Abstract
Long non-coding RNAs (lncRNAs) have emerged as critical regulators of inflammation. To further understand the interaction between inflammatory signaling pathways and lncRNAs, we characterized the function of the lncRNA, Carlr, a lncRNA expressed in both mouse and human cells of diverse tissues. Carlr expression is increased following NF-κB signaling in macrophages, with concomitant translocation to, and enrichment of, the transcript in the cytoplasm. Knockdown of Carlr results in impaired expression of NF-κB pathway genes and influences the interaction between macrophages and intestinal cells in an inflammatory environment. In human celiac disease patient samples, increased levels of the Carlr transcript were detected in the cytoplasm, alongside elevated expression of NF-κB pathway genes. These findings suggest that increased Carlr expression and/or cytoplasmic localization, is required for efficient NF-κB signaling and is associated with the inflamed tissue state observed in human celiac disease.
Introduction
Long non-coding RNAs (lncRNAs) represent a large portion of the noncoding genome, and are versatile molecules that regulate diverse cellular processes in many biological pathways, including immunity and inflammation (1, 2). Recent work has demonstrated that lncRNAs are important regulators of lymphocyte function (3), immune cell development (4),(5), and inflammatory signaling (6, 7). LncRNAs do not code for proteins but rather function in numerous other capacities, including as scaffolds for other cellular proteins/RNAs, as direct co-factors of proteins, or as sponges for other RNAs. Although the functions of some lncRNAs are well-defined, a clear mechanistic understanding of most lncRNAs remains to be established. Many lncRNAs interact with key signaling pathways to regulate cellular processes. They have also been linked to different diseases such as cancer and intestinal inflammatory diseases such as celiac disease (8, 9).
The intestine contains the largest pool of macrophages in the body, which perform many critical functions including maintenance of mucosal homeostasis and protective immunity, and are implicated in the development of inflammatory intestinal diseases (10). The NF-κB transcription factor functions as a central hub for integrating inflammatory signals and aberrant activation of the NF-κB pathway in macrophages can lead to the overproduction of pro-inflammatory cytokines, such as TNF-α, IL-1β and IL-6, leading to mucosal inflammation that contributes to intestinal inflammatory diseases (11). At the same time, induction of NF-κB signaling induces changes in the expression of lncRNAs that have been shown to be important for both cytokine production, as well as subsequent resolution of inflammation (6, 9, 12). To explore the possible involvement of lncRNAs in intestinal inflammatory diseases we screened for lncRNAs whose expression was significantly increased following LPS stimulation in macrophages. We identified one such lncRNA, Carlr (cardiac apoptosis-related lncRNA), which has recently been characterized in cardiomyocytes (13). Mouse Carlr is a single exon intergenic lncRNA located between genes Spag6 (BC061194) and Pip4k2a on mouse chromosome 2. Carlr suppresses mitochondrial fission and apoptosis by targeting miR-539 and PHB2 in cardiomyocytes. Interestingly, Carlr is known to be expressed in various tissues, including myeloid lineage cells and the intestine (13), and its expression is induced in macrophages and dendritic cells in response to LPS (11, 14, 15). However, unpublished studies from our group show that miR-539 levels do not change in response to LPS stimulation, and LPS-induced genes are not known to be targeted by this microRNA. These findings suggest that Carlr might have other cell-type/stimuli-specific functions that remain to be characterized.
Here, we describe the regulation and function of Carlr upon NF-κB activation in macrophages and intestinal cells of both mouse and human origin. Our results support a role for Carlr in promoting induction of NF-κB-stimulated genes, which may promote the pathogenesis of human inflammatory bowel diseases. These findings highlight the potential for lncRNAs to have divergent, cell-type specific functions and further emphasize their potential importance in human disease.
Material and Methods
Cell Culture, Coculture and In vitro stimulations
Primary bone marrow derived macrophages (BMDMs) were generated by culturing bone marrow from WT C57Bl/6 mice in 20% L929 cell-conditioned DMEM (Gibco BRL) medium. For lipopolysaccharide (LPS) stimulation, cells were seeded in 12-well plates at 105 cells/ml and stimulated with 10ng/ml of E. coli-derived LPS (Sigma Aldrich). 5μM and 10μM of BAY-11-7082 were used for the inhibition of NF-κB. The human THP-1 macrophage-like cell line was cultured in RPMI (Gibco BRL) supplemented with 10% FBS and antibiotics. For stimulation, cells were seeded at 105 cells/ml and stimulated with 100ng/ml of LPS. The mouse and human intestinal cell lines C26 and Caco2 (derived from colorectal carcinoma) were cultured in RPMI and DMEM (Gibco BRL) respectively, supplemented with 10% heat-inactivated fetal calf serum (Gibco BRL) and antibiotics (500 IU/ml penicillin and 100 μg/ml streptomycin; Gibco BRL). For coculture experiments Caco2 cells were plated in 6 well plates and allowed to attach overnight. THP-1 cells were added the following day and co-culture was maintained in supplemented RPMI media for the duration of the experiment. After culturing, THP-1 non-adherent cells were removed by three rounds of washing and the Caco2 adherent cells were detached and used for the RNA or protein extraction.
RNA Knockdown
For silencing experiments, macrophages immortalized by inoculation with J2 retrovirus (16) were used for the establishment of stable Carlr knockdown cell lines using the lentiviral pLKO.1-puro vector. shRNAs were cloned in the pLKO.1-puro vector according to the manufacturer’s instructions (Addgene). The same shRNAs were used for transient silencing of mouse intestinal C26 cells. For silencing of human Carlr, two siRNAs were purchased from IDT (si3:5′-CAUUUACUUUCCCAUGAUAAAACAG-3′ and si5:5′AUCACAUUUACUUUCCCAUGAUAAA-3′) and transfected into the THP-1 cells using lipofectamine RNAiMax (Invitrogen).
Patient samples
Celiac disease was diagnosed according to the ESPGHAN (European Society of Pediatric Gastroenterology Hematology and Nutrition) criteria in effect at the time of patient recruitment, including anti-gliadin (AGA), anti-endomysium (EMA) and anti-transglutaminase antibody (TGA) determinations, as well as a confirmatory small bowel biopsy. The study was approved by the Institutional Board (Cruces University Hospital code CEIC-E09/10 and Basque Clinical Trials and Ethics Committee code PI2013072) and analyses were performed after informed consent was obtained from all subjects or their parents. Biopsy specimens from the distal duodenum of each patient were obtained during routine diagnostic endoscopy. None of the patients suffered from any other concomitant immunological disease. None of the controls showed small intestinal inflammation at the time of the biopsy.
RNA extraction, RT and qPCR
Total RNA was extracted from cells using QIAGEN RNA mini/micro kits. All samples were subjected to DNAse I treatment. A total of 1ug of RNA was used for reverse transcription using SSIII enzyme (Invitrogen) and real time QPCR was carried out using 2X SYBR green fluorescent dye (Quanta Bioscience). The amplified transcripts were quantified using the comparative Ct method. All experiments were performed in triplicate. TaqMan assays and Universal Master Mix (Applied Biosystems) were used for the quantification of Carlr targets in human samples. All primer sequences are available upon request.
Cellular fractionation
For quantification of Carlr levels in nuclear and cytoplasmic compartments, nuclei were isolated using C1 lysis buffer as described previously (6) and the amount of specific nuclear RNA measured by RT-qPCR was compared to the total amount of the RNA in the whole cell. Nuclear/cytoplasmic protein fractionation was done using hypotonic and hypertonic buffers with NP-40 detergent.
RNA-protein interaction assay
Carlr was in vitro transcribed using a T7 polymerase after amplification of the region with primers harboring a T7 promoter. RNA was allowed to fold, and 80ng were mixed with protein lysates and incubated at RT for 1h. After incubation, glycerol was added and the samples were run in a 1% agarose gel with TB buffer. RNA bands were stained using Sybr Safe.
RNA immunoprecipitation (RIP)
For RIP experiments, cells were lysed in RIP buffer (150mM KCl, 25mM Tris pH 7.4, 0.5 mM DTT, 0.5% NP-40) and homogenized using a syringe. For tissue sample RIP preparation, Santa Cruz Biotechnology Protocols were followed. Briefly, tissue was immediately flash frozen and stored at −80°C. Before RIP, tissue was thawed in RIPA buffer (10 mM Tris-Cl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl), disrupted and homogenized, while maintaining temperature at 4°C throughout all procedures. Afterwards, samples were incubated on ice for 30 minutes and centrifuged to obtain cell lysates. For all samples, agarose beads were coupled with anti-p65 and anti-IκBα antibodies and incubated with lysates overnight at 4°C. Samples were then washed 8 times and bead-bound material was divided for WB and qPCR analyses.
Western blot
SDS sample buffer was added to cell lysates. Proteins were separated in 10% SDS-polyacrylamide gel and transferred to PVDF membranes. Immunoblotting was performed with the following primary antibodies: anti-beta tubulin (Sigma Aldrich), anti-Hdac1, anti-p65, anti-p50 (Santa Cruz Biotechnology), anti IκBα (Thermo Scientific). Signals were detected using Pierce ECL Western Blotting Substrate (Thermo Scientific).
Proliferation assay
Caco2 cells (1.7 × 10^4) were seeded in a 24 well plate, and THP-1-conditioned medium was added the following morning. Proliferation was measured at day 0, 1, 3 and 5. For crystal violet staining, cells were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. After staining, cells were washed and 10% acetic acid was added. Absorbance was measured at 590nm.
Results
Carlr lncRNA expression increases and translocates to the cytoplasm after NF-κB activation in mouse macrophages
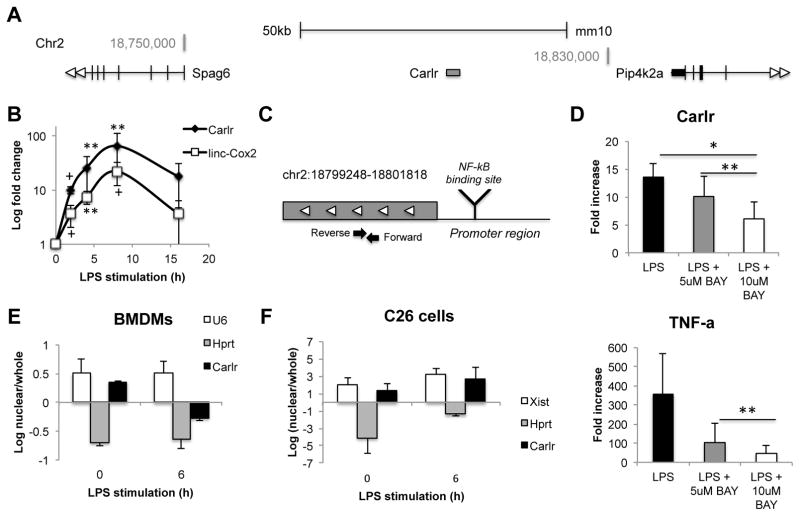
To assess the role of long non-coding RNAs in inflammation, we characterized the expression of several lncRNAs in response to LPS stimulation in mouse macrophages. The intergenic lncRNA Carlr located on mouse chromosome 2 (Figure 1A) was upregulated upon stimulation, resembling what has been described for other LPS regulated lncRNAs such as linc-Cox2 (6) (Figure 1B). These results confirmed that LPS induces dynamic changes in multiple lincRNA expression profiles with cell type-specificity. Promoter analysis using LASAGNA-Search 2.0 (http://biogrid-lasagna.engr.uconn.edu/lasagna_search/) showed an NF-κB binding site at position -327/328 (from transcription start site) (Figure 1C). To confirm whether the increase of Carlr in macrophages is due to NF-κB activation, we stimulated cells with LPS in the presence of two different concentrations of the NF-κB inhibitor BAY-11-7082. The increase of Carlr in response to LPS was diminished after NF-κB inhibition in a dose-dependent manner, suggesting that NF-κB is involved in the upregulation of this lncRNA (Figure 1D). To determine the subcellular localization, and likely site of function of Carlr, cellular fractionation and QPCR analysis were employed. Carlr was found to exhibit a primarily nuclear localization at basal conditions in mouse macrophages (Figure 1E). However, after LPS stimulation, the majority of Carlr transcript was located in the cytoplasm (Figure 1E). Interestingly, stimulation of mouse intestinal cells with LPS did not show a change of expression (Supl. Fig. 1A) or localization of this lncRNA (Figure 1F). Given the observed expression pattern, we hypothesized that Carlr could be a circular RNA (circRNA) that is transported by a nuclear export system (17), however attempts to amplify a circular form of Carlr were not successful (data not shown). These results implicate Carlr as an NF-κB-regulated lncRNA that is dynamically expressed following LPS stimulation in mouse macrophages.
Figure 1.
A) Schematic diagram of Carlr intergenic location in mice (mm10). B) Mouse macrophages stimulated with LPS for indicated time points. Data were normalized to Hprt. Data represent the mean and standard error of three independent experiments. p-values were calculated relative to basal level, +p≤0.1, *p<0.05, **p<0.01 based on unpaired Student’s t-test. C) Closer view of the Carlr region. Reverse and Forward black arrows represent the location of the RT-qPCR primers. NF-κB binding site was predicted using Lasagna-Search 2.0. D) LPS induced Carlr upregulation is diminished after NF-κB inhibition by BAY-11-7082 (right). RT-qPCR data is represented as the mean +/− standard error of three independent experiments. TNF levels (left) are shown as a control for the NF-κB inhibition. RT-qPCR analysis on whole cell and nuclear RNA to determine localization of Carlr in mouse macrophages (E) and intestinal cells (F). U6 was used as nuclear and Hprt as cytoplasmic control. Data represents the mean +/− standard error of three independent experiments
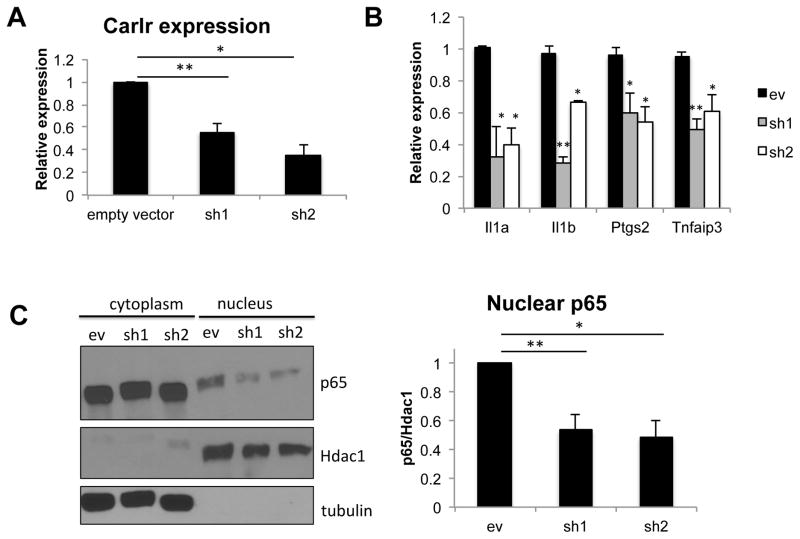
Knock down of Carlr alters the NF-κB pathway in mouse cells
To gather more information about the function of Carlr, we analyzed a panel of 92 NF-κB target genes to correlate the increased expression and cytoplasmic export of this lncRNA with the differential expression of genes in this pathway. After LPS stimulation, genes were grouped by their temporal expression-pattern using STEM (short term expression miner), a software application designed specifically to identify temporal expression profiles and the genes associated with these profiles (18). We observed that Carlr is co-expressed with a group of NF-κB target genes (including Tnfaip3, Ptgs2, NF-κB1 and NF-κB2 or Il1a and Il1b) (Sup. Fig. 1B) suggesting that it might function as a modulator similar to the NF-κB Interacting LncRNA (NKILA) (19). To address the role of Carlr in NF-κB signaling and downstream pathways, we silenced this lncRNA using two different target sequences in both macrophages origin. We first confirmed that Carlr expression was significantly reduced in the cell lines transfected with these shRNAs (Figure 2A). When silencing Carlr we observed significantly decreased expression of four (Nfkb2, Ptgs2, Il1a and Il1b) out of ten of the analyzed co-expressed NF-κB targets in both macrophage knockdown cell lines (Figure 2B). To address if Carlr silencing affects only NF-κB-dependent gene expression or is also involved in nuclear NF-κB activity, we measured the amount of NF-κB p65 subunit (a component of the most abundant NF-κB complex) in the sh-Carlr macrophages. Both silenced cell lines had less nuclear p65 than the cells transduced with the empty vector (Figure 2C). Silencing of Carlr also led to a decrease in phosphorylation of IκBα, which results in less degradation of IκBα and consequently less nuclear translocation of NF-κB (Sup. Fig. 1C). Taken together, these results suggest that Carlr is involved in the NF-κB signaling cascade in mouse macrophages and in lesser extent in intestinal cells.
Figure 2.
A) Carlr levels are significantly reduced in both sh1 and sh2 cell lines. Empty vector was used as a control. Data represents the mean and standard error of four independent experiments. **p<0.01, *p<0.05. B) RT-qPCR analysis was performed on the sh1 and sh2 cell lines to determine the effect of Carlr knockdown on the correlated genes. Genes with statistically significant decreased expression in both cell lines are shown. Data is represented as the mean and standard error of four experiments. *p≤0.05, **p<0.01 based on unpaired Students t-test. C) p65 levels were decreased in the nucleus of sh1 and sh2 cell lines. β-tubulin and Hdac1 were used as loading controls. Western blot signal was quantified using ImageJ software, *p≤0.05, **p<0.01.
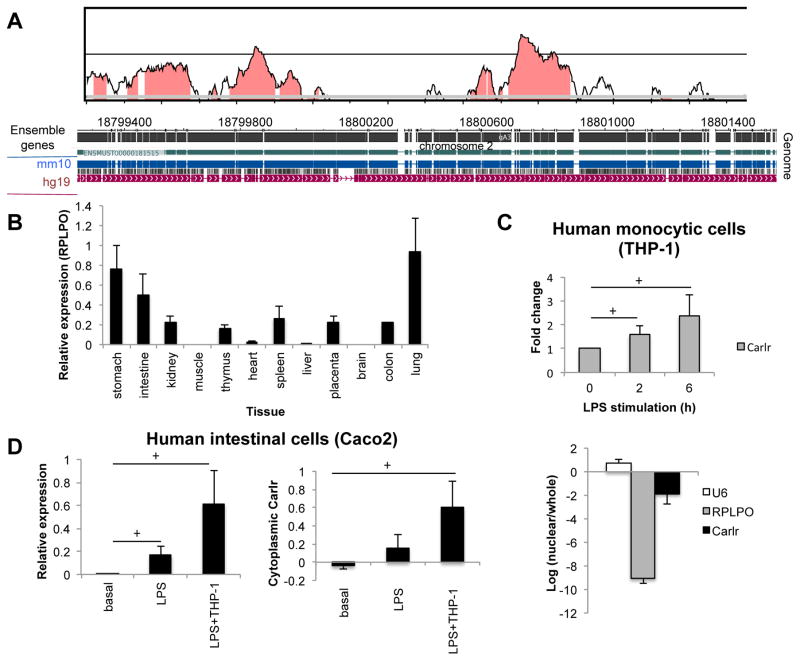
Carlr is expressed in human tissues
These observations led us to examine whether a homologous lncRNA for mouse Carlr is transcribed from the human genome. Alignment and conservation analysis using different databases and analysis tools showed that the mouse and human genomic regions exhibit significant sequence homology (Figure 3A). Moreover, data from the Epigenome Roadmap project (20) shows RNAseq signals in this region in several cell/tissue types (Sup. Fig. 2A). We designed qPCR primers and evaluated the expression of human Carlr using a pool of RNAs derived from 12 human tissues. We observed that this region is transcribed from the human genome in most of the tissues analyzed, showing higher expression in stomach, intestine and lung (Figure 3B). When RT-qPCR primers located in a conserved region upstream from the Carlr region were used, no transcript amplification was observed (Sup. Fig. 2B). We then analyzed the expression of Carlr in the human monocytic cell lines U937 and THP-1. Carlr was found to be expressed in these human cell lines, and its expression was also induced in response to LPS stimulation as seen in mouse macrophages (Figure 3C, Sup. Fig. 2C). To evaluate if the regulation of Carlr is conserved across species, we assessed the localization of Carlr in the human monocytic cell lines. Localization of Carlr in THP-1 and U937 cells showed a predominantly cytoplasmic location (Figure 3C, Sup. Fig. 2D). Interestingly, these cell lines have been described as having constitutively active NF-κB (21) which would therefore explain the cytoplasmic location of Carlr prior to LPS stimulation.
Figure 3.
A) Conservation analysis of Carlr region performed using VISTA (36). Mouse sequence is shown on the x-axis and percentage similarity to the corresponding human sequence on the y-axis. The graphical plot is based on sliding-window analysis of the underlying genomic alignment. A 100-bp sliding window is at 40-bp nucleotide increment is used. Pink shading indicates conserved noncoding genomic region (top). The image at the bottom represents pairwise alignment of the mouse and human genomes corresponding to Carlr region. Genome alignment was produced by blastz using WashU Epigenome Browser. B) Human Carlr expression (relative to stomach) measured by RT-qPCR in RNA pool of different tissues purchased from Clontech (Human total RNA master panel II) (average ± s.d; two independent RT-qPCRs). C) Expression of Carlr in basal conditions and after LPS stimulation (top) and localization of Carlr (bottom) in the human monocytic cell line THP-1. Data represents the average and standard error of three independent experiments. D) Expression (left) and cytoplasmic amount (right) of Carlr in the human intestinal cell line Caco2 at basal conditions and after incubation with LPS and with LPS stimulated THP-1 cells.
Enterocytes exist in close association with tissue macrophages that are located within the lamina propria close to the epithelial monolayer. Macrophages located in the intestinal epithelium interact with epithelial cells via soluble factors as cytokines. Increase in inflammatory cytokines can cause alterations in the epithelial cells leading to mucosal inflammation. In response to LPS, macrophages become activated, releasing proinflammatory cytokines that can disrupt the adjacent enterocyte monolayer and lead to inflammatory disease (22, 23). To evaluate the role of Carlr in the macrophage-enterocyte interaction, we assessed the localization of Carlr in the intestinal cell line Caco2 at steady state and in the presence of LPS and activated THP-1 cells. This cell line is widely used as a model for human intestinal inflammatory diseases and is able to produce NF-κB regulated cytokines such as IL-6 (24–26). Caco2 cells showed nuclear localization of Carlr at basal conditions, however, when co-culturing the intestinal cells with LPS or with activated THP-1 cells, resembling the intestinal inflammatory environment, we saw that Carlr expression was increased and at the same time translocated to the cytoplasm, mainly in the co-culture condition (Figure 3D). These results indicate that LPS induced Carlr translocation to the cytoplasm is a sign of inflammation of human macrophages and intestinal cells that should be present in the intestinal epithelia of inflammatory patients.
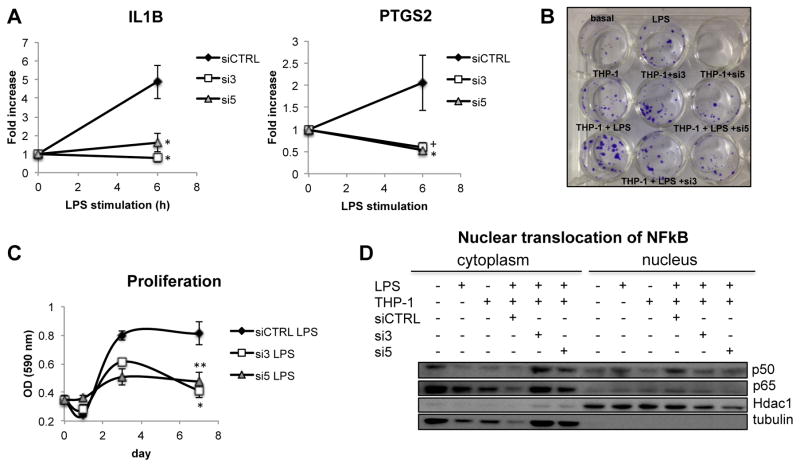
Carlr is involved in the signaling interaction between macrophages and intestinal cells during inflammation
As interactions between intestinal cells and macrophages induced cytoplasmic translocation of Carlr in the intestinal cells, we sought to evaluate the role of Carlr in the interplay between human macrophages and intestinal cells in an inflammatory setting. To assess the involvement of Carlr in this interaction, we silenced this lncRNA in THP-1 cells using two different siRNAs (Sup. Fig. 3A). Upon LPS stimulation of THP-1 cells with Carlr knockdown, we observed that two of the putative Carl-regulated genes whose expression correlates with Carlr, IL1B and PTGS2, were not induced following silencing (Figure 4A). Both of these genes are important players in inflammatory intestinal diseases and their activation induces secretion of prostaglandin and the IL1β inflammatory cytokine by THP-1 cells, soluble factors that modulate interactions between epithelial and immune cells. To further analyze the relationship between THP-1 and intestinal cells, we utilized a co-culture model of intestinal inflammation (27). After co-culturing LPS-stimulated THP-1 cells with intestinal Caco2 cells, we observed an increase in proliferation of the intestinal cells (Figure 4B) that was diminished if Carlr was previously silenced in THP-1 cells (Figure 4C). This decrease in the proliferation of Caco2 cells could be mediated by the lower levels of IL1β and PTGS2 observed in the Carlr knockdown cells. To further evaluate the effect of silencing Carlr on the interaction of macrophage and intestinal cells, we quantified the amount of NF-κB nuclear translocation in Caco2 cells. THP-1 cells release inflammatory cytokines, inducing an inflammatory cascade in intestinal cells with subsequent translocation of NF-κB to the nucleus. This inflammatory sequence resembles that which is observed in intestinal inflammatory diseases where NF-κB activity is increased (28, 29). When Carlr was silenced in THP-1 cells, we observed that translocation of NF-kB was diminished in intestinal cells following co-culture (Figure 4D), consistent with the decrease in proliferation and lower induction of IL1β and PTGS2 in THP-1 cells. Taken together, these results suggest that Carlr is an important player in the signaling between the macrophages and intestinal cells in an inflammatory environment.
Figure 4.
A) LPS induction of IL1B and PTGS2 genes is inhibited in THP-1 cells silenced for Carlr. Data represents average and standard error of three independent experiments. *p<0.05. B) Crystal violet staining of Caco2 cells incubated under different conditions for 48h. C) Proliferation of Caco2 cell after culturing with conditioned media of LPS stimulated THP-1 cells with and without Carlr silencing. Data represents the average and standard error of three independent experiments. D) p50 and p65 translocation to the nucleus in Caco2 cultured alone and with THP-1 cells under different conditions.
Carlr interacts with the active form of NF-κB
To further characterize Carlr function, we evaluated if this lncRNA is able to interact with protein complexes using a native agarose gel electrophoretic mobility shift assay. We observed changes in the migration pattern of in vitro transcribed Carlr upon mixing with protein lysates from THP-1 cells, indicating that it is able to interact with protein complexes (Figure 5A). As we are interested in the NF-κB pathway and previously saw that Carlr modulates NF-κB signaling, we performed a computational prediction using RPIseq that predicts RNA protein interaction using datasets extracted from the Protein-RNA Interface Database (30). This analysis suggested that human Carlr preferentially interacts with p65 NF-κB, rather than p50 or IκBα (Figure 5B). To experimentally evaluate the ability of Carlr to interact with these members of NF-κB family, we performed RIP (RNA immunoprecipitation) experiments in human THP-1 cells. p65 antibody immunoprecipitation showed Carlr bound to the p65 member of the NF-κB complex (Figure 5C). Interestingly, although we were able to co-IP the inactive NF-κB complex, i.e. NF-κB bound to IκBα, we did not detect significant enrichment of Carlr, suggesting that Carlr interacts with NF-κB only after it is released from the inhibitory complex. Taken together, these data suggest that Carlr is able to interact with protein complexes, and that it binds NF-κB p65 after it is released from the inhibitory complex, before translocation to the nucleus.
Figure 5.
A) Nondenaturing agarose gel for IVT Carlr incubated with different protein lysates shows that Carlr is able to interact with protein complexes. B) Interaction prediction of Carlr with NF-κB members p65 and p50 and with the inhibitory protein IκBα using RPIseq tool (30), tubulin is used as an unrelated protein. C) Levels of Carlr bound to NF-κB and IκBα after RIP performed in THP-1 cell lysates. Data is represented as average and standard error of three independent immunoprecipitations; *p=0.05 (right). Representative WB of the immunoprecipitation (left).
Carlr is associated with Celiac Disease pathogenesis
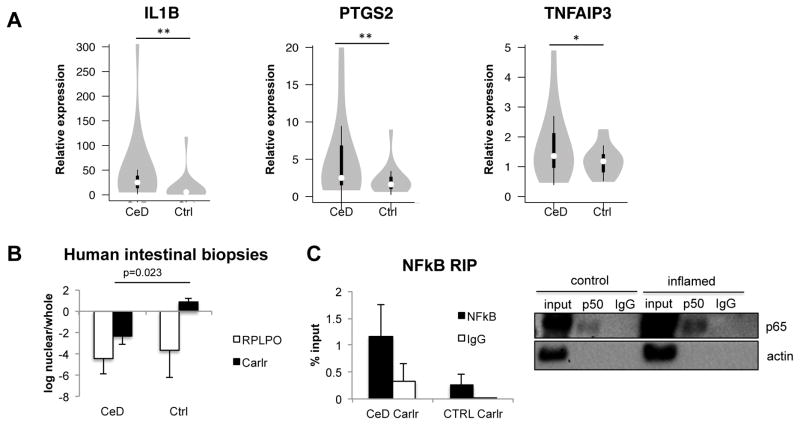
Based on its role in the NF-κB pathway and its expression in the human gastrointestinal tract, macrophages, and intestinal cells, we assessed the potential involvement of Carlr in celiac disease. Celiac disease (CeD) is an immune-mediated enteropathy that presents with constitutive NF-κB activation (31). To assess the role of lncRNAs in inflammatory bowel diseases, and specifically in CeD, we evaluated the expression of the putative Carlr targets in intestinal biopsies of celiac patients and control individuals. Three of the putative Carlr regulated targets, TNFAIP3, IL1B and PTGS2, showed statistically significant, higher expression (p ( 0.05) in celiac patients compared to control samples (Figure 6A). These results suggest that human Carlr could be involved in the dysregulation of these genes during CeD pathogenesis. Next, we measured the expression of Carlr in intestinal biopsies and, contrary to our expectations, we saw that Carlr levels were decreased (p=0.018) in celiac patients (Sup. Fig. 3C). As NF-κB is constitutively active in the celiac mucosa (28, 31), we hypothesized that this signaling pathway could make Carlr exclusively cytoplasmic in celiac patients, enabling its function as an inducer of certain disease-characteristic NF-κB pathway genes. To test this hypothesis, we isolated the nuclei of intestinal biopsy cells and quantified the fraction of Carlr in the nucleus compared to the whole cell. As predicted, celiac biopsies showed elevated cytoplasmic localization of Carlr, while in control biopsies Carlr was mainly located in the nucleus (Figure 6B). Moreover, as previously seen in vitro, Carlr was found to interact with active NF-κB complex in both control (non-inflamed) and celiac patient (inflamed) intestinal biopsies (Figure 6C) but could not bind the inactive complex (Sup. Fig. 3D). Taken together, these findings indicate that elevated Carlr cytoplasmic activity is associated with disease-characteristic elevated NF-κB signaling in CeD tissue.
Figure 6.
A) Celiac disease patient intestinal biopsies have significantly higher levels of the Carlr targets measured by RT-qPCR. RPLPO was used as housekeeping gene. **p<0.05, *p=0.05; unpaired Students t-test. White circles show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. B) Subcellular localization of Carlr in cells isolated from biopsies of five CeD patients and three controls. RPLPO was used as a control; p=0.023, unpaired Students t-test. C) Levels of Carlr bound to NF-κB after RIP performed in lysates from three control (non-inflammed) and three CeD patients (inflamed) (right). Representative WB of the immunoprecipitation (left).
Discussion
Recent studies have implicated an increasing number of lncRNA molecules in inflammation and immunity. Here, we characterize the role of the lncRNA Carlr in the NF-κB pathway in the context of intestinal inflammation. Mouse Carlr was first described in cardiomyocytes, where it exhibits decreased expression in response to anoxia (13). In the present study, we found that Carlr expression is increased in mouse and human macrophages in response to LPS stimulation and that this increase is dependent on NF-κB activation, highlighting the cell type and stimuli specificity of lncRNA expression. Indeed, we found that Carlr is translocated to the cytoplasm after NF-κB activation in mouse and human macrophages. Several lncRNAs have been described to reside within, or to be dynamically shuttled, to the cytoplasm where they regulate protein localization, mRNA translation and stability (32) so we wanted to further analyze the function of this lncRNA and its link with inflammation.
In mammalian gut tissue, enterocytes are closely associated with different types of immune cells, including tissue macrophages that are located in the lamina propia (23). For instance, macrophages exposed to LPS secrete cytokines and growth factors that can directly, or indirectly, affect epithelial function (22). Therefore macrophages play an important role in immune and inflammatory events of the intestinal mucosa. Interestingly, human Caco2 intestinal cells cultivated with activated macrophages also show a translocation of Carlr to the cytoplasm, suggesting that the cytoplasmic location of this lncRNA could be a hallmark of intestinal inflammation.
To further characterize the function of Carlr, we assessed its role in NF-κB pathway gene expression. We observed that downregulation of Carlr decreases the levels of some NF-κB pathway genes and also influences the translocation of the transcription factor to the nucleus, suggesting that it contributes to the perpetuation of the signaling cascade. Knockdown of Carlr in human THP-1 cells reduces the levels of IL1B and PTGS2 gene expression induced by LPS. These two genes are important regulators of intestinal inflammation; they are present at high levels in the inflammatory microenvironment. PTGS2 has been linked with inflammation-associated cancer development and proliferation, while IL1β is able to induce the NF-κB pathway, two major events in the development of intestinal inflammatory diseases (33, 34). Interestingly intestinal cells cultured in conditioned media from Carlr knockdown macrophages show less proliferation and a decrease in NF-κB translocation to the nucleus suggesting that Carlr is important in the interaction between macrophages and intestinal cells that occurs during the development of inflammatory diseases.
RNA EMSA and immunoprecipitation experiments revealed that Carlr lncRNA is able to interact with proteins, specifically the activated NF-κB transcription factor complex. We hypothesize that the cytoplasmic form of Carlr could recognize IκBα-free NF-κB and facilitate its transport to the nucleus, thus augmenting the signaling pathway. However, further studies are needed to decipher how and under what specific condition Carlr modulates NF-κB activation.
Finally, we wanted to assess the relevance of Carlr to human inflammation and immunity. Celiac disease is an immune-mediated enteropathy in which NF-κB is constitutively active (31), and it is a useful disease model to assess the potential involvement of this lncRNA in inflammatory diseases of the gut. When we explored Carlr expression in healthy and diseased human tissue we surprisingly found that its expression is not increased in the patient tissue. However, further analyses revealed that in celiac patients the majority of Carlr is located in the cytoplasm while in control individuals it is mainly found in the nucleus. Experiments done in vitro showed that long-term LPS stimulation does not induce such an increase of Carlr, suggesting that after induction and translocation to the cytoplasm, the levels of this lncRNA remain stable, thus explaining the lack of overexpression in the disease tissue. The phenotype of Carlr cytoplasmic localization in celiac patients resembles our results with macrophage-intestinal cell cocultures, suggesting that although Carlr expression is not elevated in CeD patients, it may still mediate an inflammatory function. Additionally, we found that the Carlr targets IL1B and PTGS2 are overexpressed in CeD patient biopsies, in line with the increased NF-κB activation and proliferation described in this disease (28, 35). Moreover, Carlr also interacts with activated NF-κB in intestinal biopsies, supporting the hypothesis that it may play a role in the development of inflammation.
Taken together, these results implicate Carlr as a potential novel player of the NF-κB inflammatory pathway and a clinically intriguing candidate associated with the pathogenesis of immune-mediated diseases.
Supplementary Material
Acknowledgments
The work was supported by grants from NIH (RO1 DK102180) and a Juan de la Cierva reincorporation fellowship (Spanish Ministry of Economy and Competitiveness)
Abbreviations
- LncRNA
long-noncoding RNA
- Carlr
cardiac apoptosis-related lncRNA
- BMDM
bone marrow derived macrophages
- LPS
lypopolysacharide
- qPCR
quantitative PCR
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- RIP
RNA immunoprecipitation
- CeD
celiac disease
Footnotes
Disclosure
The authors have no financial conflicts of interest.
References
- 1.Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, Meadows S, Montoya NR, Herrera NG, Domingos AI, Rastinejad F, Myers RM, Fuller-Pace FV, Bonneau R, Chang HY, Acuto O, Littman DR. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Kotzin JJ, Spencer SP, McCright SJ, Kumar DB, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, Harman CC, Virtue AT, Zhu S, Bailis W, Stein J, Hughes C, Raj A, Wherry EJ, Goff LA, Klion AD, Rinn JL, Williams A, Flavell RA, Henao-Mejia J. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537:239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7:39. doi: 10.1186/s13073-015-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–95. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bain CC, Mowat AM. Macrophages in intestinal homeostasis and inflammation. Immunol Rev. 2014;260:102–117. doi: 10.1111/imr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanwar JR, Kanwar RK. Gut health immunomodulatory and anti-inflammatory functions of gut enzyme digested high protein micro-nutrient dietary supplement-Enprocal. BMC Immunol. 2009;10:7. doi: 10.1186/1471-2172-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, Fitzgerald KA, Caffrey DR. Cutting Edge: A Natural Antisense Transcript, AS-IL1alpha, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1alpha. J Immunol. 2015;195:1359–1363. doi: 10.4049/jimmunol.1500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 14.Rowley SM, Kuriakose T, Dockery LM, Tran-Ngyuen T, Gingerich AD, Wei L, Watford WT. Tumor progression locus 2 (Tpl2) kinase promotes chemokine receptor expression and macrophage migration during acute inflammation. J Biol Chem. 2014;289:15788–15797. doi: 10.1074/jbc.M114.559344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pletinckx K, Stijlemans B, Pavlovic V, Laube R, Brandl C, Kneitz S, Beschin A, De Baetselier P, Lutz MB. Similar inflammatory DC maturation signatures induced by TNF or Trypanosoma brucei antigens instruct default Th2-cell responses. Eur J Immunol. 2011;41:3479–3494. doi: 10.1002/eji.201141631. [DOI] [PubMed] [Google Scholar]
- 16.Cox GW, Mathieson BJ, Gandino L, Blasi E, Radzioch D, Varesio L. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. J Natl Cancer Inst. 1989;81:1492–1496. doi: 10.1093/jnci/81.19.1492. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Skipper M, Eccleston A, Gray N, Heemels T, Le Bot N, Marte B, Weiss U. Presenting the epigenome roadmap. Nature. 2015;518:313. doi: 10.1038/518313a. [DOI] [PubMed] [Google Scholar]
- 21.Frankenberger M, Pforte A, Sternsdorf T, Passlick B, Baeuerle PA, Ziegler-Heitbrock HW. Constitutive nuclear NF-kappa B in cells of the monocyte lineage. Biochem J. 1994;304(Pt 1):87–94. doi: 10.1042/bj3040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zareie M, McKay DM, Kovarik GG, Perdue MH. Monocyte/macrophages evoke epithelial dysfunction: indirect role of tumor necrosis factor-alpha. Am J Physiol. 1998;275:C932–939. doi: 10.1152/ajpcell.1998.275.4.C932. [DOI] [PubMed] [Google Scholar]
- 23.Anand RJ, Dai S, Rippel C, Leaphart C, Qureshi F, Gribar SC, Kohler JW, Li J, Stolz DB, Sodhi C, Hackam DJ. Activated macrophages inhibit enterocyte gap junctions via the release of nitric oxide. Am J Physiol Gastrointest Liver Physiol. 2008;294:G109–119. doi: 10.1152/ajpgi.00331.2007. [DOI] [PubMed] [Google Scholar]
- 24.Al-Ghadban S, Kaissi S, Homaidan FR, Naim HY, El-Sabban ME. Cross-talk between intestinal epithelial cells and immune cells in inflammatory bowel disease. Sci Rep. 2016;6:29783. doi: 10.1038/srep29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammers KM, Lu R, Brownley J, Lu B, Gerard C, Thomas K, Rallabhandi P, Shea-Donohue T, Tamiz A, Alkan S, Netzel-Arnett S, Antalis T, Vogel SN, Fasano A. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology. 2008;135:194–204. e193. doi: 10.1053/j.gastro.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang FC. Upregulation of Salmonella-induced IL-6 production in Caco-2 cells by PJ-34, PARP-1 inhibitor: involvement of PI3K, p38 MAPK, ERK, JNK, and NF-kappaB. Mediators Inflamm. 2009;2009:103890. doi: 10.1155/2009/103890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe F, Satsu H, Mochizuki T, Nakano T, Shimizu M. Development of the method for evaluating protective effect of food factors on THP-1-induced damage to human intestinal Caco-2 monolayers. Biofactors. 2004;21:145–147. doi: 10.1002/biof.552210129. [DOI] [PubMed] [Google Scholar]
- 28.Maiuri MC, De Stefano D, Mele G, Fecarotta S, Greco L, Troncone R, Carnuccio R. Nuclear factor kappa B is activated in small intestinal mucosa of celiac patients. J Mol Med (Berl) 2003;81:373–379. doi: 10.1007/s00109-003-0440-0. [DOI] [PubMed] [Google Scholar]
- 29.Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Intern Med. 2008;263:591–596. doi: 10.1111/j.1365-2796.2008.01953.x. [DOI] [PubMed] [Google Scholar]
- 30.Muppirala UK, V, Honavar G, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Irastorza I, Elcoroaristizabal X, Jauregi-Miguel A, Lopez-Euba T, Tutau C, de Pancorbo MM, Vitoria JC, Bilbao JR. Coregulation and modulation of NFkappaB-related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet. 2014;23:1298–1310. doi: 10.1093/hmg/ddt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 33.Al-Sadi RM, Ma TY. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178:4641–4649. doi: 10.4049/jimmunol.178.7.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanayakkara M, Lania G, Maglio M, Discepolo V, Sarno M, Gaito A, Troncone R, Auricchio S, Auricchio R, Barone MV. An undigested gliadin peptide activates innate immunity and proliferative signaling in enterocytes: the role in celiac disease. Am J Clin Nutr. 2013;98:1123–1135. doi: 10.3945/ajcn.112.054544. [DOI] [PubMed] [Google Scholar]
- 36.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.