Abstract
Characterizing virus-host relationships is critical for understanding the impact of a virus on an ecosystem, but is challenging with existing techniques, particularly for uncultivable species. We present a general, cultivation-free approach for identifying phage-associated bacterial cells. Using PCR-activated cell sorting, we interrogate millions of individual bacteria for the presence of specific phage nucleic acids. If the nucleic acids are present, the bacteria are recovered via sorting and their genomes analyzed. This allows targeted recovery of all possible host species in a diverse population associated with a specific phage, and can be easily targeted to identify the hosts of different phages by modifying the PCR primers used for detection. Moreover, this technique allows quantification of free phage particles, as benchmarked against the “gold standard” of virus enumeration, the plaque assay.
Keywords: Bacteriophage, host specificity, microfluidics
Introduction
Viruses substantially impact the health and dynamics of all communities, from causing disease in individuals to influencing global biogeochemical cycles (Bohannan and Lenski, 2000; Suttle, 2005). Cyanophages, for example, play an important role in oceanic carbon fixation, since a core photosynthetic protein is of phage origin (Hurwitz et al., 2013; Thompson et al., 2011). Moreover, there are an astronomical number of viruses in the biosphere (Suttle, 2005), but it is unclear what hosts the vast majority of these viruses infect. Studying the impact of environmental viruses necessitates methods for characterizing virus-host interactions. However, the challenges involved in growing heretofore “uncultivable” bacteria means that many of these potential hosts and their viruses cannot be studied in isolation (Rappé and Giovannoni, 2003; Vartoukian et al., 2010).
Next generation sequencing (NGS) is a powerful, culture-independent method for studying virus-host interactions in diverse populations (Dhillon and Li, 2015; Labonté et al., 2015; Reyes et al., 2012; Roux et al., 2014). In this approach, metagenomic sequences are interrogated for molecular traces left by the viruses co-occurring with host genes such as small subunit ribosomal DNA sequences. These viral traces are often resistance-conferring, such as CRISPR-associated sequences (Andersson and Banfield, 2008; Weinberger et al., 2012). Identifying virus-host relationships using NGS is only possible, however, if such traces are identifiable. Moreover, even when they occur, these sequences are rare, making their identification challenging. Single cell genomics of uncultivated organisms is another powerful method for identifying virus-host pairs (Labonté et al., 2015; Roux et al., 2014). In one study, 127 single amplified genomes (SAGs) from a known clade of uncultivated organisms were screened for viruses and 69 new virus clades were found (Roux et al., 2014). In another study, bioinformatic analysis of 58 SAGs allowed identification of 30 new virus genomes (Labonté et al., 2015). To isolate these organisms for SAG analysis, fluorescence-activated cell sorting (FACS) sorts individual cells into wells based on non-specific dyes or scattering properties; once in the wells, the cells can be identified with PCR or other sequence analysis. This limits throughput to identification of just hundreds of cells, making it challenging to identify the hosts of rare viruses. A higher-throughput method for detecting infected cells is PhageFISH (Fluorescence In Situ Hybridization) which allows phage hosts to be separated based on virus infection by staining infected hosts with fluorescent probes complimentary to virus sequences (Allers et al., 2013). PhageFISH is potentially high throughput since millions of hosts can be stained in parallel and sorted at >10,000 per second with FACS. However, probe hybridization is performed in the complex milieu of a fixed cell, necessitating substantial assay optimization extremely challenging with uncultivable organisms. Viral tagging, on the other hand, uses fluorescently labeled viruses to isolate cells that they are associated with by flow cytometry (Deng et al., 2012) or for identifying new viruses that infect specific hosts (Deng et al., 2014).
A powerful, cultivation-free method for identifying specific phage hosts is single-cell PCR in individual microfluidic chambers (Tadmor et al., 2011). In this approach, TaqMan PCR interrogates individual microbes for phage sequences such that, if the sequences are present, a fluorescent signal is produced. This allows identification and recovery of phage-associated microbial genomes by interrogating positive chambers. Because this method uses PCR, which yields exponential amplification when phage sequences are present, the signal difference between phage-associated and phage-free genomes is large, permitting unambiguous identification. Moreover, harsh PCR thermocycling facilitates bacterial lysis and enhances access of hybridization primers to their targets, reducing false-negatives. The principal limitation of this method is that it lacks scalability, enabling interrogation of just thousands of bacteria; this limits its utility for studying most environments, comprising billions of microbes per milliliter (Dang and Sullivan, 2014). To allow comprehensive characterization of phage-host relationships, an optimal method would be applicable to uncultivable species, capable of analyzing millions of microbes, and enable the specific recovery of the genomes of all bacteria infected with the target phage.
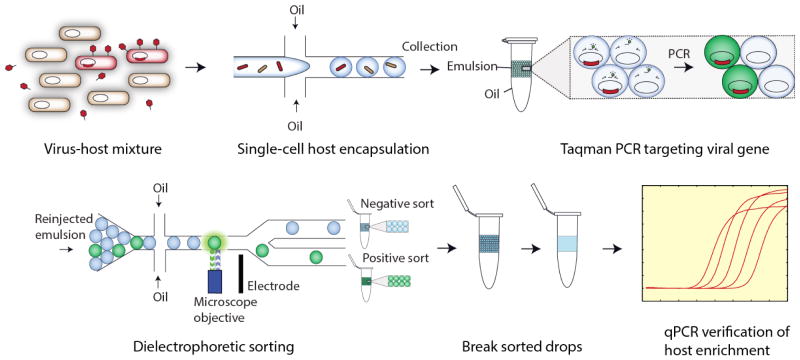
In this paper, we present a general method for identifying phage-host relationships applicable to uncultivable organisms; the method combines the scalability of FACS with the generality and precision of single-cell PCR. Using PCR-activated cell sorting (PACS) we sort bacteria based on infection by specific phages (Eastburn et al., 2014; Lim et al., 2015a). Besides applying PACS to phage sequences in infected bacteria, we also show in this paper that certain bacteriophage viral particles can be directly quantified via droplet-based PCR. This is accomplished by isolating individual bacteria from a heterogeneous population in water-in-oil droplets and performing PCR in each droplet with primers specific to the target phage genome (Fig. 1). If the phage is present in- or bound to- the host, the PCR produces a fluorescent signal, allowing the host bacterial and phage genomes to be recovered by sorting the fluorescent droplets. The sorted genomes can then be analyzed to identify the host species. Importantly, the PCR performed in the droplets is used only as a means of identifying whether the phage is present with the bacteria; the material that is recovered and sequenced is not amplified and, thus, does not need to be known in order to be recovered and sequenced.
Figure 1.
Microfluidic workflow for PACS-based viral detection and host sorting. Virus-infected hosts are first encapsulated with PCR reagent, primers and probe in picoliter-volume droplets, then thermocycled to yield fluorescent drops. These drops contain targeted genomes of viruses and their hosts, which are then sorted to yield a purified population of DNA. This material can be used for downstream sequencing analysis in this case qPCR, but any sequence analysis is possible.
PACS has a number of advantages over other methods for characterizing phage-host relationships: It does not require that the target bacteria or phage be cultivable. It is ultrahigh-throughput, allowing millions of individual microbial genomes to be sorted based on phage association, permitting detection of bacteria infected with rare phages. The TaqMan assay can be multiplexed using probes of different color allowing, for example, identification of specific micobes in a sample containing multiple, specific phage sequences, such as co-infection by distinct phages. The sorting can be easily retargeted against different species by modifying the PCR primers, while maintaining highly specific sorting. This requires minimal optimization, and as a direct comparison to antibody-based methods of cell identification, does not require access to microbial antigens. PACS is particularly applicable to the identification of putative hosts for phages whose sequences have been identified in metagenomic studies. In a recent study attempting to computationally connect viruses to their hosts, metagenomic viral sequences were associated with pathogenic species like Fusobacterium and Leptotrichia that had heretofore not been associated with any viruses (Paez-Espino et al., 2016). Our method can build on similar known short assembled segments of phage sequences to design primers that target specific hosts.
Materials and methods
2.1 Preparation of bacteriophages, plaque assays and bacterial hosts
Bacteriophage T4 (T4), bacteriophage ΦX174 (ΦX174), and E. coli hosts were obtained from Carolina Biological Supply. T4 was propagated by infection of E. coli B (ATCC 11303) (Karam, 1994) and ΦX174 by infection of E. coli C (ATCC 13706). Bacteriophage lambda (lambda cI857ts) was obtained from the lambda lysogen E. coli strain KL470 provided by R. Raghavan. Bacteriophage lambda was propagated by infection of E. coli C600 (Carolina) and plaques formed as previously described (Arber et al., 1983). A lambda lysogen of lambda cI857ts in E. coli C was prepared (Arber et al., 1983) and purified by two rounds of single colony isolation on LB agar. We use a derivative of E. coli strain BW25113, containing mCherry as an uninfected control (Lim et al., 2015b). Titers of T4, ΦX174 and lambda were determined with plaque assays as previously described (Hafenstein and Fane, 2002). Bacteriophage-containing lysates were separated from cellular debris by 5 minutes of centrifugation at 3000g, filtered through 0.2 μm filters (Sartorius, Minisart) and preserved with a single drop of chloroform in the preparation.
2.4 Microfabrication of devices
The microfluidic chips are fabricated using standard soft lithography techniques in poly (dimethylsiloxane) (PDMS) (Xia and Whitesides, 1998). To fabricate a device master from which the PDMS replicates are molded, SU-8 photoresist (MicroChem) is spun onto a 3″ silicon wafer at a thickness of 25 μm, and exposed to UV light from a UV photodiode (ThorLabs) through a UV-absorbent Mylar mask containing an inverse-image of the microfluidic chip (Fineline Imaging). The wafer is baked at 95°C on a hotplate for 1 minute, and developed in propylene glycol monomethyl ether acetate (PGMEA) to remove uncrosslinked resist, followed by post-baking in accordance with the manufacturer’s instructions. PDMS polymer and crosslinker is combined at a ratio of 11:1, poured over the master, degassed to remove trapped air bubbles, and baked at 75°C for 4 hours to crosslink the device. The device is peeled from the master and holes are punched using a 0.75 mm biopsy coring needle. The punched device is washed with isopropyl alcohol and patted with scotch tape to remove debris prior to plasma bonding to a glass slide. To render the channels hydrophobic for water-in-oil emulsification, Aquapel™ is flushed into the channels, after which the device is baked in an oven for 20 min at 65°C.
2.5 Sample encapsulation and droplet PCR
Prior to encapsulation, bacterial suspensions are washed three times by centrifugation at 3000g and the pellets are resuspended in distilled water. Phage suspensions are encapsulated without washing. The viral or bacterial suspensions are mixed with the appropriate primers, TaqMan probes, and PCR mix (2X ddPCR MasterMix, Bio-Rad). The primers and TaqMan probes are used at 1 μM and 250 nM, respectively (Primers and probes are listed in Supplemental Table 1). Droplet loading is assumed to follow Poisson statistics (Mazutis et al., 2013a). For the experiments where two strains of bacteria are mixed, E. coli C and BW25113, the bacteria are first measured for their optical density at OD600, and then mixed in appropriate volumes, so that their final concentration ratio was 1:9 or 1:999, corresponding to a spike-in ratio of E. coli C: E coli BW25113 of 10% and 0.1% respectively. The mix is loaded into an upright 1 ml syringe pre-filled with 200 μl HFE-7500 fluorinated oil (3M), connected to a PDMS flow-focus droplet generator (Supp. Data S3) through a 21 gauge needle and polyethylene tubing. Droplet generation oil for probes (Bio-Rad) is introduced into the carrier-phase inlet of the microfluidic device through another syringe and tube; the oil comes with a proprietary surfactant included to stabilize the generated droplets during the heating and cooling steps of PCR. Using computer-controlled syringe pumps, the aqueous phase is injected at 200 μlhr−1 and the oil at 400 μlhr−1 (New Era), generating 25 μm diameter droplets at ~3.6 kHz in a droplet maker with nozzle 20 μm wide and 25 μm tall. The emulsion is collected into 200 μl PCR tubes and thermocycled on a T100 thermocycler (Bio-Rad), using the following conditions: 10 min. at 95°C, 35 cycles of 10 s. at 95°C, 15 s. at 55°C and 30 s. at 70°C. To verify specificity of the PCR, the emulsions are chemically ruptured with chloroform and DI water and the aqueous fractions electrophoresed on a 2% agarose gel to confirm amplicon length.
2.6 Detection and sorting of droplets
After thermocycling the emulsions, the droplets must be sorted based on fluorescence. This is accomplished by loading the thermocycled emulsions into a syringe with 200 μl HFE-7500, maintaining the syringe vertically so that the needle faces up, and allowing the emulsion to cream for ~10 min; this ensures the droplets are at the top of the emulsion before the syringe pump is started so that they flow into the device at a controlled rate and closely packed. The droplets are injected into the detection and sorting device (Fig. S3) (Agresti et al., 2010; Eastburn et al., 2014; Lim et al., 2015a; Mazutis et al., 2013b) at a flow rate of 50 μlhr−1, with spacer oil flow rate 1000 μlhr−1. The flow rate for the second oil spacer at the sorting junction is set to 100 μlhr−1. All droplet spacing is performed with pure HFE-7500. All electrodes on the device, including the sorting electrode and moat shielding the droplets from stray field, are filled with 2M NaCl solution (Sciambi and Abate, 2015a, 2014). A 100 mW, 532 nm laser is focused upstream of the sorting junction to excite droplet fluorescence. Photomultiplier tubes (PMTs) focused on the same spot measure emitted light and output a voltage proportional to the light intensity to a computer outfitted with an FPGA card (National Instruments) programmed in LabVIEW. The card detects droplets as peaks in fluorescence over time and, when a droplet is to be sorted, outputs a 40 kHz, signal amplified to 1000 V (Trek) applied to the salt-water electrodes on the microfluidic chip. Custom LabVIEW software (available on request) allows adjustment of PMT gain, droplet fluorescence intensity thresholds for sorting, and electrode AC voltage pulse frequency and magnitude (Supplementary Data S4). An image of the optical setup can be found in the Supplementary Data (S5).
2.7 Quantitative PCR analysis of sorted droplets
Genomic material from sorted droplets is recovered by rupturing the droplets via addition of 100 μl chloroform together with 50 μl DI water and vortexing for 10 min (Lim et al., 2015a). To measure the degree of enrichment from PACS, we probe for specific genomic regions in both E. coli strains (BW25113 and C) before and after sorting. For E. coli C, we use primers specific to PRP (Primer Names: E coli C FWD & REV), a gene in that strain but not in E. coli BW25113. For E. coli BW25113, the strain has an integrated ybgF-mCherry cassette, and primers specific to mCherry are used (Primer Names: E Coli BW25113 FWD & REV). All primer sequences are listed in Supplementary Table 1. To assess primer specificity, the primers for E. coli BW25113 and C are tested against each other’s target templates; we observe no background amplification by qPCR or interrogating amplified products with gel electrophoresis. The primers are tested for linearity by constructing a serial dilution for each over a factor of one hundred thousand for the target template. qPCR measurements of genomic material yield cross-threshold (Ct) values corresponding to the number of PCR cycles needed for fluorescence levels to exceed a threshold. We obtain two Ct values for pre- and post-sorted samples, and compute an enrichment factor. The amplification reagent for all the qPCR measurements is Maxima SYBR Green Master Mix (Thermo Scientific).
Results
3.1 PACS workflow for identifying virus-host relationships
To enable the ultrahigh-throughput sorting of microbes based on the presence of phage nucleic acids, we use PACS, a droplet-based microfluidic technology (Eastburn et al., 2014; Lim et al., 2015a). In PACS, picoliter-volume aqueous droplets are used as reactors in which to perform TaqMan PCR on single bacteria. Using flow-focus emulsification (Christopher and Anna, 2007; Nie et al., 2008; Tran et al., 2013), individual phage or bacteria from a heterogeneous sample are isolated in ~107 droplets with PCR reagents and probes targeting specific phage genes (Fig. 1). At a loading rate of ~0.1 bacterial cells per droplet, we interrogate 106 cells. At that loading ratio, we can use Poisson statistics to determine at over 95% of droplets that are filled contain a single cell. After all bacterial cells are encapsulated, the emulsion is thermocycled, performing 106 parallel single-cell PCR reactions, as illustrated in Fig. 1. During thermocycling, the bacteria lyse and their nucleic acids are subjected to TaqMan amplification. If a droplet contains the nucleic acids of a phage targeted by the probes, the nucleic acids are amplified, generating a fluorescent signal that fills the encapsulating droplet. This marks the droplet as containing a bacteria associated with the target virus, allowing the putative host cell nucleic acids to be recovered by sorting the droplet, as illustrated in Fig. 1.
3.2 Specific detection and quantification of viral genomes from bacteriophage T4 and ΦX174
Detecting cells associated with a specific virus depends on the ability to reliably and specifically amplify virus nucleic acids. To investigate the robustness of this step in the PACS process, we perform TaqMan PCR in microfluidic droplets on two distinct virus species, bacteriophage T4 (T4) and bacteriophage ΦX174 (ΦX174). Pure preparations of the viruses are combined with PCR and TaqMan reagents immediately prior to microfluidic emulsification and thermocycling. After thermocycling, we observe clearly fluorescent droplets in a population of non-fluorescent droplets, as shown in Fig. 2A, indicating successful amplification of the viruses. In negative controls, we swap the TaqMan probes by including T4 probes in ΦX174 preparations and ΦX174 probes in T4 preparations. Neither of these controls yield detectable fluorescent droplets, demonstrating that the probes are specific to their target virus.
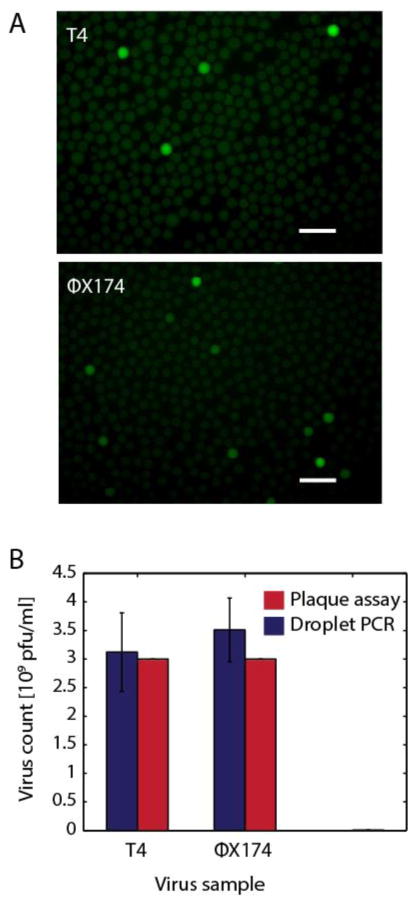
Figure 2.
A) Digital detection of phage particles after droplet PCR. Bacteriophages T4 and ΦX174 virions are partitioned into droplets for TaqMan PCR detection. Scale bars are 100μm. B) Plaque assay results closely mimic digital viral particle quantitation, suggesting that phage genomes are accurately measured with this new method. Error bars represent the standard deviation of 3 technical replicates.
An important factor when using PACS to enrich bacterial cells from a heterogeneous sample is the rate of false negatives since this limits the number of positive events detected. To characterize the sensitivity of the method, we scan the emulsions created in the previous experiment using flow dropometry (Lim et al., 2015a), recording fluorescence values for ~30,000 individual droplets (Figs. S1,S2). Using the known droplet volumes and assuming phage encapsulation is governed by Poisson statistics (Huebner et al., 2007), we estimate phage concentrations in the starting samples and compare them to estimates from plaque assays, Fig. 2B. For bacteriophage T4, plaque assays on samples used for PACS yield 3.0 * 109 pfu/ml compared to the 3.1 * 109 particles/ml for PACS. For ΦX174, the plaque assay yields 3.0 * 109 pfu/ml versus 3.5 * 109 particles/ml for PACS. The estimates using both methods are in excellent agreement indicating that the PCR conditions are sufficient to efficiently lyse the viral particles. The slightly higher values estimated by PACS may reflect that some phage genomes are incorrectly packaged, mutated, or are in non-infectious particles. In addition to validating droplet PCR for PACS, this demonstrates that droplet digital PCR using this apparatus is an alternative approach for quantitating phage genomes in a sample. Our method of directly encapsulating phage particles for digital PCR circumvents the need to pre-lyse the virus before measurement which is needed for quantitative PCR protocols (Anderson et al., 2011; Refardt, 2012).
3.3 Sorting E. coli infected with lambda bacteriophage
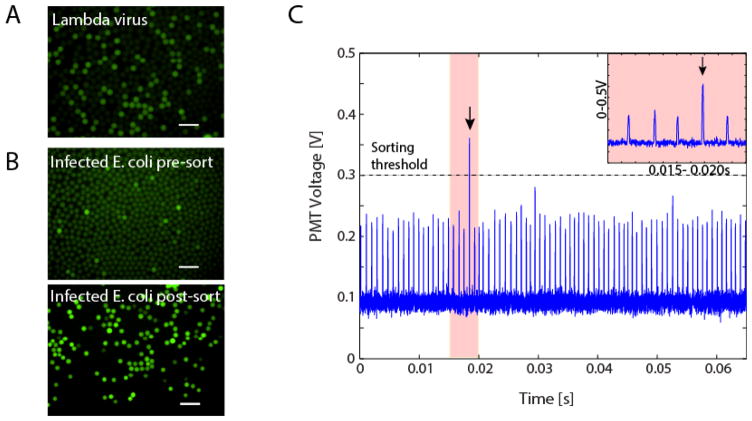
PACS enables the detection of bacterial cells associated with specific phages, including lysogens, and recovery of the host cell genomes. To illustrate this, we construct a test system comprising two E. coli strains: a lambda lysogenic C-strain, and an uninfected BW25113 strain. To validate the TaqMan probes, we analyze a sample of pure lambda virus in suspension and observe digital signals in droplets, indicating single-virion detection (Fig. 3A). We then spike E. coli C (lambda). into uninfected E. coli BW25113 in a ratio of 1:9, and wash the mixture to remove any free phage. We subject this sample to droplet PCR and again observe a digital signal corresponding to a small subpopulation of positive droplets which contain E. coli cells with the lambda genome, as shown in Fig. 3B. To verify that the digital fluorescence corresponds to droplets containing lambda genomes, we sort the emulsion to recover the positive droplets, which we accomplish using dielectrophoretic droplet sorting, Fig. 1 (Agresti et al., 2010; Eastburn et al., 2014; Lim et al., 2015a; Mazutis et al., 2013b; Sciambi and Abate, 2015a). Dielectrophoresis is a phenomenon in which a particle experiences polarization forces in a non-homogenous electric field (Pethig, 2010). The thermocycled emulsion is injected into the sorting device, which flows the droplets spaced by oil individually through the focused excitation laser. As a droplet passes through the laser, its fluorescence is excited and the resulting emitted light is measured with a photomultiplier tube (PMT), which outputs a voltage proportional to the fluorescence intensity analyzed by the computer and FPGA card. The droplets appear as peaks in voltage as a function of time, in which the amplitude of the peak is proportional to the droplet intensity, as shown in Fig. 3C. When a positive droplet passes through the laser, an abnormally tall peak is observed, as seen at t = 0.0185 seconds (Fig. 3C). Upon detection of a positive droplet, the computer outputs an alternating voltage amplified to ~1000 V applied to on-chip electrodes generating an attractive force that pulls the positive droplet into the collection channel (Agresti et al., 2010; Eastburn et al., 2014; Lim et al., 2015a). When a negative droplet passes through the laser the peak amplitude does not fall above the user-defined threshold; the electrode remains un-energized and the droplet flows passively into the waste channel. In this way, the droplet sorter separates the positive from the negative droplets. This method of sorting can separate drops at the rate of a couple of kilohertz, although modifications to the sorter design can further push sorting rates up to 30kHz (Sciambi and Abate, 2015b).
Figure 3.
A) Digital detection of lambda particles and B) pre- and post- sorted drops containing lambda and its E. coli host. Digital detection of lambda using the probes and primers from Figure 3A. C) Time trace of fluorescent droplet detection. Droplets are run through a dielectrophoretic microfluidic sorter, and droplets above a set threshold are sorted and collected each peak represents a single droplet. Inset figure shows a magnified view of the lone sorted event in the time period 0.015–0.025 seconds. All scale bars are 100μm.
3.4 Enrichment of phage-infected E. coli genomes
Sorting the bacterial cells based on presence of lambda DNA should yield primarily E. coli C cells, since only this strain was infected by lambda. To quantify the enrichment for phage-infected cells afforded by PACS, we analyze the fraction of E. coli C DNA in the sorted population using qPCR. We perform a second sort in which we lower the concentration of lysogenic E. coli C (lambda) to 0.1%, while increasing uninfected E. coli BW25113 to 99.9%. Using PACS, we recover all TaqMan positive droplets and the nucleic acids from their cells. To measure the fraction of nucleic acids corresponding to E. coli C and E. coli BW25113, we perform qPCR on pre- and post-sorted samples using primers specific to the two strains; the results for both the 10% and 0.1% mixtures for both primer sets are provided in Fig. 4. The signal from E. coli BW25113 is strongly de-enriched in the PACS-sorted material, corresponding to curve shifts by ~10 Ct values (Figure 4, upper panels). By contrast, the curves for E. coli C shift by a much smaller distance in the same direction, demonstrating that PACS for lambda virus nucleic acids enriches for these cells. To obtain a quantitative metric of enrichment, we define the enrichment factor as the ratio of host microbe purity in the pre-sorted to the post-sorted samples,
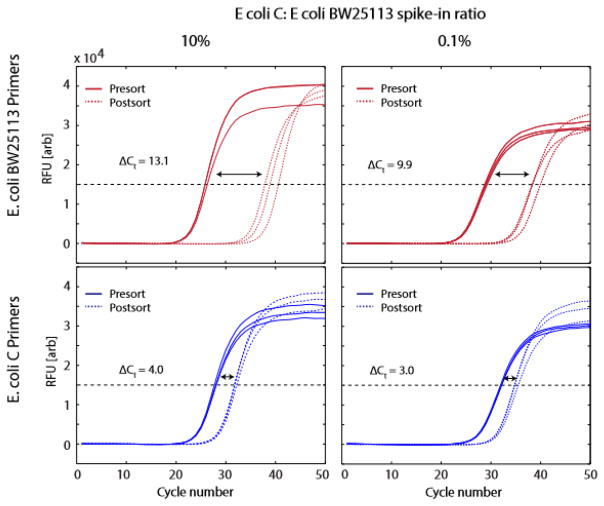
Figure 4.
qPCR detection of host genomes before and after droplet sorting based on the presence of lambda DNA. Each quadrant shows qPCR amplification curves for DNA extracted from drops before and after sorting. The shifts in the curves reflect the 2-fold change of the DNA quantity according to the specific primers being tested.
where Cpre and Cpost are the number of E. coli C cells present in the pre- and post-sorted samples, while Kpre and Kpost are the number of E. coli BW25113 present in the pre- and post-sorted samples, respectively. The relationships between Cpre and Cpost, together with Kpre and Kpost, can be determined via qPCR, which measures the log-2 fold change of gene copy numbers specific to either microbe,
where ΔCtc and ΔCtk are the differences between the qPCR cross-threshold values for E. coli C and BW25113, respectively. This is an estimation, since the actual rate of DNA amplification may be less than 2 per cycle. We know Cpre and Kpre because we begin with controlled numbers of each species before sorting, enabling us to define the ratio of the two species with respect to one another,
and to simplify the enrichment factor to,
For our initial sort, n = 9 (10% E. coli C to BW25113 ratio), we obtain 4.0 for ΔCtc and 13.1 for ΔCtk (Fig. 4.), yielding e = 9.84, indicating that the final sample is enriched to 98.4% for E. coli C from an initial concentration of 10%. For the 0.1% spike-in, we obtain 3.0 for ΔCtc and 9.9 for ΔCtk (Fig. 4.), providing e = 106, so that E. coli C is enriched by about a hundred times to 10.6% final concentration. Larger enrichment factors can be achieved by further diluting the sample prior to partitioning into droplets, which reduces the rate of co-encapsulation of the two species and false-positive recovery of off-target cells.
Discussion
When confronted with large, heterogeneous populations, cell sorting is invaluable because it allows subpopulations to be systematically isolated and studied. However, currently, specific cell sorting is only possible using affinity reagents, such as antibodies or oligomer probes that specifically bind to the cell type of interest (Mamanova et al., 2010; Turner et al., 2009); such reagents are rarely available for uncultivable targets. Nonetheless, in many of these cases, sequence data, often partial, is available. In these instances, PACS is uniquely suited for specific cell sorting since it relies on PCR to differentiate between cells based on the presence of a specific nucleic acid sequence. In this application of PACS, this allows bacterial sorting based on the presence of a phage genome within or attached to the host cell (Fig. 4). We cannot differentiate between bound phage and infected hosts using techniques presented here as intact virions also provide positive PACS signals (Fig. 2), but a stripping step could be included in the PACS workflow, if only hosts with internal viruses are desired. However, previous studies have shown that labeled viruses associated with cells are usually infectious (Deng et al., 2011; 2014). Clearly, full determination of phage-host interactions will require analysis of the specific phage-host pair and cultivation of both, but PACS allows identification of potential hosts in mixed systems, drastically narrowing the search for elusive hosts.
In its current implementation, PACS differentiates between bacterial cells based on one sequence. However, since PACS relies on TaqMan assays, probes of different color can be used to interrogate multiple sequences simultaneously (Taly et al., 2013). This should enable multi-parametric cell sorting similar to current FACS analysis with multiple antibodies. Multi-parametric cell-sorting could recover, for example, the genomes of all cells associated with specific variants of a virus, co-infection of different viral species, or the presence of specific host and virus sequences associated with the same cell.
A limitation of the current system is that the lysis of the bacteria is achieved using the detergents present in the PCR reagents and the high temperature of PCR thermocycling, which may be insufficient for lysing some bacteria or viruses. To broaden the applicability of the method, harsher lysis procedures could be implemented that allow the inclusion of enzymes to digest cellular material. Such procedures can be implemented using recently described agarose emulsion and multi-step droplet merger workflows, which enable the digestion of cell lysates with proteases (Eastburn et al., 2014). Indeed, this is essential for reliably performing PACS on mammalian cells in sub-nanoliter droplets, since undigested mammalian cell lysates can inhibit PCR (Eastburn et al., 2014).
PACS is founded on droplet-based microfluidic technologies whose intrinsic throughput is thousands of droplets per second (Tran et al., 2013). This enables facile and rapid sorting of millions of cells (Sciambi and Abate, 2015a). By implementing faster emulsification strategies using air-bubble triggered droplet generation (Abate and Weitz, 2011) or sequential droplet splitting, and using double emulsion FACS for sorting (Lim and Abate, 2013), it should be possible to enhance the throughput of the system by another order of magnitude; this will further increase the number of microbes that can be sorted for identifying rare phage-host relationships, such as identifying the host of chimeric RNA-DNA virus genomes recently discovered in numerous ecosystems (Diemer and Stedman, 2012; Stedman, 2013). PACS could also be used for screening emerging pathogenic virus sequences to identify possible reservoir hosts.
PACS paired with NGS can be a particularly potent tool for applications in microbiology and virology. PACS-sorted lysates can be recovered and subjected to deep sequencing of cellular DNA and/or RNA, enabling correlation of specific viruses with host cell genomes and their expression patterns. Virus-based PACS can be used, for example, to study how viruses modulate host cell gene expression, or if certain host genetic variants are more susceptible to virus infection. The ability to sort millions of entities based on nucleic acids is valuable for sieving through complex cell populations to determine specific phage-host interactions. Current available metagenomic datasets can be mined for phage sequences for the design of virus-specific primers.. From a complex population, one could assay the sorted genomic material for host provenance by targeted 16S or whole-genome sequencing. In addition to sorting microbes based on infection by bacteriophage, PACS can be applied to the clinical setting such as the isolation of mammalian cells latently infected by HIV. By combining PACS with sequencing, the presence of a specific pathogen can be correlated with host cell properties, such as somatic mutations or gene expression. Such investigations would be valuable for studying how different pathogens manipulate their hosts.
Supplementary Material
Highlights.
PACS (PCR-Activated Cell Sorting) for enrichment of phage-associated bacteria
Ultrahigh-throughput, cultivation-independent, microfluidic technique
Phage digital PCR without lysis step can quantitate phage virions accurately
Acknowledgments
We would like to thank Rahul Raghavan for E. coli strain KL740 and Bentley Fane for advice on ΦX174 cultivation. K. S. was supported by internal funds from Portland State University. This work was supported by the National Science Foundation through a CAREER Award [grant number DBI-1253293]; the National Institutes of Health (NIH) [grant numbers HG007233-01, R01-EB019453-01, DP2-AR068129-01]; and the Defense Advanced Research Projects Agency Living Foundries Program [contract numbers HR0011-12-C-0065, N66001-12-C-4211, HR0011-12-C-0066]. S. W. L. was supported by a fellowship from the Agency of Science, Technology and Research, Singapore. The authors would also like to thank TWiV (www.twiv.tv) for stimulating this collaboration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate AR, Weitz Da. Air-bubble-triggered drop formation in microfluidics. Lab Chip. 2011;11:1713–1716. doi: 10.1039/c1lc20108e. [DOI] [PubMed] [Google Scholar]
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz Da. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J, Amann R, Sullivan MB. Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol. 2013;15:2306–2318. doi: 10.1111/1462-2920.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B, Rashid MH, Carter C, Pasternack G, Rajanna C, Revazishvili T, Dean T, Senecal A, Sulakvelidze A. Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and NanoSight-based assays. Bacteriophage. 2011;1:86–93. doi: 10.4161/bact.1.2.15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Arber W, Enquist L, Hohn B, Murray N, Murray K. Experimental methods for use with lambda. Lambda II 1983 [Google Scholar]
- Bohannan BJM, Lenski RE. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol Lett. 2000 doi: 10.1046/j.1461-0248.2000.00161.x. [DOI] [Google Scholar]
- Christopher GF, Anna SL. Microfluidic methods for generating continuous droplet streams. J Phys Appl Phys. 2007;40:R319–R336. doi: 10.1088/0022-3727/40/19/R01. [DOI] [Google Scholar]
- Dang VT, Sullivan MB. Emerging methods to study bacteriophage infection at the single-cell level. Front Microbiol. 2014;5:1–8. doi: 10.3389/fmicb.2014.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging. mBio. 2012;3:1–8. doi: 10.1128/mBio.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- Dhillon V, Li X. Single-Cell Genome Sequencing for Viral-Host Interactions. J Comput Sci Syst Biol. 2015;8:160–165. doi: 10.4172/jcsb.1000183. [DOI] [Google Scholar]
- Diemer GS, Stedman KM. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastburn DJ, Sciambi A, Abate AR. Identification and genetic analysis of cancer cells with PCR-activated cell sorting. Nucleic Acids Res. 2014;42:1–10. doi: 10.1093/nar/gku606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafenstein S, Fane Ba. Phi X174 genome-capsid interactions influence the biophysical properties of the virion: Evidence for a scaffolding-like function for the genome during the final stages of morphogenesis. J Virol. 2002;76:5350–5356. doi: 10.1128/JVI.76.11.5350-5356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner a, Srisa-Art M, Holt D, Abell C, Hollfelder F, deMello aJ, Edel JB. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem Commun Camb Engl. 2007;2:1218–1220. doi: 10.1039/b618570c. [DOI] [PubMed] [Google Scholar]
- Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013;14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam JD. Molecular Biology of Bacteriophage T4. ASM Press; 1994. [Google Scholar]
- Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ, Sullivan MB, Woyke T, Eric Wommack K, Stepanauskas R. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015:1–14. doi: 10.1038/ismej.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SW, Abate AR. Ultrahigh-throughput sorting of microfluidic drops with flow cytometry. Lab Chip. 2013;13:4563–72. doi: 10.1039/c3lc50736j. [DOI] [PubMed] [Google Scholar]
- Lim SW, Tran TM, Abate AR. PCR-Activated Cell Sorting for Cultivation-Free Enrichment and Sequencing of Rare Microbes. Plos One. 2015a;10:e0113549. doi: 10.1371/journal.pone.0113549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SW, Tran TM, Abate AR. PCR-Activated Cell Sorting for Cultivation-Free Enrichment and Sequencing of Rare Microbes. PLOS ONE. 2015b;10:e0113549. doi: 10.1371/journal.pone.0113549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/NMETH.1419. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013a;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazutis L, Gilbert J, Ung WL, Weitz Da, Griffiths AD, Heyman Ja. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013b;8:870–91. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Seo M, Xu S, Lewis PC, Mok M, Kumacheva E, Whitesides GM, Garstecki P, Stone Ha. Emulsification in a microfluidic flow-focusing device: Effect of the viscosities of the liquids. Microfluid Nanofluidics. 2008;5:585–594. doi: 10.1007/s10404-008-0271-y. [DOI] [Google Scholar]
- Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Pethig R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics. 2010;4:022811. doi: 10.1063/1.3456626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Refardt D. Real-time quantitative PCR to discriminate and quantify lambdoid bacteriophages of Escherichia coli K-12. Bacteriophage. 2012;2:98–104. doi: 10.4161/bact.20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Hawley AK, Beltran MT, Scofield M, Schwientek P, Stepanauskas R, Woyke T, Hallam SJ, Sullivan MB. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell and environmental genomics. eLife in review. 2014:1–20. doi: 10.7554/eLife.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciambi A, Abate AR. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip. 2015a;15:47–51. doi: 10.1039/C4LC01194E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciambi A, Abate AR. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip. 2015b;15:47–51. doi: 10.1039/C4LC01194E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciambi A, Abate AR. Generating electric fields in PDMS microfluidic devices with salt water electrodes. Lab Chip. 2014;14:2605–2609. doi: 10.1039/c4lc00078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman K. Mechanisms for RNA capture by ssDNA viruses: Grand theft RNA. J Mol Evol. 2013;76:359–364. doi: 10.1007/s00239-013-9569-9. [DOI] [PubMed] [Google Scholar]
- Suttle Ca. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K, Blons H, Bouché O, Landi B, Hutchison JB, Laurent-Puig P. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW. PNAS Plus: Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci. 2011 doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TM, Lan F, Thompson CS, Abate aR. From tubes to drops: droplet-based microfluidics for ultrahigh-throughput biology. J Phys Appl Phys. 2013;46:114004. doi: 10.1088/0022-3727/46/11/114004. [DOI] [Google Scholar]
- Turner EH, Ng SB, Nickerson Da, Shendure J. Methods for genomic partitioning. Annu Rev Genomics Hum Genet. 2009;10:263–284. doi: 10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of “unculturable” bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- Weinberger AD, Sun CL, Pluciński MM, Denef VJ, Thomas BC, Horvath P, Barrangou R, Gilmore MS, Getz WM, Banfield JF. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Whitesides GM. Soft Lithography. Annu Rev Mater Sci. 1998;28:153–184. doi: 10.1146/annurev.matsci.28.1.153. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.