Abstract
We have used two approaches to search for sequence variants in the 18S coding region of amplified ribosomal DNA (rDNA) from Xenopus laevis oocytes. First, using clones derived from amplified rDNA, we compared the equivalent of a complete 18S coding region from two clones and short regions from two other clones with the 18S sequence previously determined from a "reference" clone. The respective sequences in all the clones were identical. Secondly, we examined greater than 60% of the 18S sequence in "pooled 18S genes" in uncloned amplified rDNA. The predominant sequence corresponded to that in the reference clone and no heterogeneities were apparent. Since many chromosomal rDNA units contribute to rDNA amplification the findings indicate that 18S coding sequences in X. laevis are largely homogeneous. The previously established sequence is the predominant one, thus providing a reliable basis for studies on 18S rRNA. Sequencing gels on uncloned amplified rDNA confirmed the absence of methylated cytosine in this DNA. The 18S sequence lacks major open reading frames.
Full text
PDF
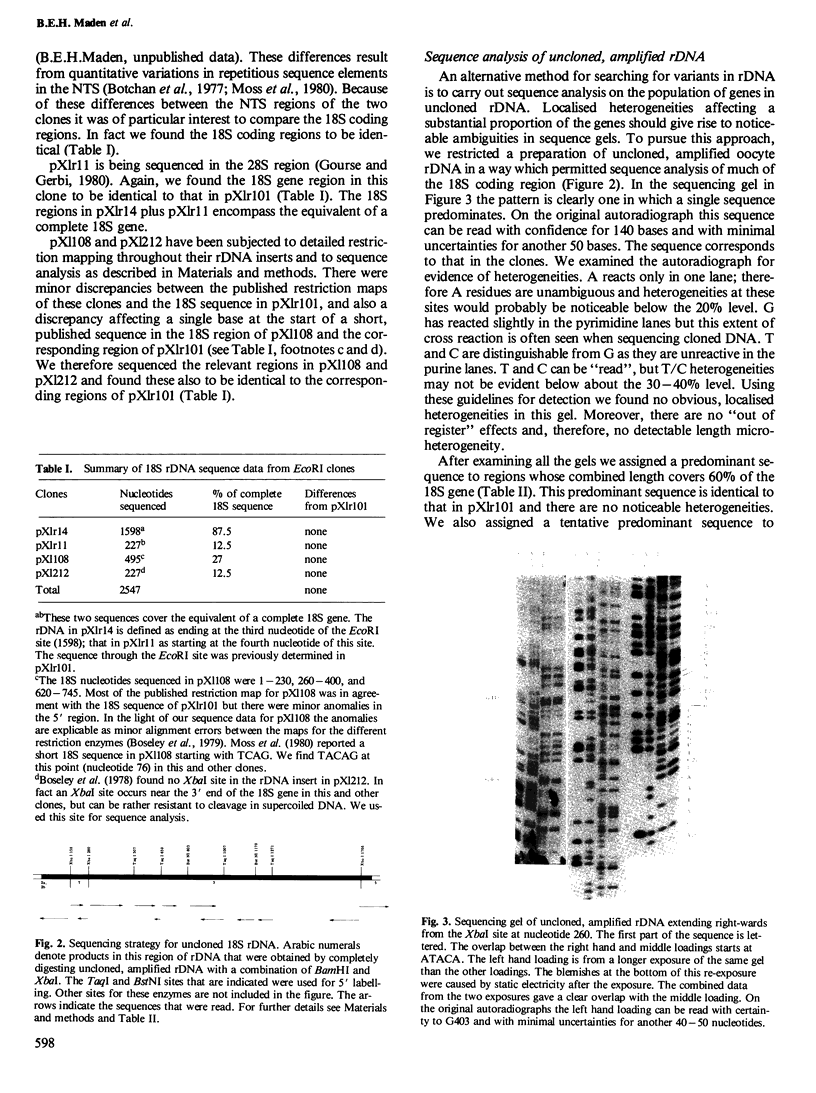



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Brown D. D., Reeder R. H. Composition and structure of chromosomal and amplified ribosomal DNA's of Xenopus laevis. J Mol Biol. 1970 Jul 28;51(2):341–360. doi: 10.1016/0022-2836(70)90147-6. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980 Jan;19(1):79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- Kumar A., Subramanian A. R. Ribosome assembly in HeLa cells: labeling pattern of ribosomal proteins by two-dimensional resolution. J Mol Biol. 1975 May 25;94(3):409–423. doi: 10.1016/0022-2836(75)90211-9. [DOI] [PubMed] [Google Scholar]
- Maden B. E. Methylation map of Xenopus laevis ribosomal RNA. Nature. 1980 Nov 20;288(5788):293–296. doi: 10.1038/288293a0. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Visualization of nucleolar genes. Science. 1969 May 23;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Cohen S. N., Chang A. C., Boyer H. W., Goodman H. M., Helling R. B. Replication and transcription of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1974 May;71(5):1743–1747. doi: 10.1073/pnas.71.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Brown D. D., Reeder R. H. The molecular basis for length heterogeneity in ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):461–486. doi: 10.1016/0022-2836(76)90229-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Carroll D., Brown D. D., Deutch A., Higashinakagawa T., Dawid I. B. Amplified ribosomal DNA from Xenopus laevis has heterogeneous spacer lengths. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2823–2827. doi: 10.1073/pnas.71.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Dawid I. B., Brown D. D. Arrangement of length heterogeneity in repeating units of amplified and chromosomal ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):487–505. doi: 10.1016/0022-2836(76)90230-8. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]