The application of single-cell genome sequencing to large cell populations has been hindered by technical challenges in isolating single cells during genome preparation. Here we present single-cell genomic sequencing (SiC-seq), which uses droplet microfluidics to isolate, fragment, and barcode the genomes of single cells, followed by Illumina sequencing of pooled DNA. We demonstrate ultra-high sequencing throughput of >50,000 cells per run in a synthetic community of Gram negative and Gram-positive bacteria and fungi. The sequenced genomes can be sorted in silico based on characteristic sequences. We use this approach to analyze the distributions of antibiotic resistance genes, virulence factors, and phage sequences in microbial communities from an environmental sample. The ability to routinely sequence large populations of single cells will enable the de-convolution of genetic heterogeneity in diverse cell populations.
Organisms are living expressions of their genomes and, hence, genome sequencing is a powerful way to study how they grow and function. Organisms are phenotypically diverse, and this diversity is mirrored by heterogeneity at the genomic level and plays important roles in populations as a whole, particularly among populations of single cells. A common challenge when applying single cell sequencing to heterogeneous systems is that they often contain massive numbers of cells: a centimeter-sized tumor can contain hundreds of millions of cancer cells1, while a milliliter of seawater can contain millions of microbes2. Moreover, each cell has a tiny quantity of DNA, making it challenging to accurately amplify and sequence single cells. Indeed, a long history of methods based on optical tweezers3, flow sorting4, microfluidics5,6, and single cell isolation using gel matrices7–9 can be used to isolate and process hundreds of single cells for sequencing, but this constitutes a minute fraction of most communities. The sparseness of the sampling limits the questions that can be addressed, with the majority of findings relating to the most abundant subpopulations. A method that could markedly increase the number of cells sequenced at the single cell level would impact a broad range of problems across biology where heterogeneity is important.
Droplet microfluidics enables millions of independent picoliter reactions, and has recently been used to deep sequence single DNA molecules10, tag nucleosomes to enable single-cell ChIP-seq11, and to profile the transcriptomes of single cells all at high throughput 12–14. However, sequencing the genomes of single cells presents unique challenges, because genomic DNA must be purified from the cellular matter and processed through a series of enzymatic steps to prepare it for sequencing. Consequently, while droplet microfluidics provides the potential for sequencing of single cell genomes at ultra high-throughput, no approach for accomplishing this has yet been described.
We describe a method for single cell genome sequencing at ultra high-throughput (SiC-seq) using droplet microfluidics. In SiC-seq, we encapsulate cells in hydrogel microspheres (microgels) that are permeable to molecules with hydraulic diameters smaller than the pore size, including enzymes, detergents, and small molecules, but sterically trap large molecules such as genomic DNA15. This allows us to use a series of “washes” on encapsulated cells, to perform the requisite steps of cell lysis and genome processing, while maintaining compartmentalization of each genome. Using a combination of microgel and microfluidic processing steps, we lyse the cells, fragment the genomes, and attach unique barcodes to all fragments, in a workflow that processes >50,000 cells in a few hours. The barcoded fragments for all cells can then be pooled and sequenced, and the reads grouped by barcode, providing a library of single cell genomes that can be subjected to additional downstream processing, including demographic characterization and in silico cytometry (Fig. 1).
Figure 1. Schematic of SiC-seq workflow.
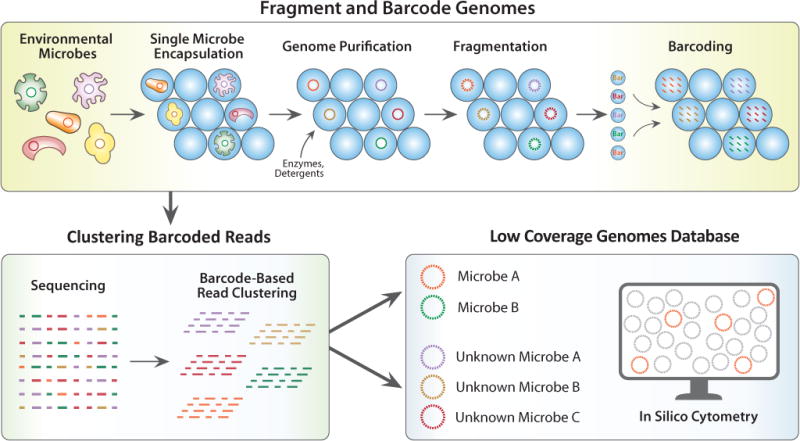
Top: Droplet workflow to generate single cell genome barcoded sequencing library. Bottom Left: Sequencing and generation of barcode groups representing reads from single cells. Bottom Right: The groups of reads comprise a database of low coverage genomes of single cells, which can be searched repeatedly in silico.
Results
SiC-seq workflow
The principal strategy of SiC-seq is to label all DNA fragments originating from the same genome with a sequence identifier (barcode) unique to that cell. The resultant products are chimeric, comprising a barcode sequence covalently linked to a random fragment of the cell genome. The barcodes allow all reads belonging to a given cell to be identified through shared sequence. We use libraries of barcode droplets containing the barcode sequences that are merged with the genome containing droplets to be barcoded10. To prepare a barcode droplet library, we encapsulate into droplets at limiting dilution, oligonucleotides comprising 15 random bases flanked by constant sequences with PCR reagents and primers complementary to the constant regions of the barcodes with one side containing the Illumina P7 flow cell adapter (Fig. 2a)16. The droplets are then thermal cycled to amplify the barcode sequences via digital droplet PCR, generating ~10 million barcode droplets in a few hours.
Figure 2. Microfluidic and biochemical workflow to generate a SiC-seq library.
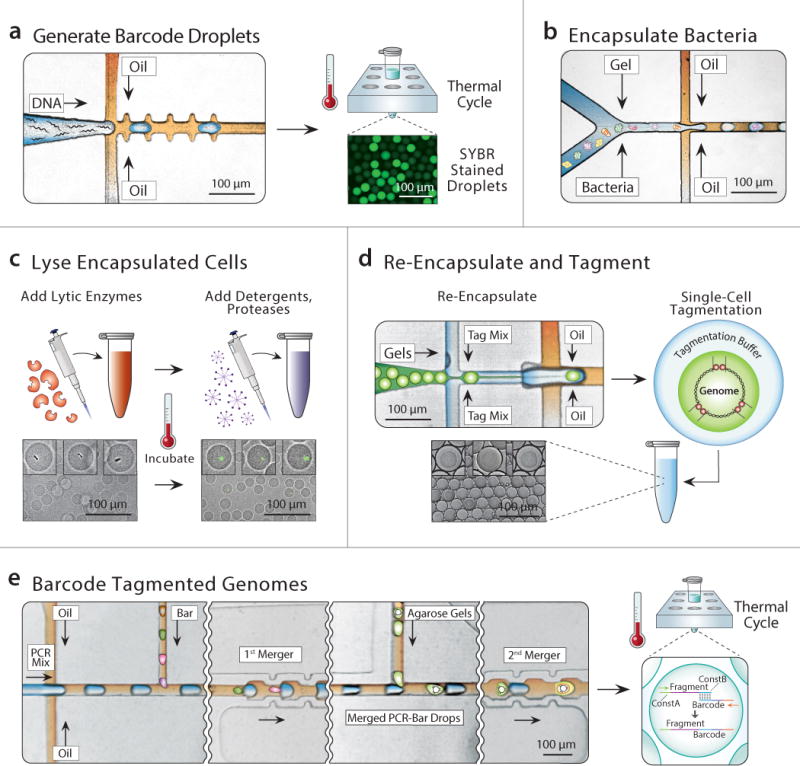
a) Generating barcode droplets by encapsulating random DNA oligos at limiting dilution and amplification by in-droplet PCR (SYBR stained for visualization). b) Cells are encapsulated at limiting dilution with molten agarose to generate single cell containing agarose microgels. c) The single cell genomes are purified through a series of bulk enzymatic and detergent lysis steps (see Online Methods). d) Microgels are re-encapsulated in droplets containing tagmentation reagents. e) The droplets containing tagmented genomes are merged sequentially with PCR reagents and barcode droplets at a 1:1 ratio, followed by PCR to splice barcodes to genomic fragments.
Before the single cell genomes can be barcoded, they must be physically isolated, purified, and fragmented. To accomplish this, we encapsulate single cells in agarose microgels using a two-stream co-flow droplet maker, which merges a cell suspension stream with a molten agarose stream, forming a droplet consisting of an equal volume of both streams (Fig. 2b and Supplementary Fig. 1a). The droplet maker runs at ~10 kHz, allowing us to generate ~10 million ~22 μm diameter droplets in ~20 minutes in a total volume of aqueous emulsion of ~60 μL. Hence, droplet generation is fast and the total volume consumed small, allowing us to load cells at a rate of 1:10 to minimize multi-cell encapsulation. After solidifying the agarose by cooling, the microgels are then transferred from oil to aqueous carrier phase to be subjected to cell lysis and genome purification. To lyse the cells, we incubate the microgels overnight in a mixture of lytic enzymes, digesting the protective microbial cell walls (Online methods). We then incubate them in a mixture of detergents and proteases for 30 minutes, solubilizing lipids and digesting proteins, preserving only high molecular weight genomic DNA, which we verify by staining with SYBR green dye (Fig. 2c). To fragment the genomes and attach the universal sequences to act as PCR handles, we re-encapsulate the gels in the Nextera® reaction (Fig 2d and Supplementary Fig. 1b). Because the transposases are dimeric, the fragmented genome remains intact as a macromolecular complex, remaining sterically encased within the hydrogel network (Supplementary Fig. 2) 17. Nevertheless, we re-encapsulate the gels into separate droplets during fragmentation to ensure that there is no cross-contamination of DNA between the gels.
After the genomes are purified and fragmented, they are barcoded for sequencing. We use a microfluidic device that merges each microgel-containing droplet with droplets containing PCR reagents and a barcode droplet (Fig. 2e and Supplementary Fig. 1c). The resulting droplets, which contain fragmented-genome and barcode DNA are collected into a PCR tube and thermal cycled, splicing the barcode sequences onto the genomic fragments via complementarity through the PCR handles added by the transposase. At this point, the spliced fragments contain both the P5 and P7 Illumina sequencing adaptor required for sequencing on the Illumina platforms. We remove droplets that coalesce during thermal cycling using a micropipette, then the remaining droplets are chemically merged and their contents pooled and prepared for sequencing (Online methods). After sequencing, the reads are filtered by quality and grouped by barcode, providing single cell genomic sequence data.
Validation of SiC-seq on an artificial microbial community
The objective of SiC-seq is to provide single cell genomic sequences bundled in barcode groups. To validate that SiC-seq generates single cell barcode groups, we applied it to an artificial microbial community containing three Gram-negative bacteria, five Gram-positive bacteria, and two yeasts, which are typically difficult cell types to lyse. We prepared a single-cell library from this community using SiC-seq and sequenced it on an Illumina MiSeq, yielding ~6 million single-end reads of 150 bp after quality filtering. We grouped reads by barcode and discard groups with < 50 reads yielding the final 48,989 barcode groups (Fig. 3a). Each barcode group represents a low-coverage genome of a cell, with a sequencing depth of ~0.1% to ~1% (Supplementary Fig. 3).
Figure 3. SiC-seq performance on an artificial microbial community consisting of ten different cell species.
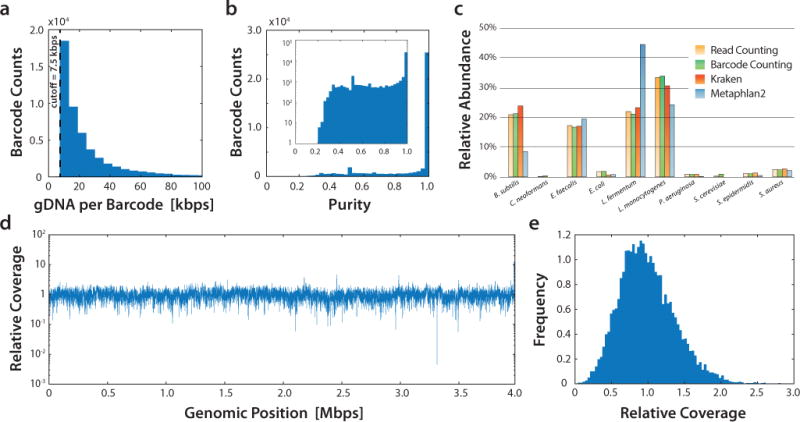
a) Distribution of sequencing yield of each barcode group. b) Histogram of the purity of each barcode group, which is defined as the fraction of reads mapping to the most mapped species for that group. The inset is plotted with the counts on a logarithmic scale. c) Relative abundance estimates of each species using read counting, barcode counting, and two different taxonomic profiling programs (Kraken and Metaphlan2). d) Relative coverage of the Bacillus subtilis genome for all Bacillus subtilis barcode groups, showing good uniformity. See Supplemental Fig. 8 for coverage maps of other species. e) Coverage histogram for the Bacillus subtilis genome binned by relative coverage.
To determine whether the barcode groups indeed correspond to single cells, we mapped all reads to the reference genomes of the ten species. If two microbes reside within the same barcode group, reads will map to two genomes. We defined a group purity score as the fraction of reads mapping to the most mapped reference (the ideal barcode group has a purity score of 1.0). The distribution of purity scores is strongly skewed to high values with the majority of purity score over 0.95 suggesting that most barcode groups represent single cells; this result is consistent even taking into account the different genome sizes of the ten species (Fig. 3b and Supplementary Fig. 4) as well as when purity is examined individually for each species (Supplementary Fig. 5). We further examined the rare barcode groups with low (<0.8) purity scores and determined that the majority of those barcode groups represent rare cases where two cells were encapsulated into one droplet or the occasional coalescence of two single-cell containing droplets (Supplementary Fig. 6).
To determine whether SiC-seq barcodes abundances reflect the organism abundances in the dataset, we compared abundance estimates calculated via short-read alignment, k-mer based sequence classification, and counting under bright-field microscopy (Fig 3 and Supplementary Fig. 7). We found that all methods are in reasonable agreement when reads are pooled and analyzed in bulk and when species identities are assigned to each barcode based on the most commonly mapped species in a group. This demonstrates that SiC-seq enables estimation of species abundance in a microbial population consistent with accepted metagenomic methods.
Sequencing the genome of a single cell typically incurs coverage distribution bias18 due to uneven amplification of DNA starting from a single genome copy. To investigate coverage distribution bias in SiC-seq, we plotted the normalized coverage distribution for reads aggregated from all barcode groups for each microbe (Fig. 3d, 3e, and Supplementary Fig. 8). With the exception of coverage gaps due to low abundances of cells of certain species within the standard microbial community, we observed no substantial coverage bias. This indicates that the sampling of each genome within a barcode group is random, so that when all groups are overlaid, a uniform distribution is obtained. We further inspected the distribution of reads in individual barcode groups and found no substantial bias (Supplementary Fig. 9). We believe that coverage bias is minimal because each genome is amplified in a tiny volume of ~65 pL, which has been shown to curtail bias-inducing runaway of exponential amplification19. Since the sequencing library is composed of ~50,000 amplified genomes, the amplification of each genome can be limited by the tiny volume while still producing sufficient total DNA for sequencing.
SiC-seq data analysis with in silico cytometry
The genomic sequences generated using SiC-seq are grouped according to single cells, which is complementary to the sequences generated from shotgun metagenomic sequencing. Existing computational tools are ill-suited to analyze these data because they do not exploit the single cell barcode information unique to SiC-seq. To address this, we utilize a sequence analysis pipeline in which reads are organized hierarchically as barcode groups, generating a Single Cell Reads database (SiC-Reads) (Supplementary Fig. 10). To build SiC-Reads, we filter raw sequences by quality, group them by barcode, and estimate a taxonomic classification of each group using phylogenetic profilers. We also estimate a purity score equal to the fraction of reads mapping to the dominant taxon within the classifiable set. Additional properties of barcode groups and reads, such as presence of sequences corresponding to antibiotic resistance genes, can be added to the database as they are discovered during analysis.
The massive set of single cell genomes present in SiC-Reads provides new opportunities for discovering associations between sequences within single cells, in a process we dub in silico cytometry. SiC-Reads comprises a collection of single cell genomes that can be sorted in silico, analogous to what is commonly done with flow cytometry on single cells. The database can be sorted repeatedly to mine for correlations between different genetic sequences and structures. Moreover, as new associations are learned, new sorting parameters can be defined, enabling discoveries without having to repeat the experiment.
Taxonomic distribution of antibiotic resistance in microbes
To demonstrate in silico cytometry, we used SiC-seq to sequence a microbial community recovered from coastal seawater of San Francisco (Online methods). We obtained ~8 million reads of 150 bp length after quality filtering (representing of ~55% of raw reads), with which we generated a SiC-Reads database (Supplementary Fig. 10). Using a phylogenetic profiler, 601,348 (6.89%) of reads were successfully classified into taxa representing 99.8% bacteria, 0.04% archaea, and 0.16% viruses (Supplementary Fig. 11a). Barcode groups were assigned a taxonomic classification based on the reads they contained, following the rule that more than 10% of reads in a barcode group must be classified, and the assigned classification is the taxon with the most supporting reads. Most barcode groups were high purity based on the classifiable sequences (~91% average), in accordance with our control sample (~94% average) (Supplementary Fig. 11b). Using this SiC-reads database, we demonstrate in silico cytometry by exploring the distribution of antibiotic resistance, virulence factors, and phage sequences in the microbial community.
Antibiotic resistance has become increasingly common and represents a significant threat to global human health20. While antibiotic resistance genes can be identified in most environments by short-read sequencing, scant information on how they are distributed among taxa is available, because obtaining this information usually requires testing or whole genome sequencing of single species; however, culture conditions for most species have not been identified, precluding such analyses.
To determine the distribution of antibiotic resistance genes among taxa in our dataset, we searched our SiC-Reads database for known antibiotic resistance genes, finding 1,081 (0.012% of reads), representing 108 (0.30%) of barcode groups. The taxonomic distribution of antibiotic resistance genes in our database has a clear structure, although it does not correlate with what is known from genomes in public databases (Fig. 4a and Supplementary Fig. 12a). This is unsurprising as differences are expected in the natural coastline environment compared to the environment of isolated and sequenced strains. The most abundant taxa associated with antibiotic resistance are not the most abundant community members overall, suggesting that in this community certain taxa tend to associate more with antibiotic resistance genes.
Figure 4. Application of SiC-seq to a marine community recovered from the San Francisco coastline.
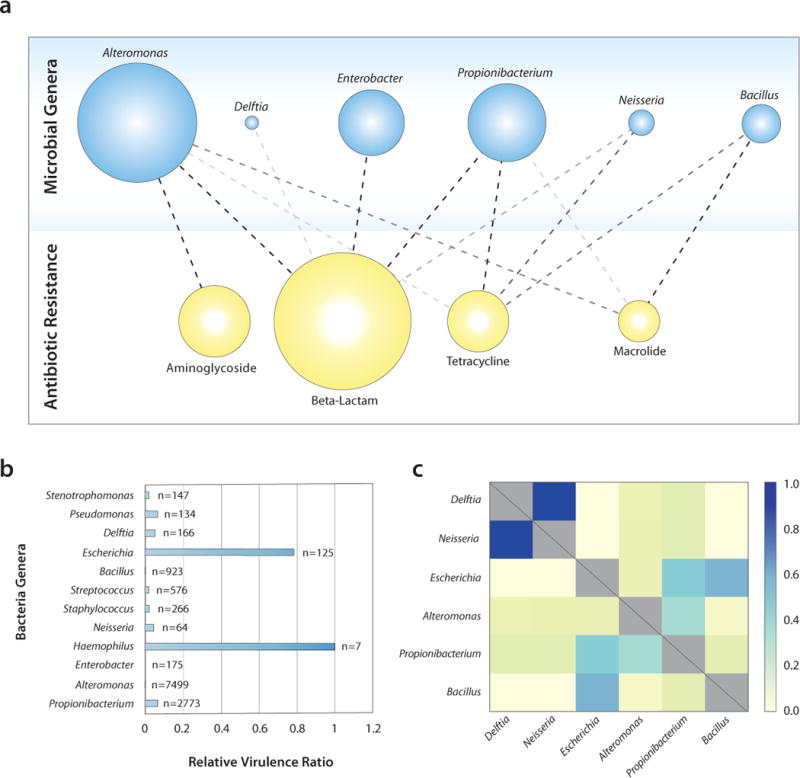
a) Distribution of antibiotic resistance genes according to genus of host microbe. The opacity of connecting lines reflects the number of interactions detected in the database. b) Relative abundance of virulence factors in each genus detected in the community. c) Relative potential for transduction between bacterial taxa, determined by the relative number of common phage sequences detected in their respective genomes, plotted as a heat map.
Association of virulence factors with host bacteria
Virulence factors, like antibiotic resistance genes, are important genetic factors in determining the threat that specific microbes pose to human health. Many opportunistic pathogens reside in natural communities in the environment and cause outbreaks when transmitted to a suitable host21. Monitoring and detecting potentially pathogenic microbes is important for public health. Like antibiotics resistance genes, traditional methods can detect the presence of these genes but not their taxonomic distribution.
To examine the taxonomic distribution of virulence factors in our dataset, we searched our coastal microbial community database for known virulence factor genes, yielding matches in 1,949 (0.022%) reads in 101 (0.28%) barcode groups consisting of 29 prevalent virulence factors distributed among 13 microbial genera. The abundances of taxa where virulence factors were found did not reflect that of the total population, suggesting that certain genera tend to carry more virulence factors than others. To quantify this, we calculated the virulence factor ratio, the ratio between the number of barcode groups containing virulence factors and the total number of barcodes in the community for that species, and normalized the results to the highest virulence factor ratio for comparison (Fig. 4b). Haemophilus and Escherichia stand out amongst all species, both of which are known opportunistic human pathogens. Comparing the virulence factor ratios of the San Francisco coastline community with ones calculated for publicly-available whole genomes, and down sampled to match our per-cell read depth (Supplementary Fig. 12b), we found that the ratios are higher for the public genomes, an expected result given that isolated and sequenced genomes are are biased towards pathogenic strains.
Determining transduction potential between bacteria
Many virulent bacterial strains are thought to arise from horizontal gene transfer aided by cross-infection of bacteriophages. Phages can modify the genomes of their hosts, leaving a copy of their own genome behind or transporting fragments of one species to another in a process known as transduction22,23. Characterizing the distribution of these mobile elements is challenging in an ecological context because confident identification of foreign genomic fragments within a specific host requires sequencing large numbers of cultures of single species or single cells. Nevertheless, this information is valuable for understanding how bacteria transfer genetic material in general, and how virulent new strains may emerge via this mechanism.
To explore transduction in the microbial community, we searched the SiC-Reads database of the San Francisco coastal community for barcode groups containing phage sequences. A phage sequence found in a bacterial genome is evidence of infection, an association that is normally extremely difficult to make for uncultivable microbes and their likely uncultivable infecting phages. We found matches in 6,805 (0.078%) reads representing 260 (0.72%) barcode groups and 106 phage genomes. Since transduction can occur between two host cells that can be infected by the same phage, the potential for transduction depends on the likelihood of phages infecting both hosts. To visualize this, we plot the normalized sum of the number of times we detect the sequences matching to the same phage in two bacterial taxa, normalized by the number of barcode groups in those taxa (Fig. 4c). According to this analysis, Delftia and Neisseria, which are closest related out of the taxa in our analysis, have the highest potential for transduction. The dearth of representative phage genomes in databases and the limited sequence information per barcode group, limits the accuracy of this approach. Therefore, higher coverage of the genomes and better phage genome databases are required to definitively identify the phages that are found in the database. Nevertheless, SiC-seq’s ability to detect these sequences and correlate them within single genomes can provide a useful approach to studying phage-host interactions.
Discussion
SiC-seq generates a metagenomic database grouped by single cell genomes amenable to repeated mining via in silico cytometry, for rapid hypothesis generation and testing. We demonstrated its use in measuring the distributions of antibiotic resistance genes, virulence factors, and transduction potential in microbial communities. The ability to sequence all cells in a sample without the need to culture is a powerful aspect of SiC-seq that should aid in our ability to characterize the ‘microbial dark matter’.
The barcoded nature of SiC-seq data necessitates additional quality control of measures for the data, in addition to the quality control measures utilized in standard sequencing. First, the barcode reads themselves must be of high quality, thus eliminating any reads containing low quality barcode sequences, regardless of the quality of the genomic sequences. Second, barcode groups must be quality controlled to remove small-sized barcode groups, which are the result of mutations in the barcode sequences and background contamination of free DNA. These quality control measures together result in a typical yield of ~55% of raw reads contributing to the SiC-reads database. Improvements in yield can be made by, for example, computationally identifying reads with mutated barcodes ‘correcting’ their sequence, but we have found only modest improvements in yields using this method alone10.
The taxonomic classification of microbes remains an integral part of studying community dynamics, from ecosystems on Earth to those residing in and on our bodies24,25. However, the taxonomic classification of short reads is error prone, due to the diversity of microbes in most communities and the high degree of horizontal gene transfer that mixes genomic elements in unpredictable ways. SiC-seq improves upon traditional metagenomics sequencing in addressing this challenge because taxonomic identification can be made based on hundreds of reads within a barcode group. Advanced strategies can be applied to estimate taxonomy of a barcode group, including Bayesian probabilistic ones based on classification of each read in the group, or ones weighted towards specific taxonomic markers. With even this improvement, accurate classification is difficult because the vast majority of sequences remain unclassifiable and the classification of sequences are biased towards well-sequenced taxa in the databases. As genome coverage improves in future iterations of SiC-seq, taxonomic classification of barcode groups should become more confident and precise, potentially arriving at strain level classifications under certain circumstances. It is worth noting that taxonomic classification with SiC-seq is also subject to the fundamental limitations of reference based classification paradigms where the classification is only as accurate as the match between the sample and the references. Hence, like traditional methods, SiC-seq phylogenetic profiling will become more reliable and complete with the expanding database of reference genomes.
The degree of genome coverage impacts the usefulness of single cell data, including the ability to generate assemblies or identify characteristic sequences for in silico cytometry. A limitation of SiC-seq is that, while the number of cells sequenced far exceeds currently described methods, the coverage per cell is significantly lower. Therefore, dropouts in coverage and false negatives can be expected in in silico cytometry analysis. For abundant organisms with a random distribution of coverage in each barcode group, the system is robust to dropouts because results are averaged over many barcode groups. For example, approximately 7,000 Alteromonas barcode groups were taken into account to determine the antibiotic resistance profile for Alteromonas bacteria. However, for less abundant species, such as Haemophilus, more dropouts can be expected because there may not be enough total sequence information to detect a specific genetic factor. For this reason, the analysis of SiC-reads databases should be limited to relative comparisons of species within the database, and the abundance of target genes within subpopulations should be normalized to the number of barcode groups in the subpopulation. It is worth noting that the dropout phenomenon is not unique to SiC-seq data, but all metagenomic sequencing data where the subpopulation to be analyzed represents a very small fraction.
Although coverage can be increased by sequencing more reads, the coverage per cell per barcode group will be below 100%. This is because the method begins with a single genome copy without amplification and losses incurred during enzymatic and microfluidic processing are irrevocable, thus limiting the maximum coverage attainable. In future iterations of SiC-seq, coverage may be increased by pre-amplifying genomes prior to processing, for example, with multiple displacement amplification in droplets7. Additionally, different strategies for barcoding genomes may yield higher coverage, such as recently described combinatorial indexing via transposase libraries40, which should be applicable to single cell genomes encapsulated in microgels.
The de novo assembly of whole genomes from metagenomics sequences is a common goal in the field of metagenomics. Mate-paired sequencing can be used to bridge contigs in a metagenomics sequencing dataset and potentially assemble whole genomes given sufficient coverage26. Though powerful, the method is limited by the required micrograms of starting DNA that can be difficult to obtain from microbial ecosystems. Furthermore, many mate-paired reads are required to assemble a whole genome, since each mate-pair bridges only two contigs. SiC-seq data improves on mate-paired sequencing in this respect by requiring minimal amounts of sample as well as enabling the bridging of multiple contigs per barcode group. Consequently, SiC-seq should allow generation of draft genomes from shotgun metagenomic data with far less DNA input requirement and sequencing effort.
While we focused on microbial communities, SiC-seq is also applicable to populations of mammalian cells, where it can have a more direct impact on human health. The grouped reads provided by SiC-seq should afford the information required to determine copy-number variations within the genome, which is relevant to cancer27. The enormous size of mammalian genomes, however, limits the number of cells that can be sequenced for a target level of coverage. Nevertheless, as the cost of sequencing continues to decrease, more cells can be sequenced to greater depth, creating opportunities for characterizing mammalian tissues, cell-by-cell.
SiC-seq method is a means to isolate and barcode large DNA molecules, irrespective of the entity from which they originate. While we have focused on cells, similar approaches can be applied to any entities whose genomes can be trapped and processed within the gel matrix. SiC-seq’s ability to build and mine large databases of genomes grouped by single cells should contribute to the characterization of heterogeneity across biology.
Online Methods
Microfluidic Devices
To fabricate the microfluidic devices, poly(dimethylsiloxane) (Dow Corning, Sylgard 184) is poured over a negative photoresist (MicroChem, catalog no. SU-8 3025) patterned on a silicon wafer (University Wafer) using UV photolithography. The PDMS devices are cured in an oven for 1 hour, extracted with a metal scalpel, and punched with a 0.75 mm biopsy core (World Precision Instruments, catalog no. 504529) to create inlets and outlets. Devices are bonded to a glass slide using an oxygen plasma cleaner (Harrick Plasma) and the channels treated with Aquapel (PPG Industries) and baked at 80°C for 10 min to render them hydrophobic.
Barcode Emulsions
Barcode emulsions are prepared through digital PCR process wherein barcode oligonucleotides are amplified as single molecules in droplets containing PCR reagents. Barcode oligonucleotides (GCAGCTGGCGTAATAGCGAGTACAATCTGCTCTGATGCCGCATAGNNNNNNNNNNNNNNNTAAGCCAGCCCCGACACT) (IDT) at 0.01 pM concentration are added to a PCR reaction mix containing 1X NEB Phusion Hot Start Flex Master Mix (NEB, catalog no. M0536L), 2% (w/v) Tween 20 (Sigma-Aldrich, catalog no. P9416), 5% (w/v) PEG-6000 (Santa Cruz Biotechnology, catalog no. sc-302016), and 400 nM primers FL128 (CTGTCTCTTATACACATCTCCGAGCCCACGAGACGTGTCGGGGCTGGCTTA) and FL129 (CAAGCAGAAGACGGCATACGAGATCAGCTGGCGTAATAGCG, contains P7 adapter sequence) (IDT). The PCR mixture and HFE-7500 fluorinated oil (3M) with 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant (008-Fluoro-surfactant, Ran Technologies) are loaded into separate 1 mL syringes (BD) and injected at 300 and 500 μL/hr, respectively, into a flow-focusing droplet maker using syringe pumps (New Era, catalog no. NE-501) controlled with a custom Python script (https://github.com/AbateLab/Pump-Control-Program). The emulsion is collected in PCR tubes, and the oil underneath the emulsion removed via pipette and replaced with FC-40 fluorinated oil (Sigma-Aldrich, catalog no. 51142-49-5) with 5% (w/w) PEG-PFPE amphiphilic block copolymer surfactant for improved thermal stability. The emulsion is thermal cycled (Bio-Rad, T100) with the following program: 98°C for 3 min, followed by 40 cycles with 2°C per second ramp rates of 98°C for 10s, 62°C for 20s, and 72°C for 20s, followed by a hold at 12°C. Fluorescent DNA staining using 10X SYBR Green I (Thermo Fisher Scientific) in HFE-7500 oil is used to quantify barcode encapsulation rate under a fluorescent microscope (Life Technologies, catalog no. AMAFD1000).
Water Sample Collection and Filtering
To obtain a natural sample of a microbial community, we collect marine water from Ocean Beach in West San Francisco, California, USA (37°44′55.6″N 122°30′33.6″W). Approximately 2 L of water is obtained by submerging two 1000 mL glass bottles below the water surface ~20 m from the shoreline. Samples are placed on ice during transport to the lab. 100 mL of the sample is passed through a 40 μm cell strainer (Corning, product no. 352340) to remove large debris, including sand. The sample is loaded into a 0.45 μm vacuum filter (Millipore, catalog no. SCHVU01RE); this filtering step separates microbes, which are captured on the membrane, and viruses, which are discarded in the filtrate. The membrane is extracted from the apparatus using a scalpel and inserted into a 15 mL centrifuge tube, to which 5 mL of PBS is added. The tube is vortexed at high speed for ~2 min to free the bacterial cells from the membrane. Finally, the cell solution is loaded into a 10 mL syringe and passed through a 5 μm syringe filter (Millipore, catalog no. SLSV025LS) to remove remaining large particulate. The marine cells are counted using the same protocol as the liquid cultures.
Cell Encapsulation in Agarose Microgels
To prepare the artificial community for processing through the SiC-seq workflow, the frozen stock of cells (Zymo Research, catalog no. D6300) is thawed gently in a room-temperature water bath. Cell concentration is determined by manual cell counting under a microscope, and diluted to an appropriate concentration for single cell encapsulation. The calculated volume of cell solution is transferred to a 1.5 mL centrifuge tube (Fisher Scientific) and washed twice in 1 mL PBS. The cells are re-suspended in a 1 mL solution of PBS containing 17% OptiPrep Density Gradient Medium (Sigma-Aldrich), 0.1 mg/mL BSA (Sigma-Aldrich, catalog no. A9418), and 1% (v/v) Pluronic F-68 (Life Technologies). The cell solution is loaded into a 1 mL syringe and placed on a syringe pump (New Era, catalog no. NE-501). 1 mL of a 3% solution of low gelling temperature agarose (Sigma-Aldrich, catalog no. A9414) and TE buffer (Teknova, catalog no. T0225) is prepared in a 1.5 mL centrifuge tube and heated on a block at 90°C for approximately 10 minutes to completely dissolve the agarose powder. The hot agarose is transferred to a 1 mL syringe and placed on a syringe pump. To keep the agarose molten during the microfluidic experiment, a personal space heater is positioned ~5 cm from the agarose syringe and set to run continuously at high heat. HFE-7500 fluorinated oil with 2% (w/w) de-protonated Krytox surfactant (DuPont, catalog no. 157FSH) is loaded into a 3 mL syringe. The cell solution, molten agarose, and oil are injected into the co-flow droplet maker at flow rates of 200, 200, and 400 μL/hr, respectively, to form the 1.5% agarose microgels. Approximately 500 μL of droplets are collected in a 15 mL centrifuge tube on ice and incubated for 30 min at 4°C to ensure complete solidification of the microgels.
Resuspending Microgels in Aqueous Buffer
The droplets are centrifuged at 300 g for 1 min to maximize separation of the emulsions from the oil. The oil layer is extracted from the tube using a 5 mL syringe and discarded. Emulsions are broken using 2 mL of a 10% (v/v) solution of perfluorooctanol (Sigma-Aldrich, catalog no. 370533) in HFE-7500; the emulsions are then mixed by pipetting and centrifuged at 300 g for 1 min. The oil is removed from the tube using a syringe and the droplet breaking step is repeated. Following droplet breaking, 2 mL of hexane containing 1% (v/v) Span 80 (Sigma-Aldrich) is added to the microgels to dissolve any remaining oil, and this solution is mixed and centrifuged at 300 g for 1 min. The hexane supernatant is removed from the tube and the hexane addition step is repeated. Finally, the microgels are washed three times in 10 mL of aqueous solution TE buffer containing 0.1% (v/v) Triton X-100 nonionic surfactant (Sigma-Aldrich). The microgels are centrifuged at 1000 g for 2 min and the supernatant aspirated between washes. The washed microgels are stored in 5 mL TE buffer at 4°C prior to cell lysis.
Cell Lysis in Microgels
To lyse the cells in the microgels, the particles are submerged in a solution of 2 mL TE buffer solution containing 10 mM DTT (manu), 2.5 mM EDTA (Teknova), and 10mM NaCl (Sigma-Aldrich). The following quantities of lytic enzymes are also included: 4 U zymolyase (Zymo Research), 10 U lysostaphin (Sigma-Aldrich, catalog no. L7386), 100 U mutanolysin (Sigma-Aldrich, catalog no. M9901), and 40 mg lysozyme (MP Biomedicals, catalog no. 195303). Cell lysis proceeds overnight in a shaking incubator at 37°C. The turbid lysate mixture is centrifuged at 1000 g for 1 min, the supernatant removed, and 3 mL of a solution containing 0.5% (w/v) lithium dodecyl sulfate (Sigma-Aldrich) and 10 mM EDTA in TE buffer is added, along with 4 U of Proteinase K (NEB) to solubilize cell debris and digest cellular proteins. The solution is incubated at 50°C on a heating block for 30 min. Following lysis, the microgels are thoroughly washed to ensure complete removal of detergents and other chemical species which may inhibit downstream molecular biology reactions. The following washes occur in 10 mL volumes with centrifugation magnitudes of 1000 g between additions of wash solutions: one wash with 2% (v/v) Tween 20 in water; one wash in 100% ethanol (Koptec) to denature any remaining Proteinase K; and five washes with 0.02% (v/v) Tween 20 in water.
Tagmentation of Genomic DNA in Microgels
Using reagents from a Nextera DNA Library Prep Kit (Illumina, catalog no. FC-121-1030), the washed and lysed gels containing high-molecular-weight genomic DNA are simultaneously fragmented and tagged with a common adapter sequence. Microgels are re-encapsulated into droplets to minimize cross-contamination during the tagmentation step. A solution of 192 uL DI water, 200 uL tagmentation buffer, and 8 uL Nextera enzyme is prepared and loaded into a 1 mL syringe. Microgels and the tagmentation solution are injected into the re-encapsulation device (Supplementary Fig. 1). The re-encapsulated microgels are incubated in a 1.5 mL tube on a heating block at 50°C for one hour.
Microfluidic Barcoding of Encapsulated Cells
Tagmented microgel droplets, barcode droplets, and 500 μL of PCR solution containing 1X Invitrogen Platinum Multiplex PCR Master Mix (Thermo Fisher Scientific, catalog no. 4464268), 400 nM primers FL127 (AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTC, contains P5 adapter sequence) and FL129 (CAAGCAGAAGACGGCATACGAGATCAGCTGGCGTAATAGCG), 50X dilution of NT buffer from the Nextera XT Kit (0.2% SDS) (Illumina, catalog no. FC-131-1024), 1% (w/v) Tween 20, 1% (w/v) PEG-6000, 2.5 U/μL Bst 2.0 WarmStart DNA Polymerase (NEB, catalog no. M0538S) are each loaded into a 1 mL syringe and injected into the sequential merger device as shown in Supplementary Fig. 1. HFE-7500 fluorinated oil with 2% (w/w) 008-Fluorosurfactant is used as the continuous phase of the emulsion. Merger of the barcode and gel droplet emulsions is achieved using an electrode connected to a cold cathode fluorescent inverter and DC power supply (Mastech). A voltage of 2.0 V at the power supply produces a ~2 kV AC potential at the electrode which causes touching droplets to merge. The emulsion is collected in a 0.5 mL thin-walled PCR tube (Applied Biosciences), and the HFE-7500 replaced with FC-40 with 5% (w/w) 008-Fluorosurfactant prior to thermal cycling with the following protocol: 65°C for 5 mins, 95°C for 2 mins, then 30 cycles at 2°C/s ramp rates of 95°C for 15s, 60°C for 1 min, 72°C for 1 min, and then 72°C for 5 mins with optional 12°C overnight hold. After thermal cycling, large (coalesced) droplets are removed using a micropipette, and the emulsion is broken by addition of 20 μL of perfluoro-octanol and brief centrifugation in a micro-centrifuge. The upper aqueous phase is collected and the DNA library is purified using a Zymo DNA Clean & Concentrator-5 kit (Zymo Research). The library is size-selected for DNA fragments in the 200–600 bp range using Agencourt AMPure XP beads (Beckman Coulter), quantified with a Bioanalyzer 2100 instrument and High Sensitivity DNA chip (Agilent), and sequenced on an Illumina MiSeq using a custom index primer (GCCCACGAGACGTGTCGGGGCTGGCTTA).
Generating the SiC-Reads Database
Raw reads from the MiSeq-generated FASTQ files are filtered by quality and grouped by barcode sequence using the Python script barcodeCleanup.py. A given read is discarded if more than 20% of its bases have a Q-score less than Q20, and all reads associated with a barcode containing less than 50 reads are discarded. This step ensures that all barcode groups, representing single cells, contain a sufficient number of high-quality reads. The resulting reads are exported to a table in a SQLite database with fields containing the barcode sequence, barcode group size, a unique read ID number, and read sequence. When the reference genomes are known, as in the case of the synthetic cell population experiment, the reads are aligned using bowtie2 v2.2.9 with default settings and the SQLite table is updated with relevant alignment information for each read. For environmental samples, the reads are classified by taxonomy using Kraken v0.10.5 with “–quick –min-hits 2” options set, and the output is exported to the SQLite database. krakenAnalysis.py assigns taxonomic identities from the Kraken database to barcode groups by a majority rule, in which barcode group is classified according to the most common taxonomic label among its classifiable reads. Barcode group purity is calculated from reference alignment data or phylogenetic labels using the script purity.py.
In Silico Cytometry
Reads from the SiC-Reads database are aligned, using bowtie2 v2.2.9 with –very-sensitive and –end-to-end settings to reference sequences of interest (antibiotic resistance database obtained from (41), virulence factor database obtained from core virulence factor genes at the virulence factor database (VFDB) 42, Phage sequence database obtained from Phage genome database accessed on May 2016 at http://www.ebi.ac.uk/genomes/phage.html. Mapping reads are then filtered for MapQ > 2 in order to remove ambiguously mapping reads. Barcode groups containing reads that map to the databases are annotated as containing the target sequence and are exported for further analysis if they are taxonomically classified with purity > 0.8. To generate the heatmap for transduction potential, all reads associated with a phage and a Kraken-classified barcode group were extracted and grouped according to phage type. Duplicate and near-duplicate reads were removed. The heatmap intensities were calculated as follows: for a given pair of bacterial hosts, the total number of host-phage-host connections in the database were counted. To normalize the data by host abundance, this number was divided by the total number of barcode groups associated with the two hosts.
Calculating the Virulence Factor Ratios
The virulence factor ratios calculations in Figure 4b of the main text was reproduced using reference genomes for the genera shown in the figure. The complete genomes of all species associated with these 12 genera were downloaded from the RefSeq database using the Perl script ncbiDownloader.pl. Genomes were pooled into FASTA files labeled by genus. From these reference files, a Python script (bargroupGenerator.py) generated simulated barcode groups of 200 reads per group, with each single-end read 150 bp long. The number of simulated bargroups generated for a given genus was equal to the number of barcode groups identified for this genus in the San Francisco Coast water sample. The simulated barcode group reads were then aligned to the original virulence factor database using bowtie2 v2.2.9 in ‘local’ alignment mode with default sensitivity settings. Unaligned sequences were removed using Samtools v1.3.1 (samtools view -b -F) and the remaining aligned reads were used to produce the data shown in Supplementary Figure 7b.
Generating the Antibiotic Resistance Network with Reference Genomes
An antibiotic resistance graph in Supplementary Fig. 7a was generated using references for the 6 genomes most commonly associated with antibiotic resistance in the SiC-Reads database of the San Francisco Coast water microbial community. The following genomes (with accession numbers) were downloaded from the NCBI RefSeq repository: Alteromonas macleodii ATCC 27126 (CP003841.1), Bacillus subtilis subsp. spizizenii strain NRS 231 (CP010434.1), Delftia acidovorans SPH-1 (CP000884.1), Enterobacter cloacae subsp. cloacae ATCC 13047 (CP001918.1), Neisseria meningitidis MC58 (AE002098.2), Propionibacterium acnes KPA171202 (AE017283.1). These genomes were combined into a single FASTA file and passed to a short read simulator, wgsim v0.3.2, which generated 10M single-end reads of 70 bp each with a base error rate of 0. These reads were aligned to the antibiotic resistant gene reference using bowtie2 in ‘local’ mode with default sensitivity settings. All unaligned sequences were removed using Samtools (samtools view -b -F). The aligned sequences in SAM format were imported into Cytoscape v3.4.0 and the network shown in Supplementary Fig. 7a was generated using the reference genus and antibiotic resistance genes as the network targets and sources, respectively. The darkness of the graph’s edges scale linearly with the total number of connections in the data, where darker lines have a greater number of associations.
Characterizing Diffusion of Genomic DNA Fragments in Agarose Microgels
A microgel sample containing encapsulated, lysed bacteria was stained with SYBR Green I and observed under a fluorescent microscope before and after tagmentation at various time points (Supplementary Fig. 2a). In another experiment, the concentration of DNA in the supernatant and contents of the gels was measured in a sample incubated at room temperature. After two days at room temperature, the beads were pelleted by centrifugation and DNA was extracted from the beads and the supernatant using a DNA gel extraction kit and DNA clean up and concentrator kit (Zymo research D4001T) and quantified using the Qubit dsDNA high sensitivity assay and Bioanalyzer High Sensitivity dsDNA chip (Supplementary Fig. 2b). As an additional experiment, microgels were incubated at 55°C with and without tagmentation enzyme to demonstrate the corresponding change in genomic DNA fragment size distribution before and after tagmentation (Supplementary Fig. 2c).
Cell Culture and Counting
To generate an additional artificial community with which to validate the SiC-seq workflow, liquid cultures of Staphylococcus epidermidis, Saccharomyces cerevisiae (strain S288c), and Bacillus subtilis (strain 168) are grown overnight in a shaking incubator. The following culture conditions are used: Staphylococcus epidermidis and Bacillus subtilis are grown in 3 mL LB broth at 37°C; Saccharomyces cerevisiae is grown in 3 mL YPD broth at 30°C. Cell concentration is determined by manually counting serial dilutions of the liquid culture on plastic slides (Thermo Fisher Scientific, catalog no. C10228) using a microscope. The cultures are kept at 4°C before being used in the microfluidic experiment. An analysis of the SiC-seq experiment for this synthetic microbial community is shown in Supplementary Fig. 7.
Code Availability
Scripts used to generate the data in this paper can be accessed at the Abatelab Github: https://github.com/AbateLab/SiC-seq.
Supplementary Material
Acknowledgments
We are grateful for K. Stedman, R. Malmstrom, R. Andino, K. Pollard, M. Fischbach for very helpful discussion and advice on the manuscript. We thank C. O’Loughlin at UCSF for providing microbial strains. This work was supported by the National Science Foundation through a CAREER Award [grant number DBI-1253293]; the National Institutes of Health (NIH) [grant numbers HG007233-01, R01-EB019453-01, 1R21HG007233, DP2-AR068129-01, R01-HG008978]; and the Defense Advanced Research Projects Agency Living Foundries Program [contract numbers HR0011-12-C-0065, N66001-12-C-4211, HR0011-12-C-0066]. Funding for open access charge: [NIH grant number DP2-AR068129-01]. F.L. is supported by a PGS-D grant from the National Science and Engineering Research Council of Canada (NSERC).
Footnotes
Accession Codes
All sequencing data is accessible at the NCBI Sequence Read Archive through the following accession numbers: SRX2516128, SRX2522025, SRX2516129.
Author Contributions
F.L. and A.R.A conceived of the SiC-seq method. F.L., B.D, and N.A. designed and performed the experiments, and analyzed data. F.L. and A.R.A wrote the manuscript.
Competing Financial Interests
Patents pertaining to this workflow may be licensed to Mission Bio, of which A.R.A is a shareholder.
ED SUM: More than 50,000 single cell genomes are sequenced in a single run using droplet barcoding.
References
- 1.Monte UD. Does the cell number 109 still really fit one gram of tumor tissue? Cell Cycle. 2009;8:505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 2.Maranger Roxane, Bird David. viral abundance in aquatic systems: a comparison between marine and fresh waters. Mar Ecol Prog Ser. 1995;121:217–226. [Google Scholar]
- 3.Zhang H, Liu KK. Optical tweezers for single cells. J R Soc Interface. 2008;5:671–690. doi: 10.1098/rsif.2008.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinke C, et al. Obtaining genomes from uncultivated environmental microorganisms using FACS–based single-cell genomics. Nat Protoc. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 5.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung K, et al. Robust high-performance nanoliter-volume single-cell multiple displacement amplification on planar substrates. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1520964113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamminen MV, Virta MPJ. Single gene-based distinction of individual microbial genomes from a mixed population of microbial cells. Microb Physiol Metab. 2015;195 doi: 10.3389/fmicb.2015.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Brito IL, Alm EJ, Blainey PC. Virtual microfluidics for digital quantification and single-cell sequencing. Nat Methods. 2016;13:759–762. doi: 10.1038/nmeth.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan F, Haliburton JR, Yuan A, Abate AR. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat Commun. 2016;7:11784. doi: 10.1038/ncomms11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein AM, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotem A, et al. High-Throughput Single-Cell Labeling (Hi-SCL) for RNA-Seq Using Drop-Based Microfluidics. PLoS ONE. 2015;10:e0116328. doi: 10.1371/journal.pone.0116328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak R, et al. Single Cell Multiplex Gene Detection and Sequencing Using Microfluidically-Generated Agarose Emulsions. Angew Chem Int Ed Engl. 2011;50:390–395. doi: 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garstecki P, et al. Formation of monodisperse bubbles in a microfluidic flow-focusing device. Appl Phys Lett. 2004;85:2649–2651. [Google Scholar]
- 17.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourcy CFA de, et al. A Quantitative Comparison of Single-Cell Whole Genome Amplification Methods. PLOS ONE. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nathan C, Cars O. Antibiotic Resistance — Problems, Progress, and Prospects. N Engl J Med. 2014;371:1761–1763. doi: 10.1056/NEJMp1408040. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz FH. Processes controlling the transmission of bacterial pathogens in the environment. Res Microbiol. 2007;158:195–202. doi: 10.1016/j.resmic.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Jiang SC, Paul JH. Gene Transfer by Transduction in the Marine Environment. Appl Environ Microbiol. 1998;64:2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochman H, Moran NA. Genes Lost and Genes Found: Evolution of Bacterial Pathogenesis and Symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 24.Consortium, T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afshinnekoo E, et al. Geospatial Resolution of Human and Bacterial Diversity with City-Scale Metagenomics. Cell Syst. 2015;1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iverson V, et al. Untangling Genomes from Metagenomes: Revealing an Uncultured Class of Marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 27.Ni X, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


