Abstract
Ribosomal biogenesis is tightly associated with cellular activities, such as growth, proliferation, and cell cycle progression. Perturbations in ribosomal biogenesis can initiate so-called nucleolar stress. The process through which ribosomal proteins (RPs) transduce nucleolar stress signals via MDM2 to p53 has been described as a crucial tumor-suppression mechanism. In this review we focus on recent progress pertaining to the function and mechanism of RPs in association with the MDM2–p53 tumor-suppression network, and the potential implications this surveillance network has for cancer development.
The Emergence of RPs in the p53 Pathway
Cancer is a collective designation for hundreds of diseases. Hallmarks of cancer include sustained proliferative signaling, evasion of growth suppressors, resistance to cell death, replicative immortality, induced angiogenesis, and activation of invasion and metastasis [1]. Despite the vast complexity of different cancers, all have at least one feature in common: uncontrolled cell proliferation, which is a consequence of enhanced protein synthesis, a process facilitated by the ribosomal machinery. Thus, as a limiting factor for protein translation, ribosome biogenesis plays a central role in oncogenic signaling processes associated with several protooncogenes such as RAS, MYC, and PI3K (reviewed in [2,3]). To counteract adverse deregulation of ribosome biogenesis, mammalian cells have developed surveillance mechanisms that monitor aberrant ribosome production through activation of a set of tumor suppressors such as p53, p14ARF (p19ARF in mice) and PTEN that suspend cellular proliferation in the event of altered protein synthesis [4]. As the ‘guardian of the genome’, p53 plays a pivotal role in cellular stress response networks. Over the past decade, escalating evidence from in vitro cell systems, animal models, and human clinical data have demonstrated that ribosomal stress inhibits p53 ubiquitination and induces p53 transactivation, consequently leading to p53-dependent cell cycle arrest or apoptosis. Mechanistically, it has been well documented that a large number of RPs are involved in the regulation of p53 via interaction with murine double minute-2 (MDM2), where RPs inhibit MDM2-mediated p53 proteosomal degradation [5–7]. This review will summarize the identification of the RP–MDM2–p53 pathway as a regulatory checkpoint for protein synthesis, the mechanisms involved in p53 regulation, and the implications for tumor surveillance based on data generated in vitro, as well as in animal models.
The RP–MDM2–p53 Pathway Is a Checkpoint for Ribosome Biogenesis
Ribosome biogenesis consists of three events in the nucleolus: ribosomal RNA (rRNA) transcription, rRNA processing, and ribosome assembly. In eukaryotes, 79 RPs have been identified, of which 33 are incorporated into the 40S subunit and 46 into the 60S [8,9]. Ribosome biogenesis is a complex, but precisely coordinated cellular process, and likely the most significant metabolic event within a proliferating cell. The entire process of ribosome biogenesis has been estimated to consume nearly 60% of cellular resources [10]. For this obvious reason, it occupies a central position in both component biosynthesis and energy homeostasis. Failure to properly regulate ribosome biogenesis is a key signal to cellular stress response pathways, collectively referred to as ribosomal stress or nucleolar stress, suggesting that the integrity of ribosome biogenesis serves as a vital checkpoint for cellular homeostasis.
Stress-induced p53 activation constitutes a major cellular response mechanism triggered by deregulation of homeostasis [11]. Metabolic perturbation, nutrient deprivation, hypoxia, DNA damage, viral infection, or even temperature change can interrupt ribosomal integrity and induce ribosomal stress [3,12]. The points of ribosome biogenesis that signal a stress response can be stratified into three categories: disruption of rRNA synthesis, disruption of rRNA processing and maturation, and imbalance of ribosome components. The primary mechanism driving the ribosomal stress response is the interaction between nucleolar proteins and MDM2, which inhibits the E3 ligase activity of MDM2 towards p53 and consequently stabilizes p53 (reviewed in [5]). The first evidence of RP interaction with MDM2 involved RPL5 binding to MDM2 in a 5S rRNA–RPL5–MDM2–p53 ribonucleoprotein complex [13]. However, the functional consequences of the RP–MDM2 interaction were not appreciated until several additional studies revealed that other RPs including RPL11, RPL23, and RPL5 could activate p53 through interaction with MDM2 [7,14–18]. To date, as many as 16 RPs have been identified to bind to MDM2, particularly to the central acidic domain, and regulate p53 activity (Table 1). Among them, RPL5 and RPL11 function as both sensor and effector of ribosomal stress, while other RPs including RPL6, RPL23, RPL26, RPL37, RPS3, RPS7, RPS14, RPS15, RPS20, RPS25, RPS26, RPS27/RPS27L, and RPS27a bear the effector function and bind to MDM2 [7,19]. Of note, almost all the aforementioned RPs exert their protective effects of p53 by antagonizing the E3 ligase activity of MDM2, providing increased weight of evidence for an RP–MDM2–p53 stress response pathway.
Table 1.
RPs and Biosynthesis Factors Involved in Regulating the MDM2-p53 Pathway.
| Protein | Binding to MDM2 | Ablation Induces p53 | L5/L11 Dependency in p53 Regulation | Mechanism of p53 Regulation | Refs |
|---|---|---|---|---|---|
| RPL5 | Yes | No | N/A | Inhibits Mdm2 E3 activity | [13,16] |
| RPL11 | Yes | No | N/A | Inhibits Mdm2 E3 ligase activity; Increases p53 acetylation and transactivity | [14,15] |
| RPS3 | Yes | No | N/A | Inhibits Mdm2 E3 activity | [96] |
| RPS15 | Yes | Yes | N/A | Inhibits Mdm2 E3 activity | [97] |
| RPS20 | Yes | Yes | N/A | Inhibits Mdm2 E3 activity | [97] |
| RPS25 | Yes | No | N/A | Inhibits Mdm2 E3 activity | [26] |
| RPS27 | Yes | No | N/A | Inhibits Mdm2 E3 activity | [22] |
| RPS27a | Yes | No | N/A | Inhibits Mdm2 E3 activity | [98] |
| RPL6 | Yes | No | N/A | Inhibits Mdm2 E3 activity | [23] |
| RPL26 | Yes | No | N/A | Enhances p53 translation | [99] |
| RPS7 | Yes | Yes | L5/L11 | Inhibits Mdm2 E3 activity | [20,35] |
| RPS14 | Yes | Yes | L5/L11 | Inhibits Mdm2 E3 activity | [51] |
| RPS26 | Yes | Yes | L11 | Inhibits Mdm2 E3 activity | [100] |
| RPS27L | Yes | Yes | L11 | Inhibits Mdm2 E3 activity | [22,66] |
| RPL23 | Yes | Yes | L5/L11 | Inhibits Mdm2 E3 activity | [17,18] |
| RPL37 | Yes | Yes | L11 | Inhibits Mdm2 E3 activity | [101] |
| RPS6 | No | Yes | L11 | Maintains ribosomal integrity | [35] |
| RPS9 | No | Yes | L11 | Maintains ribosomal integrity | [102,103] |
| RPS19a | No | Yes | L5/L11 | Maintains ribosomal integrity | [51,104] |
| RPS23 | No | Yes | L11 | Maintains ribosomal integrity | [35] |
| RPL7A | No | Yes | L11 | Maintains ribosomal integrity | [35] |
| RPL22 | No | Yes | N/A | Inhibits p53 protein synthesis | [88] |
| RPL24 | No | Yes | L11 | Maintains ribosomal integrity | [84] |
| RPL29 | No | Yes | L11 | Maintains ribosomal integrity | [87] |
| RPL30 | No | Yes | L11 | Maintains ribosomal integrity | [87] |
| SRSF1 | Yes | No | L5 | Inhibits Mdm2 E3 activity | [71] |
| NPM | Yes | No | N/A | Inhibits Mdm2 E3 activity | [105] |
| NCL | Yes | No | N/A | Inhibits Mdm2 E3 activity | [106] |
| PAK1IP1 | Yes | Yes | L5/L11 | Inhibits Mdm2 E3 activity | [107] |
| NS | No | Yes | L5/L11 | Inhibits Mdm2 E3 activity | [108] |
| PICT1 | No | Yes | L11 | Nucleolar anchor of L11 | [70] |
| MYBBP1A | No | No | L5/L11 | Facilitate p53 acetylation | [109] |
In addition to the direct binding of RP to MDM2, additional mechanisms have been ascribed. For example, RP–MDM2 interaction provides a platform for MDM2 to degrade RP substrates such as RPS7 [20,21], RPS27L [22], RPL6 [23], and RPL26 [24]. MDM2 can negatively regulate RPL26 through polyubiquitination and proteasomal degradation, attenuating the association between RPL26 and p53 mRNA, thereby repressing the RPL26-mediated augmentation of p53 gene translation [24]. In addition, RPL11 was reported to be recruited to the promoter region of p53 targeting genes to induce p53 transcriptional activity [25]. Apart from regulatory effects of RPs on p53, several RPs were identified as direct p53 target genes [22,26,27], suggesting the existence of a feedback loop that maintains a balance between activation and degradation of p53 by modulating the available pool of MDM2-interacting RPs. Other than regulating p53, MDM2 also suppresses TAp73 transcriptional activity by directly binding to its N-terminal transactivation (TA) domain. Interestingly, RPL5 and RPL11 can compete with MDM2 to bind to the TA domain of TAp73 in response to ribosomal stress, which results in elevated TAp73 transcriptional activity, suggesting an alternative mechanism underlying potential tumor inhibition by RPs via different p53 family members [28].
Ribosomal Component Imbalance Stimulates p53 Activation
A fundamental question remains as how the ribosomal stress signals communicate with MDM2 to regulate p53. Ribosome biogenesis requires the production of roughly equimolar amounts of each rRNA species and RPs to generate 60S and 40S subunits and to yield sufficient numbers of mature 80S polysomes [29]. Intensive studies on ribosomal stress have revealed that imbalance of ribosomal components is the key source of unassembled RPs. RNA polymerases (Pol) I and Pol III are responsible for the transcription of the ribosomal DNA (rDNA) genes. Inhibition of RNA Pol I by a low concentration of actinomycin D (<10 nM) or 5-fluorouracil (5-FU) can induce breakdown of nucleolar structure and activate a p53 stress response [30–32]. Infidelity in rRNA processing can lead to the accumulation of unprocessed intermediate transcripts, leading to attenuation of subunit assembly that triggers a nucleolar stress event. For example, ablation of Wrd36, a protein required for 18S rRNA processing in zebrafish, disrupts pre-rRNA processing and leads to the inappropriate accumulation of immature rRNA, which signals a stress response to p53 [33]. In addition, studies from in vitro knockdown or in vivo deletion of RPs demonstrated that the deficiency of several RPs results in the induction of p53-dependent stress responses (Table 1). It is thus argued that imbalance of ribosome components is the major reason for ribosomal stress and the consequent activation of p53. A continuous and stable supply of RPs is essential for maintaining adequate translational machinery. Otherwise, in response to ribosomal stress, the balance among ribosome components is disrupted, and unassembled RPs function as a regulatory unit, effectively constituting a quality control system to transmit cellular stress response signals through MDM2 to p53.
Unassembled RPs provides prompt the ribosome assembly machinery, mainly residing in the nucleolar compartment, to crosstalk with MDM2 and p53. Studies on RPL11 suggest two possible models that may explain the physical RP–MDM2 interaction. One model holds that disruption of the nucleolus promotes the release of RPL11 into the nucleoplasm where it can bind MDM2 and stabilize p53 [34]. This model is further supported by the finding that actinomycin D-induced RPL6 translocation from nucleolus to nucleoplasm precedes interactions with MDM2 [23]. A second model proposes that nucleolar stress induces an increase in the translation of RP-enriched 5′-terminal oligopyrimidine tract (5′-TOP) mRNA [35]. In this model, nucleolar stress causes an increase in RPL11 expression, thus raising the concentration of unassembled free RPL11 available to bind to MDM2. Sladana and colleagues demonstrated that latency in proteasomal degradation of RPL5 and RPL11 could be another source for the accumulation of free RPs [36]. Taken together, this evidence supports an RP–MDM2–p53 regulatory mechanism responsive to perturbation of ribosome biogenesis.
Oncogenic activation of ribosome biogenesis is a useful experimental approach to investigate the mechanisms and consequences of aberrant ribosome production. Overexpression of c-MYC induces ribosome biogenesis via the upregulation of both rRNA and RPs, contributing to the incidence of lymphoma [37]. On the other hand, reduced RP expression can inhibit c-MYC-induced tumorigenesis. Indeed, heterozygosity for RPL24 or RPL38 impedes the oncogenic activity of c-MYC [38]. Macias and colleagues generated a mouse model bearing a single point mutation in the zinc-finger domain of MDM2 (MDM2C305F) that inhibits the binding between MDM2 and RPL5/RPL11 and mitigates the p53 response specific to ribosomal stress. By crossing with Eμ-Myc transgenic mice, MDM2C305F mice displayed strikingly accelerated lymphomagenesis [39], indicating that the RP–MDM2–p53 pathway is triggered in response to the hyperactivation of ribosome biogenesis driven by c-MYC overexpression.
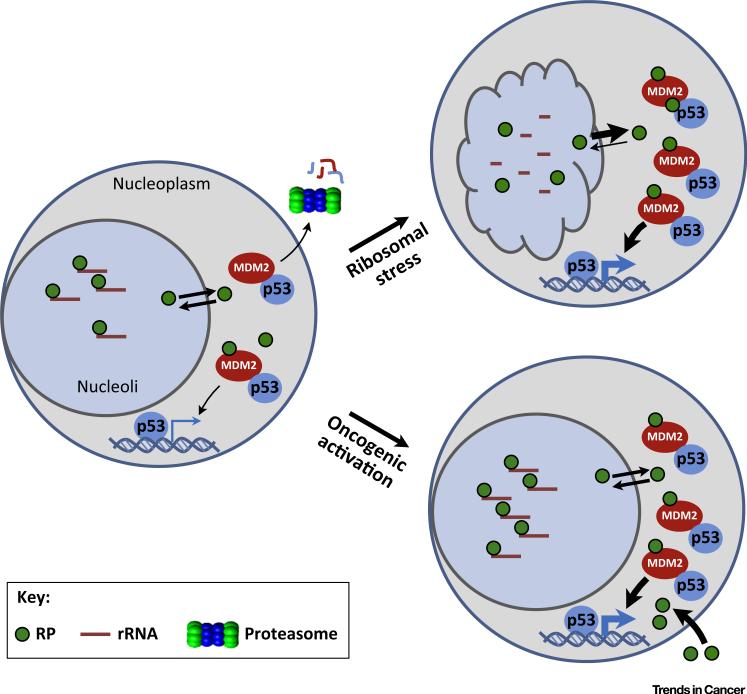
We postulate that, under normal situations, there is a dynamic balance between unassembled RPs and RPs integrated into ribosomes. In response to ribosomal stress, such as drug-induced inhibition of ribosomal biogenesis [3] or nutrient depletion [40], some RPs are released from the nucleolar pre-ribosome structure and enter into the nucleoplasm where they bind to MDM2 and stabilize p53. Upon oncogenic activation, the translation of nascent RPs is constitutively enhanced, leading to excessive expression of RPs, and the unassembled RPs accumulate in the nucleoplasm and initiate the RP–MDM2–p53 pathway [39,41]. Thus, the RP–MDM2–p53 signaling pathway acts as a sensitive surveillance network to monitor either impairment or hyperactivation of ribosome biogenesis (Figure 1).
Figure 1. Dynamic Balance of Ribosome Biogenesis Regulates the RP–MDM2–p53 Pathway.
Ribosome biogenesis is a highly dynamic cellular event that determines the shift of ribosomal proteins (RPs) between ribosome structural components or unassembled RP available for transmitting signals to p53. In response to ribosomal stressors such as chemical agents, serum starvation, or nutrient deprivation, RPs translocate from the nucleolus to the nucleoplasm where particular RPs, such as RPL5 and RPL11, bind to MDM2 to abrogate MDM2 E3 ligase activity and activate p53. Oncogenic stimulation leads to hyperactivation of RP expression and rRNA synthesis. The elevated de novo RP synthesis facilitates ribosome biogenesis in the nucleolus and simultaneously increases the nucleoplasmic concentration of free RPs which are available to bind to MDM2 and activate p53.
RPL5 and RPL11: Essential Messengers of the RP–MDM2–p53 Pathway
Given that different RPs are continuously being identified that complex with MDM2 and regulate p53, it is worth asking whether all these proteins share similar importance in p53 activation. Deficiency in a multitude of nucleolar proteins leads to p53 stabilization, however, some lack any defined association with MDM2 (Table 1). Of note, concomitant depletion of RPL5 or RPL11, but not of RPS7 and RPL23, results in abrogation of ribosomal stress-induced p53 activation and cell cycle arrest [42]. These insights imply that RPL5 and RPL11 may play a pivotal role in sensing the stress resulting from an imbalance of ribosomal components. However, what is still not completely understood from these findings is why RPL5 and RPL11 appear to be unique messengers of ribosomal stress, although there is some evidence in this direction.
The first line of evidence comes from the increased stability of RPL5 and RPL11 compared to other RPs. It is thought that cells maintain high expression levels of RPs beyond what is required for regular ribosome biogenesis. Excess unassembled RPs are actively turned over in the nucleoplasm through proteasome degradation, a process which is accelerated by the inhibition of rRNA transcription [43]. However, RPL5 and RPL11 escape the degradation machinery by mutual protection via a so far unknown mechanism, resulting in the accumulation of unassembled RPL5 and RPL11 in the nucleoplasm where they are capable of binding to MDM2 [36]. Structural analysis of the eukaryotic ribosome reveals that RPL5 and RPL11 reside on the 40S binding site of the 60S subunit together with 21 other large subunit (LSU) RPs. However, in contrast to other RPs, RPL5 binds to, and flanks, 5S rRNA to form the 5S ribonucleoprotein (RNP) in the cytoplasm. The whole complex is then imported into the nucleolus where it binds to RPL11, which sits in one of the two RNA-binding clusters of the LSU. This process is indispensable during ribosome biogenesis and places RPL5 and RPL11 in a unique structural position to facilitate proper assembly of the pre-60S subunit [44]. The key positioning may also make RPL5 and RPL11 more sensitive to disruption in subunit assembly, and thus transduce stress signaling to p53. Indeed, 5S rRNA was originally found to co-precipitate with RPL5 using anti-MDM2 antibody [13], and lately has been demonstrated to be an essential component for inhibition of MDM2 and activation of p53 [45]. More recently, several studies have revealed that the whole 5S RNP machinery, consisting of RPL5, RPL11, and 5S rRNA, are required for p53 activation triggered by Pol I inhibition, RP depletion, or oncogenic stress [44,46–48]. These findings collectively suggest that the 5S RNP complex is redirected from assembly into 60S ribosomes towards MDM2 binding under ribosomal stress conditions. Together, the cooperation of RPL5 and RPL11 in the 5S RNP, as well as their resistance to proteasomal degradation, emphasize the particular importance of RPL5 and RPL11.
A second line of evidence is highlighted by recent resolution of the crystal structure of unassembled RPL11 complex with MDM2 [49]. MDM2 is characterized to compete with 28S rRNA for binding to RPL11. The acidic domain of MDM2 forms a negatively charged surface where an electrostatic interaction with the positively charged surface of the palm of RPL11 is established. Mutation of MDM2 at the RPL11 binding domain nullifies the interaction and fails to promote RPL11 binding-induced p53 stability. This finding provides a structural basis for the development of novel strategies for therapeutic intervention by targeting the RPL11 binding domain on MDM2. The research emphasis on RPL11 and RPL5 does not necessarily minimize the contributions of other RPs. Despite the general consensus that most RPs bind to the central acidic domain of MDM2 [6], it is possible that RPs with different allosteric properties may bind to MDM2 en masse, or in a sequential order, and thus exert more powerful and rapid control over MDM2 E3 ligase activity. In addition, it is worth noting that hyperphosphorylation of MDM2 on Ser166 and Ser186 can selectively affect its binding to specific RPs (RPS14 but not RPS19, RPL5, or RPL11) in Namalwa, but not in U937 cells [50]. Moreover, RPS19 was found to be unable to bind to MDM2 when ectopically expressed in 293 cells with MDM2, but was shown to interact with MDM2 in Namalwa cells [50,51]. These findings imply that MDM2 may selectively interact with specific RPs in a stress- or tissue-specific manner.
RP Dysfunction and Cancer
Given the heterogeneous outcomes associated with ribosome biogenesis deficiency, there are questions regarding the wisdom of a universal approach to target RP–MDM2–p53. Ribosome-related disorders – ribosomopathies – have distinct pathological features, but commonly share bone marrow failure and anemia at an early age, followed by a significantly enhanced risk for cancer development. Because reduced translational capacity and increased tumor-suppressor activity of p53 are characteristics of ribosomopathies [52], it is counterintuitive to find that these patients appear to be predisposed to cancer [53–55]. In addition, in vivo experimental data from animal models also support a model whereby alterations in ribosome biogenesis may promote neoplastic transformation. In Drosophila, reduced expression of RPS6 can cause aberrant cell growth and hematopoietic neoplasias [56].
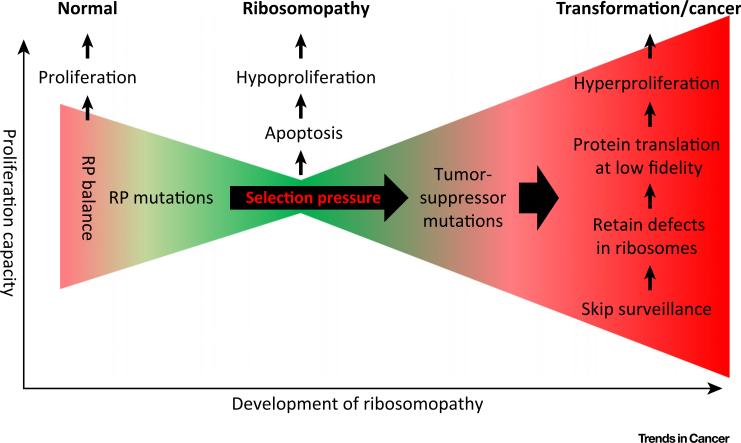
The factors governing increased predisposition to hematological malignancies in patients with ribosomopathies are unclear. A ‘selective pressure-based suppressor mutation’ model has been proposed that may yield a more plausible explanation (Figure 2). A recent study found single allele deletion of RPL22 in 10% of T cell acute lymphoblastic leukemia (T-ALL) patients. Of note, RPL22 haploinsufficiency remarkably accelerated T cell lymphoma in a MyrAkt2 T-ALL mouse model [57]. Rare somatic mutations in the RPL11 gene were also identified in pediatric T-ALL patients, although whether it functions as a disease inducer remains to be determined experimentally [58]. In addition, a separate study identi ed somatic mutations in RPL5 (1.6%) and RPL10 (8.2%) from 122 children diagnosed with T-ALL [57,59]. The exclusive mutation in arginine 98 (R98S) of RPL10 leads to the failure of the so-called ‘test drive’ of pre-60S ribosome subunit – a quality control checkpoint ensuring that only properly functioning subunits are allowed to enter the pool of translationally active ribosomes [59–61]. The shortage of 60S subunit then exerts selective pressure on cells to mutate suppressors of ribosome biogenesis to restore cell proliferation and survival. NMD3 is a primary export adaptor that serves as a checkpoint for the maturation of pre-60S subunit in yeast. In strains carrying the RPL10-R98S mutation, a second mutation in tyrosine 379 of NMD3 (NMD3-Y379D) completely rescues RPL10-R98S associated ribosome biogenesis defects and growth inhibition [62]. The NMD3-Y379D mutation allows the dysfunctional 60S subunit to enter the translational machinery, without correcting the intrinsic structural and biochemical defects of RPL10-R98S mutant ribosomes. As a result, the defective ribosome conferring decreased fidelity in protein translation potentially contributes to cell transformation. Although analysis of T-ALL patient samples from the same group failed to uncover similar mutations in those genes behaving as the quality control of the 60S LSU, it is still possible that the selection pressure can pass through ribosome biogenesis factors and inactivate the downstream ribosomal checkpoints, such as p53.
Figure 2. From Ribosomopathy to Cancer: A Model of Selective Mutation.
In healthy cells, ribosome biogenesis is tightly regulated and exists in a homeostatic balance that is determined by the overall demand for protein synthesis. Ribosomopathies develop when mutations in genes involved in ribosome biogenesis lead to hypoproliferation resulting from p53-dependent or -independent cell cycle arrest and apoptosis. Clonal selection for cells harboring mutations in tumor-suppressor genes that enables bypass of normal surveillance mechanisms can result in a population with decreased capacity for high fidelity ribosome biogenesis. Continued enrichment of this functionally deficient population gives rise to additional mutations that enable cellular transformation to cancer. This model was adapted from Sulima et al. [62].
The selection model described above (Figure 2) is supported by clinical evidence showing that p53 is frequently mutated in patients with chromosome 5 aberration-associated myelodysplastic syndrome (MDS). In a cohort of 318 MDS patients, 55 (17%) harbored chromosome 5 aberrations. Surprisingly, 47% (26/55) of them harbored TP53 mutations compared to only 1.5% (4/263) of patients without 5q aberrations [63].
Consistent with the selection model, 11 of 12 zebrafish lines with elevated rate of cancer development harbored mutations in various RP genes [64]. Of note, the distribution and penetrance of cancers in these zebrafish lines phenocopies the malignancies in p53-null zebrafish. Further analysis revealed that tumors in zebrafish with ribosomal haploinsufficiency also lost expression of p53 [65], indicating that cancer cells may utilize p53 mutation to select against surveillance by a ribosomal integrity–p53 checkpoint. In addition, tumorigenesis was directly evaluated in two recently generated RP-deficient mouse models. Xiong and colleagues found elevated spontaneous lymphoma in RPS27L null mice [66]. Knockout of RPS27L does not disrupt the ribosome profile, although it does cause a moderate decrease in rRNA maturation. Despite slightly altered ribosome biogenesis, RPS27L depletion causes p53-dependent postnatal death and hematopoietic failure. Although p53 activity is increased by RPS27L deletion, Rps27L −/− ;p53+/− mice tend to develop T lymphoblastic lymphoma and B cell lymphoma. Further analysis revealed that the wild-type p53 allele was deleted in 29 of 30 mice, strongly indicating that depletion of RPS27L imposes selective pressure towards tumor-suppressor p53. The progressive inactivation of p53 in response to RP depletion was further appreciated recently by a zebrafish study in which deletion of RPS7 or RPL11 accelerate p53 proteasome degradation through the inactivation of Akt signaling, providing a potential alternative mechanism for the predisposition of ribosomopathies to cancer [67]. Thus, while hematopoietic cells bearing ribosome deficiency are cleared by p53-dependent apoptosis, those cells with inactivated tumor suppressors such as NMD3 or p53 can escape from RP–MDM2–p53 surveillance and survive. Enrichment of these cells is plausible for neoplastic transformation, although the model has yet to be empirically tested.
In fact, there is abundant evidence suggesting a tumor-suppressive function of RPs via both p53-dependent and -independent mechanisms. For example, while homozygous depletion of RPL5 or RPL24 causes mouse embryonic lethality, spontaneous sarcoma is observed in a small portion of RPL5 or RPL24 heterozygous mice, suggesting a potential tumor-suppressor function for these two genes [68]. Similarly to RPS27L knockout mice, heterozygous knockout of RPL11 is embryonic lethal. RPL11 haploinsufficiency revealed anemia and impaired erythrocyte maturation in adult mice that correlated with remarkable G1 cell cycle arrest and apoptosis in erythroid progenitors [69]. Further examination demonstrated that partial loss of RPL11 increases mouse sensitivity to ionizing radiation-induced lymphoma. These effects are correlated with a compromised p53 response to ribosomal stress, and enhanced oncogenic activity of c-MYC, c-MYB, and TGF-β, suggesting that RPL11 may function as a tumor suppressor [69]. RPL11 can be sequestered in the nucleolus by nucleolar protein PICT1 [70]. Loss of PICT1 leads to RPL11 translocation to the nucleoplasm where it binds to MDM2 and stabilizes p53 to induce G1 cell cycle arrest and apoptosis. Analysis of patients with colorectal and esophageal cancer demonstrates that patients with lower PICT1 expression survive longer, supporting a notion that the RPL11–MDM2–p53 pathway is a potential tumor-suppressing pathway in some cancers [70].
The RP–MDM2–p53 pathway is a key cellular defense mechanism against aberrant cell proliferation stemming from enhanced oncogenic activity. As mentioned, disruption of RPL11–MDM2 complexes accelerates c-MYC-induced carcinogenesis via the abrogation of p53 activity [39]. A recent study demonstrated that oncogenic splicing factor SRSF1 directly binds to RPL5 in complex with MDM2, and is necessary for the stabilization and activation of p53 in response to ribosomal stress. Increased SRSF1 expression activates the RPL5–MDM2–p53 pathway, decreases cell proliferation, and ultimately induces tumor suppression and oncogene-induced senescence [71]. Thus, it appears that the RP–MDM2–p53 pathway is an integral component of a self-monitoring system that maintains robust cell growth and proliferation. When RPs are incorporated into the ribosome, they promote growth, a process upregulated by proto-oncogenic signals. This is supported by observations that numerous RPs are upregulated at either the mRNA or protein levels in various human tumors [72]. However, when unassembled as a free pool, RPs are capable of binding to MDM2 to activate p53-dependent tumor suppression. Therefore, a delicate balance is struck between the rate of RP synthesis, production, and incorporation into functional units as a means to modulate the rate of cell growth and subsequent proliferation. Although the RP–MDM2–p53 pathway is likely the primary mechanism of RP-directed tumor suppression, alternative mechanisms are intertwined with cancer signaling processes. Therefore, the interpretation that dysfunctional ribosomal biogenesis is either oncogenic or tumor-suppressive requires caution, and may depend on the context of specific cancer types.
Animal Models for Human Ribosomopathies
Phenotypes associated with RP deficiency are heterogeneous. In humans, ribosomopathies include Diamond–Blackfan anemia (DBA), 5q-syndrome, Shwachman–Diamond syndrome (SDS), Treacher Collins syndrome (TCS), cartilage-hair dysplasia (CHH), and dyskeratosis congenita (DKC) (reviewed in [73,74]). Although each ribosomopathy displays unique characteristics, they do share some common features. The most notable is the presence of hypoplastic behavior, characterized by decreased cell proliferation and increased apoptosis, that strongly correlates with p53 activation [75].
Several mouse models with genetic manipulation of RPs were generated to dissect the relationship between RP deficiency and ribosomopathy, and the outcome ranges from lethality to manifestations of tissue-specific defects (Table 2). Human DBA is characterized by anemia, impaired erythropoiesis, and bone marrow failure. Mutations in several RPs including RPS24, RPS17, RPL35A, RPS7, RPS15, RPS27A, RPL36, RPL5, and RPL11 have been identified in approximately 50% of human DBA patients. Genetic manipulation of RPS6, RPS7, RPS19, RPS20, RPS27A, RPS27L, and RPL11 in mice was used to study DBA. RPS6 was the rst RP to be deleted in mice, and homozygous deletion led to embryonic lethality [76]. Surprisingly, tissue-specific deletion of RPS6 in liver demonstrated deficiency in cell proliferation but not in cell growth [76]. Further studies showed that RPS6 haploinsufficiency phenocopies human DBA, and concomitant deletion of p53 rescues the DBA phenotype, providing evidence that ribosomal integrity is monitored by p53 [77,78]. Likewise, haploinsuffiency of RPS7 [79], RPS19 [80–82], RPS20 [78], RPS27A [83], RPS27L [66], and RPL11 [69] all phenocopy human DBA. The disease phenotype can be corrected by p53 deletion in almost all these mouse models, with the exception of mice with RPL11 heterozygosity where the contribution of p53 to DBA-related anemia was not specifically addressed.
Table 2.
Knockout/Mutant Mouse Models of Ribosomal Proteins.
| Protein | Genotype | Phenotype | Mechanism | Ribosome Biogenesis | p53 Rescue | Cancer Connection | Refs |
|---|---|---|---|---|---|---|---|
| RPS6 | +/− | Perigastrulation lethal | Cell cycle arrest | 40S ribosome biogenesis abolished | Yes | Upregulation in lymphoma | [76-78] |
| DBA phenotype | Yes | ||||||
| Inducible liver-KOa | Liver regeneration failed after partial hepatectomy | N/Aa | |||||
| RPS19 | −/− | Embryonic lethal | Apoptosis and cell cycle arrest | 40S ribosome subunit defect | N/A | [78,80] | |
| Dsk3/+ | DBA phenotype | Yes | |||||
| Dsk3/Dsk3 | Embryonic lethal | N/A | |||||
| shRNA-Tg | DBA phenotype | Yes | |||||
| RPS20 | Dsk4/+ | DBA phenotype | Apoptosis and cell cycle arrest | Yes | [78] | ||
| Dsk4/Dsk4 | Embryonic lethal | N/A | |||||
| RPS27L | −/− | Postnatal death and hematopoietic failure | Apoptosis | Moderate decrease in rRNA processing | Yes | Spontaneous lymphoma in p53+/− background | [66] |
| RPL11 | +/− | Embryonic lethal and hematopoietic failure | Cell cycle arrest | Impaired rRNA processing | N/A | Increased lymphomagenesis in L11+/− | [69] |
| RPS7 | Zma/+ or Mtu/+ | Reduced body size, abnormal skeletal morphology and mid-ventral white spotting | Neuronal apoptosis | Defect in 18S pre-rRNA processing | Yes | Suppresses ovarian cancer | [79] |
| Zma/Zma or Mtu/Mtu | Embryonic lethal | N/A | |||||
| RPS27a | Sfa/+ | Cerebellar ataxia, pancytopenia and epidermal hyperpigmentation | Neuronal apoptosis | Yes | [83] | ||
| Sfa/Sfa | Embryonic lethal | N/A | |||||
| RPL22 | −/− | Deficiency in αβ T cell development | T cell apoptosis | Yes | Downregulation/mutation in lung cancer | [88,89] | |
| RPL29 | −/− | Postnatal lethal, growth retardation Delayed osteogenesis |
Cell cycle arrest | Impaired protein synthesis and reduced RPs | N/A | Promotes angiogenesis | [85,86] |
| RPS14 | +/−b | Phenocopy 5q- syndrome: anemia and bone marrow failure | Apoptosis in hematopoietic progenitor cells | Defect in 18S rRNA processing | Yes | [110] | |
| RPL24 | Bst/+ | Defect in pigmentation and congenital dysfunction | Cell cycle arrest | Impaired protein synthesis | Yesc | Haploinsufficiency abrogates Myc-induced lymphoma | [38,84] |
| Bst/Bst | Embryonic lethal | ||||||
| RPL38 | Heterozygous mutation | Skeletal patterning defects like kinky tail | Impaired translation of Hox mRNAs | Global protein synthesis is unchanged | No | Haploinsufficiency abrogates Myc-induced lymphoma | [38,111] |
Abbreviations: N/A, data not available; KO, knockout.
Haploinsufficiency of the Cd74-Nid67 interval (containing RPS14).
p53 deletion rescue the Bst phenotype however cause mouse death.
The current mouse models of ribosomopathies imply that RP deficiency-induced p53 predominantly triggers apoptosis in hematopoietic cells. p53 can also promote the survival of RPL24-deficient mice during the embryonic stage by delaying the cell cycle to induce a Bst (belly spot and tail) phenotype in adults [84]. A less extreme phenotype is observed in RPL29 null mice, which are viable but exhibit postnatal lethality due to low birth weight, immature organ development, and delayed osteogenesis associated with reduced expression of RPs and decreased rates of proliferation and protein synthesis [85,86].
More recently, Sun and colleagues reported that deletion of RPL29 facilitates RPL11–MDM2 binding and activation of p53 in human cancer cells [87], suggesting that p53 signaling is involved in the growth retardation observed in RPL29-null mice. p53 appears to have conflicting roles in determining cell survival or cell death in response to RP insufficiency. One explanation might be the differential expression of RPs in a tissue-specific or developmental stage-dependent manner. Indeed, homozygous deletion of RPL22 in mice does not cause a lethal phenotype, but selectively suppresses αβ-lineage T cell development. Mechanistically, deletion of RPL22 triggers selective activation of p53 in the αβ-lineage progenitor cells, resulting in increased levels of p21 and BAX, and subsequent apoptosis [88,89]. Overall, mutant RP phenotypes in mice appear to be the result of two distinct mechanisms: (i) ribosome-dependent suppression of protein synthesis, and (ii) extra-ribosomal functions that activate p53. To our surprise, the latter mechanism seemingly dominates the cellular response to RP deficiency. This suggests that restoration of normal p53 turnover could be a therapeutic approach for treating ribosomopathies. Indeed, Lenalidomide (Len), an inhibitor of PP2Ac, causes phosphorylation of MDM2, leading to the release of MDM2 from RPS14, the stabilization of MDM2, and degradation of p53 [50]. By targeting the RPS14–MDM2–p53 axis, Len has successfully the extended survival of patients with lower risk MDS [90].
Concluding Remarks
As knowledge pertaining to extra-ribosomal function of RPs accumulates, the potential for unassembled RPs to interact with non-nucleolar components has opened doors to an unexpected new field where RPs play potentially opposing functions independent of their classical function as structural units of the ribosome. As an important defense mechanism against uncontrolled cell proliferation, the RP–MDM2–p53 pathway stands out as one of the better-characterized pathways that fulfill a role as a sensor of ribosomal integrity. Transmission of ribosome processing defects to p53 enables a stopgap mechanism that inhibits further cell cycle progression and protects genome stability. This discovery highlights the potential for pharmaceutical intervention in the RP–MDM2–p53 pathway in cancer. Several molecules targeting the RP–MDM2–p53 pathway have already been developed, but yielded adverse effects owing to severe cytotoxicity. Among them, two inhibitors of RNA Pol I, CX-3543, and CX-5461, are in Phase II clinical trials and are demonstrating clinical efficacy with promising tolerance and safety profiles [91,92].
Many open questions remain to be addressed to fully understand the RP–MDM2–p53 pathway (see Outstanding Questions). For example, it is unclear why a redundant number of RPs bind to MDM2 to elicit a seemingly similar p53 response. There may be nuanced regulation of ribosome production and function that could further lead to novel areas for therapeutic intervention. On a different note, there is a gap in experimental evidence to suggest whether RPL11, or the RPL5–RPL11–5S rRNA complex, is the dominant switch in this pathway. The types and magnitudes of cellular stressors that can transmit signals though the RP–MDM2–p53 pathway, particularly endogenous signals, are largely unexplored. Finally, increasing evidence reveals that induction of p53 following ribosomal stress does not necessarily require catastrophic genomic damage but may instead respond to more subtle and transient fluctuations in genome fidelity. Given that p53 target genes are differentially regulated and tailored to stress-specific responses that dictate cell fate between life and death [93–95], it will be interesting to understand whether there is any specificity in ribosomal stress-induced p53 signaling, especially in terms of tumorigenesis. If this is the case, how does ribosomal stress induce divergence between life or death events? A greater depth of knowledge regarding RP–MDM2–p53 regulation will advance our understanding of RP signaling in both health and disease.
Trends.
As one hallmark of cancer, uncontrolled cellular proliferation is sustained by enhanced ribosome biogenesis. Disruption of ribosome synthesis triggers a nucleolar stress response that transmits signals to p53 to maintain genomic and cellular homeostasis. The central mediators of this signaling process are MDM2 and RPs. In response to nucleolar stress, RPs bind to MDM2 to promote p53 stabilization and activation. The resulting checkpoint is defined as the RP–MDM2–p53 pathway.
In addition to monitoring the fidelity of ribosomal biogenesis, the RP–MDM2–p53 surveillance network is important for inhibiting oncogenic activity. Hyperactivity of the c-Myc oncogene contributes to lymphomagenesis. A mouse model harboring a mutation in MDM2 (MDM2C305F) bypasses RP signaling to p53 and accelerates lymphoma development. The findings provide key mechanistic insight between oncogene-induced cell growth and p53 regulation of cell cycle dynamics.
Failure to synthesize specific RPs leads to a collection of pathological conditions known as ribosomopathies. Many are characterized by hematological deficiencies and a predisposition to cancer development. Emerging evidence implicates the RP–MDM2–p53 pathway as a key mechanism contributing to these endpoints. Aberrant activation of the RP–MDM2–p53 pathway may provide selective pressure for loss of p53-dependent cell cycle suppression, providing a causal link to ribosomal deficiency-mediated cancers.
Outstanding Questions.
What are the natural triggers of the RP–MDM2–p53 pathway under physiological conditions? Accumulating evidence reveals that induction of p53 following ribosomal stress does not necessarily require catastrophic genomic damage but may instead respond to more subtle and transient environmental fluctuations. Defining the perturbations that lead to nucleolar stress represents a key data gap for future investigations.
Does RP–MDM2–p53 signaling provide a protective effect through promotion of pro-survival or pro-death signals? The transcriptional programs activated by p53 may vary depending on the nature of the stress input. Classifying these potentially differential responses is important for defining the specificity of RP–MDM2–p53 signaling in the context of tumorigenesis.
Why do a redundant number of RPs bind to MDM2 to elicit a seemingly similar p53 response? As many as 16 RPs have been found to bind to MDM2 and stabilize p53, but it is not well understood whether these RPs work independently or synergistically. Future work exploring the complementary effects of RPs on the magnitude, duration, and specificity of the p53 response is warranted.
What are the regulatory mechanisms that govern RP translocation to the nucleus following nucleolar stress? Movement of free RPs from the nucleolus to the nuclear space is important for transmitting stress signals to p53. How RPs primed for ribosomal synthesis are redirected toward MDM2 to elicit a p53 response remains to be clarified.
Is the RP–MDM2–p53 pathway a druggable target for cancer treatment? Most RPs bind to the central zinc-finger domain of MDM2 to prevent interaction with p53. Small molecules with a strong affinity for the zinc-finger domain of MDM2 may provide a viable means for activating p53 tumor suppression in susceptible cancer types. Active investigation into screening and application of novel drug candidates is a crucial gap to translate these research findings into novel cancer therapeutics.
Acknowledgments
The authors thank members of the laboratory of Y.Z. for critical discussions during the preparation of this review. This work was supported by grants from the National Institutes of Health (CA127770, CA100302, and CA167637) and the Natural Science Foundation of China (81272207).
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013;5:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James A, et al. Nucleolar stress with and without p53. Nucleus. 2014;5:402–426. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein–Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TH, et al. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget. 2014;5:860–871. doi: 10.18632/oncotarget.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2014;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisvert FM, et al. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 9.Kressler D, et al. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 11.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulon S, et al. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marechal V, et al. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohrum MA, et al. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 17.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin A, et al. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, et al. Scission of the p53–MDM2 loop by ribosomal proteins. Genes Cancer. 2012;3:298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D, et al. Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong X, et al. Ribosomal protein S27-like and S27 interplay with p53–MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30:1798–1811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai D, et al. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014;42:1799–1811. doi: 10.1093/nar/gkt971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofir-Rosenfeld Y, et al. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahata B, et al. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Identification of ribosomal protein S25 (RPS25)–MDM2-p53 regulatory feedback loop. Oncogene. 2013;32:2782–2791. doi: 10.1038/onc.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, et al. Ribosomal proteins L11 and L5 activate TAp73 by overcoming MDM2 inhibition. Cell Death Differ. 2015;22:755–766. doi: 10.1038/cdd.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry RP. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose–response of different RNA species. J. Cell Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 31.Iapalucci-Espinoza S, Franze-Fernandez MT. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 1979;107:281–284. doi: 10.1016/0014-5793(79)80390-7. [DOI] [PubMed] [Google Scholar]
- 32.Longley DB, et al. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 33.Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum. Mol. Genet. 2008;17:2474–2485. doi: 10.1093/hmg/ddn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat KP, et al. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fumagalli S, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bursac S, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Riggelen J, et al. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 38.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macias E, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein– Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Ribosomal protein–Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng X, et al. Oncogenic c-Myc-induced lymphoma-genesis is inhibited non-redundantly by the p19Arf–Mdm2-p53 and RP–Mdm2-p53 pathways. Oncogene. 2015;34:5709–5717. doi: 10.1038/onc.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fumagalli S, et al. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam YW, et al. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klinge S, et al. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 45.Donati G, et al. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sloan KE, et al. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura K, et al. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 2015;10:1310–1323. doi: 10.1016/j.celrep.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 48.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 49.Zheng J, et al. Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Genes Dev. 2015;29:1524–1534. doi: 10.1101/gad.261792.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei S, et al. Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32:1110–1120. doi: 10.1038/onc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, et al. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakhoul H, et al. Ribosomopathies: mechanisms of disease. Clin. Med. Insights Blood Disord. 2014;7:7–16. doi: 10.4137/CMBD.S16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donadieu J, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90:45–53. [PubMed] [Google Scholar]
- 54.Taskinen M, et al. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am. J. Med. Genet. Part A. 2008;146A:2370–2375. doi: 10.1002/ajmg.a.32478. [DOI] [PubMed] [Google Scholar]
- 55.Alter BP, et al. Cancer in dyskeratosis congenita. Blood. 2009;113:6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson KL, et al. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao S, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tzoneva G, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bussiere C, et al. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sulima SO, et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42:2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sulima SO, et al. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5640–5645. doi: 10.1073/pnas.1400247111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kulasekararaj AG, et al. TP53 mutations in myelodys-plastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br. J. Haematol. 2013;160:660–672. doi: 10.1111/bjh.12203. [DOI] [PubMed] [Google Scholar]
- 64.Amsterdam A, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacInnes AW, et al. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiong X, et al. Ribosomal protein S27-like is a physiological regulator of p53 that suppresses genomic instability and tumorigenesis. Elife. 2014;3:e02236. doi: 10.7554/eLife.02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antunes AT, et al. Ribosomal protein mutations result in constitutive p53 protein degradation through impairment of the AKT pathway. PLoS Genet. 2015;11:e1005326. doi: 10.1371/journal.pgen.1005326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kazerounian S, et al. Development of soft tissue sarcomas in ribosomal proteins L5 and S24 heterozygous mice. J. Cancer. 2016;7:32–36. doi: 10.7150/jca.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morgado-Palacin L, et al. Partial loss of Rpl11 in adult mice recapitulates Diamond–Blackfan anemia and promotes lymphomagenesis. Cell Rep. 2015;13:712–722. doi: 10.1016/j.celrep.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 70.Sasaki M, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat. Med. 2011;17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fregoso OI, et al. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol. Cell. 2013;50:56–66. doi: 10.1016/j.molcel.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou X, et al. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell. Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teng T, et al. Growth control and ribosomopathies. Curr. Opin. Genet. Dev. 2013;23:63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Ellis SR. Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim. Biophys. Acta. 2014;1842:765–768. doi: 10.1016/j.bbadis.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 75.Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Semin. Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 77.Panic L, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGowan KA, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watkins-Chow DE, et al. Mutation of the Diamond–Blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9:e1003094. doi: 10.1371/journal.pgen.1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matsson H, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol. Cell. Biol. 2004;24:4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devlin EE, et al. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond–Blackfan anemia. Blood. 2010;116:2826–2835. doi: 10.1182/blood-2010-03-275776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaako P, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond–Blackfan anemia. Blood. 2011;118:6087–6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 83.Terzian T, et al. Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J. Pathol. 2011;224:540–552. doi: 10.1002/path.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barkic M, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol. Cell. Biol. 2009;29:2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirn-Safran CB, et al. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn. 2007;236:447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- 86.Oristian DS, et al. Ribosomal protein L29/HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J. Orthop. Res. 2009;27:28–35. doi: 10.1002/jor.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun XX, et al. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J. Biol. Chem. 2010;285:25812–25821. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson SJ, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 89.Stadanlick JE, et al. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J. Immunol. 2011;187:664–675. doi: 10.4049/jimmunol.1100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.List AF, et al. Extended survival and reduced risk of AML progression in erythroid-responsive lenalidomide-treated patients with lower-risk del(5q) MDS. Leukemia. 2014;28:1033–1040. doi: 10.1038/leu.2013.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drygin D, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 92.Bywater MJ, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 95.Kruiswijk F, et al. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 96.Yadavilli S, et al. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair. 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daftuar L, et al. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53–MdmX network. PLoS ONE. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun XX, et al. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J. Biol. Chem. 2011;286:22730–22741. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takagi M, et al. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 100.Cui D, et al. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 101.Llanos S, Serrano M. Depletion of ribosomal protein L37 occurs in response to DNA damage and activates p53 through the L11/MDM2 pathway. Cell Cycle. 2010;9:4005–4012. doi: 10.4161/cc.9.19.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lindstrom MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lindstrom MS, Nister M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS ONE. 2010;5:e9578. doi: 10.1371/journal.pone.0009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dutt S, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kurki S, et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 106.Saxena A, et al. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- 107.Yu W, et al. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53–MDM2 loop. Nucleic Acids Res. 2011;39:2234–2248. doi: 10.1093/nar/gkq1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai MS, et al. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuroda T, et al. RNA content in the nucleolus alters p53 acetylation via MYBBP1A. EMBO J. 2011;30:1054–1066. doi: 10.1038/emboj.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barlow JL, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q-syndrome. Nat. Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]