medical concepts in the EMRs, we were able to obtain a binary vector for each medical concept in the sketch indicating which EMRs mentioned that concept. The sum of these binary vectors, which we denote as m, contains, for each EMR, the number of concepts in common with the CPT of the EMR and the sketch. A single iteration over m allowed us to compute the number of EMRs with CPTs within each level of similarity denoted as n0…n|z|. The smoothed likelihood of a clinical sketch was then calculated by interpolating the number of EMRs at each similarity level (ni):
where α ∈ [0,1] is a scaling factor such that when α = 0 no smoothing is performed and when α=1, the smoothed likelihood is the sum of vector n, or the total number of EMRs whose CPT shares each level of similarity with the medical sketch.
Thus, we have considered three approaches for inferring the distribution of CPTs given by Equation 7 which is used by our system not only to rank extracted medical answers, but also to rank the documents for each answer.
5. EXPERIMENTAL RESULTS
We evaluated the role of our approach for question answering towards clinical decision support in terms of (1) the quality of answers returned for each topic as well as (2) the quality of scientific articles retrieved for each topic. In addition to the quality of answers and retrieved scientific articles, we also analyzed (3) the quality of the clinical picture and therapy graph. In these evaluations, we used the set of 30 topics (numbered 31–60) used for the 2015 TREC-CDS evaluation [26].
5.1 Medical Answer Evaluation
To determine the quality of answers produced by our system, we relied on a list of “potential answers” produced by the authors of the 2015 TREC-CDS topics and distributed by the TREC CDS organizers after the conclusion of the evaluation. These potential answers were not provided to the relevance assessors for judging document retrieval, nor were they provided to participating teams until after the evaluations were performed. Although the 2015 TREC-CDS evaluation focused only the ability to retrieve and rank scientific articles, the potential answers provided after the conclusion of the evaluation allowed us to cast the TREC-CDS task as a question-answering problem. To our knowledge, we are the first to publish any results based on the potential answers to the 30 topics used in the 2015 TREC-CDS evaluation. The potential answers indicate a single possible answer that the topic author had in mind when designing the topic; as such they do not represent the “best” answer, nor are they guaranteed to be represented in the document collection. Nevertheless, we evaluated the ranked list of answers produced by our system by using the potential answers as the gold-standard. We computed the Mean Reciprocal Rank (MRR) used in previous TREC Q/A evaluations [33], which is the average of the reciprocal (multiplicative inverse) of the rank of the first correct answer retrieved for each topic Table 1 lists the results for evaluating the inferred medical answers when considering each answer inference technique described in Section 4 for each medical sketch z ∈ {Z(t),EZ(t,l)} described in Section 2.
Table 1.
The Mean Reciprocal Rank (MRR) obtained when using each of the 3 inference techniques and each medical sketch.
| Z(t) | ⋆EZ(t,l) | |
|---|---|---|
| ⋆Bethe Approximation | 0.125 | 0.694 |
| Pair-wise Variational | 0.083 | 0.502 |
| Interpolated Smoothing | 0.124 | 0.601 |
When evaluating the answers inferred from each possible medical sketch z and answer inference method, the highest performance was obtained using the Bethe Approximation method for answer inference and relying on the extended medical sketch (EZ(t,l)). Note that the difference between Bethe Approximation and the Interpolated Smoothing method for probabilistic inference was not statistically significant (p<0.001, N =30, Wilcoxon signed-ranked test), however both methods significantly outperformed the pair-wise variational method which strongly suggests that modeling the clinical picture and therapies of patients requires more than pair-wise information about medical concepts. Overall, the answers obtained using Z(t) were of significantly poorer quality than those obtained using EZ(t,l). We observed that the answers produced by Z(t) typically included the most common diseases, tests, or treatments indicated in the EMR collection. This confirms our belief that incorporating the medical knowledge discerned from relevant scientific articles into the sketch yields substantially higher-quality answers. The results of EZ(t,l), from Table 1 show that the inferred answers from the clinical picture and therapy graph correspond reasonably-well to the possible answers generated by the TREC topic creators.
In addition to the Mean Reciprocal Rank shown in Table 1, Figure 7 shows the reciprocal rank of the gold-standard answer for each topic used in the 2015 TREC-CDS evaluation when using Interpolated Smoothing on the answers produced by EZ(t,l). As shown, for the majority of topics, our top-ranked answer was equivalent to the gold-standard answer. We obtained the correct answer for nearly all of the treatment topics (topics 51–60), but for some of the diagnosis topics (31–40), and many of the test topics (41–50) we did not rank the gold answers on the first position. For example, Table 2 illustrates the ten highest-ranked answers produced by our system against the gold answer provided by the TREC topic authors, along with the held-out diagnosis for each topic previously shown in Figure 1.
Figure 7.
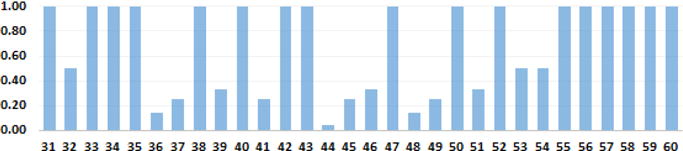
Reciprocal Rank for each topic evaluated in TREC-CDS 2015 based on probabilistic ranking using Interpolated Smoothing applied to the extended medical sketch EZ(t,l)
Table 2.
Examples of answers discovered for the medical cases illustrated in Figure 1
| Topic 32 |
EMAT:
DIAGNOSIS Answers: cytomegalovirus; leishmania donovani; kala-azar; mycobacterium; columbiense; salmonella; interferon-gamma; pneumonitis; lymphocytic alveolitis; pulmonary infection Gold Answer: cytomegalovirus |
| Topic 44 |
EMAT:
TEST Diagnosis: paroxysmal nocturnal hemoglobinuria Answers: Hb electrophoresis; stability tests; genetic workup; renal biopsy; laboratory evaluation; ham test; sugar water tests; phosphatase; cd55; cd59; ultrasonography Gold Answer: flow cytometry |
| Topic 53 |
EMAT:
TREATMENT Diagnosis: Dengue Answers: nonsteroidal anti-inflammatory drugs; fluid replacement; methylprednisolone; acetaminophen; bed rest; isotonic fluids; starch; dextran; albumin; physiotherapy; methotextrate; analgesics Gold Answers: supportive care, analgesics, fluid management |
As shown, for Topic 32, we obtain the correct answer at the highest position. This is because the gold answer, cytomegalovirus was frequently mentioned in relevant scientific articles and had a strong association to the medical case in the CPTG. Topic 44, however, was more difficult and produced the answers with the lowest MRR of any topic processed by our system. In fact, the gold answer, flow cytometry was ranked as the 20-th most likely answer by our system. In analyzing this behavior, we compared the answers produced by our system against a literature review of PubMed articles about paroxysmal nocturnal hemoglobinuria, the diagnosis for the topic. Many of the answers we proposed represent alternative tests recommended by Medline Plus for patients diagnosed with the disease, such as the sugar water test, and Hb electrophoresis. In contrast, the high rank of genetic workup highlights an area for future improvement: certain highly general tests, such as a genetic workup, are likely to have already been considered by the physician. As such, future work may benefit by narrowing the resultant answers to favor rarer diseases, tests, or treatments. For Topic 53, we obtained the correct answer as the second-highest ranked answer. This highlights the ability of our system to discover the gold answer, fluid management, through mentions of the synonymous concept fluid replacement. Unfortunately, the top-ranked answer, nonsteroidal anti-inflammatory drugs (NSAIDs) produced by our system was actually counter-indicated for the diagnosis of Dengue in most articles. That is, although the concept was mentioned with a present assertion in the article, the context indicated that the concept should not be used to treat patients with Dengue. This indicates that the belief values expressed by assertions may not be sufficient for answer inference in all cases.
5.2 Medical Article Retrieval Evaluation
To evaluate the quality of ranked scientific articles, we relied on the relevance judgments produced for the 2015 TREC-CDS topics by Oregon Health and Science University (OHSU). Physicians provided relevance judgments for each of the participating systems by manually reviewing the twenty top-ranked articles as well as a 20% random sample of the articles retrieved between ranks 21 and 100 for each topic. A total of 37,807 topic-article judgments were produced for the 2015 topics. These judgments indicate whether retrieved scientific articles were (1) relevant, (2) partially relevant or (3) non-relevant. In our evaluations, as in the official TREC-CDS evaluations, we did not distinguish between relevant and partially relevant documents, considering only the binary relevance of each article. This allowed us to measure the quality of articles retrieved in terms of four information retrieval metrics also used by TREC: (1) the inferred Average Precision (iAP), wherein retrieved articles were randomly sampled and the Average Precision was calculated as in [37]; (2) the inferred Normalized Discounted Cumulative Gain (iNDCG), wherein retrieved articles were randomly sampled and the NDCG was calculated as per [38]; (3) the R-Precision, which measures the precision of the highest R-retrieved documents, where R is the total number of relevant documents for the topic; and (4) the Precision of the first ten documents retrieved (P@10) [19].
We compared the quality of ranked scientific articles produced by our system when considering the medical sketch (Z(t)) or the extended medical sketch (EZ(t,l)), (2) each of the three answer inference methods reported in Section 4, and relying on (3) five relevance models. BM25 relied on the Okapi-BM25 [24] (k1 =1.2 and b=0.75) relevance model; TF-IDF used the standard term frequency-inverse document frequency vector retrieval relevance model; LMJM and LMDir leveraged language-model ranking functions using Jelinek-Mercer (λ=0.5) or Dirichlet (μ=2,000) smoothing [40], respectively; and DFR considered the Divergence from Randomness framework [1] with an inverse expected document frequency model for information content, a Bernoulli-process normalization of information gain, and Zipfian term frequency normalization. We also compare our performance against the top-performing systems for the 2015 TREC-CDS evaluation for both Task A (in which no explicit diagnoses was provided) and Task B (in which an explicit diagnoses was given for each topic focusing on a medical test and treatment). It should be noted that our system did not incorporate the gold-standard diagnoses given in Task B (i.e. our system was designed for Task A). Moreover, our system relies on only basic query expansion (described in Section 3.1) and a standard relevance model (BM25) while the top-performing systems submitted to the TREC-CDS task relied on significantly more complex methods for query expansion and often incorporated additional information retrieval components (e.g. pseudo-relevance feedback, rank fusion) which were not considered in our architecture [23] As in the official evaluation, we distinguish between automatic systems which involved no human intervention, and manual systems in which arbitrary human intervention was allowed. Table 3 illustrates these results. Clearly, the best performance obtained by our system (denoted with a ‘⋆’) relies on (1) the extended medical sketch (EZ(t,l)), (2) the interpolated-smoothing method for answer inference, and (3) the BM25 ranking function. Note that just as with the answer evaluation, there was no statistically significant difference in the performance obtained when using the Interpolated Smoothing or the Bethe Approximation methods for answer inference. As shown, our Q/A-informed relevance approach yields significantly improved performance to top reported systems for each task [26]. In task A, we obtained a 49% increase in inferred NDCG compared to the best reported automatic system [4] and a 40% increase to the best reported manual system [4]. In task B, in which participants were given the gold-standard diagnosis for every topic (except topics 31–40 in which the purpose was to retrieve documents describing possible diagnoses), we obtained a 14% increase in inferred NDCG compared to the best reported automatic [27] and manual system [39]. This suggests that much of the increased performance obtained by our system in Task A was based on our ability to infer the correct diagnosis. Moreover, it suggests that the ability to infer multiple related medical concepts (beyond the gold-standard diagnosis) can improve the relevance of retrieved scientific articles. This suggests that the relevant articles in the TREC-CDS task considered more answers than only those in the gold-standard set. Moreover, the high performance of our approach clearly demonstrates the impact of medical Q/A for medical clinical decision support.
Table 3.
Performance results obtained for the system reported in this paper (Q/A-CDS) when using each type of medical sketch, method for answer inference, and relevance model as well as the iNDCG obtained by the State-of-the-Art (SotA) automatic and manual systems submitted to TREC.
| iAP | iNDCG | R-Prec | P@10 | ||
|---|---|---|---|---|---|
| Q/A-CDS | Z(t) | .006 | .010 | .020 | .062 |
| ⋆EZ(t,l) | .147 | .434 | .344 | .722 | |
|
| |||||
| Bethe Approximation | .140 | .432 | .336 | .701 | |
| Pair-wise | .128 | .382 | .330 | .610 | |
| ⋆Interpolated | .147 | .434 | .344 | .722 | |
|
| |||||
| ⋆BM25 | .042 | .204 | .163 | .387 | |
| TF-IDF | .041 | .197 | .169 | .350 | |
| LMJM | 0.040 | .193 | .151 | .357 | |
| LMDir | 0.043 | .203 | .170 | .360 | |
| DFR | .039 | .197 | .167 | .333 | |
|
| |||||
| SotA | Task A Automatic | – | .294 | – | – |
| Task A Manual | – | .311 | – | – | |
|
| |||||
| Task B Automatic | – | .382 | – | – | |
| Task B Manual | – | .381 | – | – | |
5.3 Medical Knowledge Evaluation
The clinical picture and therapy graph (CPTG) that we have automatically generated contained 634 thousand nodes and 13.9 billion edges with 31.2% of all nodes being diagnoses, 21.84% being signs or symptoms, 23.62% encoding medical tests, and 23.34% of nodes encoding medical treatments. The distribution of assertions that we obtained were: 13.1% were absent, 0.01% were ASSOCIATED-WITH-SOMEONE-ELSE, 1.13% were CONDITIONAL, 33.31% were CONDUCTED, 17.05% were HISTORICAL, 0.72% were HYPOTHETICAL, 8.37% were ONGOING, 1.04% were ORDERED, 0.55% were POSSIBLE, 1.12% were PRESCRIBED, 22.34% were PRESENT, and 0.89% were SUGGESTED. Using the 2010 i2b2/VA shared-task annotations, medical concept detection achieved an F1-score of 79.59%, while assertion classification obtained 92.75%. We evaluated our new assertion types using 10-fold cross validation against the 2,349 annotations we created and obtained an accuracy of 75.99%. Evaluating the quality of the edges contained in the CPTG was prevented by the fact all nodes in the CPTG are qualified by their assertions while no medical ontologies capture relations between concepts with these types of beliefs.
6. CONCLUSIONS
In this paper, a novel medical Q/A framework is presented in which answers are probabilistically inferred from an automatically derived medical knowledge graph. We experimented with three probabilistic inference methods, which enabled the identification of answers with surprisingly high MRR scores when evaluating the questions from the 2015 TREC-CDS task. Although the questions were related to complex medical cases, the results that were obtained rivaled the performance of Q/A results obtained for simpler, factoid questions. To our knowledge, the medical Q/A framework presented in this paper is the first to address the feasibility of identifying the answers to the TREC-CDS questions instead of providing a ranked list of articles from PubMed where the answers can be found. We also explored the quality of the answers obtained to the medical questions from an automatically derived medical knowledge graph in two cases: (1) when considering the medical topic by itself and (2) when considering both the medical topic and a relevant scientific article. The second case proved to be far more successful than the first one, indicating that successful medical Q/A from knowledge bases need to combine three sources of knowledge: (1) knowledge of the medical case; (2) knowledge from scientific articles (reflecting knowledge developed in medical research); and (3) knowledge from a large EMR collection (reflecting knowledge acquired during medical practice). Moreover, when the answers of a medical question are known, they inform the ranking of relevant articles from PubMed with 40% increased inferred Average Precision to current state-of-the-art systems evaluated in the most recent TREC-CDS.
Acknowledgments
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
All participants in the 2015 TREC-CDS track were provided with the correct answers to the topics a few months after the evaluation.
Although we could have considered every possible PubMed article, we limited our experiments to only the top one-thousand articles to improve computational efficiency.
Although we have used stochastic gradient descent, any method for convex optimization may be used.
References
- 1.Amati G, Van Rijsbergen CJ. Probabilistic models of information retrieval based on measuring the divergence from randomness. Transactions on Information Systems, TOIS. 2002;20(4):357–389. [Google Scholar]
- 2.Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. In AMIA, page. 2001:17. [PMC free article] [PubMed] [Google Scholar]
- 3.Auer S, Bizer C, Kobilarov G, Lehmann J, Cyganiak R, Ives Z. Dbpedia: A nucleus for a web of open data Springer. 2007 [Google Scholar]
- 4.Balaneshin-kordan S, Kotov A, Xisto R. Wsu-ir at trec 2015 clinical decision support track: Joint weighting of explicit and latent medical query concepts from diverse sources. Text Retrieval Conference, TREC. 2015 [Google Scholar]
- 5.Bao J, Duan N, Zhou M, Zhao T. Knowledge-based question answering as machine translation. Cell. 2014;2(6) [Google Scholar]
- 6.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollacker K, Evans C, Paritosh P, Sturge T, Taylor J. International Conference on Management of Data, SIGMOD. ACM; 2008. Freebase: a collaboratively created graph database for structuring human knowledge; pp. 1247–1250. [Google Scholar]
- 8.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 9.Dong L, Wei F, Zhou M, Xu K. Question answering over freebase with multi-column convolutional neural networks. Association for Computational Linguistics, ACL. 2015;1:260–269. [Google Scholar]
- 10.Fellbaum C. WordNet: An Electronic Lexical Database. The MIT press; 1998. [Google Scholar]
- 11.Garg AX, Adhikari NK, McDonald H, Rosas-Arellano MP, Devereaux P, Beyene J, Sam J, Haynes RB. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: a systematic review. Journal of the American Medical Association, JAMA. 2005;293(10):1223–1238. doi: 10.1001/jama.293.10.1223. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin T, Harabagiu SM. Graphical induction of qualified medical knowledge. International Journal of Semantic Computing, IJSC. 2013;7(04):377–405. [Google Scholar]
- 13.Goodwin T, Harabagiu SM. Clinical data-driven probabilistic graph processing. LREC. 2014:101–108. [Google Scholar]
- 14.Kim J-D, Ohta T, Tateisi Y, Tsujii J. Genia corpus – a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl 1):i180–i182. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 15.Kingsbury P, Palmer M. From treebank to propbank. In LREC Citeseer. 2002 [Google Scholar]
- 16.Koller D, Friedman N. Probabilistic graphical models: principles and techniques. MIT press; 2009. [Google Scholar]
- 17.Lee J, Scott DJ, Villarroel M, Clifford GD, Saeed M, Mark RG. Engineering in Medicine and Biology Society, EMBC. IEEE; 2011. Open-access mimic-ii database for intensive care research; pp. 8315–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipscomb CE. Medical subject headings (mesh) Bulletin of the Medical Library Association. 2000;88(3):265. [PMC free article] [PubMed] [Google Scholar]
- 19.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. Vol. 1. Cambridge university press; Cambridge: 2008. [Google Scholar]
- 20.Pearl J. Fusion, propagation, and structuring in belief networks. Artificial intelligence. 1986;29(3):241–288. [Google Scholar]
- 21.Recht B, Re C, Wright S, Niu F. Hogwild: A lock-free approach to parallelizing stochastic gradient descent. Advances in Neural Information Processing Systems, NIPS. 2011:693–701. [Google Scholar]
- 22.Roberts K, Harabagiu S. A flexible framework for deriving assertions from electronic medical records. Journal of the American Medical Informatics Association. 2011;18(5):568–573. doi: 10.1136/amiajnl-2011-000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts K, Simpson M, Demner-Fushman D, Voorhees E, Hersh W. State-of-the-art in biomedical literature retrieval for clinical cases: a survey of the trec 2014 cds track. Information Retrieval Journal. 2014:1–36. [Google Scholar]
- 24.Robertson SE, Walker S, Jones S, Hancock-Beaulieu MM, Gatford M, et al. Okapi at trec-3. Text Retrieval Conference, TREC. 1995:109–109. [Google Scholar]
- 25.Scheuermann RH, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. Proceedings of the 2009 AMIA Summit on Translational Bioinformatics. 2009;2009:116–120. [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson MS, Voorhees E, Hersh W. Overview of the trec 2014 clinical decision support track. Text Retrieval Conference, TREC. 2014 [Google Scholar]
- 27.Song Y, He Y, Hu Q, He L. Ecnu at 2015 cds track: Two re-ranking methods in medical information retrieval. Proceedings of the 2015 Text Retrieval Conference. 2015 [Google Scholar]
- 28.Stone PJ, Dunphy DC, Smith MS. The general inquirer: A computer approach to content analysis. 1966 [PubMed] [Google Scholar]
- 29.Surdeanu M, Turmo J. Proceedings of the Ninth Conference on Computational Natural Language Learning. Association for Computational Linguistics; 2005. Semantic role labeling using complete syntactic analysis; pp. 221–224. [Google Scholar]
- 30.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association, JAMIA. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varmus H, Lipman D, Brown P. Pubmed central: An nih-operated site for electronic distribution of life sciences research reports. Vol. 24. Washington DC: National Institutes of Health Retrieved December; 1999. p. 1999. [Google Scholar]
- 32.Vontobel PO. Counting in graph covers: A combinatorial characterization of the bethe entropy function. Information Theory, IEEE Transactions on. 2013;59(9):6018–6048. [Google Scholar]
- 33.Voorhees EM, et al. The trec-8 question answering track report. In Text Retrieval Conference, TREC. 1999;99:77–82. [Google Scholar]
- 34.Woods WA. In Proceedings of the June 4–8, 1973, national computer conference and exposition. ACM; 1973. Progress in natural language understanding: an application to lunar geology; pp. 441–450. [Google Scholar]
- 35.Yao X, Van Durme B. ACL. Citeseer; 2014. Information extraction over structured data: Question answering with freebase; pp. 956–966. [Google Scholar]
- 36.Yedidia JS, Freeman WT, Weiss Y. Constructing free-energy approximations and generalized belief propagation algorithms. Information Theory, IEEE Transactions on. 2005;51(7):2282–2312. [Google Scholar]
- 37.Yilmaz E, Aslam JA. Proceedings of the 15th ACM international conference on Information and knowledge management. ACM; 2006. Estimating average precision with incomplete and imperfect judgments; pp. 102–111. [Google Scholar]
- 38.Yilmaz E, Kanoulas E, Aslam JA. Proceedings of the 31st annual international ACM SIGIR conference on Research and development in information retrieval. ACM; 2008. A simple and efficient sampling method for estimating ap and ndcg; pp. 603–610. [Google Scholar]
- 39.You R, Zhou Y, Peng S, Zhu S, China R. Fdumedsearch at trec 2015 clinical decision support track. In Text Retrieval Conference, TREC. 2015 [Google Scholar]
- 40.Zhai C, Lafferty J. Special Interest Group on Information Retrieval, SIGIR. ACM; 2001. A study of smoothing methods for language models applied to ad hoc information retrieval; pp. 334–342. [Google Scholar]


