SUMMARY
Developing renewable energy sources is critical to maintaining the economic growth of the planet while protecting the environment. First generation biofuels focused on food crops like corn and sugarcane for ethanol production, and soybean and palm for biodiesel production. Second generation biofuels based on cellulosic ethanol produced from terrestrial plants, has received extensive funding and recently pilot facilities have been commissioned, but to date output of fuels from these sources has fallen well short of what is needed. Recent research and pilot demonstrations have highlighted the potential of algae as one of the most promising sources of sustainable liquid transportation fuels. Algae have also been established as unique biofactories for industrial, therapeutic, and nutraceutical co-products. Chlamydomonas reinhardtii’s long established role in the field of basic research in green algae has paved the way for understanding algal metabolism and developing genetic engineering protocols. These tools are now being utilized in C. reinhardtii and in other algal species for the development of strains to maximize biofuels and bio-products yields from the lab to the field.
Keywords: Chlamydomonas reinhardtii, recombinant proteins, biofuels, bio-products, molecular engineering
INTRODUCTION
Access to affordable and environmentally sustainable fuels and energy sources may be the greatest challenge of this century. With demand continuing to increase and new supplies costing ever more to extract, the availability of fossil fuels will inevitably shrink, resulting in rising energy prices worldwide. With the rising cost of energy also comes the rising cost of food, as food and fuel prices are closely linked. Recently, algae-based biofuels have been highlighted as one of the best current alternative source of renewable energy (Merchant et al., 2011; Georgianna and Mayfield, 2012; Leite et al., 2013; Oncel, 2013). Algae do not compete for arable land, have fast generation times, can grow in salt and waste water, and have the potential to produce more oil per acre than land plants (Dismukes et al., 2008; Demirbas and Demirbas, 2011). While most focus has been on the production of biodiesel, algae can also be a source of other fuels, such as bioethanol, biohydrogen, and biogas (Jones and Mayfield, 2011; Oncel, 2013).
While algal biofuels hold significant promise to meet future energy demands, improvements are needed at all levels of production in order to realize this potential. Algal biofuels are not economically viable on their own at current production levels (Brownbridge et al., 2013; Nagarajan et al., 2013). One near-term solution is to couple biofuel production with high-value co-products to increase the commercial value of the entire algal biomass. Current high-value bio-products produced in algae include industrial and therapeutic proteins as well as nutraceuticals and other high-value small molecules (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Rasala and Mayfield, 2014).
As the most characterized algal species with the largest set of genetic tools and techniques, Chlamydomonas reinhardtii is an excellent model organism to understand and improve biofuels and bio-products production in algae. Chlamydomonas reinhardtii has led the field in the development of molecular tools for strain selection and engineering for green alga. By far, more recombinant proteins have been expressed in C. reinhardtii than all other algal species combined (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Rasala and Mayfield, 2014). Studies in C. reinhardtii have also helped elucidate the molecular mechanisms behind algal lipid and hydrogen metabolism (Merchant et al., 2011; Torzillo and Seibert, 2013). C. reinhardtii is also among the first of the engineered algal species to be studied in commercial settings, which allows academic researchers to begin to understand the challenges of bringing transgenic algae to commercial-scale production (Scoma et al., 2012; Gimpel et al., 2014; Schoepp et al., 2014).
BIOENGINEERING ALGAE
Chloroplast engineering
Over the last 70 years, C. reinhardtii has become the flagship alga for laboratory studies and genetic manipulation. The eukaryotic green alga has three modifiable genomes and is capable of producing a wide variety of protein products (Rosales-Mendoza et al., 2012; Barrera and Mayfield, 2013; Rasala et al., 2013b). The efficient manufacture of these products at commercial viability will require a myriad of genetic tools to enhance protein accumulation and bioactivity (Figure 1). To date, the chloroplast genome has been the primary target for engineering protein production, predominantly because it readily performs homologous recombination and is easily transformed (Boynton et al., 1988). Typically, recombinant protein expressed from the chloroplast accounts for 1–10% of total protein (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Rasala and Mayfield, 2014).
Figure 1.
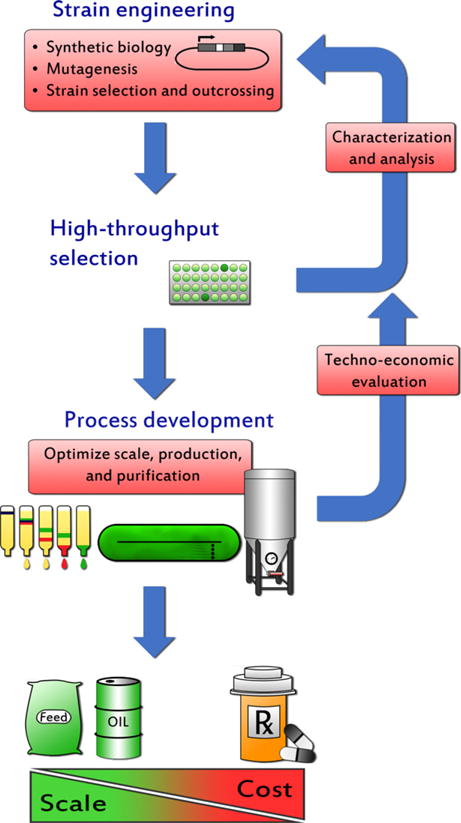
Steps to engineering a production strain of green algae. Genetic manipulation using advanced tools will result in a desired phenotype. The isolated strain will be tested for scale-up and reanalyzed for characteristics such as growth rate, population density, recombinant protein accumulation, and lipid profile. If these traits do not overcome economic constraints of production, additional genetic manipulation in the laboratory is required. Once an ideal strain is identified, fuels and co-products can be extracted from scaled-up cultures.
One current limitation of chloroplast engineering in C. reinhardtii is the use of native promoters to achieve high levels of protein accumulation. The best exogenous gene expression achieved utilizes the endogenous psbA promoter (Manuell et al., 2007). However, the effective use of the psbA promoter requires that the endogenous psbA regulatory region be removed due to a phenomenon known as auto-attenuation (Minai et al., 2006; Rasala and Mayfield, 2011). This renders the organism non-photosynthetic, eliminating one of the unique attributes of algae. One way to alleviate this issue is to reintroduce the psbA gene under the control of a different endogenous promoter (Gimpel et al., 2014). This method results in photosynthetic C. reinhardtii that can produce recombinant proteins, albeit at reduced levels compared with the non-photosynthetic versions. Exogenous transcriptional regulation machinery have been used successfully in C. reinhardtii (Kato et al., 2007), however exogenous promoters remain an underutilized resource for chloroplast engineering. Synthetic regulatory elements, such as UTRs, that modify gene expression have also been used in the C. reinhardtii chloroplast (Specht and Mayfield, 2012), although the successful use of fully synthetic promoters has yet to be described. A full understanding of synthetic promoter synthesis would allow for fine-tuning of gene expression, which is particularly useful for metabolic engineering.
In addition to engineering strains to produce maximal amounts of recombinant protein, chloroplast engineering is necessary for developing strains for robust growth, particularly in outdoor settings. Light intensity and availability is a major concern for maximizing growth rates and cell density. Excessive solar irradiation leads to photoinhibition, while low light flux due to weather or cell culture density can lead to reduced photosynthetic activity and slower growth. Chloroplast engineering for improved function under varying light conditions would allow maximum production of algae throughout the year. Early data have shown that the light antenna of C. reinhardtii can be modified to alter chlorophyll content and subsequently adjust its absorbance spectrum, altering photosynthetic productivity (Kirst et al., 2012).
An alternative method of chloroplast engineering to alter photosynthetic activity has focused on the protein subunits of the photosystem itself. Photosystem II is a highly conserved protein complex, which performs the initial rate limiting step of photosynthesis. It is known that PSII is in part responsible for enhanced fitness in various light conditions (Mulo et al., 2012). Recently, the D1 protein of C. reinhardtii was replaced with the homologous D1 proteins from Synechococcus sp. 7942 (Vinyard et al., 2013, 2014). These studies showed that complementation of the native D1 protein with natural variants from other species could increase photosynthetic efficiencies under varying light conditions, and also that this increased efficiency led to improved overall growth. In addition, due to C. reinhardtii’s unique low fluorescence background, these studies were the first to elucidate the mechanisms behind the optimized photosynthetic activity. Thus, not only were improved strains generated, but C. reinhardtii proved to be an ideal model for understanding PSII function.
Nuclear engineering
Although the chloroplast can effectively accumulate high levels of recombinant protein, nuclear transformation technology is required to fully engineer algae. Nuclear expression allows for organelle targeting or secretion of proteins, and enables more complex post-translational modifications of proteins such as glycosylation. In particular, proper protein localization is required for metabolic engineering, which relies on compartmentalization for some steps in syntheses. The nuclear genome of C. reinhardtii has been much more difficult to develop for recombinant protein production than the chloroplast. Low protein accumulation levels due in part to transgene silencing and insertion positional effects have limited the use of the nuclear genome from a protein manufacturing standpoint (Cerutti et al., 1997; De Wilde et al., 2000; Wu-Scharf et al., 2000). However, recently it was shown that fusing recombinant proteins to the Sh Ble antibiotic resistance gene, which requires high protein accumulation, could substantially increase recombinant protein accumulation (Rasala et al., 2012). In addition, C. reinhardtii lacks strong constitutive promoters comparable to the viral promoters used in plants. While hybrid promoters have had some success in increasing transgene expression in C. reinhardtii (Schroda et al., 2000), truly synthetic promoters may hold great promise for tight regulation of robust recombinant protein expression (Venter, 2007).
Homologous recombination
Homologous recombination (HR) allows for targeted gene knock-outs as well as targeted insertion for reducing positional effects for recombinant gene expression. This mechanism has enabled sophisticated engineering in the C. reinhardtii chloroplast; however at present, HR does not occur at sufficient levels in the nuclear genome to be of practical use. Knockdowns have been created by artificial microRNAs, which is also a valuable tool for genetic engineering (Molnar et al., 2009; Zhao et al., 2009; Moellering and Benning, 2010; Schmollinger et al., 2010). Alternatively, a knockout strain library is currently being developed by insertional mutagenesis. Because of the scale of this project, a high-throughput screen known as ChlaM-meSeq has been established to identify genes disrupted by insertional mutagenesis. In a pilot screen, a pool of insertional mutants was shown to cover 39% of known protein coding genes (Zhang et al., 2014). Although each of these methods have been valuable for progress of the field, a rapid method for developing novel knock-outs and targeted gene introduction is essential for future development of C. reinhardtii.
To date, methods of identifying HR in C. reinhardtii have not been reliable, nor have there been ways to significantly increase the low basal rate of HR in Chlamydomonas (Sodeinde and Kindle, 1993; Gumpel et al., 1994; Nelson and Lefebvre, 1995). However, there have been many advances in targeted gene delivery across other platforms in the past decade. Transcription-activator-like effectors (TALEs) allow one to design a unique DNA-binding target sequence and the addition of a nuclease can achieve targeted restriction digestion and subsequent homology directed repair (Li et al., 2011). Successful gene targeting by a TALE has been shown by a subsequent increase in endogenous ARS expression (Gao et al., 2014). However, to this point, TALENs have not successfully been utilized for targeted gene disruption in C. reinhardtii. The CRISPR/Cas9 system is an alternative method for inducing targeted gene delivery. Here, a guide RNA will direct restriction by the Cas9 nuclease to a specific sequence of DNA. The CRISPR/Cas9 system has been successful in mammals, fish, fungus, and plants (tobacco, Arabidopsis, sorghum and rice; Jiang et al., 2013). Cas9 has recently been transiently expressed in C. reinhardtii, although stable cell lines have not been established (Jiang et al., 2014a). Successful transient expression shows that Cas9 is capable of modifying the C. reinhardtii genome.
The C. reinhardtii nuclear genome generally performs repairs of double-stranded breaks almost exclusively by non-homologous end joining (NHEJ) and errors are often introduced during the process. Unfortunately, because Chlamydomonas relies so heavily on NHEJ as a repair mechanism, even if TALENs or the CRISPR/Cas9 systems are developed in C. reinhardtii, it is unlikely that a large percentage of gene delivery will occur by targeted homology repair. NHEJ machinery is comprised of three main subunits: Ku70, Ku80, and DNA ligase IV. In yeast, Ku70 and Ku80 form a heterodimer that binds to double stranded breaks and recruits DNA ligase IV to mediate NHEJ (Kanaar et al., 1998). The C. reinhardtii genome contains homologues to each of these genes. Based on a study in Pichia pastoris, one strategy may be to stifle expression of these repair proteins in Chlamydomonas in order to increase the level of HR activity (Näätsaari et al., 2012).
High-throughput screening
High-throughput screening allows detection of rare phenotypes including those following gene insertion or mutagenesis. One of the fastest quantitative methods of screening mutants is fluorescence-activated cell sorting (FACS). This technique has been utilized for detection of high lipid accumulating mutants by a process utilizing Nile Red, a lipid dye, to analyze lipid levels in a set of mutants generated using EMS (Xie et al., 2014). This method successfully isolated C. reinhardtii strains that accumulated 23–58% higher fatty acid content by dry mass when compared to their parent strain. Alternatively, Terashima et al. (2014) developed a high lipid-accumulating mutant pool by insertional mutagenesis, a strategy termed Chlamydomonas High Lipid Sorting (CHiLiS). This method generates a detectable insertion that can be used to rapidly identify the disrupted gene(s). Characterization of the strains isolated by this method showed a significant increase in triacylglycerol accumulation. These papers demonstrate that rapid screening can identify mutants with altered metabolism for the production of biofuels, which can easily be translated beyond C. reinhardtii. In addition to fluorescent stains, fluorescent protein reporters in C. reinhardtii have been shown to effectively sort using FACs (Rasala et al., 2013a). By tagging recombinant proteins with fluorescent protein tags, production strains can be rapidly screened and genes that affect recombinant protein accumulation can be identified. Fluorescent reporters fused to antibodies have also been successfully used to sort for wild isolates with specific cell wall components (Jiang et al., 2014b). Development of techniques using these reporters will be necessary to rapidly isolate and engineer production strains.
Ultimately, high-throughput screening technologies will be an essential tool for generating strains for outdoor production. High-throughput screening allows one to select exclusively based on desirable traits, rather than validating transformation with linkage to an antibiotic marker. Horizontal gene transfer of antibiotic resistance cassettes to environmental microbes poses a theoretical threat to human health and safety. Therefore, high-throughput screening mechanisms that avoid generation of transgenic algae with drug resistant markers, and that can be scaled up to outdoor ponds for economically feasible biofuels production, are an area where more research is required.
ALGAL BIOFUELS
Biodiesel
Chlamydomonas reinhardtii has been a key model in understanding complex algae lipid metabolism. Current efforts have focused on the use of triacylglycerols (TAGs) as a first generation biodiesel. TAGs from algae are also of interest as alternatives to plant-based edible oils (Klok et al., 2014). Traditionally, C. reinhardtii has not been seen as an oleaginous species (James et al., 2011). However, studies have shown that natural levels of lipids can vary widely in C. reinhardtii strains (Siaut et al., 2011). In addition, wild-type and mutant C. reinhardtii can accumulate significant amounts of TAGs in response to nitrogen or salt stress (Wang et al., 2009; Li et al., 2010; Work et al., 2010; Goodson et al., 2011; James et al., 2011; Siaut et al., 2011). In fact, C. reinhardtii studies have helped elucidate the mechanisms behind lipid accumulation in response to nitrogen deprivation (Boyle et al., 2012; Msanne et al., 2012; Goodenough et al., 2014). Chlamydomonas reinhardtii has also been an excellent model to identify algal genes involved in TAG metabolism (Merchant et al., 2011; Klok et al., 2014).
Targeted overexpression of putative TAG metabolic genes in C. reinhardtii has been met with mixed success. Overexpression of type 2 diacylglycerol acyltransferases (DGATs) DGAT2–1 and DGAT2–5 led to increased lipid content, while DGAT2-a,b,c overexpression had no effect (Deng et al., 2012; La Russa et al., 2012). Overexpression of acyl-ACP (acyl carrier protein) esterase (AAE) led to an altered lipid profile but not an increase in lipid content (Blatti et al., 2011). It is clear that even in C. reinhardtii, a better understanding of lipid metabolism is required in order to fully utilize algae’s potential as a source of TAGs for biodiesel or edible oil production.
Biohydrogen
As an alternative to liquid fuels, C. reinhardtii has also been extensively studied as a model for the photoproduction of biohydrogen. Renewed interest in C. reinhardtii biohydrogen production began when significant and sustained H2 production was demonstrated in sulfur starved strains (Melis et al., 2000). The theoretical maximum light conversion efficiency for H2 production is 13.4%, which is higher than the 11.2% limit for biodiesel (Torzillo and Seibert, 2013). In addition, purities of up to 98% can be achieved without the need of extraction (Torzillo et al., 2009). However, several factors significantly limit the actual production of H2 in algae. The most significant limitation is the sensitivity of hydrogenase to O2, which inhibits hydrogenase activity by affecting transcription and protein maturation (Ghirardi et al., 2007). Additional constraints include competition for electrons from alternative pathways, inefficient light conversion of large light-harvesting antennae, and inherent limitations of anaerobic growth (Torzillo and Seibert, 2013).
Studies in C. reinhardtii to address these limitations have included the generation of improved strains through either mutagenesis or targeted genetic engineering (Melis et al., 2007; Esquível et al., 2011; Torzillo and Seibert, 2013; Baltz et al., 2014; Xu et al., 2014). Studies have also shown significantly improved H2 production through the optimization of growth conditions including periodic sulfur starvation (Melis et al., 2000) and optimized light, media and photobioreactor conditions (Torzillo and Seibert, 2013; Oncel et al., 2014). However, despite these optimizations, the best light conversion efficiency reached has been approximately 3%. Even if these optimal conditions could be perfectly scaled to outdoor growth, the cost of algal H2 would be well over US$8 per gallon gasoline equivalent (gge) (James et al., 2009). However, continued research holds promise to optimize algae strains and growth conditions to make biohydrogen a competitive fuel in the future.
HIGH-VALUE BIO-PRODUCTS
Production of recombinant proteins in C. reinhardtii has been a fruitful area of research in recent years. Several recent articles have thoroughly reviewed the plethora of protein products that have been successfully expressed within C. reinhardtii (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Rasala and Mayfield, 2014). To date, 34 different protein co-products have been produced in algae, 29 of which were produced in C. reinhardtii.
The vast majority of targeted proteins have been therapeutic proteins. With a current market value of US$140 billion, therapeutic proteins provide an attractive co-product target to increase the economic value of algal biomass (Walsh, 2014). Therapeutic proteins produced include the UV-protectant metallothionein (Zhang et al., 2006), antibody mimics 14FN3 and SAA-10FN3 (Rasala et al., 2010), and anti-cancer proteins TNF-Related Apoptosis Inducing Ligand (TRAIL; Yang et al., 2006), and allophycocyanin (Su et al., 2005). The glycoprotein hormone erythropoietin has also been produced from the C. reinhardtii nucleus, demonstrating the ability to generate recombinant proteins with glycosylation, the most common post-translational modification found on protein therapeutics (Eichler-Stahlberg et al., 2009; Walsh, 2014).
The largest class of therapeutic proteins that have been produced in C. reinhardtii are subunit vaccines (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Rasala and Mayfield, 2014). Vaccines are a particularly attractive target because algae are generally regarded as safe (GRAS) by the FDA. Therefore, if orally available vaccines can be produced, these vaccines may be stored and administered as lyophilized algal pellets. Edible algal vaccines would reduce the cost of vaccines by orders of magnitude and vastly increase their availability since algal vaccine pellets do not need to purified, do not require needle injection, and can be stored for up to 20 months at RT (Dreesen et al., 2010; Gregory et al., 2013).
Chlamydomonas reinhardtii has also been shown to be able to produce a novel type of antibody therapeutic. Monoclonal antibodies (mAbs) are the largest growing segment of protein therapeutics with almost 27% of the new approvals and six of the top 10 selling biotherapeutics products in 2013 (Walsh, 2014). In particular, antibody-drug conjugates (ADCs) are gaining in popularity as more effective cancer treatment. The most significant drawback of ADCs is the requirement for chemical linkage of the toxic molecule, which leads to increased production costs and potential off-target toxicity of free toxins. In contrast, the unique environment of the C. reinhardtii chloroplast is able to produce immunotoxins in which toxic proteins are genetically linked to the targeting antibody (Tran et al., 2013a,b). This reduces the cost of production since the immunotoxin is produced as a single protein and this production method can also help eliminate off-target toxicity.
Protein co-products expressed in C. reinhardtii also include a range of nutraceuticals for use as feed additives and/or human nutritional supplements. Feed additives include phytase, xylanase, and flounder growth hormone (Kim et al., 2002; Yoon et al., 2011; Georgianna et al., 2013). Mammary-associated serum amyloid A (MAA) is one of the best expressed bio-products in C. reinhardtii, accumulating to over 5% of total soluble protein (Manuell et al., 2007). MAA can be used as a prophylaxis against enteric bacterial infections in humans as well as replacement for antibiotics in animal feed (Larson et al., 2003; Mack et al., 2003). Recombinant proteins have also been engineered to increase high-value nutritional compounds in C. reinhardtii such as organic selenium (Hou et al., 2013), carotenoids (Baldo et al., 2011; Couso et al., 2011), and triacylglycerols (La Russa et al., 2012). Carotenoids and triacylglycerols have the added benefit of being used in biofuels formulations (Merchant et al., 2011; Peralta-Yahya et al., 2011).
OUTDOOR CULTIVATION SYSTEMS
Large-scale microalgae cultivation can occur in either open or closed systems (Ugwu et al., 2008; Singh et al., 2011). Open systems range from small artificial ponds to large open bodies of water; however the most productive open systems currently implemented are shallow raceway ponds (Singh et al., 2011). Although these open systems require large areas of land, they can be constructed in arid environments and avoid competition with preexisting agriculture (Georgianna and Mayfield, 2012). Raceways can be constructed relatively cheaply and require little maintenance (Singh et al., 2011; Schoepp et al., 2014). Open systems are inherently susceptible to contamination and thus are predominantly used to culture only robust photoautotrophic species (Ugwu et al., 2008; Rasala and Mayfield, 2014). Heterotrophic growth in open systems is not a viable option due to the heightened susceptibility of contamination. Greenhouses can be used to cover smaller open systems in order to deter contamination and potentially increase productivity. Additionally, greenhouses allow for the growth of genetically engineered strains in contained ‘open’ systems, but these structures will add to the initial cost of a facility.
Closed systems are highly variable in design but overall provide a more productive environment while protecting the culture from contamination (Ugwu et al., 2008; Singh et al., 2011). More strains of algae can be cultivated in these closed systems, including mixotrophic and hydrogen-producing species of microalgae (Scoma et al., 2012; Gimpel et al., 2014). The productivity and sterility benefits of a closed system are countered by their heightened costs as they may cost up to 10 times that of open systems due to the additional infrastructure and operational costs (Del Campo et al., 2007).
Recently, pilot facilities containing a variety of these systems have been built to test strains in a setting more comparable to conditions encountered at commercial facilities (Scoma et al., 2012; Gimpel et al., 2014; Schoepp et al., 2014). Experiments with C. reinhardtii have been conducted in these outdoor systems to test the viability of the strain as a producer of biomass, biohydrogen and even high-value bio-products. These studies are invaluable for understanding and addressing the challenges of bringing laboratory strains into commercial production.
OUTDOOR CULTIVATION OF C. REINHARDTII
Researchers at the University of California, San Diego have developed one of the first systems to measure research-scale production of algal strains in closed bags and open ponds. Biomass production (g L−1 day−1) was reported for several algae and cyanobacteria species (Table 1; Schoepp et al., 2014). Chlamydomonas reinhardtii (CC-1690) out produced eight other species in a 100-L closed system, but production was reduced by 80% when grown in an open 800-L pond. Lipid content was not measured in this study; however, reported average lipid content in C. reinhardtii is comparable to other species (Table 1; Griffiths and Harrison, 2009). This suggests that C. reinhardtii is competitive with other algal species in closed systems. While closed systems may be economical for high-value products, the cost and size limitations of closed systems may hinder sufficient production of lipid-accumulating strains for biofuel production (Figure 1).
Table 1.
Biomass production of various algal species in an outdoor setting compared to average reported lipid content
| Species | Closed system production (g L−1 day−1) |
Open system production (g L−1 day−1) |
Lipid content (nutrient-replete, % dry weight) |
|---|---|---|---|
| Scenedesmus dimorphus | 0.095 | 0.090 | 26 |
| Chlorella vulgaris | 0.047 | 0.035 | 25 |
| Chlamydomonas reinhardtii | 0.078 | 0.015 | 21 |
| Arthrospira platensis | 0.040 | 0.018 | 13 |
| Anabaena sp. | 0.055 | – | 5 |
| Porphyridium purpureum | 0.074 | 0.036 | 11 |
| Nannochloropsis salina | 0.043 | 0.028 | 27 |
| Dunaliella tertiolecta | 0.039 | 0.031 | 15 |
| Phaeodactylum tricornutum | 0.021 | – | 21 |
Biomass production data from Schoepp et al. (2014) and lipid content from Griffiths and Harrison (2009).
Chlamydomonas reinhardtii was also tested in an outdoor system for its ability to produce biohydrogen gas. Scoma et al. cultivated C. reinhardtii heterotrophically in a 50-L tubular closed system. By starving the culture of sulfur, photosystem II was down-regulated to the point where oxygen consumption was greater than oxygen evolution, creating the necessary anaerobic conditions for biohydrogen production. However, the combination of uncontrolled solar intensities and nutrient deprivation can lead to photo-damage and the production of reactive oxygen species in microalgae, thus hindering production. To address this photoinhibition, Scoma et al. acclimated their cultures to natural light for 1 week before experimentation to induce physiological changes such as reduced chlorophyll, higher levels of xanthophylls, and increased photosynthetic and respiratory rates. When cells were allowed to acclimate to solar light, a more robust photoprotection system was observed, but biohydrogen production was still only equal to 20% of the laboratory results.
Recombinant protein production in C. reinhardtii has been demonstrated in a pilot commercial setting. Bovine Milk Amyloid A (MAA), a protein with anti-microbial properties, has previously been produced to high levels in C. reinhardtii in the laboratory (Manuell et al., 2007). High accumulation of MAA in this strain required removal of the psbA gene, thus making the strain non-photosynthetic and unfit for outdoor growth. Recently, psbA was rescued using the alternative regulatory regions to eliminate issues of auto-attenuation (Gimpel et al., 2014). The best rescued strain was then grown in 100-L hanging bags and accumulated MAA at approximately 3 mg L−1 of culture for a maximum yield of 11.8 g MAA kg−1 dry weight biomass. Even with lower photosynthetic efficiency and lower total soluble protein production versus the knockout laboratory strain, the production of MAA in C. reinhardtii has the potential to be highly profitable since it can be administered as a solid algal pellet at a cost 60 times lower than traditional purified bovine colostrum MAA.
CONCLUSION
Chlamydomonas reinhardtii with its extensive research history and genetic tool infrastructure has been an excellent system to begin to understand algal metabolism and strain development. Chlamydomonas reinhardtii’s ease of transformation, large toolset, and natural variety has led to strains with significantly increased biofuels and bio-product yields. The biggest successes with C. reinhardtii have been the production of high-value products at laboratory scale in closed systems (Almaraz-Delgado et al., 2013; Barrera and Mayfield, 2013; Gimpel et al., 2014; Rasala and Mayfield, 2014). However, C. reinhardtii remains limited by its naturally low oil content, sensitivity to high solar irradiation and its weak growth in outdoor ponds, thus making it a relatively poor choice for large-scale biofuels production (James et al., 2011; Scoma et al., 2012; Schoepp et al., 2014). While genetic engineering may help to overcome some of C. reinhardtii’s limitations, it will be essential to also expand the knowledge and tools developed in C. reinhardtii to other algal species.
Algae are the most diverse organisms on the planet, able to grow in almost any environment, from marine systems to hot springs to desert soil crusts and even sewer drains (Norton et al., 1996; Dufresne et al., 2008; Parker et al., 2008; Tirichine and Bowler, 2011; Blunt et al., 2012). Through these adaptations, strains have naturally developed favorable traits for specific manufacturing applications. Many wild-type species of algae will be economically viable as production platforms for bio-products with minimal strain optimization. For instance, cold-tolerant microorganisms have to maintain higher levels of polyunsaturated fatty acids to maintain membrane fluidity at colder temperatures. These extremophiles make obvious choices for sources of omega-3 fatty acids. To date, several natural species have already demonstrated robust growth in commercial-scale systems, mostly for nutraceuticals (Oncel, 2013). In addition, over the past 20 years, we have expanded the use of genetic tools to a wide variety of algal species, albeit with varying success. These tools will be needed to engineer an arsenal of pest-resistant strains with optimal growth rates and production capabilities as well as to utilize algae as a protein biomanufacturing platform. The combination of improved strain selection and engineering in more diverse species is our best chance of developing algal strains for large-scale renewable energy and sustainable recombinant protein production in the future.
Acknowledgments
This work is supported by the US Department of Energy (DEEE0003373) and the California Energy Commission Initiative for Large Molecule Sustainable Fuels (CILMSF 500-10-039).
References
- Almaraz-Delgado AL, Flores-Uribe J, Pérez-España VH, Salgado-Manjarrez E, Badillo-Corona JA. Production of therapeutic proteins in the chloroplast of Chlamydomonas reinhardtii. AMB Express. 2013;4:57. doi: 10.1186/s13568-014-0057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo FC, Inmaculada C, Rosa L, Herminia R, Vargas MÁ. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl Microbiol Biotechnol. 2011;91:341–351. doi: 10.1007/s00253-011-3262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz A, Dang KV, Beyly A, Auroy P, Richaud P, Cournac L, Peltier G. Plastidial expression of type II NAD(P)H dehydrogenase increases the reducing state of plastoquinones and hydrogen photoproduction rate by the indirect pathway in Chlamydomonas reinhardtii. Plant Physiol. 2014;165:1344–1352. doi: 10.1104/pp.114.240432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera DJ, Mayfield SP. High-value recombinant protein production in microalgae. In: Richmond A, Hu Q, editors. Handbook of Microalgal Culture: Applied Phycology and Biotechnology. Oxford, UK: John Wiley & Sons, Ltd; 2013. pp. 532–544. [Google Scholar]
- Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS One. 2011;7:e42949. doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Marine natural products. Nat Prod Rep. 2012;29:144–222. doi: 10.1039/c2np00090c. [DOI] [PubMed] [Google Scholar]
- Boyle N, Page M, Liu B, et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem. 2012;287:15811–15825. doi: 10.1074/jbc.M111.334052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, et al. Chloroplast transformation in Chlamydomonas with high-velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Brownbridge G, Azadi P, Smallbone A, Bhave A, Taylor B, Kraft M. The future viability of algae-derived biodiesel under economic and technical uncertainties. Bioresour Technol. 2013;151:166–173. doi: 10.1016/j.biortech.2013.10.062. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Johnson AM, Gillham NW, Boynton JE. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso I, Vila M, Rodriguez H, Vargas MA. Overexpression of an exogenous phytoene synthase gene in the unicellular alga Chlamydomonas reinhardtii leads to an increase in the content of carotenoids. Biotechnol Prog. 2011;27:54–60. doi: 10.1002/btpr.527. [DOI] [PubMed] [Google Scholar]
- De Wilde C, Van Houdt H, De Buck S, Angenon G, De Jaeger G, Depicker A. Plants as bioreactors for protein production: avoiding the problem of transgene silencing. Plant Mol Biol. 2000;43:347–359. doi: 10.1023/a:1006464304199. [DOI] [PubMed] [Google Scholar]
- Del Campo JA, Garcia-Gonzalez M, Guerrero MG. Outdoor cultivation of microalgae for carotenoid production: current state and perspectives. Appl Microbiol Biotechnol. 2007;74:1163–1174. doi: 10.1007/s00253-007-0844-9. [DOI] [PubMed] [Google Scholar]
- Demirbas A, Demirbas FM. Importance of algae oil as a source of biodiesel. Energy Convers Manage. 2011;52:163–170. [Google Scholar]
- Deng XD, Gu B, Li YJ, Hu XW, Guo JC. The roles of acyl-CoA: diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii. Mol Plant. 2012;5:945–947. doi: 10.1093/mp/sss040. [DOI] [PubMed] [Google Scholar]
- Dismukes CG, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Dreesen IA, Charpin-El Hamri G, Fussenegger M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J Biotechnol. 2010;145:273–280. doi: 10.1016/j.jbiotec.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Dufresne A, Ostrowski M, Scanlan DJ, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler-Stahlberg A, Weisheit W, Ruecker O, Heitzer M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta. 2009;229:873–883. doi: 10.1007/s00425-008-0879-x. [DOI] [PubMed] [Google Scholar]
- Esquível MG, Amaro HM, Pinto TS, Fevereiro PS. Efficient H2 production via Chlamydomonas reinhardtii. Trends Biotechnol. 2011;29:595–600. doi: 10.1016/j.tibtech.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Gao H, Wright DA, Li T, Wang YJ, Horken K, Weeks DP, Yang B, Spalding MH. TALE activation of endogenous genes in Chlamydomonas reinhardtii. Algal Res. 2014;5:52–60. [Google Scholar]
- Georgianna DR, Mayfield SP. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 2012;488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- Georgianna DR, Michael JH, Marina M, Shuiqin W, Kyle B, Alex JL, James H, Michael M, Stephen PM. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta. Algal Res. 2013;2:2–9. [Google Scholar]
- Ghirardi ML, Posewitz MC, Maness PC. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms*. Annu Rev Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- Gimpel JA, Hyun JS, Schoepp NG. Production of recombinant proteins in microalgae at pilot greenhouse scale. Biotechnol Bioeng. 2014;112:339–345. doi: 10.1002/bit.25357. [DOI] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell. 2014;13:591–613. doi: 10.1128/EC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson C, Roth R, Wang ZT, Goodenough U. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell. 2011;10:1592–1606. doi: 10.1128/EC.05242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JA, Topol AB, Doerner DZ, Mayfield SP. Alga-produced cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl Environ Microbiol. 2013;79:3917–3925. doi: 10.1128/AEM.00714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Phycol. 2009;21:493–507. [Google Scholar]
- Gumpel NJ, Rochaix JD, Purton S. Studies on homologous recombination in the green alga Chlamydomonas reinhardtii. Curr Genet. 1994;26:438–442. doi: 10.1007/BF00309931. [DOI] [PubMed] [Google Scholar]
- Hou QT, Qiu S, Liu Q, Tian J, Hu ZL, Ni JZ. Selenoprotein-transgenic Chlamydomonas reinhardtii. Nutrients. 2013;5:624–636. doi: 10.3390/nu5030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James BD, Baum GN, Perez J, Baum KN. Technoeconomic boundary analysis of biological pathways to hydrogen production. U.S. Dept. of Energy, Washington, DC; 2009. p. 207. (US Department of Energy, Office of Energy Efficiency & Renewable Energy ed. Subcontract report NREL/SR-560-46674). [Google Scholar]
- James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price DG, Djordjevic MA. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour Technol. 2011;102:3343–3351. doi: 10.1016/j.biortech.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. Successful transient expression of Cas9 and gingle guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell. 2014a;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Cossey S, Rosenberg JN, Oyler GA, Weeks D. A rapid live-cell ELISA for characterizing antibodies against cell surface antigens of Chlamydomonas reinhardtii and its use in isolating algae from natural environments with related cell wall components. BMC Plant Biol. 2014b;14:244. doi: 10.1186/s12870-014-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CS, Mayfield SP. Algae biofuels: versatility for the future of bioenergy. Curr Opin Biotechnol. 2011;23:346–351. doi: 10.1016/j.copbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JHJ, van Gent DC. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kato K, Marui T, Kasai S, Shinmyo A. Artificial control of transgene expression in Chlamydomonas reinhardtii chloroplast using the lac regulation system from Escherichia coli. J Biosci Bioeng. 2007;104:207–213. doi: 10.1263/jbb.104.207. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim YT, Cho JJ, Bae JH, Hur SB, Hwang I, Choi TJ. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar Biotechnol. 2002;4:63–73. doi: 10.1007/s1012601-0070-x. [DOI] [PubMed] [Google Scholar]
- Kirst H, Garcia-Cerdan JG, Zurbriggen A, Melis A. Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol. 2012;158:930–945. doi: 10.1104/pp.111.189910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok AJ, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. Edible oils from microalgae: insights in TAG accumulation. Trends Biotechnol. 2014;32:521–528. doi: 10.1016/j.tibtech.2014.07.004. [DOI] [PubMed] [Google Scholar]
- La Russa M, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH. Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J Biotechnol. 2012;162:13–20. doi: 10.1016/j.jbiotec.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Larson MA, Wei SH, Weber A, Mack DR, McDonald TL. Human serum amyloid A3 peptide enhances intestinal MUC3 expression and inhibits EPEC adherence. Biochem Biophys Res Commun. 2003;300:531–540. doi: 10.1016/s0006-291x(02)02901-7. [DOI] [PubMed] [Google Scholar]
- Leite GB, Abdelaziz AEM, Hallenbeck PC. Algal biofuels: challenges and opportunities. Bioresour Technol. 2013;145:134–141. doi: 10.1016/j.biortech.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, Hu Q. Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng. 2010;12:387–391. doi: 10.1016/j.ymben.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack DR, McDonald TL, Larson MA, Wei S, Weber A. The conserved TFLK motif of mammary-associated serum amyloid A3 is responsible for up-regulation of intestinal MUC3 mucin expression in vitro. Pediatr Res. 2003;53:137–142. doi: 10.1203/00006450-200301000-00023. [DOI] [PubMed] [Google Scholar]
- Manuell AL, Beligni MV, Elder JH, Siefker DT, Tran M, Weber A, McDonald TL, Mayfield SP. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol J. 2007;5:402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- Melis A, Zhang L, Forestier M, Ghirardi ML. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2000;122:127–136. doi: 10.1104/pp.122.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Seibert M, Ghirardi ML. Hydrogen fuel production by transgenic microalgae. In: Léon R, Galván Cejudo A, Fernández E, editors. Transgenic Microalgae as Green Cell Factories. New York: Springer New York; 2007. pp. 110–121. [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J. TAG, You’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol. 2011;23:352–363. doi: 10.1016/j.copbio.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman F. Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell. 2006;18:159–175. doi: 10.1105/tpc.105.037705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Msanne J, Xu D, Konda AR, Casas-Mollano JA. Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry. 2012;75:50–59. doi: 10.1016/j.phytochem.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta. 2012;1817:247–257. doi: 10.1016/j.bbabio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Näätsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Chou SK, Cao S, Wu C, Zhou Z. An updated comprehensive techno-economic analysis of algae biodiesel. Bioresour Technol. 2013;145:150–156. doi: 10.1016/j.biortech.2012.11.108. [DOI] [PubMed] [Google Scholar]
- Nelson JA, Lefebvre PA. Targeted disruption of the NIT8 gene in Chlamydomonas reinhardtii. Mol Cell Biol. 1995;15:5762–5769. doi: 10.1128/mcb.15.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TA, Melkonian M, Andersen RA. Algal biodiversity. Phycologia. 1996;35:308–326. [Google Scholar]
- Oncel SS. Microalgae for a macroenergy world. Renew Sustain Energy Rev. 2013;26:241–264. [Google Scholar]
- Oncel SS, Kose A, Faraloni C, Imamoglu E, Elibol M, Torzillo G, Sukan FV. Biohydrogen production using mutant strains of Chlamydomonas reinhardtii: the effects of light intensity and illumination patterns. Biochem Eng J. 2014;92:47–52. [Google Scholar]
- Parker MS, Mock T, Armbrust EV. Genomic insights into marine microalgae. Annu Rev Genet. 2008;42:619–645. doi: 10.1146/annurev.genet.42.110807.091417. [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Ouellet M, Chan R. Identification and microbial production of a terpene-based advanced biofuel. Nat Commun. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Mayfield SP. The microalga Chlamydomonas reinhardtii as a platform for the production of human protein therapeutics. Bioeng Bugs. 2011;2:50–54. doi: 10.4161/bbug.2.1.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Mayfield SP. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth Res. 2014;123:227–239. doi: 10.1007/s11120-014-9994-7. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Lee PA, Jager M, Cardoso RMF, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8:719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Lee PA, Shen ZX, Briggs SP, Mendez M, Mayfield SP. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A Peptide. PLoS One. 2012;7:e43349. doi: 10.1371/journal.pone.0043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B, Barrera D, Ng J, Plucinak T, Rosenberg J, Weeks D, Oyler G, Peterson T, Haerizadeh F, Mayfield S. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013a;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Gimpel JA, Tran M, Hannon MJ. Genetic engineering to improve algal biofuels production. In: Borowitzka MA, Moheimani NR, editors. Algae for Biofuels and Energy. Dordrecht, Netherlands: Springer Netherlands; 2013b. pp. 99–113. [Google Scholar]
- Rosales-Mendoza S, Paz-Maldonado LM, Soria-Guerra RE. Chlamydomonas reinhardtii as a viable platform for the production of recombinant proteins: current status and perspectives. Plant Cell Rep. 2012;31:479–494. doi: 10.1007/s00299-011-1186-8. [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Strenkert D, Schroda M. An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr Genet. 2010;56:383–389. doi: 10.1007/s00294-010-0304-4. [DOI] [PubMed] [Google Scholar]
- Schoepp NG, Stewart RL, Sun V, Quigley AJ. System and method for research-scale outdoor production of microalgae and cyanobacteria. Bioresour Technol. 2014;166:273–281. doi: 10.1016/j.biortech.2014.05.046. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Scoma A, Giannelli L, Faraloni C, Torzillo G. Outdoor H2 production in a 50-L tubular photobioreactor by means of a sulfur-deprived culture of the microalga Chlamydomonas reinhardtii. J Biotechnol. 2012;157:620–627. doi: 10.1016/j.jbiotec.2011.06.040. [DOI] [PubMed] [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011;11:7. doi: 10.1186/1472-6750-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Nigam PS, Murphy JD. Mechanism and challenges in commercialisation of algal biofuels. Bioresour Technol. 2011;102:26–34. doi: 10.1016/j.biortech.2010.06.057. [DOI] [PubMed] [Google Scholar]
- Sodeinde OA, Kindle KL. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1993;90:9199–9203. doi: 10.1073/pnas.90.19.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht E, Mayfield SP. Synthetic oligonucleotide libraries reveal novel regulatory elements in Chlamydomonas chloroplast mRNAs. ACS Synth Biol. 2012;2:34–46. doi: 10.1021/sb300069k. [DOI] [PubMed] [Google Scholar]
- Su ZL, Qian KX, Tan CP, Meng CX, Qin S. Recombination and heterologous expression of allophycocyanin gene in the chloroplast of Chlamydomonas reinhardtii. Acta Biochim Biophys Sin. 2005;37:709–712. doi: 10.1111/j.1745-7270.2005.00092.x. [DOI] [PubMed] [Google Scholar]
- Terashima M, Freeman ES, Jinkerson RE, Jonikas MC. A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 2014;81:147–159. doi: 10.1111/tpj.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Bowler C. Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant J. 2011;66:45–57. doi: 10.1111/j.1365-313X.2011.04540.x. [DOI] [PubMed] [Google Scholar]
- Torzillo G, Seibert M. Hydrogen production by Chlamydomonas reinhardtii. In: Richmond A, Hu Q, editors. Handbook of Microalgal Culture: Applied Phycology and Biotechnology. 2nd. Oxford, UK: John Wiley & Sons, Ltd; 2013. pp. 417–432. [Google Scholar]
- Torzillo G, Scoma A, Faraloni C, Ena A. Increased hydrogen photoproduction by means of a sulfur-deprived Chlamydomonas reinhardtii D1 protein mutant. Int J Hydrogen Energy. 2009;34:4529–4536. [Google Scholar]
- Tran M, Henry RE, Siefker D, Van C, Newkirk G, Kim J, Bui J, Mayfield SP. Production of anti-cancer immunotoxins in algae: ribosome inactivating proteins as fusion partners. Biotechnol Bioeng. 2013a;110:2826–2835. doi: 10.1002/bit.24966. [DOI] [PubMed] [Google Scholar]
- Tran M, Van C, Barrera DJ, Pettersson PL, Peinado CD, Bui J, Mayfield SP. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc Natl Acad Sci USA. 2013b;110:E15–E22. doi: 10.1073/pnas.1214638110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwu CU, Aoyagi H, Uchiyama H. Photobioreactors for mass cultivation of algae. Bioresour Technol. 2008;99:4021–4028. doi: 10.1016/j.biortech.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Venter M. Synthetic promoters: genetic control through cis engineering. Trends Plant Sci. 2007;12:118–124. doi: 10.1016/j.tplants.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Vinyard DJ, Gimpel J, Ananyev GM, Cornejo MA, Golden SS, Mayfield SP, Dismukes GC. Natural variants of photosystem II subunit D1 tune photochemical fitness to solar intensity. J Biol Chem. 2013;288:5451–5462. doi: 10.1074/jbc.M112.394668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinyard DJ, Gimpel J, Ananyev GM. Engineered photosystem II reaction centers optimize photochemistry versus photoprotection at different solar intensities. J Am Chem Soc. 2014;136:4048–4055. doi: 10.1021/ja5002967. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- Wang ZT, Ullrich N, Joo S, Waffenschmidt S. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work VH, Radakovits R, Jinkerson RE, Meuser JE. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7–10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell. 2010;9:1251–1261. doi: 10.1128/EC.00075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- Xie B, Stessman D, Hart JH, Dong H, Wang Y, Wright DA, Nikolau BJ, Spalding MH, Halverson LJ. High throughput fluorescence activated cell sorting for lipid hyperaccumulating Chlamydomonas reinhardtii mutants. Plant Biotechnol J. 2014;12:872–882. doi: 10.1111/pbi.12190. [DOI] [PubMed] [Google Scholar]
- Xu L, Wang Q, Wu S, Li D. Improvement of hydrogen yield of lba-transgenic Chlamydomonas reinhardtii caused by increasing respiration and impairing photosynthesis. Int J Hydrogen Energy. 2014;39:13347–13352. [Google Scholar]
- Yang ZQ, Li YN, Chen F, Li D, Zhang ZF, Liu YX, Zheng DX, Wang Y, Shen GF. Expression of human soluble TRAIL in Chlamydomonas reinhardtii chloroplast. Chin Sci Bull. 2006;51:1703–1709. [Google Scholar]
- Yoon SMM, Kim SY, Li KF, Yoon BH, Choe S, Kuo MM. Transgenic microalgae expressing Escherichia coli AppA phytase as feed additive to reduce phytate excretion in the manure of young broiler chicks. Appl Microbiol Biotechnol. 2011;91:553–563. doi: 10.1007/s00253-011-3279-2. [DOI] [PubMed] [Google Scholar]
- Zhang YK, Shen GF, Ru BG. Survival of human metallothionein-2 transplastomic Chlamydomonas reinhardtii to ultraviolet B exposure. Acta Biochim Biophys Sin (Shanghai) 2006;38:187–193. doi: 10.1111/j.1745-7270.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell. 2014;26:1398–1409. doi: 10.1105/tpc.114.124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]


