Abstract
The kidney is one of the body’s main filtration organs, and hence, opportunity exists for designing nanomedicine that can naturally accumulate in the kidneys for renal diseases. In addition to traditional physiochemical properties for kidney accumulation, such as size and charge, synthesized nanoparticles can be conjugated with targeting ligands which further home the nanocarriers to cell types of interest In this review, we highlight key studies that have shown success in utilizing peptide- or antibody-based ligands in nanoparticles to target the glomerulus, podocytes, or renal tubule cells in the kidney. In addition, other ligand candidates which have shown renal affinity, but have not yet been integrated into a nanoparticle are also presented. These studies can provide insight into the design of novel clinical solutions for improved detection, prevention, and treatment of renal diseases using nanomedicine efforts
1. Introduction
Medications that are administered systemically have potential for adverse side effects. The kidney and liver are especially susceptible, as together they receive approximately 45% of the cardiac output, and are the main organs involved in the excretion and metabolism of wastes in the body.1,2 For instance, common antibiotics or non-steroidal, anti-inflammatory drugs may cause tubulointerstitial nephritis and acute tubular necrosis in the kidney when high doses are administered systemically.3 Therefore, nanomedicine aims to exploit properties of matter at the nanoscale to improve traditional therapeutics or create new opportunities for treatment.4 For example, utilizing nanoparticles to encapsulate or conjugate therapeutics can improve pharmacokinetics, increase potency to a specific tissue type, or deliver molecules to tissues which are normally considered “undruggable” due to the body’s immune and excretory systems.5
In this review, we discuss key studies in nanomedicine towards kidney applications. We specifically focus on peptide-and antibody-based targeting ligands that have been used in nanoparticle studies. We also compile additional peptide and antibody ligands that show renal specificity, which may be used in nanoparticle systems in future efforts. Amino acid-based ligands are selected due to their biocompatibility and relatively safe degradation products.6,7 For an overview of kidney targeting strategies encompassing antibody mimetics, proteins, nucleic acids, and small-molecule ligands, refer to the review by Kamaly et al.8 Here, we first summarize the fundamental physical properties of nanoparticle size and charge that affect biodistribution, which provide a “passive” targeting effect toward the kidneys without specific ligand interaction with cells. Subsequently, targeting ligands reviewed will be categorized by the specific kidney cell type they target, which includes glomerular endothelial and epithelial cells, tubule cells, podocytes, or mesangium cells. This intra-kidney specificity is desirable to focus therapeutics specifically to diseased cell types, and minimize off target toxicity.
1.1. Renal anatomy and common pathologies
The main repeating functional unit of the kidney is the nephron. Nephrons are organized into layers within the kidney, where the glomerulus, proximal and distal tubules residing mostly in the cortex, and the loop of Henle and collecting duct form deeper layers in the medulla (Fig. 1).9 The first renal structure unfiltered blood encounters is the glomerulus. The glomerulus is comprised of a cluster of capillaries surrounded by Bowman’s capsule, which together form the renal corpuscle. This structure is lined with parietal epithelial cells, thought to serve as a reservoir for renal progenitor cells.10 The interstitial spaces within the glomerular capillaries are occupied with mesangial cells, which provide the maintenance of capillary organization and support filtration processes for glomerular endothelial cells.11 The glomerular filtration barrier surrounds the endothelial cells and mesangial cells, which consists of the glomerular basement membrane as well as associated podocytes. The podocytes form interdigitating foot processes along the glomerular basement membrane, creating 4–11 nm slits that prevent the passage of large macro-molecules. In sum, the kidney is an organ with many cell types exposed to ever changing external fluid compositions and concentrations.
Fig. 1.
Structure and cellular makeup of the nephron. Adapted and reprinted from ref. 8 with permission from Nature Publishing Group.
Acute kidney injury (AKI) is a broad term and historically assessed by the magnitude of serum creatinine concentration increase, a waste product generated by the muscles.12 Normal levels for a healthy individual range between 0.6–1.2 milligram per deciliter of blood.13 Common causes of acute pathologies include hypotension/sepsis, trauma, acute tubular necrosis, contrast injury, and urinary obstruction.14 Genetic predispositions may instead cause chronic conditions such as polycystic kidney disease or lupus nephritis. Repeated exposure to harmful substances and secondary chronic conditions such as diabetes or high blood pressure can also lead to glomerulone-phritis. For a review of nanoparticles used for kidney applications categorized by disease type, refer to the review by Brede et al.15
1.2. Nanoparticle size and charge
Most studies have administered nanoparticles intravenously, as this delivery route offers the highest bioavailability.16 One of the most studied factors in nanoparticle design is size, as differing diameters of the same nanoparticle composition can cause vastly different biodistribution profiles after injection.17 In healthy kidneys, nanoparticles with hydrodynamic diameters up to 7 nm are able to pass through the glomerular membrane barriers and are cleared via renal excretion.18,19 The vast majority of studies indicate that both rigid metal20 or flexible polymer21 nanoparticles in this size range are cleared by the kidney (Fig. 2).22,23 In diseased states, the breakdown of the glomerular endothelial cell lining and podocyte architecture can lead to the passage of much larger substances, leading to proteinuria.15
Fig. 2.
Typical size relation to the distribution of nanoparticle accumulation in the lungs, liver, spleen, and kidneys. Adapted and reprinted from ref. 23 with permission from Nature Publishing Group.
Deviations to this general size trend, however, have been reported. Choi et al. utilized Au-PEG nanoparticles with varying core sizes between 5 and 98 nm and a polyethylene glycol (PEG) “brush border” with the molecular weight between 5000 and 20 000 g mol−1 to investigate the size dependence of nanoparticles on kidney accumulation.24 Optimal targeting to the mesangium, or a thin membrane of cells that supports glomerular capillaries, of healthy BALB/c mice kidneys was seen for nanoparticles approximately 75 nm in diameter. Particle accumulation inside renal corpuscles revealed a strong function of size: particles on the order of 10 nm are suggested to enter the mesangium only briefly, where they are not retained due to an absence of phagocytosis. Particles larger than 75 nm are too large to pass through the glomerular endothelium pores. Interestingly, accumulation in peritubular capillaries did not seem to correlate with size, demonstrating distinct uptake characteristics in different portions of the nephron. Williams et al. constructed “mesoscale” nanoparticles, approximately 400 nm in diameter, which accumulated seven times more efficiently in the kidney compared to the heart, lung, spleen, and liver (Fig. 3).25 Poly(lactic-co-glycolic acid) conjugated to polyethylene glycol (PLGA-PEG) nanoparticles were synthesized in several forms, including anionic (ζ-potential = −19.5 ± 0.6 mV), cationic (ζ-potential = −19.5 ± 0.6 mV), and neutral (ζ-potential = 0.38 mV) forms. No significant difference was observed regarding organ distribution among all surface charge variations.26 They were observed to localize in the basolateral region of proximal tubule epithelial cells under histological analysis. However, studies on the opposite end of the size spectrum, with ultrasmall nanoparticles <5.5 nm, indicate that charge is a primary determinant of kidney uptake or excretion. Negatively-charged quantum dots (QDs) (~3.7 nm) as a model system were used by Liang et al. and found to accumulate in mesangial cells, with little found in urine.27 In contrast, cationic QDs of a similar size (~5.67 nm) were found to be readily excreted into urine shortly after injection.
Fig. 3.
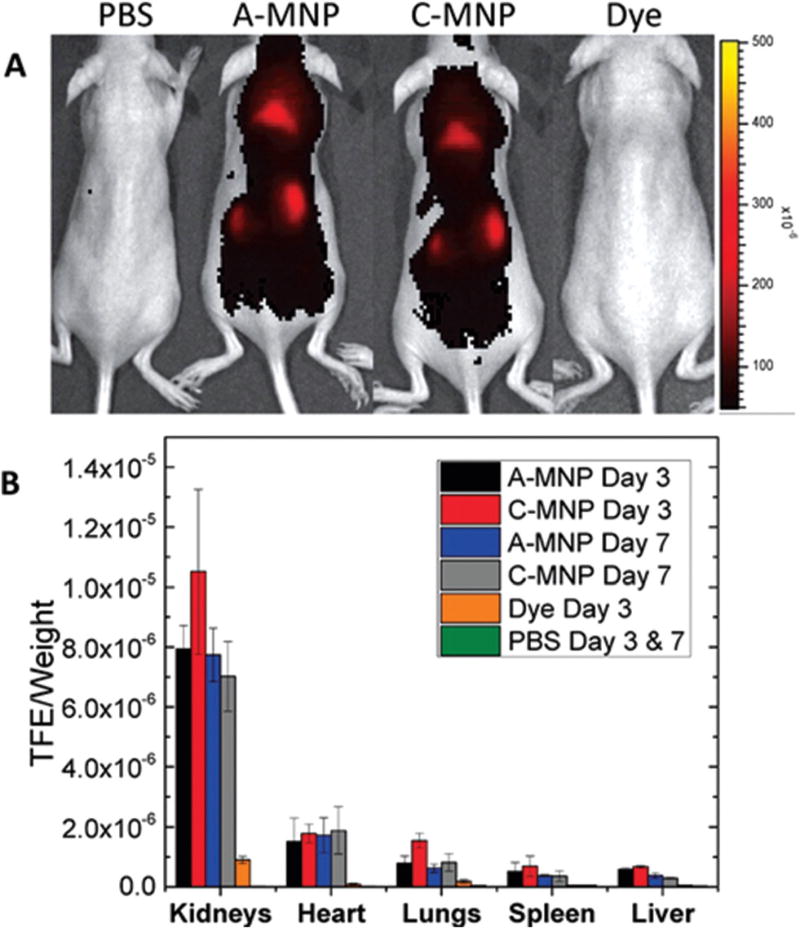
In vivo biodistribution of mesoscale nanoparticles, approximately 400 nm in diameter, showing heightened accumulation in the kidneys compared to other organs. (A) Dorsal image of mice treated with PBS, 50 mg kg−1 anionic-MNP (A-MNP), 50 mg kg−1 cationic-MNP (C-MNP), and an equal molar weight of free dye. (B) Ex vivo organ fluorescence from mice injected with MNPs, dye, or PBS (mean ± SD). Adapted and reprinted from ref. 25 with permission from Nature American Chemical Society.
In addition, deviations from the general size trend can be achieved using glutathione, a naturally occurring tri-amino acid peptide present in both oxidized and reduced forms in human kidneys, when coated on copper nanoparticles of 2 nm average diameter. Yang et al. expected that dissociated components of this particle would be cleared by the kidney faster, as is the case with most nanoparticles that dissociate.28,29 However, these glutathione copper nanoparticles were found to clear faster compared to their dissociated components. This surprising result indicates that the surface modification can also dictate particle clearance. The authors discuss that the coated nanoparticles may have a lower affinity to serum protein adsorption compared to their fragments, allowing faster urinary system excretion.
Recently, the density of the nanoparticle has also been shown to affect biodistribution. By again utilizing glutathione as a coating for gold or silver core nanoparticles of varying densities, Tang et al. found that the renal clearance efficiency exponentially increased with a decrease in particle density two hours post-injection.30 This behavior likely originates from the density-dependent margination effect in blood circulation, where higher density nanoparticles circulate more slowly, thus leading to slower renal clearance.
2. Peptide ligands
The active targeting of nanoparticles typically involves ligands bound to the surface which enables a higher degree of specificity compared to untargeted particles of the same physical characteristics such as size and charge.31 Peptide ligands take advantage of highly specific interactions between the ligand and the target site to promote accumulation of nanoparticles. Peptides are naturally degradable, easily synthesized, and custom tunable with a variety of linker chemistries. By employing targeting peptides to nanoparticles, drugs have the potential to reduce side effects and toxicity associated with current therapies for kidney disease, and can generate a higher intrarenal drug concentration compared to that of free drug.32 Notably, many potential candidates for kidney targeting peptides from phage display technologies have been identified,33 but relatively few nanoparticle studies for targeted drug delivery applications have been attempted in recent years. We present potential peptides, organized by cell target, that have shown renal specificity and can be used as active targeting ligands for nanomedicine. Compiled in Table 1 is a summary of these targeting peptides.
Table 1.
Kidney targeting peptide sequences
| Target cell | Peptide sequence | Ref. |
|---|---|---|
| Podocytes | Cyclo(RGDfC) | 35 |
| Glomerular endothelium/basement membrane | PKNGSDP, DSHKDLK | 39 |
| CYFQNCPRG | 30 and 41 | |
| CLPVASC | 33 | |
| Tubule cells | MCLPVASCGGPGVG (VPGxG)160VPGWPGSGGC | 43 |
| ANTPCG-PYTHDCPVKR | 32 | |
| ELRGD(R/M)AX(W/L) | 44 | |
| GVKGVQGTL, HGVRGNLIS, and GVRGQLATP | 45 | |
| LTCQVGRVH | 46 | |
| (KKEEE)3K | 49 |
2.1. Podocytes
Podocytes surround the capillaries and mesangium, and possess unique foot-like structures which aid in selectively filtering blood components. Many common genetic diseases affect podocytes such as the Alport syndrome, membranous nephropathy, and segmental glomerular sclerosis. Secondary conditions such as hypertension and diabetic neuropathy can also cause podocyte dysfunction which results in kidney disease.34 Pollinger et al. developed cyclo(RGDfC)-modified quantum dots (Qdots), which are rod-shaped constructs with a length of 10 nm to 15 nm and a width of 5 nm.35 Cyclo (RGDfC) was hypothesized to demonstrate ligand-specific binding for the αvβ3 integrin expressed on podocytes. This particular integrin is discussed to play a role in kidney viral infection, making it a candidate for facilitating cellular uptake.36 Highly specific, cell and receptor binding was observed in an ex vivo study of primary podocytes. Confocal microscopy showed cellular Qdot uptake into vesicle-like structures within glomerular podocytes. The receptor mediated nature of nanoparticles binding to podocytes was further confirmed with a competition assay using free peptides.
2.2. Glomerular endothelial cell barrier and basement membrane
The glomerular endothelial cells are adjacent to the glomerular capillaries, and form 60–80 nm intercellular pores, integral to the function of filtration.8 Impairment of this endothelial surface layer leads to albuminuria and glomerulosclerosis.37 The glomerular endothelial cells and podocytes secrete a layer of extracellular matrix which forms the glomerular basement membrane, composed of collagen IV, laminin, and proteoglycans.38 Recently, Jung et al. took a different approach to traditional phage display studies by performing several machine learning models to rapidly predict the tissue-specific targeting capabilities of peptides based on sequence information.39 Several machine learning algorithms utilized training data gathered from previous phage display studies.33 Possible peptide sequences were given a prediction score for different organ affinities. Two sequences showed significantly higher prediction scores for the kidney: PKNGSDP and DSHKDLK. The authors conclude that PKNGSDP and DSHKDLK can recognize the glomerular endothelial markers of the kidney based on predictions of their model.
To enhance the tissue-specific interactions of targeting peptides, Suzuki et al. linked the carbohydrate-modified Arg-vaso-pressin (AVP) peptide, or CYFQNCPRG, to a variety of sugars via an octamethylene spacer group.40 They proposed that the carbohydrate-modified peptides would act as the targeting moiety while the sugar would act as a ligand to deliver drugs by attaching to sugar recognition molecules previously investigated.41 To determine the kidney targeting capabilities of carbohydrate-modified AVP, Suzuki et al. injected radiolabeled carbohydrate-modified AVP, as well as unmodified AVP as control, into Sprague-Dawley ex vivo. They found that unmodified AVP distributed in the medulla, which contain the majority of nephron structures. Glycosylated (Glc-O-C8-AVP) and manno-sylated (Man-O-C8-AVP) forms distributed mainly in the cortex, which contains support blood vessels and portions of the proximal convoluted tubule. Suzuki et al. found that clearance of Glc-O-C8-AVP (1.7 mL min−1 g−1) was 13 times greater than glomerular filtration clearance (0.13 mL min−1 g−1). The authors note that there are two main routes of renal uptake, which include circulating blood through the basolateral membrane, and from the luminal side after glomerular filtration. They reason that since clearance via the circulating blood cannot be greater than the rate of glomerular filtration, the results indicate that the renal uptake of Glc-O-C8-AVP takes place from blood via the basolateral membrane.
Historically, to determine organ-selective targeting peptides, namely to the kidney and brain, Pasqualini et al. utilized phage display to find that the peptide sequence CLPVASC demonstrated about seven-fold greater preferential binding to the kidney relative to other organs.33 A closer examination via immunohistochemistry revealed that CLPVASC was found in both the glomeruli and between tubules of the kidney.
2.3. Tubular cells
Tubular epithelial cells in the proximal tubule gather the urine to the collecting ducts, and exchange substances throughout the length of the tubule. This epithelium is highly susceptible to injury, and will be an important cellular target for therapies intended to treat proinflammatory and fibrogenic tubular injury.42 Bidwell et al. modified the sequence suggested by Pasqualini et al., CLPVASC, with an elastin-like polypeptide (ELP), to specifically target the tubule.43 The authors discuss that fusing small-molecules and peptide therapeutics to relatively large, nonimmunogenic protein-based carrier ELPs can increase their stability for enhanced renal accumulation. This modified ELP peptide was hypothesized to behave like an extended rod-like protein, suggesting that it is filtered by the glomerulus and reaches the tubules by protein re-uptake. One mechanism suggested to underlie this phenomenon is the hydrostatic forces orienting these rod-like structures perpendicularly to the basement membrane, enabling the insertion of their <10 nm axis through the membrane.8 The kidney-selective biopolymer demonstrated significantly higher renal accumulation (over 15-fold higher than in other organs), and a five-fold increase in plasma half-life over untargeted ELPs upon administration in rats. Fluorescence slide scanning of the rat kidney sections showed that the CLPVASC-ELP primarily localized in the renal cortex. To show that the effects of CLPVASC-ELP are not species-specific, Bidwell et al. also injected the biopolymer in swine and found localization in the tubule cells, vascular endothelial, and smooth muscle cells. The authors suggest that the free COOH-terminal thiol group on CLPVASC-ELP will allow thiolated drugs or other small-molecule drugs with thiol cleavable linkers to be attached for kidney-specific delivery.43
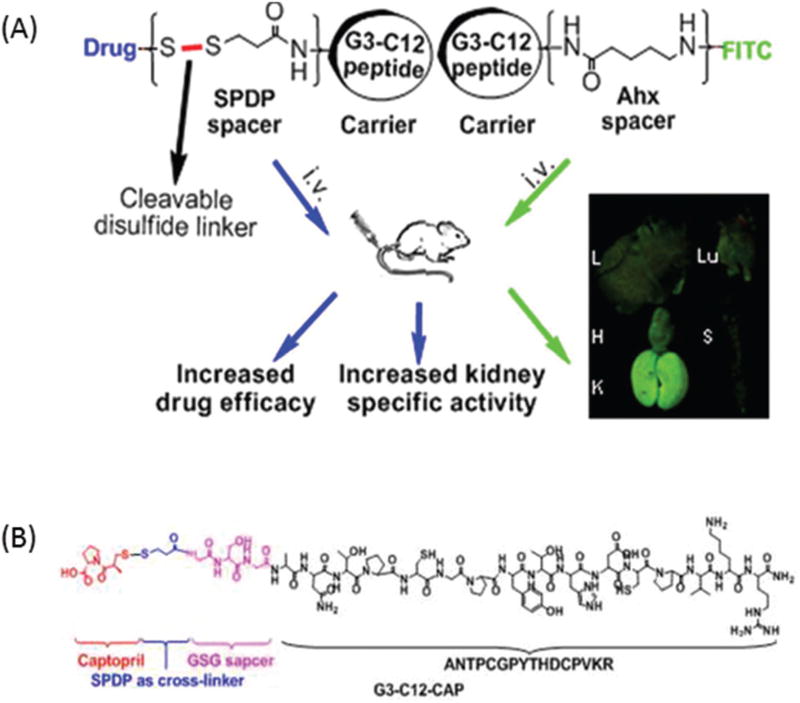
A study by Geng et al. showed that the targeting peptide identified by phage display, G3-C12 (ANTPCG-PYTHDCPVKR) was able to accumulate in the kidneys via renal proximal tubule cell reabsorption only three minutes post-injection (Fig. 4A and B).32 A small molecule drug model captopril (CAP), which acts to reduce glomerular hypertension and renal injury, was coupled to the targeting peptide with a disulfide bond. The bond was demonstrated to be cleavable in the kidney, releasing free CAP into the renal proximal tubule cells. CAP release from the carrier–drug conjugate was caused by the abundance of reduced glutathione in the kidney, which cleaved the disulfide bond and allowed CAP to be separated from G3-C12 at the proximal tubule cells. Following this process, the G3-C12 peptides were filtered by the glomerulus and reabsorbed by the proximal tubule cells; they were then transferred to the lysosomal compartment to be metabolized. The fact that the carrier itself was internalized by the tubule cells is important, as it generated a higher drug concentration in the kidney compared to other parts of the body such as the liver, lung, heart, and spleen. Geng et al. explain that G3-C12 was likely taken up by the renal proximal tubule cells via endocytosis because three out of its 16 total amino acids are positively charged. This allows the peptide to interact with and be reabsorbed by megalin, a negatively charged receptor highly expressed in the renal proximal tubules.
Fig. 4.
(A) Geng et al. successfully developed the peptide–drug conjugate G3-C12 for the kidney-targeted delivery of CAP. The disulfide bond serves as a covalent linker that can be cleaved by the reduced gluta-thione in the kidney, releasing free CAP. (B) The structure of G3-C12-CAP, which contains a GSG spacer (to allow for conformational freedom and low steric hindrance), a N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP) cross-linker, and CAP. Adapted and reprinted from ref. 32 with permission from Nature American Chemical Society.
Odermatt et al. were the first to report a new method for screening phage-display libraries ex vivo on microdissected intact kidney tubules instead of plated cells.44 They found that the linear peptide ligand with the sequence ELRGDMAAL (or a similar sequence with the motif ELRGD(R/M)AX(W/L)) selectively binds to the basolateral cell surface of cortical collecting ducts (CCD) compared to other parts of the kidney such as the proximal convoluted tubules (PCT). Peptide sequences containing this consensus motif exhibited a 16-fold higher binding to CCD compared with PCT in rats, and a 39-fold higher binding compared with the control phage. Using this same method, Odermatt et al. found that phages with the consensus motif K(X3)TNHP bound preferentially to PCT. Peptide sequences with this motif showed a 2-fold higher binding to PCT compared to CCD, and a 10-fold higher binding compared with the control phage. Through screening phage display libraries ex vivo on microdissected renal tubular segments, Audigé et al. identified two distinct motifs, GV(K/R) GX3(T/S) (GV-phage) and RDXR (RD-phage), that bind specifically to receptors expressed at the basolateral membrane of PCT.45 Specifically, the peptide sequences of the GV-phages are GVKGVQGTL, HGVRGNLIS, and GVRGQLATP and the sequences of the RD-phages are GMRDHRMTI, ETMQRDVRA, YRDFRDIWA, SLRDRGFT, HLNMWRDGG, and GGAIKDTQN. The GV-phage or RD-phage exhibited 15-fold and 13-fold higher binding to PCT compared to the control phage, respectively.
Kim et al. coupled the kidney targeting peptide, LTCQVGRVH, identified by phage display, to a biodegradable poly(ester amine) (PEA) via an amide bond between the amine group of PEA and the carboxyl group of the peptide.46 PEA is a gene carrier with a high transfection efficiency. The PEA developed in this study was based on glycerol dimethacrylate (GDM) and low molecular weight polyethylenimine (LMW PEI), which is a non-viral vector system with high transfection efficiency but also high toxicity. PEA based on GDM and LMW PEI was developed to create a carrier with reduced cytotoxicity while maintaining high transfection efficiency. The kidney targeting and gene delivery potential of LTCQVGRVH-PEA was evaluated both in vitro using the 293T cell line and in vivo with Sprague-Dawley rats with induced unilateral ureteral obstruction (UUO) kidney fibrosis models. Results from both types of studies indicate that the peptide-conjugated PEA achieved low cytotoxicity, as well as high transfection efficiency and high specificity delivering the hepatocyte growth factor (HGF) gene which is involved in promoting tubular repair and to inhibit tissue fibrosis.47 More specifically, Kim et al. showed that rats treated with peptide-conjugated PEA delivering HGF had recovered renal function and reduced collagen.46
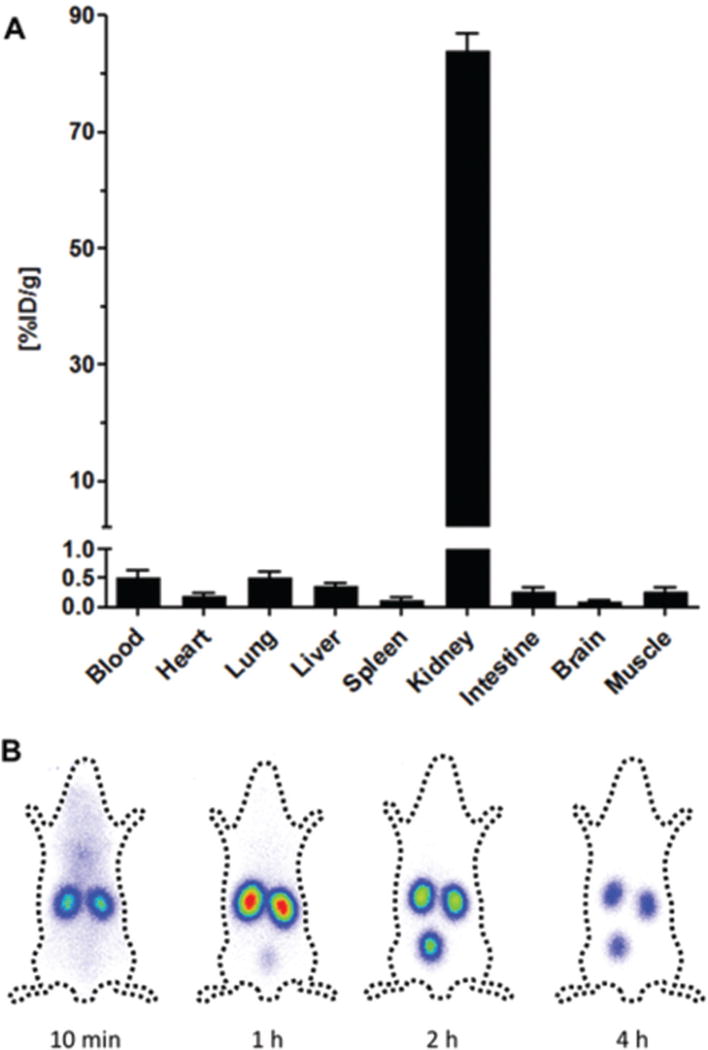
Building on previous work regarding lysine interactions with receptors on the apical side of proximal renal tubule cells,48 Wischnjow et al. developed the peptide sequence (KKEEE)3K.49 They showed that the targeting peptide had high renal specificity and accumulation in proximal tubule cells, making it an ideal kidney-specific carrier for drug delivery and the treatment of kidney diseases (Fig. 5). Immunohistochemistry showed that (KKEEE)3K specifically accumulated in the renal cortex. Like the G3-C12 peptide, (KKEEE)3K was absorbed via megalin-mediated endocytosis as demonstrated by the lack of (KKEEE)3K accumulation in mice with megalin-deficient kidneys. The peptide remained stable in serum after 24 hours with no signs of degradation. In addition to having exceptional kidney accumulation and stability, (KKEEE)3K demonstrated renal clearance within a few hours to prevent toxicity due to long kidney retention.
Fig. 5.
Organ distribution study of labeled (KKEEE)3K in mice at 1 hour post-injection. (A) γ-Scintigraphy of (KKEEE)3K time-course. Adapted and reprinted from ref. 49 with permission from Nature American Chemical Society.
3. Antibody ligands
Antibodies are typically 200–300 nm in size proteins and are part of the adaptive immune system. Much like peptide-conjugated nanoparticles, antibody-conjugated nanoparticles can generate a product that delivers potent therapeutic or imaging agents that are highly specific to a certain tissue or antigen.31,50 We present antibody–nanoparticle systems that show renal accumulation for distinct cell types (Table 2).
Table 2.
Kidney targeting antibodies
3.1. Glomerular endothelial cells and basement membrane
An antibody-conjugated nanoparticle strategy was used by Hultman to target the rat major histocompatibility class II (RT1 anti-MHC II).51 MHC II is a transmembrane macromolecular marker found typically on antigen-presenting cells, and is constitutively expressed in the healthy medulla of the human kidney, specifically in the renal peritubular and glomerular capillaries, with minimal expression in the renal cortex. Superparamagnetic iron oxide nanoparticles (SPIO) were coated with a phospholipid monolayer of PEG methyl ether (mPEG), to reduce the nonspecific binding of proteins in vivo. These nanoparticles were conjugated to RT1 anti-MHCII antibodies via a maleimide linkage, and antibody-conjugated nanoparticles showed an elimination half-life in the kidney of 255 minutes, compared to the 45 minutes of the unconjugated nanoparticles as well as nanoparticles conjugated to nonspecific antibodies. This large increase in the plasma half-life indicates that the particle is interacting in a target-selective fashion with MHC II, as suggested by the observed trend of receptor and plasma protein binding being directly proportional to the plasma half-life.52 This may provide a quantifiable vehicle for the kidney delivery of imaging agents or preventative therapeutics associated with MHC II-related disorders.
Serkova et al. utilized coated iron oxide nanoparticles, with a mean diameter of 9.7 nm, to target a membrane protein expressed in the inflamed kidney disease state.53 Complement receptor type 2 (CR2) is a transmembrane protein that binds to the circulating complement protein C3 fragments, which increases in activity when inflammation is present in the kidneys. MRL/lpr mice were used in this study; they are a mouse model that spontaneously develops lupus-like renal disease characterized by glomerular immune complex deposition and complement fragment activation. Moreover, this model has demonstrated C3 deposition along the Bowman capsule and tubular basement membrane. SPIO nanoparticles were conjugated with anti-CR2 monoclonal antibodies (CR2-Fc) on the surface, verified with flow cytometry. Iron from the nanoparticle core was detectable via magnetic resonance imaging (MRI) 72 hours after injection in some glomeruli and tubules of MRL//lpr mice injected with the CR2-Fc iron oxide nanoparticles, while not seen in normal kidneys. This strategy can be used to noninvasively detect inflammation in the kidneys due to lupis nephritis, with wider applications to several types of autoimmune and inflammatory kidney diseases which show characteristic C3 deposition.54
Vascular cell adhesion molecule-1 (VCAM-1) is an important regulatory molecule for leukocyte adhesion to tissues and is upregulated in the kidney ischemia reperfusion injury. Studies in animal models indicate that such ischemia activates endo-thelial cells, which upregulates the expression of VCAM-1.55 The anti-VCAM 1 antibody conjugated to iron oxide microparticles 1 µm in diameter has been explored as a diagnostic tool for renal endothelial cell inflammation.56 Anti-VCAM-1-conjugated particles were found to bind to the endothelium rapidly, occurring within minutes, which has potential as a fast-acting delivery vehicle for therapeutic or imaging agents. MRI contrast utilizing the iron composition of the microparticles was observed within 30 minutes of injection into mice, and persisted for the entire 90-minute imaging session. Accumulation of microparticles was evident in the renal cortex and medulla, and did not show any signs of local infarction or hemorrhage, confirming the potential of anti-VCAM-1-conjugated iron oxide as a diagnostic tool for renal inflammation.
Asgeirsdottir et al. focused on another inflammatory marker exploited for kidney targeting properties.57,58 The site-selective delivery of E-selectin targeting liposomes (AbEsel) on the order of 114 nm was shown to deliver encapsulated dexa-methasone which reduces endothelial cell activation and counteracts glomerulonephritis progression. In an induced accelerated glomerulonephritis (GN) model generated from C57bl/6 mice, liposomes reduced glomerular proinflammatory gene expression but did not affect blood glucose levels, which is typically elevated to severe levels upon administration of free dexamethasone, demonstrating a classic benefit of nanoparticle drug delivery. Dexa-AbEsel liposomes reduced renal injury as shown by a reduction of blood urea nitrogen levels, decreased glomerular crescent formation, and down-regulation of disease-associated genes in C57bl/6 mice. Immunohistochemistry confirmed that uptake corresponded to the glomerular surface.
Liposomes were formulated and conjugated with an anti-α8 integrin antibody by Scindia et al., which allows for specific accumulation and targeting to mesangial cells.59 α8 integrin, which belongs to the family of membrane proteins which allow for cell adhesion, was confirmed by the authors to be expressed in both normal and GN glomerular mesangial cells in mice via immunofluorescence. α8 integrin is not present on human kidney endothelial cells, but was confirmed in this study to be characteristic of mesangial cells.60 Immunoliposomes modified with the anti-α8 integrin antibody were found to be approximately 100 nm in diameter and entered the mesangial space from the circulation through the 130–170 nm endothelial fenestrations and are ultimately taken up by mesangial cells. It was suggested that the slit pores of 30–70 nm between podocyte foot processes prevented rapid excretion of the liposomes into the urinary space. A similar study was performed with immunoliposomes containing mycophenolate mofetil (MMF), which showed targeting to the Thy1.1 antigen in rat mesangial cells expressing GN.61 An established model of Thy-1 nephritis was used in which a single intravenous injection of an anti-Thy-1 antiserum is known to induce lysis of mesangial cells.62 Proteinuria and creatinine levels were successfully prevented to reach pathological levels in treated nephritic Wistar rats, while healthy control rats were not affected by treatment. Therapies which involve only mesangial cells can benefit from a targeted approach which spares endothelial cells from off-target drug interaction.
3.2. Tubular cells
Shirai et al. investigated silica nanoparticles (SiNPs) conjugated to anti-CD11b antibodies in a UUO mouse model characterized by inflammation in tubular cells, as well as interstitial inflammation and fibrosis.63,64 Renal inflammation imaging was successfully achieved by the intravenous injection of SiNP with fluorescent-labeled anti-CD11b. After antibodies were immobilized on the SiNP surface such that specificity to the Fc region was preserved, nanoparticles were found to have a high affinity for macrophages at the site of renal damage. Significantly higher affinity was observed for the orientation specific antibody SiNP compared with the antibody immobilized in a random orientation, or free antibody, indicating the importance of antibody orientation with respect to the particle for targeting.
Liposomes with antibodies against human renal cancer markers have also been investigated.65 Dal K29, a murine IgGl monoclonal antibody against human renal cell carcinoma, was conjugated to unilamellar liposomes on the order of 600 to 800 nm in diameter to deliver methotrexate, a common chemotherapy and immunosuppressant. In vitro studies took place on the Caki-1 cell line, which is a human clear cell renal cell carcinoma that expresses characteristic features of the proximal tubule epithelium. Endocytosis of Dal K29-linked liposomes was observed, with some antibody-conjugated lipo-somes found in clathrin coated pits.66 The liposome constructs were endocytosed more effectively and provided more potent inhibition of cancer cell growth compared to equimolar amounts of free methotrexate, as wells as liposomes conjugated to nonspecific antibodies. This study demonstrates that nanoparticle conjugation with a targeting ligand has the potential to increase specificity as well as potency compared to systemic free drug administration.
In a mouse model of rhabdomyolysis-induced AKI, which releases myoglobin into the bloodstream to cause kidney damage, Rubio-Navarro et al. incorporated the antibody targeting the injury marker CD163, which is associated with apoptosis of proximal tubule cells.67 Renal dysfunction was induced in mice through glycerol administration, which increased CD163 expression. Rhabdomyolysis-induced kidney injury in humans was also found to have enhanced CD163 expression compared to healthy tissues. Gold-coated iron oxide particles on the order of 6 nm were conjugated to anti-CD163 IgG antibodies, and electron microscopy confirmed the presence of these nanoparticles within interstitial macrophages in the kidney tubules in vivo.68 Such molecular specificity to disease cell and tissue types can lead to early detection, along with more efficient therapeutic delivery.
4. Conclusion
A variety of targeting peptides and antibodies have been utilized with nanoparticles to target the kidney and kidney cells, including the glomerular mesangial cells, glomerular endothelial cells, and tubule cells. This strategy can give researchers a wide selection of potential targets when developing diagnostic or therapeutic strategies to renal diseases that affect particular kidney cell types. Targeting nanoparticles may offer a more predictable trend in particle behavior compared to nanoparticles with a specific size, charge, and other physiochemical properties. A case by case evaluation of each nanoparticle system is needed, as a combination of different physiochemical factors may cause deviation from the size and charge trends in kidney uptake. While many studies presented herein only investigate the targeting properties of the nanoparticle system in a diseased or healthy kidney model, simultaneous comparison in both states is needed to elucidate the specific cellular differences that are interacting with nanoparticles. Nonetheless, nanoparticle systems with the targeting ligands show great promise for the treatment of kidney disease in a clinical setting and can be an effective strategy in future studies.
Acknowledgments
The authors thank Christopher Poon, Sarah Milkowski, and Timothy Chang for assisting in the literature search and revision process, and Khosrow Khodabandehlou for helpful discussions. The authors would like to acknowledge the financial support from the University of Southern California Provost Fellowship and the National Heart, Lung, and Blood Institute (NHLBI), R00HL124279.
Biography

Eun Ji Chung
Eun Ji Chung is a Gabilan Assistant Professor in the Department of Biomedical Engineering at the University of Southern California. She received her Ph.D. from the Interdisciplinary Biological Sciences Program and the Department of Biomedical Engineering from Northwestern University, and finished her postdoctoral training at the Institute for Molecular Engineering at the Chung is a recipient of the NIH Pathway to Independence Award, and her research interests include molecular design, nano-medicine, regenerative medicine, and theranostics towards clinical applications including cardiovascular and kidney diseases.
References
- 1.John R, Herzenberg AM. J. Clin. Pathol. 2009;62:505. doi: 10.1136/jcp.2008.058271. [DOI] [PubMed] [Google Scholar]
- 2.Henderson JM, Millikan WJ, Hooks M, Noe B, Kutner MH, Warren WD. Hepatology. 1989;10:288–291. doi: 10.1002/hep.1840100306. [DOI] [PubMed] [Google Scholar]
- 3.Ravnskov U. Br. J. Clin. Pharmacol. 1999;47:203–210. doi: 10.1046/j.1365-2125.1999.00869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pourgholi F, Hajivalili M, Farhad J-N, Kafil HS, Yousefi M. Biomed. Pharmacother. 2016;77:98–107. doi: 10.1016/j.biopha.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Wu SY, Lopez-Berestein G, Calin GA, Sood AK. Sci. Transl. Med. 2014;6:240. doi: 10.1126/scitranslmed.3008362. ps247-240 ps247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung EJ. Exp. Biol. Med. 2016;241:891–898. doi: 10.1177/1535370216640940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards DA, Maruani A, Chudasama V. Chem. Sci. 2017;8:63–77. doi: 10.1039/c6sc02403c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamaly N, He JC, Ausiello DA, Farokhzad OC. Nat. Rev. Nephrol. 2016;12:738–753. doi: 10.1038/nrneph.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurts C, Panzer U, Anders HJ, Rees AJ. Nat. Rev. Immunol. 2013;13:738–753. doi: 10.1038/nri3523. [DOI] [PubMed] [Google Scholar]
- 10.Romagnani P. Experimental Models for Renal Diseases. 2011;169 [Google Scholar]
- 11.Scholondorff D, Banas B. J. Am. Soc. Nephrol. 2009;20:1179–1187. doi: 10.1681/ASN.2008050549. [DOI] [PubMed] [Google Scholar]
- 12.Perrone RD, Madias NE, Levey AS. Clin. Chem. 1992;38:1933. [PubMed] [Google Scholar]
- 13.Kellum JA, Sileanu FE, Murugan R, Lucko N, Shaw AD, Clermont G. J. Am. Soc. Nephrol. 2015;26:2231–2238. doi: 10.1681/ASN.2014070724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta RL, Kellum JA, Shah SV, Molitoris BA, Ronco C, Warnock DG, Levin A. Crit. Care. 2007;11:R31. doi: 10.1186/cc5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brede C, Labhasetwar V. Adv. Chronic Kidney Dis. 2013;20:454–465. doi: 10.1053/j.ackd.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller RH, Keck CM. J. Biotechnol. 2004;113:151–170. doi: 10.1016/j.jbiotec.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 17.De Jong WH, Hagens WI, Krystek P, Burger MC, Sips AJAM, Geertsma RE. Biomaterials. 2008;29:1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 18.Lankveld DP, Oomen AG, Krystek P, Neigh A, Troost-de Jong A, Noorlander CW, Van Eijkeren JC, Geertsma RE, De Jong WH. Biomaterials. 2010;31:8350–8361. doi: 10.1016/j.biomaterials.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Dehaini D, Fang RH, Luk BT, Pang Z, Hu CM, Kroll AV, Yu CL, Gao W, Zhang L. Nanoscale. 2016;8:14411–14419. doi: 10.1039/c6nr04091h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenblum LT, Kosaka N, Mitsunaga M, Choyke PL, Kobayashi H. Mol. Membr. Biol. 2010;27:274–285. doi: 10.3109/09687688.2010.481640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi H, Brechbiel MW. Mol. Imaging. 2003;2:1–10. doi: 10.1162/15353500200303100. [DOI] [PubMed] [Google Scholar]
- 22.Longmire M, Choyke PL, Kobayashi H. Nanomedicine. 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco E, Shen H, Ferrari M. Nat. Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi CHJ, Zuckerman JE, Webster P, Davis ME. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6656–6661. doi: 10.1073/pnas.1103573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams RM, Shah J, Ng BD, Minton DR, Gudas LJ, Park CY, Heller DA. Nano Lett. 2015;15:2358–2364. doi: 10.1021/nl504610d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He C, Hu Y, Yin L, Tang C, Yin C. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 27.Liang X, Wang H, Zhu Y, Zhang R, Cogger VC, Liu X, Xu ZP, Grice JE, Roberts MS. ACS Nano. 2016;10:387–395. doi: 10.1021/acsnano.5b05066. [DOI] [PubMed] [Google Scholar]
- 28.Zhou C, Long M, Qin Y, Sun X, Zheng J. Chem. Angew. Int. Ed. 2011;50:3168–3172. doi: 10.1002/anie.201007321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Sun S, Zhou C, Hao G, Liu J, Ramezani S, Yu M, Sun X, Zheng J. Bioconjugate Chem. 2015;26:511–519. doi: 10.1021/acs.bioconjchem.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang S, Peng C, Xu J, Du B, Wang Q, Vinluan RD, 3rd, Yu M, Kim MJ, Zheng J. Chem. Angew. Int. Ed. 2016;55:16039–16043. doi: 10.1002/anie.201609043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol. Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng Q, Sun X, Gong T, Zhang Z-R. Bioconjugate Chem. 2012;23:1200–1210. doi: 10.1021/bc300020f. [DOI] [PubMed] [Google Scholar]
- 33.Pasqualini R, Rouslahti E. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 34.Lal MA, Young KW, Andag U. Drug Discovery Today. 2015;20:1228–1234. doi: 10.1016/j.drudis.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Pollinger K, Hennig R, Breunig M, Tessmar J, Ohlmann A, Tamm ER, Witzgall R, Goepferich A. Small. 2012;8:3368–3375. doi: 10.1002/smll.201200733. [DOI] [PubMed] [Google Scholar]
- 36.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 37.Leeuwis JW, Nguyen TQ, Dendooven A, Kok RJ, Goldschmeding R. Adv. Drug Delivery Rev. 2010;62:1325–1336. doi: 10.1016/j.addr.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Groffen AJ, Ruegg MA, Dijkman H, van de Velden TJ, Buskens CA, van den Born J, Assmann KJ, Monnens LA, Veerkamp JH, van den Heuvel LP. J. Histochem. Cytochem. 1998;46:19–27. doi: 10.1177/002215549804600104. [DOI] [PubMed] [Google Scholar]
- 39.Jung E, Lee NK, Kang S-K, Choi S-H, Kim D, Park K, Choi K, Choi Y-J, Jung DH. J. Comput.-Aided Mol. Des. 2012;26:1267–1275. doi: 10.1007/s10822-012-9614-6. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki K, Susaki H, Okuno S, Yamada H, Watanabe HK, Sugiyama Y. J. Pharmacol. Exp. Ther. 1999;288:888. [PubMed] [Google Scholar]
- 41.Susaki H, Suzuki K, Ikeda M, Yamada H, Watanabe HK. Chem. Pharm. Bull. 1994;42:2090–2096. doi: 10.1248/cpb.42.2090. [DOI] [PubMed] [Google Scholar]
- 42.Ramos AM, González-Guerrero C, Sanz A, Sanchez-Niño MD, Rodríguez-Osorio L, Martín-Cleary C, Fernández-Fernández B, Ruiz-Ortega M, Ortiz A. Expert Opin. Drug Discovery. 2015;10:541–556. doi: 10.1517/17460441.2015.1033394. [DOI] [PubMed] [Google Scholar]
- 43.Bidwell GL, Mahdi F, Shao Q, Logue OC, Waller JP, Reese C, Chade AR. Am. J. Physiol. Renal Physiol. 2017;312:F54. doi: 10.1152/ajprenal.00143.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odermatt A, AudigÉ A, Frick C, Vogt B, Frey BM, Frey FJ, Mazzucchelli L. J. Am. Soc. Nephrol. 2001;12:308–316. doi: 10.1681/ASN.V122308. [DOI] [PubMed] [Google Scholar]
- 45.Audigé A, Frick C, Frey FJ, Mazzucchelli L, Odermatt A. Kidney Int. 2002;61:342–348. doi: 10.1046/j.1523-1755.2002.00120.x. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y-K, Kwon J-T, Jiang H-L, Choi Y-J, Cho M-H, Cho C-S. J. Nanosci. Nanotechnol. 2012;12:5149–5154. doi: 10.1166/jnn.2012.6372. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Dai C, Liu Y. J. Am. Soc. Nephrol. 2002;13:2464–2477. doi: 10.1097/01.asn.0000031827.16102.c1. [DOI] [PubMed] [Google Scholar]
- 48.Kaunitz JD, Cummins VPS, Mishler D, Nagami GT. J. Clin. Pharmacol. 1993;33:63–69. doi: 10.1002/j.1552-4604.1993.tb03905.x. [DOI] [PubMed] [Google Scholar]
- 49.Wischnjow A, Sarko D, Janzer M, Kaufman C, Beijer B, Brings S, Haberkorn U, Larbig G, Kubelbeck A, Mier W. Bioconjugate Chem. 2016;27:1050–1057. doi: 10.1021/acs.bioconjchem.6b00057. [DOI] [PubMed] [Google Scholar]
- 50.Arruebo M, Valladares M, Gozalez-Fernandez A. J. Nanomater. 2009;2009:1–24. [Google Scholar]
- 51.Hultman KL, Raffo AJ, Grzenda AL, Harris PE, Brown TR, O’Brien S. ACS Nano. 2008;2:477–484. doi: 10.1021/nn700400h. [DOI] [PubMed] [Google Scholar]
- 52.Scheife RT. DICP, Ann. Pharmacother. 1989;23:S27–S31. doi: 10.1177/106002808902300706. [DOI] [PubMed] [Google Scholar]
- 53.Serkova NJ, Renner B, Larsen BA, Stoldt CR, Hasebroock KM, Bradshaw-Pierce EL, Holers VM, Thurman JM. Radiology. 2010;255:517–526. doi: 10.1148/radiol.09091134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopel T, Salant DJ. C3 glomerulopathies: Dense deposit disease and C3 glomerulonephritis. [accessed 3/21, 2017]. [Google Scholar]
- 55.Seron D, Cameron JS, Haskard DO. Nephrol., Dial. Transplant. 1991;6:917–922. doi: 10.1093/ndt/6.12.917. [DOI] [PubMed] [Google Scholar]
- 56.Akhtar AM, Schneider JE, Chapman SJ, Jefferson A, Digby JE, Mankia K, Chen Y, McAteer MA, Wood KJ, Choudhury RP. PLoS One. 2010;5:e12800. doi: 10.1371/journal.pone.0012800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asgeirsdottir SA, Kamps JA, Bakker HI, Zwiers PJ, Heeringa P, van der Weide K, van Goor H, Petersen AH, Morselt H, Moorlag HE, Steenbergen E, Kallenberg CG, Molema G. Mol. Pharmacol. 2007;72:121–131. doi: 10.1124/mol.107.034140. [DOI] [PubMed] [Google Scholar]
- 58.Asgeirsdottir SA, Zwiers PJ, Morselt HW, Moorlag HE, Bakker HI, Heeringa P, Kok JW, Kallenberg CG, Molema G, Kamps JA. Am. J. Physiol. Renal Physiol. 2008;294:F554–F561. doi: 10.1152/ajprenal.00391.2007. [DOI] [PubMed] [Google Scholar]
- 59.Scindia Y, Deshmukh U, Thimmalapura PR, Bagavant H. Arthritis Rheum. 2008;58:3884–3891. doi: 10.1002/art.24026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madsen KM, Tisher CC. Anatomy of the kidney. Elements of Normal Renal Structure and function. 2004 [Google Scholar]
- 61.Suana AJ, Tuffin G, Frey BM, Knudsen L, Muhlfeld C, Rodder S, Marti HP. J. Pharmacol. Exp. Ther. 2011;337:411–422. doi: 10.1124/jpet.110.176222. [DOI] [PubMed] [Google Scholar]
- 62.Wenzel UO, Wolf G, Jacob I, Thaiss F, Helmchen U, Stahl RAK. Kidney Int. 2015;61:2119–2131. doi: 10.1046/j.1523-1755.2002.00354.x. [DOI] [PubMed] [Google Scholar]
- 63.Shirai T, Kohara H, Tabata Y. J. Drug Targeting. 2012;20:535–543. doi: 10.3109/1061186X.2012.693500. [DOI] [PubMed] [Google Scholar]
- 64.Ucero AC, Benito-Martin A, Izquierdo MC, Sanchez-Nino MD, Sanz AB, Ramos AM, Berzal S, Ruiz-Ortega M, Egido J, Ortiz A. Int. Urol. Nephrol. 2014;46:765–776. doi: 10.1007/s11255-013-0520-1. [DOI] [PubMed] [Google Scholar]
- 65.Singh M, Ghose T, Faulkner G, Kralovec J, Mezei M. Cancer Res. 1989;49:3976. [PubMed] [Google Scholar]
- 66.Singh M, Ghose T, Mezei M, Belitsky P. Cancer Lett. 1991;56:97–102. doi: 10.1016/0304-3835(91)90082-s. [DOI] [PubMed] [Google Scholar]
- 67.Rubio-Navarro A, Carril M, Padro D, Guerrero-Hue M, Tarín C, Samaniego R, Cannata P, Cano A, Villalobos JMA, Sevillano ÁM, Yuste C, Gutiérrez E, Praga M, Egido J, Moreno JA. Theranostics. 2016;6:896–914. doi: 10.7150/thno.14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tarin C, Carril M, Martin-Ventura JL, Markuerkiaga I, Padro D, Llamas-Granda P, Moreno JA, Garcia I, Genicio N, Plaza-Garcia S, Blanco-Colio LM, Penades S, Egido J. Sci. Rep. 2015;5:17135. doi: 10.1038/srep17135. [DOI] [PMC free article] [PubMed] [Google Scholar]