Abstract
Small-molecule inhibitors of DNA repair pathways are being intensively investigated as primary and adjuvant chemotherapies. We report the discovery that cardiac glycosides, natural products in clinical use for the treatment of heart failure and atrial arrhythmia, are potent inhibitors of DNA double-strand break (DSB) repair. Our data suggest that cardiac glycosides interact with phosphorylated mediator of DNA damage checkpoint protein 1 (phospho-MDC1) or E3 ubiquitin–protein ligase ring finger protein 8 (RNF8), two factors involved in DSB repair, and inhibit the retention of p53 binding protein 1 (53BP1) at the site of DSBs. These observations provide an explanation for the anticancer activity of this class of compounds, which has remained poorly understood for decades, and provide guidance for their clinical applications. This discovery was enabled by the development of the first high-throughput unbiased cellular assay to identify new small-molecule inhibitors of DSB repair. Our assay is based on the fully automated, time-resolved quantification of phospho-SER139-H2AX (γH2AX) and 53BP1 foci, two factors involved in the DNA damage response network, in cells treated with small molecules and ionizing radiation (IR). This primary assay is supplemented by robust secondary assays that establish lead compound potencies and provide further insights into their mechanisms of action. Although the cardiac glycosides were identified in an evaluation of 2366 small molecules, the assay is envisioned to be adaptable to larger compound libraries. The assay is shown to be compatible with small-molecule DNA cleaving agents, such as bleomycin, neocarzinostatin chromophore, and lomaiviticin A, in place of IR.
INTRODUCTION
DNA is an established target for chemotherapeutic intervention; approximately 70% of small-molecule anticancer agents target DNA.1 Among the many different DNA lesions, DNA double-strand breaks (DSBs) are the most deleterious.2 It has been estimated that a single unrepaired DSB is sufficient to induce apoptosis.3 DNA DSBs are resolved by the non-homologous end joining (NHEJ)4 and homologous recombination (HR)5 repair pathways, which are part of the cellular DNA damage response (DDR) network. Sporadic and hereditary DDR mutations are widespread in many tumors,6 and while these mutations drive tumorigenesis, they also provide a context in which to obtain selectivity, as inhibition of a functional DDR pathway in transformed cells is selectively toxic because of decreased genetic buffering (synthetic lethality).7 Healthy cells, which are less reliant on the DDR (partly because of lower rates of proliferation), are not sensitized to the same degree.3,8 Consequently, small-molecule inhibitors of NHEJ or HR repair (and other DDR pathways) are of great interest and are in clinical development.8,9
Although the potential of DDR inhibitors as primary or adjuvant chemotherapies is now widely appreciated, only a single DNA repair inhibitor, olaparib,10 has been approved for clinical use. The slow progress in this area may be due to the nature of prior discovery efforts, which have focused on identifying molecules that inhibit specific factors in vitro. For example, poly(ADP-ribose) polymerase 1 (PARP1) inhibitors,11 which ushered in the era of DNA repair as a therapeutic target,12 and DNA protein kinase catalytic subunit (DNA-PKcs) inhibitors13 have been discovered using enzymatic assays, but permeability, toxicity, and solubility limitations have impeded their clinical use.14 A small number of whole-cell assays have been reported, but these have focused on specific factors, such as ataxia telangiectasia and Rad3-related (ATR)15 or ataxia telangiectasia mutated (ATM)16 kinases. To our knowledge, an unbiased high-throughput cellular assay to discover small-molecule DNA repair inhibitors has not been reported.
Here we disclose a new high-content, high-throughput cellular assay for the unbiased discovery of NHEJ and HR repair inhibitors. Our approach is enabled by the development of a time-resolved method to automatically and concurrently monitor the production and resolution of phospho-SER139-H2AX (γH2AX) and p53 binding protein 1 (53BP1) foci in cells treated with ionizing radiation (IR) and candidate DDR inhibitors. γH2AX17 and 53BP118 are key DDR components that recruit many other mediator and effector proteins and chromatin-modifying complexes to DSBs. γH2AX is formed early in the DDR, amplifies the DNA damage signal, and localizes several NHEJ and HR proteins, including 53BP1, onto the damaged DNA.17d 53BP1 recognizes a unique DSB-specific histone code and acts in conjunction with factors downstream of ATM to promote NHEJ and suppress HR repair.18d γH2AX and 53BP1 form cytologically detectable foci that can be visualized by immunofluorescence microscopy.19 As DSBs are ameliorated, feedback mechanisms terminate the DDR, leading to dissipation of these foci.20 Consequently, cellular levels of NHEJ or HR repair activity can be determined indirectly by measuring the kinetics of 53BP1 and γH2AX foci formation and resolution. As γH2AX is formed early in the DDR, delays in γH2AX foci resolution correlate with decreased DNA repair. On the other hand, as 53BP1 foci are formed later in the DDR, modulation of DNA repair activity may be expected to impede the formation of 53BP1 foci or their resolution. The concurrent monitoring of two distinct repair factors can provide insight into the point at which the pathways are disrupted. These studies have led to the discovery of cardiac glycosides, natural products in clinical use for the treatment of heart failure and atrial arrhythmia, as lead compounds for modulation of NHEJ and HR activity. This work provides an explanation for the longstanding but poorly understood anticancer activity of these compounds and suggests their application to treat DDR-deficient tumor types.
MATERIALS AND METHODS
Cell Lines and Reagents
U2OS and T98G cell lines were obtained from American Type Culture Collection (ATCC). U2OS cells were cultured in McCoy’s 5A medium (Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Life Technologies). T98G cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies) with 10% FBS. All cells were maintained at 37 °C with 5% CO2. NU7441 and BEZ-235 were purchased from Fisher Scientific and Santa Cruz Biotechnology, respectively. Strophanthidin (1) (Sigma-Aldrich), ouabain (2) (MP Biomedicals), lanatoside C (3) (Sigma-Aldrich), digoxin (4) (Sigma-Aldrich), ouabagenin (5) (Sigma-Aldrich), and digoxigenin (6) (Sigma-Aldrich) were purchased as dry powders and diluted in dimethyl sulfoxide (DMSO) for dose–response studies.
Library Compounds
We employed the NIH Clinical Collection (BioFocus DPI), the FDA Approved Drug Library (ENZO Life Sciences), and a Yale Center for Molecular Discovery Yale Procured Drugs custom collection, which total 2366 compounds. All of the compounds were stored as 10 mM solutions in DMSO at −20 °C before use.
Assay Protocol
Cells were seeded at 4000 cells per well to achieve total well volumes of 20 µL in 384-well plates (black with optically clear bottom, PerkinElmer) using a Thermo Combidrop liquid dispenser. Cells were grown for 72 h, followed by the addition of library compounds using a 384-head pin tool with quills (V&P Scientific, Inc.) on the Aquarius liquid transfer robot (Tecan). A 20 nL aliquot of each compound stock solution (10 mM in DMSO) was added to 20 µL of cells to provide final compound and DMSO concentrations of 10 µM and 0.1%, respectively. Each plate contained 16 negative vehicle control wells (0.1% DMSO) and 16 positive control wells (1 µM BEZ-235 or 5 µM NU7441). The cells were incubated with the compounds for 1 h and then irradiated with 10 Gy IR using an X-RAD KV irradiator (Precision X-ray). Following irradiation, the cells were incubated for an additional 24 h (or shorter time points for time course experiments) and then fixed and subjected to immunofluorescence.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in the presence of 0.02% Triton X-100 at room temperature for 20 min and then incubated in permeabilization/blocking solution (10% FBS, 0.5% Triton X-100 in phosphate-buffered saline (PBS)) at room temperature for 1 h. Primary antibodies [phospho-specific H2AX (Upstate 05-636), 53BP1 (Novus Biologicals NB100-904SS), phospho-specific DNA-PKcs (Abcam ab4194), and breast cancer 1 (BRCA1) (Santa Cruz sc-6954)] were diluted 1:500 in permeabilization/blocking solution and used to stain cells at 4 °C overnight. The secondary antibodies used were Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (IgG) and Alexa Fluor 647-conjugated goat anti-rabbit IgG (Molecular Probes). Cells were costained with the nucleic acid dye Hoechst 33342 (HOECHST) (λem = 460 nm) to visualize the nuclei. Immunofluorescence staining was performed in 384-well plates using liquid handlers.
Imaging and Image Analysis
Cells were imaged using the InCell 2200 Imaging System (GE Corporation). The automated image analysis protocol for the quantitative assessment of γH2AX and 53BP1 foci was developed using the InCell Analyzer software (GE Corporation). Nuclei were segmented on the basis of the nuclear HOECHST staining channel, and foci were identified on the basis of the 53BP1 or γH2AX fluorescence channel. Nuclei and foci were then linked, and the number of foci per nucleus was counted. Cell-level distributions of number of foci per nucleus were generated from at least 450–600 cells/well. A threshold of ≥15 foci per nucleus was set as defining γH2AX- and/or 53BP1-positive cells.
Data Analysis
To evaluate the robustness of the screen, Z′ factors were calculated from the mean signals of the positive and negative controls (μc+ and μc−) and their standard deviations (σc+ and σc−) for each plate using the formula Z′ = 1 − [3(σc+ + σc−)/|μc+ − μc−|].21 Primary screening data were analyzed using the commercial software and database package ActvityBase (IDBS). The effect of each library compound was calculated as the normalized percent effect using the following formula: percent effect = [(sample − μ)/(μ − μ)] × c− c+ c− 100%. Histograms of normalized data for the entire screening population were plotted using the JMP software (SAS). The hit threshold was defined as 3 standard deviations beyond the mean of the normalized percent effect values of all screened library compounds. Compounds satisfying the 3 standard deviation cutoff were selected as screen actives, arrayed into a hitpick plate, and retested in four replicates. Selected hits were purchased as powders and assayed in dose–response studies in 384-well plates.
Secondary DSB Repair Assays
The green fluorescent protein (GFP)/red fluorescent protein (RFP)-based DSB repair assays were performed using a U2OS cell line with stably integrated NHEJ and HR repair reporters (termed the EJ-DR assay) combined with a ligand-dependent I-SceI (ddSceGR), as previously described.22 Ligand-induced DNA cleavage by ddSceGR was performed by adding the Shield1 and triamcinolone acetonide (TA) ligands at concentrations of 0.5–1 µM and 100 nM, respectively, to the cell cultures arrayed in 96-well microplates. Compounds were then added at the indicated concentrations. Ligands and compounds were incubated in the cells for 24 h, followed by 1–2 washes with DMEM containing 10% FBS without ligands. The NHEJ and HR repair activities were assessed by quantification of the percentages of DsRed+ and GFP+ cells, respectively, using a FACScan flow cytometer (Becton, Dickinson and Company; BD) at the indicated times. Selected experiments were analyzed with an automated fluorescence imager (Cytation3, Biotek Instruments) as previously described.23 Standard compensation techniques were used when GFP and DsRed were analyzed simultaneously in order to minimize spectral overlap. DsRed+, GFP+, and parental cells were used as controls for optimization. The data were analyzed using FloJo (Tree Star, Inc.). Experiments were performed in either triplicate or quadruplicate, and error bars represent standard errors of the mean (SEM).
Cell-Cycle Profiling
Cell-cycle phase distributions were assessed by analysis of the DNA content using HOECHST staining of cell nuclei as previously described.22,23 In these experiments, cells were fixed in 70% ethanol at the indicated time points in 96-well microplates, washed with PBS, and then stained with 1 µg/mL HOECHST dye. HOECHST fluorescence was quantified using the Cytation3 automated fluorescence imager. Data analysis was performed using standard FloJo cell-cycle profiling tools.
RESULTS
Development of an Automated γH2AX and 53BP1 Staining and Imaging Protocol
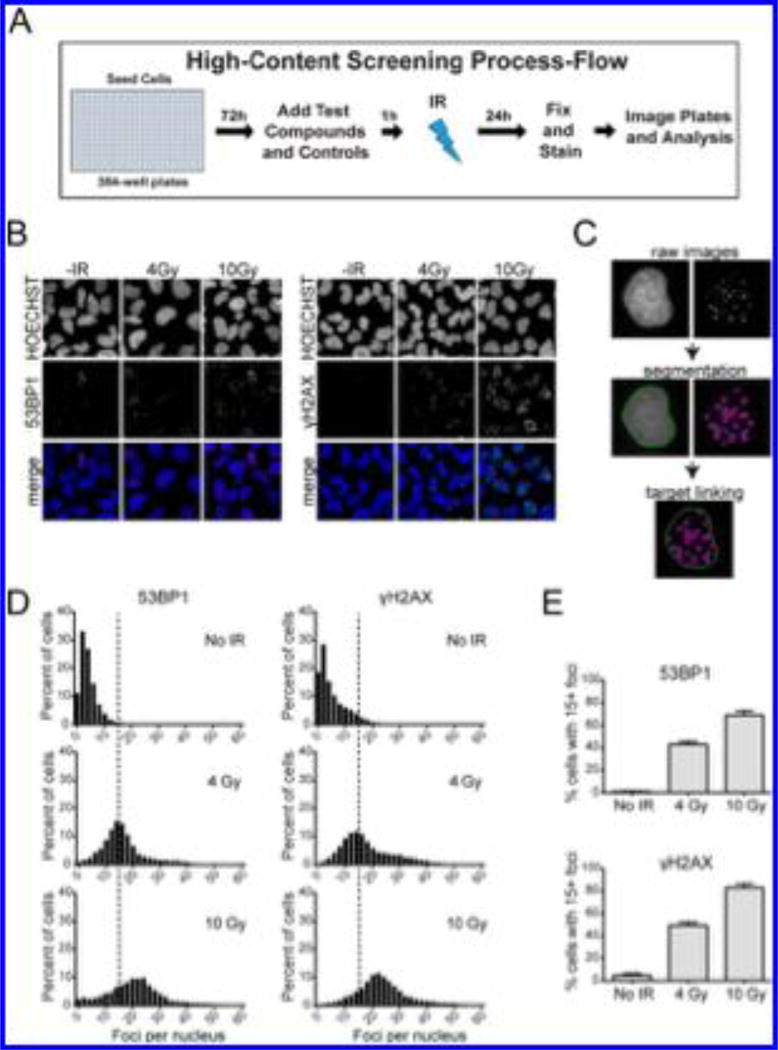
Figure 1A depicts a schematic of the primary assay. Cells were seeded in 384-well plates and grown in log phase for 72 h. Test compounds were then added, and the cells were incubated for an additional 1 h, after which the plates were irradiated. The irradiated plates were incubated for 24 h, followed by fixing, staining, and automated imaging and analysis.
Figure 1.
Development of the high-throughput primary assay for quantification of 53BP1 and γH2AX foci in U2OS cells. (A) Schematic depicting the workflow of the assay. (B) InCell 2200 images of 53BP1 and γH2AX foci in cells exposed to 4 or 10 Gy IR and analyzed 5 h postirradiation. HOECHST dye was used to stain nuclei. (C) Example of segmentation and linking of nuclei and foci using image analysis algorithm developed using the InCell Analyzer software. (D) Distribution of cell population as a function of 53BP1 and γH2AX foci number. Histograms for control cells (no IR) and cells subjected to 4 or 10 Gy IR, and analyzed 5 h postirradiation are shown. Dashed line represents 15 foci threshold described in the main text. (E) Percent of cells with ≥15 53BP1 or γH2AX foci after treatment with 4 or 10 Gy IR.
Optimization studies employed the human osteosarcoma cell line U2OS on the basis of the experience of our group and others using these cells in studies of DDR.22,24 Confirmatory studies using a second cell line are described below. A protocol to simultaneously detect IR-induced 53BP1 and γH2AX foci in a 384-well format using an automated high-content microscope was first developed. Cell density, timing and dose of IR, fixation and permeabilization conditions, and γH2AX and 53BP1 antibody staining conditions were optimized (data not shown). Exposure of U2OS cells to 0, 4, or 10 Gy IR followed by staining and imaging under optimized protocols revealed that the levels of γH2AX and 53BP1 foci increased with increasing IR (Figure 1B).
To enable automated image analysis (Figure 1C), the nuclei were first segmented on the basis of the HOECHST staining channel. Foci were identified on the basis of the 53BP1 and γH2AX fluorescence channels. The nuclei and foci were then linked, and the image features were quantified. The percentages of cells containing 0–60 53BP1 or γH2AX foci were then plotted to generate cell-level histograms from a large statistically robust population (at least 450–600 cells/well). As expected, the number of γH2AX and 53BP1 foci per cell increased with increasing IR (Figure 1D). A threshold of ≥15 foci per nucleus (dashed vertical lines in Figure 1D) was used to define cells as γH2AX- or 53BP1-positive. The γH2AX threshold excludes cells with low levels of DNA damage foci resulting from active replication and metabolism.25 By this approach the percentages of γH2AX- and 53BP1-positive cells could be determined with high precision, and robust and reproducible differences between irradiated and control cells were observed (Figure 1E).
Identical experiments employing the human glioblastoma cell line T98G provided precise and reproducible data (Supplementary Figure 1). Alternatively, use of the molecular DNA-cleavage agents neocarzinostatin,26 etoposide,26 bleomycin,26 and lomaiviticin A30,31 in place of IR led to dose-dependent increases in γH2AX and 53BP1 foci (Supplementary Figures 2–4). Pan-γH2AX staining was observed without an attendant increase in 53BP1 foci at high concentrations of these agents, which is suggestive of induction of apoptosis.27 This result underscores the utility of analyzing both γH2AX and 53BP1 foci simultaneously to distinguish DSB induction from apoptosis.
Characterization of Foci Production and Resolution Kinetics
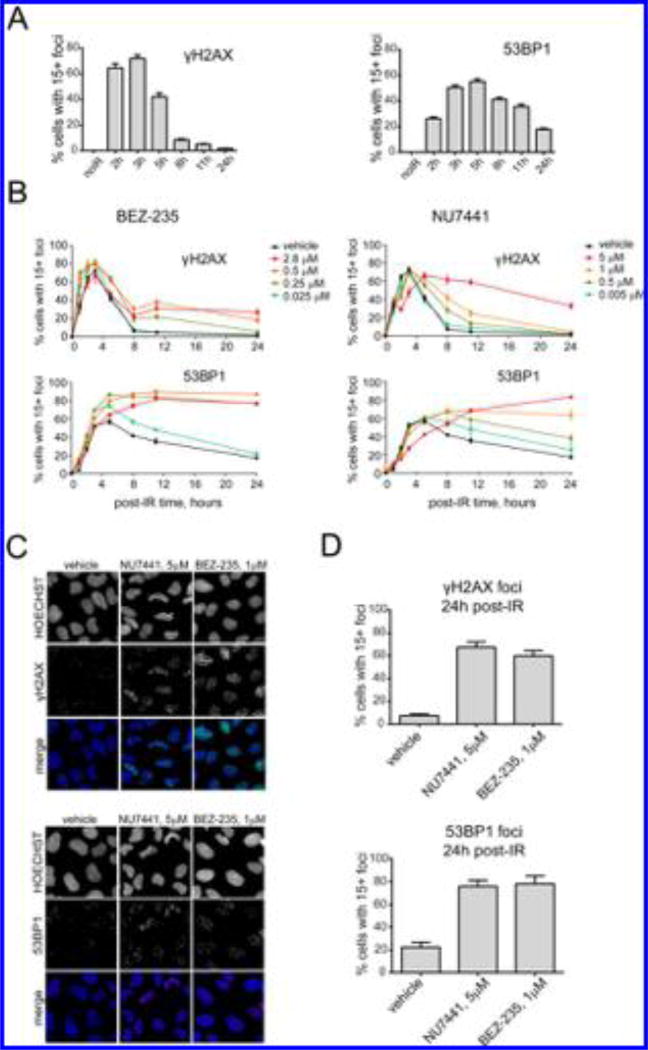
The rates of DNA damage foci production and resolution were characterized to identify the optimal time point to assay for activity. Cells were exposed to 10 Gy IR and analyzed for 53BP1 and γH2AX foci at 2–24 h after irradiation. The numbers of foci-positive cells peaked at 3 and 5 h for γH2AX and 53BP1, respectively, and then decreased over 5–24 h (Figure 2A). 53BP1 foci were more persistent, and resolution was incomplete after 24 h. Histogram data for each of the data points shown in Figure 2A are presented in Supplementary Figure 5A. These data illustrate the wide distribution in the number of foci per nucleus at each time point, emphasizing the utility of using a 15+ foci threshold and scoring percent of positive cells (i.e., cells with 15+ foci) as opposed to the mean number of foci per nucleus. Representative raw images of γH2AX and 53BP1 foci for each time point postirradiation are shown in Supplementary Figure 5B. As the numbers of DNA damage foci-positive cells reached a maximum between 3 and 5 h, we reasoned that later time points would be most optimal to detect small-molecule-induced changes in foci resolution.
Figure 2.
Validation of the foci-based screening assay using known DSB repair inhibitors. (A) Percent of cells with ≥15 γH2AX (left) or 53BP1 (right) foci as a function of time after exposure to 10 Gy IR. (B) Effects of BEZ-235 (left) and NU7441 (right) on resolution of γH2AX and 53BP1 foci. Cells were pretreated with BEZ-235 (0.025–2.8 µM) or NU7441 (0.005–5 µM) for 1 h and then subjected to 10 Gy irradiation. The percentages of cells with ≥15 foci are shown as functions of time postirradiation. (C, D) Impaired foci resolution in cells treated with DNA repair inhibitors. Shown are (C) immunofluorescence images and (D) percentages of cells with ≥15 53BP1 or γH2AX foci in cell populations pretreated with 5 µM NU7441 or 1 µM BEZ-235 for 1 h, exposed to 10 Gy IR, and analyzed 24 h postirradiation.
Assay Validation Using Known DSB Repair and DNA Damage Checkpoint Inhibitors
We then sought to test whether delays in γH2AX and 53BP1 foci resolution using known DSB repair and DNA damage checkpoint inhibitors could be detected. The DNA-PKcs inhibitors NU744128 and BEZ-23529 were employed, since they are known to block NHEJ repair after exposure to IR.30 Both drugs are chemo- and radiosensitizers in vitro and in tumor xenografts in vivo,13,14,28,29,31 and studies have demonstrated the persistence of 53BP1 and γH2AX foci as a result of impaired DNA repair in cells treated with NU7441.32 BEZ-235 (0.025–1 µM) and NU7441 (0.05–5 µM) induced dose-dependent delays in 53BP1 and γH2AX foci resolution after exposure to 10 Gy IR (Figure 2B,C), and 60–80% of the cells treated with 5 µM NU7441 or 1 µM BEZ-235 displayed ≥15 foci per nucleus after 24 h, compared with 5–20% observed in control cells (Figure 2D). These data suggested 24 h as a suitable point to probe for modulation of repair activity. Images of foci at 24 h postirradiation are shown in Supplementary Figure 6A.
BEZ-235 and NU7441 also delayed DNA-PKcs foci resolution (Supplementary Figure 6), providing further confirmation that the delays in γH2AX and 53BP1 foci resolution were due to impairment of DSB repair. We also evaluated several other known DSB repair and checkpoint inhibitors in analogous time-dependent dose–response experiments, including mirin (Mre11 inhibitor),33 VE-821 (ATR inhibitor),34 SAHA (HDAC inhibitor),35 KU55933 (ATM inhibitor),36 and TCS2312 (CHK1 inhibitor).37 All of these compounds affected γH2AX and/or 53BP1 foci clearance after irradiation (Supplementary Figure 7). Collectively, these data supported the ability of this screening approach to identify DSB modulators, led to the selection of BEZ-235 and NU7441 as positive controls, and identified 24 h after irradiation as the optimal time point to assay for delays in repair.
Small-Molecule Screen for Identification of Novel DSB Repair Inhibitors
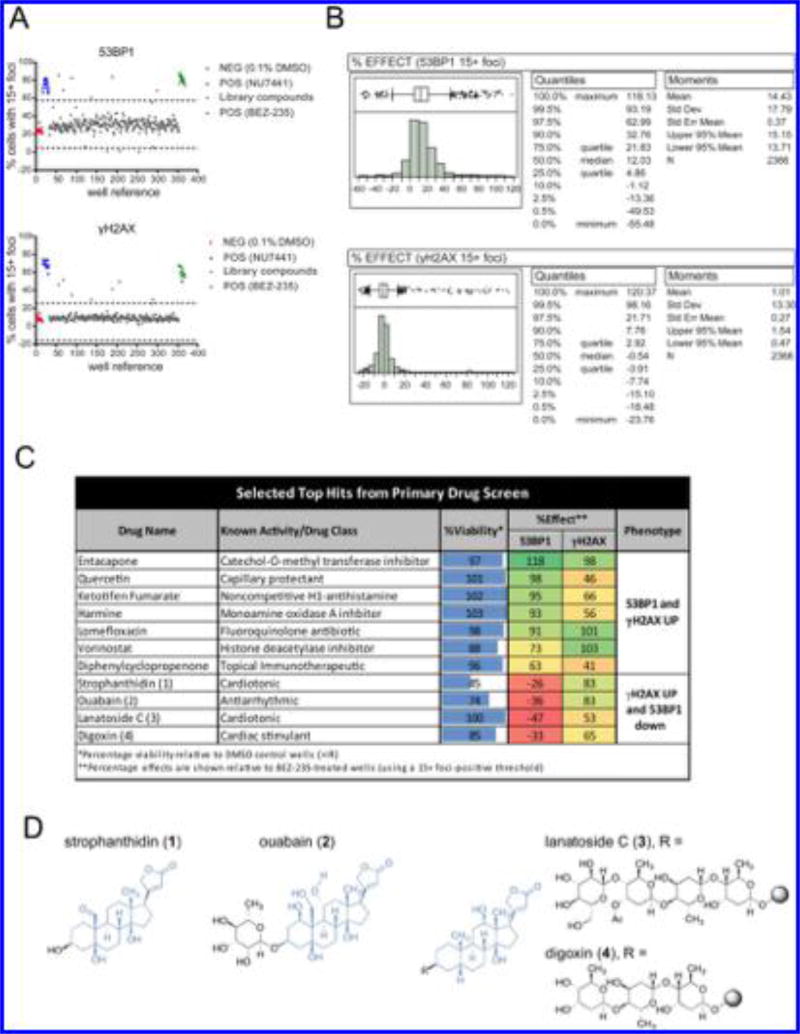
The assay parameters developed above were applied to a screen of 2366 structurally diverse small molecules. U20S cells were seeded in 384-well plates, cultured for 72 h, and treated with library compounds (10 µM). After 1 h incubation, the cells were exposed to 10 Gy IR, incubated for an additional 24 h, fixed, stained, and analyzed as above. Scatter plots of raw data (Figure 3A) showed low variability and significant separation of the signals for positive (BEZ-235 or NU7441) and negative (DMSO) control populations. The majority of compounds had negligible activity. The means and standard deviations of the control samples were used to calculate signal-to-background (S/B), coefficient of variation (CV), and Z′ factors for each screening plate (Supplementary Figure 8).21 In cells treated with NU7441, an average CV of 5% (range = 3–10%) and an average Z′ of 0.6 (range = 0.47–0.69) were observed for the 53BP1 phenotype. The γH2AX phenotype had an average CV of 7% (range = 5–9%) and an average Z′ of 0.68 (range = 0.56–0.8). In cells treated with BEZ-235, an average CV of 5.1% (range = 4–6%) and an average Z′ of 0.61 (range = 0.46–0.72) were observed for the 53BP1 phenotype. The γH2AX phenotype had an average CV of 11% (range = 6–14%) and an average Z′ of 0.5 (range = 0.39–0.73).
Figure 3.
Results of a screen of 2366 bioactive compounds. (A) Scatter plots of raw data (percentages of cells with ≥15 53BP1 or γH2AX foci) from a representative screening plate. Blue and green data points correspond to wells treated with 5 µM NU7441 or 1 µM BEZ-235, respectively. Gray data points represent the screening population, and red points represent vehicle (negative control). (B) Histograms of normalized data (percent effect relative to 1 µM BEZ-235) for the entire screening population. Quantiles and moments for all of the tested compounds (n = 2366) are shown on the right. (C) Table of selected top hits satisfying the 3 standard deviation cutoff. Normalized viabilities and effects on 53BP1 or γH2AX foci observed in the primary screen are shown. (D) Chemical structures of cardiac glycosides 1–4.
To enable data comparison across the entire screen, the raw data from each plate were normalized to the control data from the same plate, where the mean of 16 BEZ-235 wells was set as 100% effect and the mean of 16 DMSO vehicle control wells was set as 0% effect. The percent effect values across all of the compounds were nearly normally distributed for γH2AX and 53BP1 foci (Figure 3B), with a handful of compounds displaying activity comparable to that of the positive controls. A hit threshold of 3 standard deviations beyond the mean of the normalized values of the entire population was defined. This threshold corresponds to 67.8% effect relative to BEZ-235 for the 53BP1 foci phenotype and 41% effect relative to BEZ-235 for the γH2AX foci phenotype. With this threshold, 47 compounds that inhibited 53BP1 foci resolution and 46 compounds that inhibited γH2AX foci resolution were identified. Nineteen compounds delayed resolution of both γH2AX and 53BP1. Seventeen hits suppressed 53BP1 foci levels. (All of the primary screen hits are presented in Supplementary Data File 1.) The hits that suppressed 53BP1 foci and simultaneously delayed γH2AX foci were deemed to be of special interest as DSB repair modulators. As noted in the Introduction, 53BP1 foci are formed downstream of γH2AX, so suppression of 53BP1 foci with concomitant delay of γH2AX foci resolution suggests a target between these two factors (discussed further below). Several known DNA-damaging agents, such as bleomycin, mitomycin, camptothecin, and etoposide, were identified as screen actives (Supplementary Figure 9). While these compounds induce DNA damage rather than inhibit DSB repair, their detection serves as further confirmation of the accuracy of the assay.
Hit Validation
Fifteen γH2AX hits and 15 53BP1 hits showed reproducible effects upon retesting (~30% validation rate; see Supplementary Data File 2), and several compounds delayed both γH2AX and 53BP1 foci resolution at levels comparable to that observed for BEZ-235. Primary hits that showed reproducible effects and are not known DNA-damaging agents are shown in Figure 3C. Representative images for selected hits are shown in Supplementary Figure 10.
Vorinostat38 and quercetin39 are known to possess DDR inhibitory activity, and the identification of these compounds supports the sensitivity of the approach. The cardiac glycosides strophanthidin (1), ouabain (2), lanatoside C (3), and digoxin (4) (Figure 3D) uniquely elevated γH2AX foci with concomitant suppression of 53BP1 foci levels. These hits were of great interest because they suggested a functional suppression of proximal DNA damage signaling (53BP1 foci suppression) and disruption of DSB rejoining (delayed γH2AX foci resolution). The structural similarity of the aglycon residues of 1–4 (shown in blue) suggests specific recognition with a DDR target. As 3 and 4 contain an identical aglycon substructure, we focused on 3 for further studies.
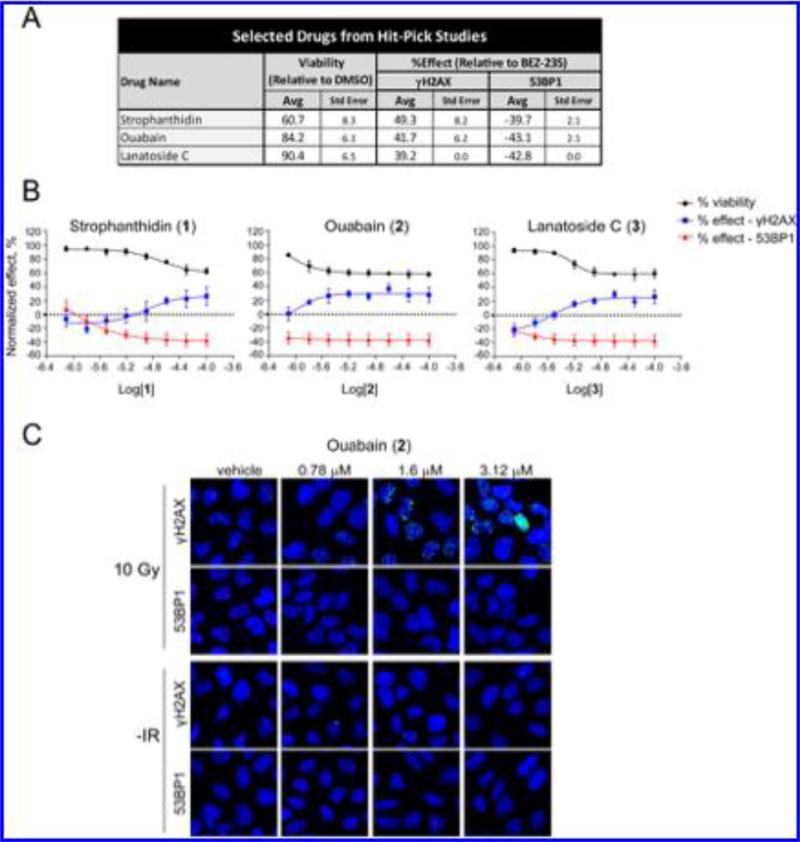
Averages from four replicate assays using 1–3 demonstrated that 1 and 2 displayed minimal effects on cell viability while 3 decreased cell viability by ~40% (Figure 4A). Moreover, reproducible dose-dependent modulation of 53BP1 and γH2AX foci was observed (Figure 4B,C). These differences did not correlate with adverse effects on cell viability, which was calculated by dividing the cell numbers in the drug-versus DMSO-treated wells. Ouabain (2) suppressed 53BP1 foci at nanomolar concentrations (780 nM), while the effects on γH2AX foci levels were apparent at higher doses (Figure 4B,C). Importantly, increases in DNA damage foci levels were not observed in the absence of IR (Supplementary Figure 11), suggesting that these effects are not related to direct DNA damage. Evaluation of foci formation at earlier time points (Supplementary Figure 12) showed that 1–3 did not affect the induction of 53BP1 foci within 3–5 h postirradiation, but dose-dependent suppression of 53BP1 foci levels was observed between 8–24 h postirradiation. These findings suggest that these hits modulate repair activity or pathway choice rather than directly inhibiting the initial recruitment of this factor to the lesion (which occurs at near normal levels in the first 4 h for cells treated with 1–3). The significance of these findings is discussed further below in the Discussion.
Figure 4.
Hitpick and dose–response analysis of selected screen actives. (A) Hitpick table showing average and standard error values from DNA damage foci assays performed in quadruplicate for 1–3. The percent effect values shown are relative to BEZ-235-treated wells, as described in Materials and Methods. (B) Dose-dependent effects of 1–3 on cell viability and 53BP1 or γH2AX foci numbers in U2OS cells subjected to 10 Gy IR. Cells were pretreated with 1–3 for 1 h, irradiated, and then analyzed 24 h postirradiation. Percent viability relative to DMSO control wells and normalized foci effects relative to BEZ-235-treated wells are shown. The X axis is the logarithm of the molar concentration of 1, 2, or 3. (C) 53BP1 and γH2AX foci images of cells 24 h postirradiation after treatment with 2.
We then tested whether 1–3 induced changes in the patterns of foci induction and resolution of other key DSB repair proteins. We chose to analyze DNA-PKcs40 and BRCA141 foci kinetics, as these factors are involved in canonical NHEJ and HR repair, respectively. In the absence of 1–3, DNA-PKcs and BRCA1 foci formation peaked at 4 and 6 h, respectively, which is consistent with previous reports by our group and others using this cell line.22–24 1–3 induced marked dose-dependent suppression of BRCA1 foci induction immediately after irradiation; this was followed by suppression of BRCA1-foci levels 8–24 h postirradiation (Supplementary Figure 13). In contrast, DNA-PKcs foci levels were moderately diminished 0–4 h after irradiation, but substantial suppression of foci levels was noted 8–24 h postirradiation (Supplementary Figure 13). Among 1–3, 2 demonstrated the greatest response; for example, a nearly 2-fold reduction in foci-positive cells was observed in cells treated with 390 nM 2 8 h postirradiation. For all three drugs, we observed nearly complete loss of BRCA1 and DNA-PKcs foci (along with suppression of 53BP1 foci levels), which corresponded to elevated γH2AX levels, suggestive of unrepaired DSBs. These data indicate a profound DSB repair defect in cells treated with these drugs. As will be presented below, we did not observe any significant changes in cell-cycle phase distribution, suggesting that these observations do not arise from cell-cycle arrest.
Analysis of the Effects of 1–3 on DSB Repair Activity Using a GFP/RFP Reporter Assay
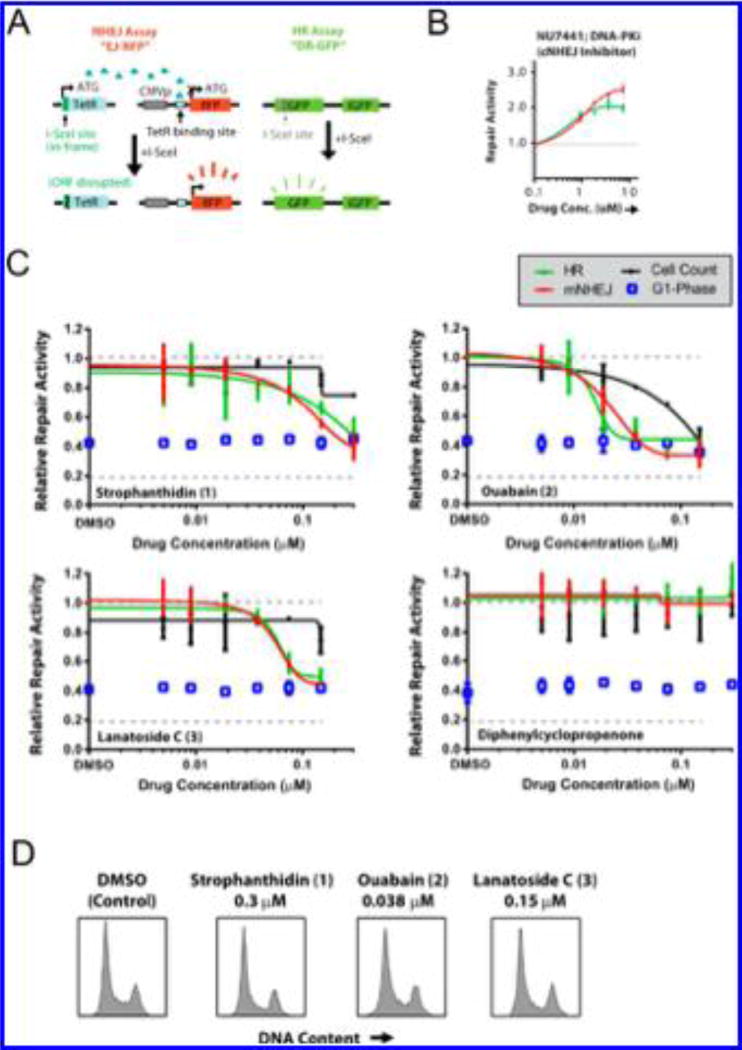
We recently developed a method to study NHEJ and HR repair at site-specific DSBs tethered to red fluorescent protein (RFP)- and green fluorescent protein (GFP)-based reporter assays in human tumor cell lines.22 A schematic of this assay is shown in Figure 5A. This NHEJ assay measures mutagenic DSB repair, which is predominantly driven by noncanonical pathways. The site-specific DSBs are created by the rare-cutting endonuclease I-SceI, which has a 23 base pair recognition sequence integrated into the cell genome.42 We also created a novel ligand-dependent I-SceI that can be stably integrated into these cells.22 In this manner, DSB cleavage kinetics can be precisely controlled, which is ideal for testing the activity of putative DSB repair inhibitors that may have short half-lives. This assay has been validated using small interfering RNAs targeting several key DSB repair genes22 and known DNA repair inhibitors such as NU7441.23 Representative data for NU7441 using this assay are presented in Figure 5B. Treatment with NU7441 induced HR and mutagenic NHEJ repair, indicating that we could detect an expected shift away from canonical NHEJ repair.22,43 These assays are based on the repair of a single cohesive-end DSB with a GFP- and RFP-based functional readout. As such, these assays and their readouts are distinct from the IR-induced DNA damage foci assays presented earlier.
Figure 5.
Secondary assay platform for validation of DSB repair inhibition. (A) Schematic of the GFP/RFP-based DSB reporter assay to measure HR and mutagenic NHEJ repair in cells at induced site-specific DSBs. (B) Validation of this reporter assay using the DNA-PK inhibitor NU7441. (C) Effects of 1–3 on HR and mutagenic NHEJ repair at a range of drug doses; the fractions of cells in the G1 phase, along with total cell counts, are shown for each dose. (D) Representative cell-cycle phase histograms for the corresponding active compound concentrations from the DSB repair assays presented in (C) are shown for each compound.
Cells were counted in parallel at each dose to assess toxicity
Cell death associated with DSB-repair-inhibitor-induced radiosensitization would not be expected in the 24 h postirradiation time period since IR-associated cell killing typically takes a minimum of 48–72 h to manifest.44 This phenomenon forms the basis for the use of clonogenic survival assays to assess the effects of DNA-damaging and other agents on cell viability.45 Similarly, inhibition of DSB repair itself typically would not be expected to induce substantial adverse effects on cell viability, on the basis of the finding that numerous cell lines have been created with homozygous null mutations in key DSB repair genes. Thus, any effects on cell viability in these foci-based assays can be attributed to nonspecific toxicity. HOECHST staining was used to measure DNA content and assess the effects of each compound on the cell cycle distribution. Since DNA DSB repair pathways are cell-cycle-dependent,22,46 these studies were necessary to exclude indirect effects on DSB repair arising from cell-cycle arrest.
The effects of 1–3 at concentrations of 0.300–5 µM on DSB repair were evaluated in these assays. As shown in Figure 5C, 1–3 suppressed both NHEJ and HR repair independent of cell toxicity, and no significant alterations in the cell cycle distribution were observed. Representative cell cycle distribution plots are shown in Figure 5D, and the corresponding plots for each dose tested here are shown in Supplementary Figure 14A. Ouabain (2) displayed the most potent phenotype, with approximately 60% reductions in both NHEJ and HR repair activity at a concentration of 38 nM. Substantial DSB repair inhibitory activity was observed for 2 even at <20 nM. DSB repair inhibition was correlated with cell toxicity for this drug, but there were no changes in the G1 phase fraction under these same conditions. Similarly, minimal changes in cell doubling were observed for these compounds at the doses where DSB repair was suppressed (Supplementary Figure 14B). Thus, the toxicity appeared to be cell-cycle-independent and could not explain the phenotype of this drug on DSB repair. Diphenylcyclopropenone was also included in these experiments since it was validated as an initial hit (Figure 3C), but it was excluded because it was found to induce DNA damage in the absence of IR (data not shown). As expected, this compound did not demonstrate any activity in these assays, which detect DSB repair specifically at two site-specific induced DSBs. These data highlight the specificity of this assay for DSB repair activity.
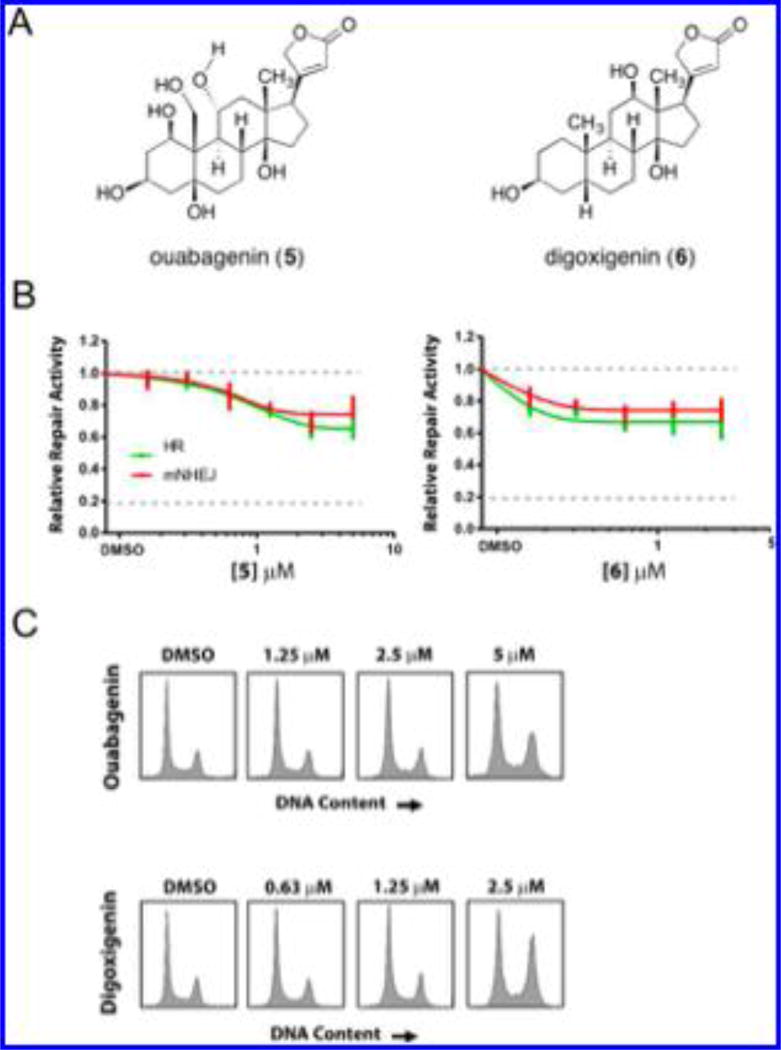
We also evaluated the activities of the isolated aglycones of 2 and 3 [ouabagenin (5) and digoxigenin (6), respectively; Figure 6A] in this repair assay to ascertain the influence of the carbohydrate residues of 2 and 3 and obtain preliminary structure–function data on the aglycon substructures. Both 5 and 6 displayed comparable, albeit less robust, NHEJ and HR inhibitory activities (Figure 6B). Negligible effects on cell toxicity were noted at the doses observed (data not shown). Cell-cycle analysis did not provide any evidence of cell-cycle arrest (Figure 6C).
Figure 6.
Analysis of the effects of aglycon substructures on DSB repair activity. (A) Structures of the isolated aglycones of 2 and 3 [ouabagenin (5) and digoxigenin (6), respectively]. (B) Effects of 5 and 6 on DSB repair activity in the GFP/RFP-based reporter assays (the assay schematic shown in Figure 5A). (C) Representative cell-cycle phase histograms for each drug at selected concentrations from the assays presented in (B).
DISCUSSION
DNA repair is being intensively investigated as a chemotherapeutic strategy.8,9 Because of deficiencies in genetic buffering, tumor cells are often sensitized toward DDR inhibitors, creating the potential to obtain selectivity in systemic treatments.3,8 However, while over 100 proteins are known to be directly involved in the DDR network, only a handful of these have been targeted by small molecules,47 and nearly all of these were discovered using target-based approaches.15,16 A whole-cell unbiased approach promises to provide inhibitors of known and unknown protein targets, including those that are not readily obtained in purified form. This approach addresses issues of solubility, uptake, and metabolism in the primary screen and provides an opportunity to identify novel essential factors in the DDR through follow-up target identification studies. A recent review has highlighted the utility of phenotypic screening in cancer drug discovery.48
The assay we have reported employs γH2AX17 and 53BP118 as markers of repair activity in the primary screen. While 53BP1 alone is often regarded as a reliable indicator of DSB repair,18d its use in conjunction with γH2AX and a time-dependent analysis provides further confirmation of phenotype and preliminary insight into the mechanism of action of the compounds. This assay is fully automated and conducted in a microwell-based format, rendering it amenable to larger compound collections. Our secondary assays comprise a unique platform of experiments to rigorously exclude false positives and gain further insight into the repair pathways targeted by each compound. All of the hits were tested for γH2AX and 53BP1 foci formation in the absence of IR in order to exclude false positive results arising from direct DNA damage induced by the compounds. The restriction-enzyme-based DSB repair assay we employed allows us to concurrently analyze mutagenic NHEJ and HR repair activities and to determine the effect of each compound on either pathway separately.22,23 Furthermore, this assay represents an orthogonal validation of activity as a DSB repair modulator for each identified hit. In addition, high-throughput cell-cycle analysis and cell counting allow us to probe for changes in cell cycle and viability, which provide insight into off-target effects such as direct DNA damage, cell-cycle arrest, and toxicity. These latter assays are essential since it has been shown that the rates of γH2AX and 53BP1 foci resolution can be influenced by the cell-cycle phase in which DSB repair is occurring.20 Consequently, there is a possibility that a compound could indirectly impair resolution of 53BP1 and γH2AX foci by inducing cell-cycle arrest.27 While such a phenotype may lead to radio- or chemosensitization, we are focused on the identification of molecules that directly modulate DSB repair. By means of these secondary assays, lead compounds identified in the primary screen can be rapidly validated as direct DSB repair inhibitors and rank-ordered for further studies.
By this approach, cardiac glycosides 1–4 emerged as inhibitors of NHEJ and HR repair. This result was particularly intriguing given the prior reported anticancer activities of these compounds.49 Several discrete mechanisms of anticancer activity have been proposed for these compounds, including inhibition of Na+/K+-ATPase, which is overexpressed in many cancers, poisoning of topoisomerase complexes, or inhibition of glycolysis.49 Our data conclusively establish that these compounds modulate NHEJ and HR repair; moreover, we did not observe toxicity or DNA damage upon treatment with the cardiac glycosides alone, which excludes Na+/K+-ATPase and topoisomerase inhibition as underlying their mechanism of action. Interestingly, these compounds prolonged the lifetime of γH2AX foci while inhibiting the formation of 53BP1 foci. These observations suggest that the cardiac glycosides may interact with a factor in the DDR between γH2AX and 53BP1. The comparable activities of the aglycones ouabagenin (5) and digoxigenin (6) suggest a common target for these compounds. A recent study showed that ATM-mediated phosphorylation of mediator of DNA damage checkpoint protein 1 (MDC1) promotes recruitment of the E3 ubiquitin–protein ligase ring finger protein 8 (RNF8) to the site of DSBs. RNF8 in turn recruits 53BP1 and BRCA1 to the damaged DNA.50 Thus, it is possible that MDC1 or RNF8, which act downstream of γH2AX but upstream of 53BP1, may be targets of the cardiac glycosides. This hypothesis is consistent with the observation that BRCA1 foci levels were also suppressed upon treatment with cardiac glycosides. In addition, 1–3 did not affect the induction of 53BP1 foci 3–5 h postirradiation, but dose-dependent suppression of 53BP1 foci levels was observed 8–24 h postirradiation. These data suggest a defect in 53BP1 foci retention rather than recruitment. Previous studies have suggested complex mechanisms that can independently regulate the retention 53BP1 after its initial recruitment to DSBs.51 We thus hypothesize that these drugs primarily affect the retention of this molecule at DSBs. Future studies will focus on evaluating
Several cardiac glycosides are in clinical use for the treatment of heart failure and atrial arrhythmia. For example, digoxin and digitoxin are used for the treatment of cardiac failure, and the latter drug is also used for the treatment of supraventricular arrhythmias, including atrial fibrillation. Peak digoxin levels of 8–12 µg/kg typically are required for a therapeutic effect. Cardiac glycosides appear to have IC50 values against their known target, Na,K-ATPase, in the range of 10–100 nM in vitro.52 We observe an altered DNA damage response phenotype at concentrations well within this in vitro range (e.g., Figure 5C, where DSB repair inhibitory activity is observed even at <20 nM). In addition, UNBS1450 is a steroid cardiac glycoside with structural similarities to digoxin53 that has been tested in a phase I clinical trial against nonsmall cell lung cancer. Taken together, these findings suggest that we can achieve effective concentrations in an oncology setting. On the basis of our studies, the facile repurposing54 of cardiac glycosides as DDR inhibitors could be pursued. Recent advances in the chemical synthesis of cardiac glycosides,55 including methods to produce libraries of glycosylated derivatives,56 provide methods to further optimize their structures and control their polypharmacology.
CONCLUSION
We have developed the first fully automated whole-cell assay for the discovery of DNA DSB repair inhibitors. Our assay employs two well-established markers of DSB repair activity, γH2AX and 53BP1, allowing us to confirm phenotype in the primary screen and gain insight into their mechanism of action. We have established a series of robust secondary assays to validate lead compounds and exclude compounds that directly damage DNA, decrease cell viability, or induce cell-cycle arrest. These studies have led to the discovery that cardiac glycoside natural products are potent modulators of NHEJ and HR repair. This observation provides an explanation for the anticancer activity of these compounds, which has remained unresolved. Our data suggest that the cardiac glycosides may target a factor in the DDR downstream of γH2AX and upstream of 53BP1, potentially either MDC1 or RNF8, and that this interaction may inhibit retention of 53BP1 at the site of DSBs. We envision that these clinically approved agents could be readily repurposed as radio- or chemosensitizers for the treatment of a broad range of cancers. More broadly, the assay we have reported is likely to enable the discovery of new DDR inhibitors and new biological targets in these clinically important pathways.
Supplementary Material
Acknowledgments
Financial support from the National Institute of General Medical Sciences (R01 GM090000 to S.B.H.), CureSearch for Children’s Cancer (R.S.B.), and the Yale Center for Molecular Discovery is gratefully acknowledged. We thank Dr. Laureen Colis for procurement of antibodies and helpful discussions during the course of these studies.
Footnotes
ASSOCIATED CONTENT
*Supporting Information The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b00162.
Supplementary Figures 1–14 (PDF) Supplementary Data File 1 (XLSX) Supplementary Data File 2 (XLSX)
The authors declare no competing financial interest.
References
- 1.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. Nucleic Acids Res. 2011;39:D1035. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schipler A, Iliakis G. Nucleic Acids Res. 2013;41:7589. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hühn D, Bolck HA, Sartori AA. Swiss Med Wkly. 2013;143 doi: 10.4414/smw.2013.13837. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. Annu. Rev. Biochem. 2010;79:181. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung P, Klein H. Nat. Rev. Mol. Cell Biol. 2006;7:739. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 6.Hoeijmakers JHJ. Nature. 2001;411:366. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 7.Hartman JL, IV, Garvik B, Hartwell L. Science. 2001;291:1001. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 8.(a) Aziz K, Nowsheen S, Pantelias G, Iliakis G, Gorgoulis VG, Georgakilas AG. Pharmacol. Ther. 2012;133:334. doi: 10.1016/j.pharmthera.2011.11.010. [DOI] [PubMed] [Google Scholar]; (b) Curtin NJ. Nat. Rev. Cancer. 2012;12:801. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 9.(a) Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. Nat. Rev. Cancer. 2008;8:193. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]; (b) Ljungman M. Chem. Rev. 2009;109:2929. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]; (c) Reed E. Clin. Cancer Res. 2010;16:4511. doi: 10.1158/1078-0432.CCR-10-0528. [DOI] [PubMed] [Google Scholar]
- 10.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui- Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JHM, de Bono JSN. Engl. J. Med. 2009;361:123. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 11.Skalitzky DJ, Marakovits JT, Maegley KA, Ekker A, Yu X-H, Hostomsky Z, Webber SE, Eastman BW, Almassy R, Li J, Curtin NJ, Newell DR, Calvert AH, Griffin RJ, Golding BT. J. Med. Chem. 2003;46:210. doi: 10.1021/jm0255769. [DOI] [PubMed] [Google Scholar]
- 12.(a) Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Nature. 2005;434:913. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]; (b) Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NMB, Jackson SP, Smith GCM, Ashworth A. Nature. 2005;434:917. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 13.Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Bioorg. Med. Chem. Lett. 2004;14:6083. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Cancer Res. 2006;66:5354. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 15.(a) Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, Cliby WA, Sarkaria J, Beale G, Edmondson RJ, Curtin NJ. Br. J. Cancer. 2011;105:372. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. Nat. Struct. Mol. Biol. 2011;18:721. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo K, Shelat AA, Guy RK, Kastan MB. J. Biomol. Screening. 2014;19:538. doi: 10.1177/1087057113520325. [DOI] [PubMed] [Google Scholar]
- 17.(a) Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. J. Biol. Chem. 1998;273:5858. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]; (b) Rogakou EP, Boon C, Redon C, Bonner WM. J. Cell Biol. 1999;146:905. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. J. Biol. Chem. 2000;275:9390. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]; (d) Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. Nat. Rev. Cancer. 2008;8:957. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. J. Cell Biol. 2000;151:1381. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Anderson L, Henderson C, Adachi Y. Mol. Cell. Biol. 2001;21:1719. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rappold I, Iwabuchi K, Date T, Chen J. J. Cell Biol. 2001;153:613. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Panier S, Boulton S. J. Nat. Rev. Mol. Cell Biol. 2014;15:7. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 19.(a) Colis LC, Woo CM, Hegan DC, Li Z, Glazer PM, Herzon SB. Nat. Chem. 2014;6:504. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Colis LC, Hegan DC, Kaneko M, Glazer PM, Herzon SB. J. Am. Chem. Soc. 2015;137:5741. doi: 10.1021/ja513117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polo SE, Jackson SP. Genes Dev. 2011;25:409. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. J. Biomol. Screening. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Bindra RS, Goglia AG, Jasin M, Powell SN. Nucleic Acids Res. 2013;41:e115. doi: 10.1093/nar/gkt255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goglia AG, Delsite R, Luz AN, Shahbazian D, Salem AF, Sundaram RK, Chiaravalli J, Hendrikx PJ, Wilshire JA, Jasin M, Kluger HM, Glickman JF, Powell SN, Bindra RS. Mol. Cancer Ther. 2015;14:326. doi: 10.1158/1535-7163.MCT-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge S. J. Nat. Cell Biol. 2012;14:318. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redon CE, Nakamura AJ, Martin OA, Parekh PR, Weyemi US, Bonner WM. Aging. 2011;3:168. doi: 10.18632/aging.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cragg GM, Kingston DGI, Newman DJ, editors. Anticancer Agents from Natural Products. 2. Taylor and Francis; Boca Raton, FL: 2011. [Google Scholar]
- 27.Cleaver JE. Photochem. Photobiol. 2011;87:1230. doi: 10.1111/j.1751-1097.2011.00995.x. [DOI] [PubMed] [Google Scholar]
- 28.Hardcastle IR, Cockcroft X, Curtin NJ, El-Murr MD, Leahy JJ, Stockley M, Golding BT, Rigoreau L, Richardson C, Smith GC, Griffin RJ. J. Med. Chem. 2005;48:7829. doi: 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- 29.(a) Mukherjee B, Tomimatsu N, Amancherla K, Camacho CV, Pichamoorthy N, Burma S. Neoplasia. 2012;14:34. doi: 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Maira S-M, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Cheǹe P, DePover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, García- Echeverría C. Mol. Cancer Ther. 2008;7:1851. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 30.Davidson D, Amrein L, Panasci L, Aloyz R. Front. Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gil del Alcazar CR, Hardebeck MC, Mukherjee B, Tomimatsu N, Gao X, Yan J, Xie XJ, Bachoo R, Li L, Habib AA, Burma S. Clin. Cancer Res. 2014;20:1235. doi: 10.1158/1078-0432.CCR-13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Tavecchio M, Munck JM, Cano C, Newell DR, Curtin NJ. Cancer Chemother. Pharmacol. 2012;69:155. doi: 10.1007/s00280-011-1662-4. [DOI] [PubMed] [Google Scholar]; (b) McLuckie KI, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, Tannahill D, Lowe C, Balasubramanian S. J. Am. Chem. Soc. 2013;135:9640. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. Nat. Chem. Biol. 2008;4:119. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charrier J-D, Durrant SJ, Golec JMC, Kay DP, Knegtel RMA, MacCormick S, Mortimore M, O’Donnell ME, Pinder JL, Reaper PM, Rutherford AP, Wang PSH, Young SC, Pollard JR. J. Med. Chem. 2011;54:2320. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 35.Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D. Gynecol. Oncol. 2014;133:599. doi: 10.1016/j.ygyno.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Cancer Res. 2004;64:9152. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 37.Teng M, Zhu J, Johnson MD, Chen P, Kornmann J, Chen E, Blasina A, Register J, Anderes K, Rogers C, Deng Y, Ninkovic S, Grant S, Hu Q, Lundgren K, Peng Z, Kania RS. J. Med. Chem. 2007;50:5253. doi: 10.1021/jm0704604. [DOI] [PubMed] [Google Scholar]
- 38.DuBois SG, Groshen S, Park JR, Haas-Kogan DA, Yang X, Geier E, Chen E, Giacomini K, Weiss B, Cohn SL, Granger MM, Yanik GA, Hawkins R, Courtier J, Jackson H, Goodarzian F, Shimada H, Czarnecki S, Tsao-Wei D, Villablanca JG, Marachelian A, Matthay KK. Clin. Cancer Res. 2015;21:2715. doi: 10.1158/1078-0432.CCR-14-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min K, Ebeler SE. Food Chem. Toxicol. 2009;47:2716. doi: 10.1016/j.fct.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 40.Jette N, Lees-Miller SP. Prog. Biophys. Mol. Biol. 2015;117:194. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy R, Chun J, Powell SN. Nat. Rev. Cancer. 2012;12:68. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukacsovich T, Yang D, Waldman AS. Nucleic Acids Res. 1994;22:5649. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunn A, Bennardo N, Cheng A, Stark JM. J. Biol. Chem. 2011;286:42470. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.(a) Thompson LH, Suit HD. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1968;13:391. doi: 10.1080/09553006814550371. [DOI] [PubMed] [Google Scholar]; (b) Thompson LH, Suit HD. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1969;15:347. doi: 10.1080/09553006914550571. [DOI] [PubMed] [Google Scholar]
- 45.(a) Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Nat. Protoc. 2006;1:2315. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]; (b) Sumantran VN. Methods Mol. Biol. 2011;731:219. doi: 10.1007/978-1-61779-080-5_19. [DOI] [PubMed] [Google Scholar]
- 46.(a) Radhakrishnan SK, Jette N, Lees-Miller SP. DNA Repair. 2014;17:2. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Singh SK, Wu W, Zhang L, Klammer H, Wang M, Iliakis G. Int. J. Radiat. Oncol., Biol., Phys. 2011;79:540. doi: 10.1016/j.ijrobp.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava M, Raghavan SC. Chem. Biol. 2015;22:17. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Moffat JG, Rudolph J, Bailey D. Nat. Rev. Drug Discovery. 2014;13:588. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 49.Prassas I, Diamandis EP. Nat. Rev. Drug Discovery. 2008;7:926. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 50.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Science. 2007;318:1637. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. J. Cell Biol. 2005;170:201. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lelievre LG, Crambert G, Allen PD. Cell. Mol. Biol. 2001;47:265. [PubMed] [Google Scholar]
- 53.Mijatovic T, Op De Beeck A, Van Quaquebeke E, Dewelle J, Darro F, de Launoit Y, Kiss R. Mol. Cancer Ther. 2006;5:391. doi: 10.1158/1535-7163.MCT-05-0367. [DOI] [PubMed] [Google Scholar]
- 54.Pantziarka P, Bouche G, Meheus L, Sukhatme V, Sukhatme VP. Future Oncol. 2015;11:181. doi: 10.2217/fon.14.244. [DOI] [PubMed] [Google Scholar]
- 55.(a) Renata H, Zhou Q, Dünstl G, Felding J, Merchant RR, Yeh C-H, Baran PS. J. Am. Chem. Soc. 2015;137:1330. doi: 10.1021/ja512022r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Renata H, Zhou Q, Baran PS. Science. 2013;339:59. doi: 10.1126/science.1230631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12305. doi: 10.1073/pnas.0503270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.