Abstract
Single cell RNA sequencing (scRNA-seq) is an exciting new technology allowing the analysis of transcriptomes from individual cells, and is ideally suited to address the inherent complexity and dynamics of the central nervous system. ScRNA-seq has already been applied to the study of molecular taxonomy of the brain. This work has paved the way to expanding our understanding of nervous systems and to providing insights into cellular susceptibilities and molecular mechanisms in neurological and neurodegenerative diseases. Here, we discuss the recent progress and the challenges in applying this technology to advance our understanding of the brain. We favor the application of scRNA-seq in the discovery of targets and biomarkers as a new approach in developing novel therapeutics for the treatment of neurodegenerative diseases.
Single Cell RNA Sequencing: A Valuable Tool
Neuroscience research has been bolstered by recent technological advances, including high resolution imaging approaches [1], optogenetic modulation (see Glossary) [2], genome engineering [3], and next generation sequencing (NGS) [4]. These techniques have enabled us to interrogate the physiology and pathophysiology of the nervous system with unprecedented precision and depth. Emerging advances using single cell RNA sequencing (scRNA-seq) in the central nervous system (CNS) have already begun to provide exciting molecular insights into the complexity of the brain by identifying novel cellular subtypes based on transcriptional profiles as well as possible disease-relevant mechanisms [5–8] (Box 1). An extension of these studies will be to apply scRNA-seq to compare different cell populations and cellular states in the context of neurological disease. Transcriptomic analyses at single cell levels using pathological samples from human brains and animal models of neurological diseases are likely to provide an remarkable opportunity to understanding disease mechanisms.
Box 1. Pioneering Transcriptomic Studies in Neuroscience.
The systematic description of cellular structures in the brain based on their morphological characteristics and localization, pioneered by Ramon y Cajal, has been guiding neuroscience studies for more than a century [9]. In this regard, translating the morphological features of the nervous system into molecular and functional terms and understanding their formation during development represent important goals of modern neuroscience. Importantly, such studies have identified transcriptional factors involved in the determination of cell fates for all cell types in the CNS, underscoring the importance of transcriptional regulation in the development and maintenance of the nervous system [10–12]. The systematic bulk RNA-seq analyses and in situ mapping of brains by researchers at the Allen Brain Institute and the laboratory of Ben Barres have provided important molecular definitions of the cell types and structures in the brain and highlighted the role of transcriptional regulation in its physiology. On the one hand, the Allen Institute for Brain Science systematically characterized the genome-wide gene expression patterns in molecularly defined cell types and anatomically defined regions in the CNS of both human and animal models [13]. On the other hand, Ben Barres’ laboratory conducted RNA sequencing analyses on isolated CNS cell populations, including neurons, oligodendrocytes, astrocytes, microglia and endothelial cells [13]. This cell-type specific bulk RNA sequencing approach has provided a baseline classification system for major cell types and an important foundation for studying the cellular landscape in the CNS [14, 15]. These and other studies have demonstrated the power of understanding the complexity of the CNS through a transcriptional viewpoint, and have also provided an excellent methodology to process large datasets in an integrative and user-friendly environment.
In this review, we discuss scRNA-seq studies as a means of assessing the dynamics of differential gene expression patterns in different cell types. We review promising research directions made possible by the application of this novel technology in the field of neurobiology and highlight studies that bear direct relevance to the discovery of neurodegeneration mechanisms and which might aid in identifying new drug targets and biomarkers. In addition, we discuss potential hurdles that need to be overcome when conducting scRNA-seq in the intact brain.
Transcriptomic Studies Unveil Therapeutically Relevant Targets for Neurodegeneration
While many early studies focused on understanding transcriptomics in the CNS in the context of development, here, we highlight the possibility that differential expression patterns can be utilized to understand disease mechanisms in the field of neurodegenerative and neurological diseases (Box 2). One important application is the understanding of the transcriptional basis of disease vulnerability. For example, two studies explored the transcriptional profiles in rat models of transient global ischemia in an attempt to understand why pyramidal neurons of the CA1 are highly vulnerable to ischemic insult and degeneration [16, 17]. By contrast, adjacent CA3 pyramidal neurons appear to be largely spared after this type of insult [16, 17]. These studies provide insights into disease pathogenesis in animal models, by identifying several novel molecules induced after ischemia such as SMN1/Smn1, a gene that is dysregulated in spinal muscular atrophy. In addition, transcriptional profiling has also been used to explore the mechanism(s) by which a lesion in multiple sclerosis (MS) occurs, and how a chronic active and an inactive lesion may differ at transcriptomic levels in MS [18, 19]. This work and that of others identified the upregulation of immune markers in active lesions in MS. Despite the fact that both of these studies were conducted before NGS, they were important attempts to align temporal and spatial localization with transcriptional states in order to better define neuronal disease pathology. However, these reports were limited by their statistical power. Indeed, many early microarray studies were technically limited because of the small number of patients amenable to this type of experiment. Multiplexing (i.e. simultaneously sequencing multiple patient barcode-labeled samples) by using NGS has enabled the quantitative assessment of transcriptional patterns in large cohorts of patients.
Box 2. Transition to scRNA-seq in the Nervous System.
While the cells in the nervous system have been traditionally classified according to their molecular markers, location, morphology, target specificity, and electrophysiological characteristics, scRNA-seq provides an opportunity to explore and understand the molecular mechanisms that define their characteristics. The dynamic changes in gene expression patterns at the single cell level in response to environmental or disease conditions may provide important insights into the ongoing cellular responses and mechanisms under relevant physiological and pathological processes. However, one should be cautious in interpreting whether a molecular profile corresponds to a cell type or a cell state [33, 37, 38]. The former refers to a unique, molecular signature that remains stable under different conditions, throughout development and into adulthood, while the latter corresponds to a more dynamic set of genes that shift their expression in response to different signaling stimuli. It is therefore not difficult to imagine that molecular profiling by scRNA-seq will adequately inform us on the properties of a vast array of neurons and glia within the nervous system, including electrophysiological properties, connectivity patterns or activation states, and under various conditions of interest.
Such methodological improvement occurred in parallel with the development of RNA sequencing technologies and the ability to gather and interpret large data sets. Indeed, the NIH has commenced several large initiatives to understand human diseases through an “omics” approach. One successful example has been the effort to understand Alzheimer’s Disease (AD) pathology by investigating the transcriptional profiles of brains from large cohorts of patients [20]. While the results from these studies are just beginning to emerge, two outcomes of this research approach are evident: one is the discovery of novel disease related targets and pathways, and the other is the identification of biomarkers for disease. Indeed, a study investigating the transcriptional profile of more than 1500 late-onset AD patients identified a pathogen phagocytosis molecular signature, revealing Tyro receptor tyrosine kinase binding protein TYROBP as a key regulator of many genes important for microglial function [21]. These findings not only point to a novel microglial-specific molecular signature in AD, but also suggest potential biomarkers in connection with TYROBP that can be further explored. Moreover, the microglial-specific signature was identified within the overall RNA profile of AD patient brain samples using a bulk sequencing approach, suggesting a robust immune signature in AD samples that can be masked by other cell types [21]. This conclusion is consistent with a recent report suggesting that bulk-sequencing changes detectable in pathological samples may be primarily driven by the changes in the composition and abundance of cell types, rather than that of gene transcription per se [22] (Box 3). This interpretation was reached based on the RNA-seq data from multiple human neurodegenerative diseases such as amyotrophic laterals sclerosis (ALS), frontotemporal dementia (FTD), and AD human post-mortem tissues as well as AD mouse models[22]. In this work, increased transcripts were mostly enriched in non-neuronal cell types including endothelial cells, astrocytes and microglia, whereas the noted down-regulated transcripts came from neuronal cells. Such evidence highlights the critical need to investigate and understand the transcriptomic changes at single cell level in human pathology and animal model studies.
Box 3. Limitations of Bulk Transcriptomic Studies.
RNA samples for bulk sequencing are often derived from a diverse population of cells, either isolated from whole brain or from a specific region of the CNS, with or without fluorescence-activated cell sorting (FACS). Thus, cell-type specific transcriptomic changes may often be lost in such studies as the final dataset represents an average of many cells from different regions and/or different cell types. Given the high degree of heterogeneity and complexity of the CNS, cell types and transcripts of low abundance are common. In particular, with bulk RNA-seq, it may be difficult to differentiate if a transcript of low abundance is expressed at low levels in a common cell type or highly expressed in a rare cell type. Furthermore, with disease, low abundance transcriptional changes in a rare cell type close to a lesion may be undetectable, or underestimated, when using a bulk RNA-seq approach. Additionally, while FACS sorting allows selection via particular biomarker(s), the intrinsic heterogeneity within cell populations might still be missed, potentially resulting in a loss of critical pieces of information, which in the context of neurodegenerative diseases, may correspond to signaling events triggering pathological cascades or, facilitating their amplification. As such, scRNA-seq techniques have the potential to overcome these limitations.
Development of scRNA-seq Technologies
Understanding the nervous system at a single cell level has been an exciting area of interest in the neuroscience community for some time [23, 24]. Early studies used elegant -- yet low throughput -- techniques to identify transcriptional differences among rat hippocampal neurons [23] as well as among cells from the mouse olfactory epithelium [24]. A parallel approach utilized laborious cell suctioning after single cell electrophysiological recordings (whole cell patch clamp) followed by semi-quantitative PCR, to couple neuronal firing rates with the expression of calcium-binding molecules and neuropeptide receptors [25]. While these studies identified specific differences in neuronal populations, aligning their functions with gene expression patterns, they were not quantitative enough, nor did they provide an unbiased assessment of global gene expression patterns. NGS has radically changed our ability to understand the transcriptional landscape in any given cellular system by quantitatively assessing gene expression patterns, distinguishing splicing events and identifying rare transcripts. The power of NGS is so immense that it has rendered microarrays obsolete[26]. While microarrays detect robust, homogeneous changes in cell populations[27], they are limited in identifying low abundance transcriptional events, and rare cell populations. In this regard, the development of the scRNA-seq approach yields the promise of overcoming such challenges (Figure 1).
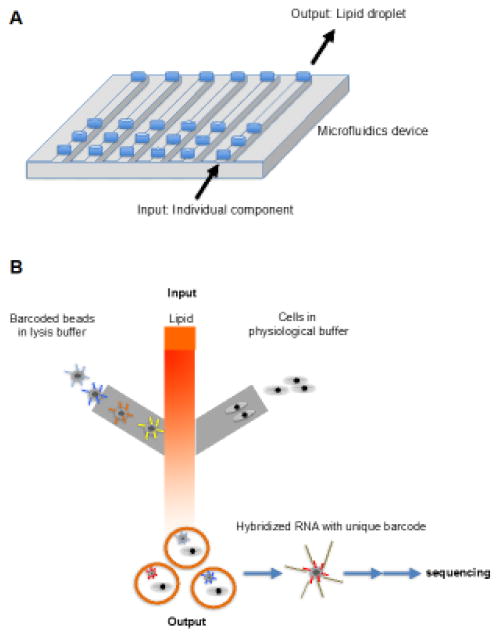
Figure 1. Single Cell Sequencing Technology.
A) schematic representation of a microfluidics chip with multiple devices (microfluidics chambers that are necessary to perform an individual experiment) on a single chip (i.e. usually a single chip can be used for multiple experiments. B) A single device has three input ports (oil, barcoded beads in lysis buffer, and cells of interest) and a single output port used for collecting bead- cell containing lipid droplets. Then each cell (or RNA in the cell) is marked by the unique barcode and processed on the bead for sequencing.
Plate-Based Techniques
Understanding biology at a single cell level has become an exciting area in biomedical research [28, 29], with various robust technological platforms developed for this purpose. The individual utility of these methods has been reviewed elsewhere [30, 31]; however, it is important to note that some scRNA-seq techniques, including Smart-seq2, Cell-seq, MATR-seq, SCRB-seq etc. rely on capturing or sorting individual cells into tubes or multi-well plates. Studies using this type of technology were the first to explore the brain at single cell levels in a high throughput format [5, 32]. However, one clear obstacle of these plate-based techniques is the limitation in cell numbers collected. An additional caveat is that these methodologies require FACS sorting, or require that cells to be separated based on size (i.e. Fluidigm/C1)[30, 33]. As a result, maintaining the native heterogeneity of CNS constituents is almost impossible. By contrast, lipid droplet/microfluidics-based cell capture techniques allow the capture and sequencing of thousands of cells simultaneously from an intact tissue. Because of the inherent variability in scRNA-seq data, it is important to ensure -- with sufficient confidence -- that sufficient numbers of cells are obtained so as to identify changes in any given cell population, on at an individual transcript level for each condition. While the exact number of cells required to understand transcriptional changes within a cellular population has to be determined experimentally, it’s clear that understanding pathological changes in cellular responses requires adequate statistical power. An additional consideration, is the depth of sequencing obtained; when sequencing resources are limiting, the greater the numbers of cells that are sequenced will reduce the number of reads for each individual cell. A critical challenge is to understand for any given experiment, what the optimal number of cells to capture is, while maintaining sufficient sequencing depth to understand comparative differences between biological groups.
Microfluidics and Lipid Droplet Techniques
A number of scRNA-seq platforms, including Drop-seq and inDrop platforms, have been developed to allow efficient multiplex RNA isolation and cDNA preparation after single cell separations using microfluidics and lipid droplet technologies [34, 35]. These platforms are flexible in the number of cells captured and sequenced. Microfluidics devices have been developed to efficiently mix the principal components, and then separate individual cells for RNA processing. Barcoded beads, each attached with a unique sequencing primer, are pumped into a chamber within the microfluidics device to mix with the cell suspension. Each bead and cell are then encapsulated in a lipid droplet. On the one hand, in the inDrop technology, the cell is lysed and the RNAs in this cell are reverse-translated into cDNAs in each Droplet, which are then captured by the unique primer on the bead [34]. On the other hand, in the Dropseq platform, the cell is lysed and RNAs are captured by the uniquely barcoded poly-dT primer on the bead, and cDNA libraries are generated after the lipid droplet is broken to produce STAMPS (single-cell transcriptomes attached to microparticles)[36]. Because each bead contains its own specific barcode that uniquely identifies transcriptomes from individual cells, these techniques allow the simultaneous analyses of transcriptomes of thousands of cells in a single reaction.
Critical differences between the Drop-seq and inDrop platforms include the number of barcodes used, and the physical properties of the beads. The inDrop method has fewer barcodes and is thus limited in the number of cells that it can process; yet it has a higher capture efficiency of cells (~10%), due to the use of soft large hydrogel beads [34]. Comparatively, the Drop-seq technology may allow the capture of a larger number of cells from complex tissue samples because a larger number of barcodes are available for sequencing, but yet, it exhibits lower capture efficiency (~1%) due to the hard- and smaller- sized methacrylic beads [35]. The laboratories that developed this technology have quickly and widely shared their experiences and expertise to disseminate this technology, enabling the greater scientific community to both validate the method, and to apply it to various fields of research. One clear limitation to both Drop-seq, InDrop, as well as other methods utilizing a unique sequencing primer, is that the DNA sequencing is carried out only from the 3′ end of the DNA. As a consequence, this method cannot detect alternative splice forms or examine allele-specific expression using single nucleotide polymorphisms (SNPs). In this regard, methods such as Smart-seq2 (a plate-based cellular collection and sequencing method) that do not use a unique sequencing primer, are able to sequence full-length transcripts [30, 31].
Challenges of Data Analysis in scRNA seq
Data obtained from scRNA-seq are technically identical to those from a traditional bulk expression profile analysis; however, they result in complex expression distributions and increased variability. These are in part caused by the heterogeneity in cell types as well as reduced abundance of transcripts, which poses new challenges demanding new statistical and computational methods for analysis. In this section, we discuss the general processes and considerations when analyzing data from scRNA-seq (Figure 2). While several methodologies exist for scRNA-seq, microfluidics-based platforms enable the capture of a large number of diverse cell types, with an ability to process them in a semi-automated way. For this main reason, we focus here on this method, as it allows to capture the inherent cellular heterogeneity of the CNS.
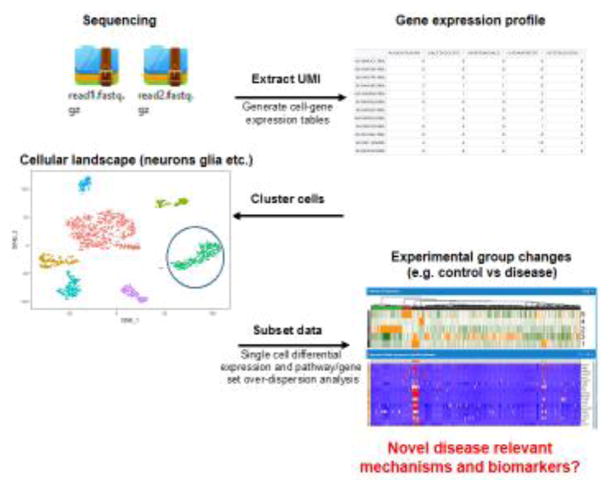
Figure 2. Single Cell Sequencing Data Analysis Workflow.
The Unique molecular identifier (UMI) is extracted from raw data, facilitating efficient filtering of duplicate transcripts from the PCR and sequencing steps. Cell-gene expression tables are then generated. Cells are clustered based on different gene expression profiles. With known cell-type specific gene markers, cell clusters can be identified as individual cell populations. Single cell differential gene expression or pathway overdispersion analysis can be applied to specific cell populations.
The data analysis associated with scRNA-seq typically can be grouped into three parts: (1) obtaining cell-gene expression tables, (2) identifying cell types, and/or (3) pursuing question-specific answers. The last two steps do not necessarily have to come in that order. The first part of the analysis is to generate a table showing the relative expression level of each gene per cell. A Microfluidics-based scRNA-seq platform requires paired-end sequencing: ‘Read1’ contains a cell-specific barcode and ‘unique molecular identifier’ (UMI)[36, 39] which is a random barcode that allows the tracking of unique transcripts, while ‘Read2’ contains a cDNA sequence near the poly-A tail of transcripts. Read2s are mapped to the genome with standard aligners (e.g, Tophat [40], STAR [41]) and they are quantified per gene and per cell by processing UMIs and cell-barcodes provided by Read1. Upon quantification, single-cell specific quality control measures are monitored, including total reads per cell, total number of genes detected per cell, and fraction of mapped reads. Based on these values, real cells are selected from empty droplets or cells of poor-quality, e.g., dying/dead cells. This provides a preliminary cell-gene expression table. Confounding factors such as batch effect and cell-cycle status should be corrected if necessary or possible, and certain cells can be also discarded.
The last procedure for generating a cell-gene expression table is to normalize -- either by dividing each UMI count with total UMI counts per cell [34], or by randomly down-sampling UMI counts in each cell such that all cells will have identical total UMI counts [42, 43]. By contrast, for plate-based scRNA-seq methods, external RNA controls consortium (ERCC) spike-in controls have been used as a known standard for normalization [44, 45]. However, this is not applicable for lipid-droplet/microfludics based methods, because the majority of droplets will not contain cells, and when added, ERCC will be amplified instead, appearing as a major RNA species at the expense of the relevant transcripts from cells. Another normalization method consists of converting the measurement of transcripts per kilobase million (TPM) into relative transcript counts, under the assumption that detected gene expression levels that are at, or below the peak of TPM distribution per cell, correspond to a single transcript [46]. However, researchers recommend the use of UMI, if possible; this is in agreement with a recent comparative report of scRNA-seq methods [31].
It is important to note that annotations of the 3′UTR are often incomplete and truncated early, resulting in the underestimation of UMI counts per gene [47]. In this regard, alternative sequencing methods, such as SMART-seq which do not rely on UMIs and can sequence full length transcripts, provide greater accuracy [31]. Extending annotations of 3′UTR thus gives more reads per gene [43], but currently there is no systematic research reporting optimal 3′UTR length for each gene in model organisms.
One critical limitation of any scRNA-seq platform is that the detected transcript levels exhibit high variability, especially for low abundant transcripts. For example, the capture rates of transcripts using inDrop and Drop-seq, have been reported as 7.1% [34] and 12.8% [35], respectively. Hence, histograms of expression levels of most genes present a large peak at 0 counts, termed ‘dropouts’ [31]. There are a variety of statistical methods developed to accommodate this problem, reviewed in [48]. This aspect of single cell data is one of the biggest hurdles of scRNA-seq; future technological improvement to increase the number of gene counts per cell might help to overcome this challenge.
The second part of the analysis identifies cell types from the cell-gene expression table based on similarity in gene expression patterns among cells. The difficulties in this process are quite variable and depend on the complexity of the population structure and the aim of the study [49]. If the cell types are very different from each other, principal component analysis (PCA) and its variants such as independent component analysis (ICA) and t-distributed stochastic neighbor embedding (tSNE), when combined with conventional k-means or hierarchical clustering would be sufficient to distinguish the different subpopulations [34, 35]. However, if the cell types are similar, or if higher resolution is required, more sophisticated methods such as Louvain-Jaccard clustering [50] and Infomap [51] can be used [52]. In addition, multiple iterations of simultaneous selection of relevant gene subsets and clustering of cell types might be necessary [37]. Thus, validation of detected cell types using histological methods is also necessary, especially when novel cell types are potentially observed in a given scRNA-seq experiment [53].
The third part of the analysis is specific to the question of interest. Standard statistical methods based on t-test, f-test, or Wilcoxon rank sum test can be used for differential expression analysis among cell types. Weighted gene co-expression network analysis (WGCNA) has also been adopted for network analysis [34, 54]. Moreover, ICA and the ‘pseudo-timing’ method have been used to reconstruct a temporal order of changes during cell differentiation [55]. In addition, Bayesian frameworks have been used to construct cell-type lineage trees [43]. Note that the former and latter methods are based on different premises– the former assumes continuity, while the latter assumes discreteness of cell-states during cell-fate transition represented in gene-expression space. In-situ hybridization data have also been a useful complement to scRNA-seq, facilitating the construction of a spatial expression map, as for example, in a developing embryo [56].
Most scRNA-seq analyses have focused on the characterization of systems of interest, which is an important first step. Similar to other high-throughput profiling methods, further challenges lie in developing algorithms that can generate experimentally testable predictions by inferring causality from the data generated from scRNA-seq, especially when attempting to understand disease mechanisms. Additionally, defining how scRNA-seq can be most effectively integrated with parallel bulk RNA-sequencing will also enhance our understanding of each individual cell within the whole network, and hopefully add to our knowledge of pathophysiological disease mechanisms.
Molecular Taxonomy of the Brain at the Single Cell Level
Since the complexity of the CNS is due, at least in part, to the presence of diverse cell types and their distinct properties governed by specific gene expression patterns, an accurate understanding of transcriptomics at the single cell level should provide the molecular taxonomy of distinct cell types in the CNS. Thus, improved resolution when characterizing gene expression patterns at the single cell level in the CNS may be translated into a better understanding of the brain in both health and disease. The desired, extensive gene expression lists are now technologically possible to obtain via single cell transcriptomics, as evidenced by recent scRNA-seq studies of the mouse cortex [57, 58].
While the field of high throughput scRNA-seq is in its infancy, these early forays using this technology suggest that it is feasible to define different cell types in the CNS by their transcriptomes at the single cell level. The power of scRNA-seq was first demonstrated in the analysis of the retina; the study, which pioneered the development of Drop-seq technology, analyzed transcriptomes from 44,808 mouse retinal cells and identified 39 transcriptionally distinct cell populations, creating a molecular atlas of gene expression for known retinal cell classes and novel candidate cell subtypes [35]. Similarly, another study used scRNA-seq to systematically examine the molecular diversity of retina bipolar cells (BCs) and identified 15 types of BCs, including all known subtypes and two novel types [52]. In addition, other researchers have used scRNA-seq to construct a cellular taxonomy of the mouse primary visual cortex, identifying multiple GABAergic, glutamatergic and non-neuronal cell subtypes [37]. These transcriptomically distinct cell types displayed specific and differential electrophysiological, and axon projection properties, which suggests that the single-cell transcriptomic signatures might be associated with specific cellular properties [37]. The report identified 49 transcriptomic cell types using ~ 1,600 cells from the mouse cortex. Of note, this study utilized a non-lipid-droplet/fluidics based approach and was able to obtain a robust dataset.
A recent large-scale scRNA-seq analysis of mouse somatosensory cortex (S1) and hippocampal CA1 region identified 47 molecularly distinct subclasses, comprising all known major cell types in the cortex, including 7 subclasses of pyramidal neurons distributed in different layers, 16 subclasses of interneurons, 2 classes of astrocytes, as well as 6 classes of oligodendrocytes, possibly representing different differentiation stages of these cells [5]. The functional relevance of such classification is supported by the fact that a subtype of interneuron, marked by the expression of Pax6 and Htr3a, showed very specific electrophysiological and morphological characteristics of late-spiking neurogliaform cells [5]. These results provide insights into the transcriptional regulation of cortical layer heterogeneity by providing an important template for future studies of disease. In addition, such studies demonstrate the region-specific heterogeneity in the continuous maturation process of cells, including neurons and glial cells in the CNS. Accordingly, distinct oligodendrocyte populations expressing molecular signatures of the whole lineage spectrum – from oligodendrocyte precursor cells to mature oligodendrocytes -- were found in different regions of the mouse brain [59]. However, the functional implications of such regional specificities of oligodendrocytes transcriptomes at different maturation stages will require further investigation.
Finally, while most of single cell transcriptome analyses have used experimental model organisms such as the mouse, an scRNA-seq analysis on 466 cells captured from healthy adult and fetal human brains (obtained during surgical procedures) was able to capture major cell types, including neurons, glia and vascular cells in the adult and fetal human brain [60]. A complement to this study was a report showing the ability to culture biopsy tissues from adult human patient brains, subjected to scRNA-seq analysis and identifying 5 classes of cells including oligodendrocytes, microglia, neurons, endothelial cells, and astrocytes [61]. Consequently, we can expect that studies such as these may eventually lead to a comprehensive cellular atlas of the human brain at the single cell level [60].
In conjunction studies defining human brain taxonomy, a recent study demonstrated the ability to perform RNA-seq on single nuclei isolated from post-mortem human neurons [8]. This work was conducted on 3,227 single neuronal nuclei derived from various regions of the cerebral cortex, leading to the identification of 16 neuronal subtypes. The method has been validated by others [62] but is thus far only amenable for isolation of nuclear RNA, which is clearly different from total cellular RNA. Nonetheless, the ability to detect different neuronal cell types in post-mortem tissue is a significant advance that may enable the characterization of a transcriptomic basis of neurodegenerative diseases at the single cell level. Furthermore, a recent report has described a method to cryopreserve human peripheral blood derived mononuclear cells and mouse tissue, successfully subjecting these to scRNA-seq [63]. This method also corroborates that conducting scRNA-seq in post-mortem brain tissue is thus possible.
Exploring the Mechanisms of Neurodegeneration at the Single Cell Level
In addition to being a molecular classification tool, scRNA-seq is being used to ask important questions relevant to disease mechanisms, as well as to provide information on possible new therapeutic targets and biomarkers. Two different studies using mouse models employed scRNA-seq to understand the regulation of serotonergic neurons with relevance to anxiety and mood disorders [64, 65]. One report used an elegant combination of genetic fate mapping, fluorescent cell sorting and genome-wide RNA-seq, both at population- and at single cell- levels, characterizing the diversity of serotonergic (5HT) neurons in mouse brains [64]. This study revealed profound differences in the patterns of gene expression both between and within anatomical domains and 5-HT neuron sub-lineages. In addition, using scRNA-seq on isolated serotonergic nuclei, another study revealed the expression of >500 G-protein coupled receptors (GPCRs) in the dorsal raphe serotonergic neurons [65]. Since GPCRs are excellent drug targets, this study might provide potential new targets for modulating DR neurons in multiple mood disorders [65]. Others used scRNA-seq to identify multiple subtypes of dopaminergic neurons, demonstrating the differential localization of various subtypes in distinct anatomic regions of the mouse brain [66]. Moreover, the authors identified a subtype of Aldh1a1+ dopaminergic neurons in the substantia nigra that were especially vulnerable in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) toxin-induced mouse model of Parkinson’s disease (PD).
The striatum is involved in controlling multiple aspects of cognitive function as well as in translating cortical activity into adaptive motor actions in the brain. Striatal dysfunction contributes to multiple neurodegenerative or psychiatric disorders, including PD, Huntington’s disease, schizophrenia and addiction [67]. The GABAergic medium spiny neurons (MSNs), the principal neurons of the striatum, are traditionally classified into D1- and D2-subtypes based on the expression of the D1-dopamine or the D2-dopamine receptors, respectively [68]. A recent study analyzed 1208 cells from the mouse striatum using a plate-based scRNA-seq approach (Fluidigm/C1, SMARTseq2) and documented that both D1 and D2-MSNs could be subdivided into additional discrete populations [69]. Since the function of dopamine as a key neurotransmitter is linked to both neurodegenerative and neuropsychiatric disorders, the characterization of distinct populations of dopaminergic neurons may facilitate our understanding of the molecular underpinning and pathogenesis of such diseases.
ScRNA-seq has also been used to investigate the regenerative potential of the brain. One study isolated 130 cells from the subventricular zone of adult mice and characterized the existence of a pool of dormant neural stem cells (NSCs) expressing distinct combinations of lineage-specific genes [70]. Furthermore, the study also examined the response of NSCs to brain ischemia, identifying the transition of stem cells from dormant, to activated state following injury, and when influenced by interferon gamma (IFN) signaling [70]. The report was the first to compare NSCs in vivo in a pathological context, identifying signaling modules that could be targeted for regeneration [70]. A challenge now will be to extend such studies into human systems and translating relevant findings into therapeutic interventions.
An alternative avenue to investigating the mechanisms of CNS diseases may be to use peripheral cells as surrogates of pathology [71]. Various research groups have examined transcriptional changes in patient derived blood cells, looking for signatures of neurological diseases and identifying potential disease mechanisms [71, 72]. These studies are particularly relevant because an emerging concept in the field of neurodegeneration includes the role of immune dysfunction in driving disease pathology [73] [74]. Thus, understanding the transcriptional landscape of peripheral cells such as monocytes may enable the discovery of disease biomarkers, but also inform on mechanistic pathways. Patient derived blood cells are easily accessible and do not require post-mortem analysis, which is a critical limitation in studying human neurodegenerative diseases [75, 76]. These cells are also amenable for scRNA-seq analysis[75, 76]. It is intriguing to imagine whether a subtype of peripheral blood cells characterized through scRNA-seq could be used as a ‘window’ into the brain. ScRNA-seq analysis performed on peripheral blood cells, coupled with RNA-seq data obtained directly from the CNS, such as post-mortem tissue from patients, could be exploited to potentially elucidate critical mechanisms contributing to human neurodegeneration.
The studies described above depict the vast potential of utilizing scRNA-seq in our quest to better understand the physiology and pathophysiology of the CNS. Because of the relative novelty of this technique and the lack of a standardized protocol in the CNS, it is important to compare different methods used to prepare samples for the single cell workflow. Some of the earlier scRNA-seq studies utilized cells that were dissociated and then FACS- enriched [5]. This methodology has two inherent caveats. First, the dissociation of cells should be conducted in a manner that faithfully recapitulates the cellular distribution of all cell types present in the CNS and where the transcriptional landscape is maintained throughout this process. Second, FACS should be conducted using multiple cell surface marker since FACS may alter the transcriptional cellular profile and may also bias the cellular collection [37]. Because of the development of new data analysis tools and optimization of dissociation protocols, some of the more recent studies have been able to conduct scRNA-seq analyses on multiple cell types of whole brain, simultaneously [34, 35]. This important transition signifies a shift in our ability to study the brain.
ScRNA-seq can be applied to understand how a genetic perturbation affects the CNS at high resolution. The application of NGS has enabled the identification of novel genetic association and risk factors associated with human diseases, such as the involvement of OPTN, TBK1, SQSTM1 and C9ORF72 in ALS [77]. The expression pattern of these genes suggests an involvement in microglial function in both mouse models and in human neurodegenerative disease [74]. Thus, performing scRNA-seq may not only help us identify the cell type(s) in which these genes are most highly expressed, but also reveal the specific cell type(s) in the CNS which is/are most affected by the perturbation of these genes. Additionally, understanding the mechanism of action (MOA) of potential therapeutic molecules and current drugs on the market remains a big challenge[78]. ScRNA-seq can be utilized both in vitro (e.g. brain slice culture) and in vivo (e.g. injection into a rodent species), allowing the assessment of cell type and pathways that may be altered by a molecule of interest. One report described the effect of drug treatment on human brain tissue, retrospectively examined; human biopsied hippocampal tissue was cultured and subject to scRNA-seq and the effects of treatment with Keppra and Dexamethasone (compounded together) were significantly associated with the expression levels of GFAP, CD14, HTRA3, and PDPN genes in endothelial cells[61]. This represents an example of testing the effect(s) of compounds or drugs of interest on animal and human brain tissues using scRNA-seq.
Concluding Remarks
ScRNA-seq may be applied to achieve three major goals in neuroscience research. The first goal is to provide a molecular atlas of the brain with unparalleled resolution. This atlas extends our knowledge on the structural organization of the brain and helps us infer connectivity patterns and functional integration. In addition, it may facilitate the design of novel tools that can be used to target with notable, spatiotemporal precision, specific subpopulations and lineages in the brain. The second goal is to provide detailed molecular profiling capability to explore the molecular vulnerability of specific neuronal subpopulations in diseases and to identify candidate target molecules for therapeutic intervention (see Outstanding Questions and Box 4). Discovering novel drug targets localized to particular cells of the CNS using scRNA-seq is an exciting direction in therapeutic research. Furthermore, the ability to link a pathology- or lesion-specific signatures to cells may paves the way in the identification of putative novel therapeutic pathways and biomarkers. An attractive scenario includes the microdissection of a lesion within the brain analyzed by scRNA-seq to identify a cell-specific signature. Such information could then be used to guide drug discovery in association with specific lesions of the CNS (see Box 2). The recent introduction of automated platforms for scRNA-seq may thus provide an opportunity not only to fully standardize this method, but also to enable a routine workflow of single cell isolation and sequencing of animal and human tissues, that may include biopsy or post-mortem brain pathological samples. The third goal is to define specific cellular targets at transcriptomic levels with pharmacological manipulations. We predict that future applications of scRNA-seq in neuroscience research might provide an exciting prospective in understanding physiological structure-function changes in human brains as well as new opportunities in therapeutics approaches to treat neurological pathologies.
Outstanding Questions Box.
What can be done to improve the number of reads per cell in scRNA-seq?
A direct comparison between different single cell methods will enable us to focus on method optimization to increase the depth of single cell runs. This will not only include optimization of dissociation methods but also provide a better delineation of an ideal sequencing protocol followed by data analysis. A critical challenge will also be to understand the optimal number of cells to be captured for a given experiment, while maintaining sufficient sequencing depth to understand differences between two or more biological groups.
What will be the best way to integrate scRNA-seq data into the discovery pipepline?
As the field of scRNA-seq matures, the way in which we use this information will need to be standardized. In particular, experimental definition of how bulk RNA sequencing approaches in conjunction with single cell data in the brain are utilized needs to occur. Additionally, how scRNA-seq is partnered with single cell genomics and proteomics technology will need to be established
What are the differences between single cell preparations in aged and/or diseased human tissue as compared to mouse tissue?
While studies have shown the ability to isolate different cells in the CNS in various animal models of neurodegenerative disease, it remains to be seen whether single cell dissociation protocols can be used to effectively isolate different cell types in the aged human brain. These studies will also have to examine whether cryoprotection of human tissue is indeed amenable to scRNA-seq.
What are some of the new applications for scRNA-seq in the CNS?
Recent studies have shown that scRNA-seq can be successfully paired with genome-wide CRISPR/Cas9 genome editing screens. One intriguing idea may be to utilize this type of approach in the intact brain in an animal model of neurodegenerative disease. One can envision the use of these experiments as a novel unbiased semi-high throughput discovery platform for drug discovery targets in CNS diseases.
Box 4. Clinician’s Corner.
Single cell RNA sequencing (scRNA-seq) provides valuable insight into the complexity of the nervous system by enabling the identification of molecular signatures at unprecedented resolution. Thus, the relative representation of different cell types within the nervous system can be inferred in health and disease.
The significance of scRNA-seq extends to being able to correlate structural and functional properties of cell types with distinct molecular signatures. Such analytical power reveals new dimensions of cell identity, and allows a deeper understanding of the relationship between disease state and cellular response. For example, the vulnerability of specific neuronal cell types in different neurodegenerative diseases might be explained, in part, in terms of gene expression. Ultimately, novel molecules and pathways might be identified as candidate therapeutic targets.
The identification of cell-type specific signatures at the resolution of a single cell can uncover a multitude of relevant biomarkers, which might be used to monitor disease progression and assess the efficacy of applied therapeutic strategies.
Future developments of scRNA-seq aim to improve the efficiency of the method by increasing coverage of the transcriptome, preserve the physiological context of cells and couple the information retrieved from RNA-seq with methods such as cell imaging or proteomics. Thus, scRNA-seq provides a platform for a truly holistic understanding of cellular physiology in health and disease.
Trends Box.
In the context of the CNS, scRNA-seq technology lies at the interface of neuroscience, computational biology and systems biology, enabling a novel and unbiased understanding of the CNS.
scRNA-seq in the brain can discriminate between different cell types in the CNS, facilitating the direct comparison of different cell types between samples on a population level.
The ability to identify individual cells in the CNS opens possibilities of finding pathogenic and/or regenerative cells and pathways that can be exploited for drug discovery.
scRNA-seq has been carried out in fresh and frozen human brain tissue. This paves the way towards a more precise understanding of neurological and neurodegenerative diseases.
Acknowledgments
This work was supported in part by grants (to JY) from the NINDS (1R01NS082257) and the NIA (1R01AG047231).
Glossary
- optogenetic modulation
genetic stimulation of cells (usually neurons), using light. This technique has been used to control the activity of neurons both in vitro and in vivo (i.e. awake free-moving mice).
- next generation sequencing (NGS)
high-throughput nucleic acid sequencing performed on multiple commercial platforms. It has replaced traditional Sanger sequencing, and utilizes a parallel sequencing process, producing many simultaneous sequences.
- Bulk RNA sequencing
Uses a ‘bulk’ population of cells as starting material; traditionally referred to as RNA-seq.
- In-situ hybridization
method where nucleic acids (RNA, DNA or modified nucleic acids strands) hybridize to complementary sequences, identifying the presence and localization of a particular RNA or DNA species within a cell or tissue.
- transient global ischemia
here, a transient event that affects the entire brain. Certain neurons of the CNS are more vulnerable to this type of insult. This includes the main excitatory neurons of the CA1 region of the hippocampus, the pyramidal cells. Adjacent CA3 pyramidal neurons of the hippocampus are relatively resistant to this type of injury.
- Multiple sclerosis
demyelinating disease of the CNS which leads to a disruption of neuronal function.
- Barcodes
Here, Labeled Nucleotides that are added to RNA or DNA, that can be used during sequencing to identify a specific sample source.
- Paired-end sequencing
refers to the two ends of the same DNA molecule that are both sequenced. Often performed to obtain additional information about the origin of the DNA molecule being sequenced (i.e. barcode, UMI etc.)
- standard aligners
software used to align sequencing data to a genome of reference (e.g human genome or mouse genome). This is a critical step in the use and quantification of sequencing data.
- batch effect
inherent variation in sequencing data from 2 or more replicate experiments.
- Cell-gene expression table
a table whose columns are cells, and rows are genes, with each element representing a relative gene expression value.
- Unique molecular identifier (UMI)
short sequence of random DNA oligomer attached to cDNA during reverse transcription; it allows tracking of unique transcripts.
- principal component analysis (PCA)
A linear transformation algorithm that embeds high-dimensional data to low-dimension (e.g., 2D), capturing the most varying direction of data points. It is frequently used for RNA-seq (bulk and single-cell) analysis to determine and visualize key changes between data sets.
- Independent component analysis (ICA)
An algorithm to separate mixed signals into independent components, which can be used as a dimensionality reduction tool.
- t-distributed stochastic neighbor embedding (tSNE)
A non-linear transformation algorithm that embeds high-dimensional data to low-dimension, preserving local structure. This method has frequently been used to visualize different cell states or different cell types in scRNA-seq.
- k-means clustering
clustering algorithm that designates each data point into a cluster with the nearest mean.
- hierarchical clustering
clustering algorithm that successively merges data according to their similarities.
- Louvain-Jaccard clustering
algorithm that clusters cells by keeping densely connected weighted edges among cells, while discarding sparse edges between clusters.
- Infomap
algorithm that decomposes a molecular network into modules by compressing a description of the probability flow on a graph.
- Network neighborhood analysis
method that finds a set of genes (the neighborhood) that is similar to an initial ‘seed’ set of genes. It defines neighborhoods on the basis of a robust interconnectedness measure, e.g. the topological overlap measure. It can be used to define multi-node similarity measures.
- weighted gene co-expression network analysis (WGCNA)
algorithm that clusters genes whose expressions are correlated by applying high power to the correlation coefficients and by using a topological overlap matrix.
- GABAergic medium spiny neurons (MSNs)
key neuronal type within the basal ganglia of the striatum; uses GABA as its main neurotransmitter, while responding to dopamine. MSNs are critical for movement initiation, or motivation and reward, depending on the expression of the dopamine receptor (Dopamine-like receptor 1 vs. Dopamine-like receptor 2).
- dorsal raphe
anatomical region of the Raphe nucleus located at the midline of the brainstem; responsible for various functions including sleep and depression. It has a concentration of serotonergic neurons which use serotonin as their main neurotransmitter, impacting sensorimotor function.
- mouse somatosensory cortex (S1)
distinct region in the mouse neocortex responsible for sensory input, especially tactile input. Much of the input comes from the mouse whiskers.
- Aldh1a1+ dopaminergic neurons
Aldehyde dehydrogenase 1A1 (Aldh1a1): enzyme facilitating the detoxification of catecholamines including dopamine (DA) and norepinephrine; found in a subset of dopaminergic neurons and helps dopamine metabolism-associated toxicity in those neurons.
- Keppra
anti-epileptic drug (aka levetiracetam).
- Dexamethasone
corticosteroid; potent anti-inflammatory drug.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162(2):246–57. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lerner TN, et al. Communication in Neural Circuits: Tools, Opportunities, and Challenges. Cell. 2016;164(6):1136–50. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 2016;17(1):36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poulin JF, et al. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci. 2016;19(9):1131–41. doi: 10.1038/nn.4366. [DOI] [PubMed] [Google Scholar]
- 5.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–42. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 6.Foldy C, et al. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc Natl Acad Sci U S A. 2016;113(35):E5222–31. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Manno G, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167(2):566–580. e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–90. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramón y Cajal S, et al. Texture of the nervous system of man and the vertebrates. 1, 3 Springer; Wien; New York: 1999. [Google Scholar]
- 10.Chizhikov VV, Millen KJ. Mechanisms of roof plate formation in the vertebrate CNS. Nat Rev Neurosci. 2004;5(10):808–12. doi: 10.1038/nrn1520. [DOI] [PubMed] [Google Scholar]
- 11.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468(7321):214–22. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 12.Zuchero JB, Barres BA. Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 2013;23(6):914–20. doi: 10.1016/j.conb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry AM, Hohmann JG. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mamm Genome. 2012;23(9–10):539–49. doi: 10.1007/s00335-012-9406-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89(1):37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin K, et al. Microarray analysis of hippocampal gene expression in global cerebral ischemia. Ann Neurol. 2001;50(1):93–103. doi: 10.1002/ana.1073. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara N, et al. Genome-wide gene expression analysis for induced ischemic tolerance and delayed neuronal death following transient global ischemia in rats. J Cereb Blood Flow Metab. 2004;24(2):212–23. doi: 10.1097/01.WCB.0000106012.33322.A2. [DOI] [PubMed] [Google Scholar]
- 18.Mycko MP, et al. Microarray gene expression profiling of chronic active and inactive lesions in multiple sclerosis. Clin Neurol Neurosurg. 2004;106(3):223–9. doi: 10.1016/j.clineuro.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Dutta R, Trapp BD. Gene expression profiling in multiple sclerosis brain. Neurobiol Dis. 2012;45(1):108–14. doi: 10.1016/j.nbd.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett DA, et al. Selected findings from the Religious Orders Study and Rush Memory and Aging Project. J Alzheimers Dis. 2013;33(Suppl 1):S397–403. doi: 10.3233/JAD-2012-129007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153(3):707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivasan K, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberwine J, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89(7):3010–4. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tietjen I, et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38(2):161–75. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 25.Cauli B, et al. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 1997;17(10):3894–906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong X, et al. Building an RNA Sequencing Transcriptome of the Central Nervous System. Neuroscientist. 2016;22(6):579–592. doi: 10.1177/1073858415610541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 29.Tischler J, Surani MA. Investigating transcriptional states at single-cell-resolution. Curr Opin Biotechnol. 2013;24(1):69–78. doi: 10.1016/j.copbio.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–6. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziegenhain C, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65(4):631–643. e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–53. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 33.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32(10):1053–8. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–6. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 37.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19(2):335–46. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodworth MB, et al. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat Rev Genet. 2017;18(4):230–244. doi: 10.1038/nrg.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hug H, Schuler R. Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J Theor Biol. 2003;221(4):615–24. doi: 10.1006/jtbi.2003.3211. [DOI] [PubMed] [Google Scholar]
- 40.Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–9. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Z, et al. A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell Stem Cell. 20(1):120–134. doi: 10.1016/j.stem.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding B, et al. Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics. 2015;31(13):2225–2227. doi: 10.1093/bioinformatics/btv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vallejos CA, et al. BASiCS: Bayesian Analysis of Single-Cell Sequencing Data. PLoS Comput Biol. 2015;11(6):e1004333. doi: 10.1371/journal.pcbi.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu X, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirion OB, et al. Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet. 2016;7:163. doi: 10.3389/fgene.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heimberg G, et al. Low Dimensionality in Gene Expression Data Enables the Accurate Extraction of Transcriptional Programs from Shallow Sequencing. Cell Syst. 2016;2(4):239–50. doi: 10.1016/j.cels.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blondel VD, et al. Fast unfolding of communities in large networks. Journal of Statistical Mechanics-Theory and Experiment 2008 [Google Scholar]
- 51.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 2008;105(4):1118–23. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shekhar K, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166(5):1308–1323. e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trimarchi JM, et al. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502(6):1047–65. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- 54.Luo Y, et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161(5):1175–86. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satija R, et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson MB, Walsh CA. Cerebral cortical neuron diversity and development at single-cell resolution. Curr Opin Neurobiol. 2016;42:9–16. doi: 10.1016/j.conb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar P, et al. Understanding development and stem cells using single cell-based analyses of gene expression. Development. 2017;144(1):17–32. doi: 10.1242/dev.133058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–9. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112(23):7285–90. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaethling JM, et al. Primary Cell Culture of Live Neurosurgically Resected Aged Adult Human Brain Cells and Single Cell Transcriptomics. Cell Rep. 2017;18(3):791–803. doi: 10.1016/j.celrep.2016.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishnaswami SR, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nature Protocols. 2016;11(3):499–U281. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillaumet-Adkins A, et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 2017;18(1):45. doi: 10.1186/s13059-017-1171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okaty BW, et al. Multi-Scale Molecular Deconstruction of the Serotonin Neuron System. Neuron. 2015;88(4):774–91. doi: 10.1016/j.neuron.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spaethling JM, et al. Serotonergic neuron regulation informed by in vivo single-cell transcriptomics. FASEB J. 2014;28(2):771–80. doi: 10.1096/fj.13-240267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poulin JF, et al. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 2014;9(3):930–43. doi: 10.1016/j.celrep.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Deurwaerdere P, et al. Expanding the repertoire of L-DOPA’s actions: A comprehensive review of its functional neurochemistry. Prog Neurobiol. 2017;151:57–100. doi: 10.1016/j.pneurobio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Kreitzer AC, Malenka RC. Striatal plasticity and basal ganglia circuit function. Neuron. 2008;60(4):543–54. doi: 10.1016/j.neuron.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gokce O, et al. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016;16(4):1126–37. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Llorens-Bobadilla E, et al. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17(3):329–40. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 71.Bradshaw EM, et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16(7):848–50. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satoh J, et al. Microarray analysis identifies a set of CXCR3 and CCR2 ligand chemokines as early IFNbeta-responsive genes in peripheral blood lymphocytes in vitro: an implication for IFNbeta-related adverse effects in multiple sclerosis. BMC Neurol. 2006;6:18. doi: 10.1186/1471-2377-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jay TR, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med. 2015;212(3):287–95. doi: 10.1084/jem.20142322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–83. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 75.Chan G, et al. Trans-pQTL study identifies immune crosstalk between Parkinson and Alzheimer loci. Neurol Genet. 2016;2(4):e90. doi: 10.1212/NXG.0000000000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167(7):1883–1896. e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 77.Taylor JP, et al. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nutt DJ, Attridge J. CNS drug development in Europe - Past progress and future challenges. Neurobiology of Disease. 2014;61:6–20. doi: 10.1016/j.nbd.2013.05.002. [DOI] [PubMed] [Google Scholar]