Abstract
Cancer cell invasion through the extracellular matrix is associated with metastatic spread and therapeutic resistance. In carcinomas, the detachment and dissemination of individual cells has been associated with an epithelial-mesenchymal transition, but tumors can also invade using collective, multicellular phenotypes. This malignant tumor progression is also associated with alignment and stiffening of the surrounding extracellular matrix. Historically, tumor invasion has been investigated using 2D monolayer culture, small animal models or patient histology. These assays have been complemented by the use of natural biomaterials such as reconstituted basement membrane and collagen I. More recently, engineered materials with well-defined physical, chemical and biomolecular properties have enabled more controlled microenvironments. In this review, we highlight recent developments in multicellular tumor invasion based on microfabricated structures or hydrogels. We emphasize the role of interfacial geometries, biomaterial stiffness, matrix remodeling, and co-culture models. Finally, we discuss future directions for the field, particularly integration with precision measurements of biomaterial properties and single cell heterogeneity, standardization and scale-up of these platforms, as well as integration with patient-derived samples.
Introduction
Tumor invasion and metastasis occurs in the context of molecular and mechanical cues from the extracellular matrix (ECM), and ultimately results in over 90% of cancer-related fatalities.1 In particular, the detachment and dissemination of individual cells from the periphery of carcinomas is reminiscent of the epithelial-mesenchymal transition (EMT) in embryonic development and wound healing.2 Moreover, tumor cells can exhibit multicellular collective invasion as loosely or tightly coordinated groups.3 Such malignant tumor progression is accompanied by a dramatic remodeling and stiffening of the ECM (i.e. desmoplasia).4 These microenvironmental changes can bias tumor cells towards individual or collective invasion, a phenomenon known as phenotypic plasticity. This reciprocity between tumor invasion and ECM has previously been investigated in cancer research using 2D monolayer culture, animal models, and patient histology. Biomimetic materials can complement these existing approaches by recapitulating important features of the tumor microenvironment.5
Biomaterials can be fabricated or synthesized into microstructural architectures that mimic the ECM. For instance, semiconductor fabrication techniques have enabled exquisitely detailed geometric features, comparable in size and spacing to matrix topography.6 Moreover, hydrogels based on hydrated, crosslinked polymer networks can mimic the biochemical composition, mechanical stiffness and degradability of ECM in tissues and tumors.7 Indeed, natural hydrogels have enabled key biological insights into EMT and tumor invasion. For instance, Hay and colleagues first observed what they termed an epithelial-to-mesenchymal transformation after embedding corneal epithelial tissue in a reconstituted matrix of fibrillar collagen I.8 Subsequently, Bissell and colleagues showed that epithelial morphogenesis can be recapitulated using reconstituted basement membrane in a 3D context, resulting in the self-organization of individual cells into gland-like structures with hollow lumens and differentiated cell-cell junctions (acini).9,10 The disorganization and dissemination of cells from these acini into the surrounding ECM recapitulates many key features of tumor progression.11 Recent advances in biomaterials have enabled new physical insights into the tumor microenvironment12 and may facilitate preclinical models of cancer with greater physiological relevance.13
In this review, we highlight recent developments in cancer cell invasion and EMT enabled by new biomaterial platforms. We focus on multicellular tissues as in vitro and ex vivo models of cancer, with emphasis on results published within the last several years. In the following sections, we consider 1) microfabricated geometries that promote EMT from 2D monolayers, 2) the spreading of 3D multicellular aggregates onto planar surfaces, 3) epithelial morphogenesis and dissemination in 3D biomaterials, and 4) co-culture of tumor and stromal cells. We conclude with a discussion of future directions for the field.
Background: Multicellular Tumor Invasion and the ECM
Cancer can be defined as a disease in which “abnormal cells proliferate in an uncontrolled fashion and spread throughout the body.”14 Most human cancers are carcinomas derived from epithelial tissues, which line the walls of surfaces and cavities of human organs. The most common types of cancers arise in the skin, prostate, breast, lung and colon.15 Nevertheless, many types of skin cancer can be diagnosed early and treated. Thus, cancer-related fatalities are most frequently due to lung, colorectal, breast, prostate, and pancreatic cancers.15
Tumor progression may be largely influenced by tissue-specific physiology. For instance, the functional unit of the breast is the lobule, which is lined by luminal and myoepithelial cells interfaced with intralobular stroma, further surrounded by interlobular stroma.16 These stroma include extensive adipocytes (fat cells), as well as fibroblasts, immune cells, stem cells, and endothelial cells. Breast carcinomas are often classified as ER (estrogen receptor) positive, HER2 (human epidermal growth factor receptor 2) positive, or triple negative. In contrast, the functional unit of the lung is the alveolus, which is lined by alveolar epithelium that consists of flattened platelike type I pneumocytes and rounded type II pnuemocytes.16 Alveolar macrophages are associated with these epithelial cells, as well as small numbers of fibroblast-like cells, smooth muscles cells, mast cells, and lymphocytes. Carcinomas of the lung are histologically classified as adenocarcinomas, squamous cell carcinomas, small cell carcinomas and large carcinomas. It should be noted that the lung is also the most common site for secondary metastases, which may be partially explained by hematogenous spread and arrest of cancer cells in the capillary bed of the lung.17 Finally, the skin includes squamous epithelial cells (keratinocytes), melanocytes, dendritic cells, and lymphocytes. Common skin cancers include melanomas, sqamous cell carcinomas and basal cell carcinomas.16 The details of tissue-specific pathophysiology are reviewed extensively elsewhere.16 Benam et al. have also recently reveied tissue-specific in vitro cancer models.18
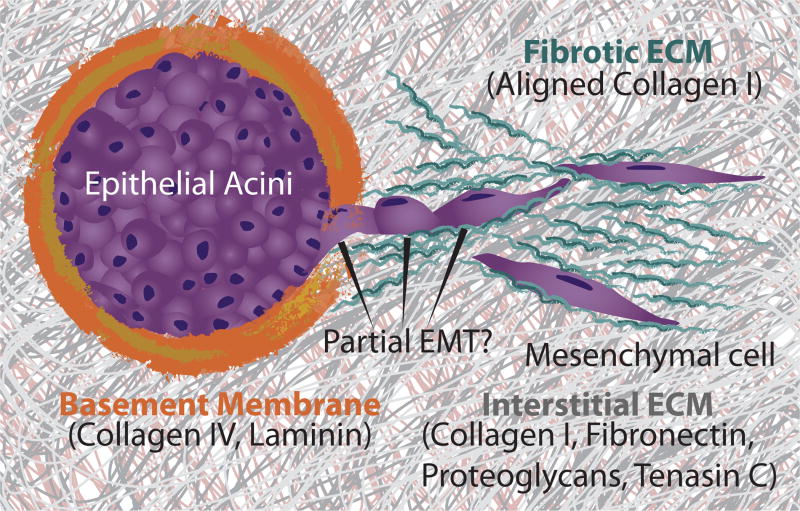
In general, epithelial tissues transition to invasive carcinomas through multicellular disorganization and dissemination (Figure 1).11 As this malignant progression occurs, cells encounter distinct ECM with dramatically different biochemical and physical properties. Furthermore, tumor cells can actively deposit and remodel ECM, which can further alter the extracellular cues that govern molecular and mechanical phenotype.4 In general, tumor cells display extraordinary phenotypic heterogeneity and plasticity in response to these dynamic microenvironmental conditions, enabling them to migrate with a variety of distinct collective and individual phenotypes.1 In particular, various aberrant stimuli may trigger an EMT, resulting in a weakening of cell-cell adhesions, as well as enhanced motility and cell-matrix adhesion.2
Fig. 1.
Multicellular tumor invasion and the ECM. Epithelial tissues are enclosed by basement membrane. During tumor invasion and EMT, tissues disorganize and disseminate into the surrounding interstitial matrix. Tumor cells can further deposit and reorganize the matrix into a more fibrotic architecture.
Overall, tumor cells are highly efficient in invasion processes for infiltration of the ECM, and subsequent intravasation to enter the bloodstream. However, successful extravasation, or exit from circulation to a secondary site, occurs with very low frequency due to the harsh nature of the bloodstream (e.g. mechanical shear stress) and susceptibility to immune cell surveillance.19 Expression of the T-cell marker, CD44, on tumor cells may favor extravasation by enhancing the adhesive interactions between cancer cells and the endothelium. Subsequently, the colonization of secondary metastatic sites often depends on the permissiveness of the microenvironment. As a consequence, primary tumors originating from certain tissues often preferentially metastasize to other tissues, a phenomenon known as organ tropism.17 For instance, primary breast tumors primarily metastasize to bone, lungs, liver, and brain, while primary lung tumors primarily metastasize to brain, bones, adrenal gland, and liver. Indeed, the primary tumor may prime a metastatic niche for colonization by reprogramming stromal cells to structurally rearrange the ECM, recently reviewed elsewhere.20 Although not a primary focus of this review, it should be noted that metastatic homing could be investigated by biomimetic materials that recapitulate the microenvironment of metastatic sites.21
From Basement Membrane to Interstitial Matrix
Under physiological conditions, the ECM presents mechanical and biochemical cues that are instructive for cellular function and the maintenance of tissue architecture.4 The ECM consists of two major subtypes known as the basement membrane and interstitial matrix. Epithelium and endothelium that support biofluid adsorption and secretion are adherent to the basement membrane (Figure 1).22 This thin, mesh-like network provides a structural framework for maintaining epithelial and endothelial tissue architecture, as well as a barrier to adjacent connective tissue. The basement membrane consists of specialized ECM proteins including collagen IV and glycoproteins such as laminin, nidogen, and perlecan. In particular, laminin contains multiple domains that mediate the binding interactions between basement membrane components and present sites for cell adhesion.23 Importantly, laminin plays a crucial role in instructing epithelial phenotype.24 Epithelial cells on the basement membrane typically exhibit a highly geometric, cobblestone-like morphology with tight junctions, which enables control over tissue barrier properties such as permeability and secretion.
The adjacent connective tissue (stroma) contains interstitial matrix, which is a three-dimensional hydrogel composed of fibrillar collagens, fibronectin, tenascin-C, elastin, glycosaminoglycans, proteoglycans, and others. The exact composition of interstitial matrix varies with tissue type and often undergoes diverse changes during carcinoma progression (Figure 1).25 The protein components that comprise the ECM have been recently characterized in terms of a “core matrisome,” which includes the structural ECM components described previously, as well as “matrisome-associated” proteins, which include ECM-affiliated proteins (galectins, semaphorins, etc.), secreted proteins (growth factors, cytokines, etc.), and ECM regulators (matrix metalloproteinases, cross-linking enzymes), etc.26 Together, these ECM constituents provide sites for cell adhesion and migration, and grant essential mechanical properties to the tissue including tensile strength, elasticity, and resistance to compressive forces. Typically, brain tissue is one of the softest tissues (E ~ 0.1 kPa), while breast and liver are slightly stiffer (E ~ 0.4 – 1 kPa).27 Cartilage and bone tend to be relatively stiff (E ~ 10 – 20 kPa).27 Interestingly, tissue stiffness scales roughly with collagen I concentration. 28
During tumor progression, the ECM is dynamically remodeled to a fibrotic-like state, which has analogies with wound healing processes, described as “a wound that does not heal.”29 Stromal cells such as fibroblasts can synthesize, reorganize and degrade the ECM, resulting in a macroscopic stiffening of the stroma (desmoplasia).30 In particular, there is often increased deposition, cross-linking and alignment of fibrillar proteins such as collagen I, resulting in an aberrant matrix topography and biomechanical properties that affect cellular phenotype (Figure 1). These aligned, track-like structures can facilitate the directional migration of tumor cells toward lymphatic and blood vessels for metastatic spread.31 Additionally, excess ECM degradation can result in the release of sequestered growth factors and other bioactive molecules which are bound to proteoglycans.32 Finally, altered ECM can enhance the recruitment and infiltration of immune cells as well as induce angiogenesis, which is reviewed elsewhere.4
Integrins and other Cell-Matrix Adhesions
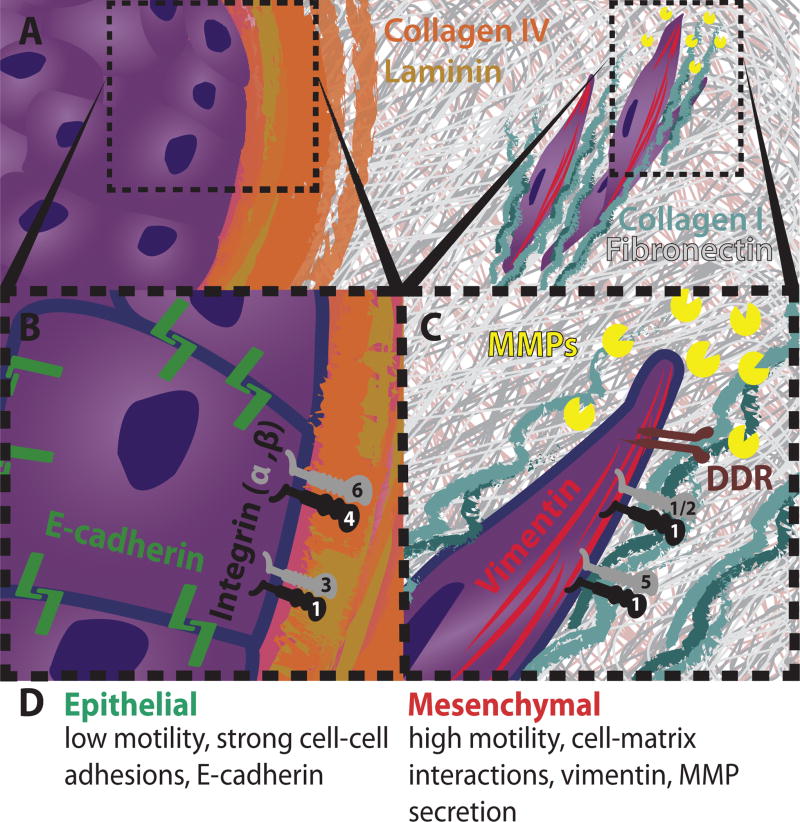
Cell adhesion and migration along the ECM are primarily mediated by integrins, a family of transmembrane glycoprotein receptors (Figure 2). Each integrin consists of an alpha and beta subunit, with 18 and 8 types respectively, yielding at least 24 distinct heterodimers.33 These diverse integrin heterodimers permit some specificity in ligand binding to direct cell-matrix interactions in a context dependent manner, although there also exists some promiscuity. In particular, epithelial cells utilize integrins α6β4 and α3β1 to attach to collagen IV and laminin in the basement membrane (Figure 2B). During the initial stages of carcinoma progression, tumor cells can locally degrade the basement membrane through MMP (matrix metalloproteinase) and ADAM (a disintegrin and metalloproteinase) protease families.34. As a consequence, tumor cells can interact with the surrounding interstitial matrix through integrins α5β1 and α1/2β1, which bind to collagen I and fibronectin (Figure 2C). Furthermore, matrix degradation can coincide with additional ECM deposition and crosslinking. In particular, lysyl oxidase expression leads to further collagen I crosslinking and subsequent clustering of β1 integrin, which further drives cell survival via the PI3K pathway and invasion through focal adhesion kinases.30 In addition to integrin binding, collagen also interacts with receptor Tyr kinases discoidin domain-containing receptors 1 and 2 (DDR1/2), which have been shown to be critical for collective cell migration and stimulate the induction of EMT, respectively.32
Fig. 2.
Cellular and molecular features of EMT. (A) Epithelial and mesenchymal cells in ECM. (B) Epithelial cell polarity is governed by cell-cell junctions and integrins. (C) Mesenchymal cell polarity is governed by integrins and DDR, but modulated by matrix metalloproteinases (MMPs) and vimentin. (D) Phenotypic features of EMT.
EMT Pathways
Features of EMT have been observed in patient tumor histology and are often associated with poor prognosis.35 These include a loss of tissue architecture with dedifferentiated phenotypes, diminished expression of E-cadherin, and invasive protrusions.36 Cell-ECM interactions may lead to the expression of EMT regulatory genes, and reciprocally, several transcription factors that regulate the induction of EMT are also involved in ECM remodeling. For instance, ECM can participate in the initiation of signal transduction pathways by serving as a reservoir for soluble signals (e.g. growth factors). As an example, transforming growth factor beta (TGFβ) binds to glycoproteins and proteoglycans in the ECM, fibrillin and heparin sulfate, respectively, and can be mobilized in response to cleavage of the complex37 to drive downstream signaling including the induction of EMT.38
Snail, Zeb, and Twist are prominent transcription factors, or master regulatory genes, involved in EMT induction.39,40 A common theme is the downregulation of E-cadherin to destabilize adherens junctions between cells. In addition, apical tight junctions and desmosomes are weakened by the repression of genes that encode claudin and occludin, as well as desmoplakin and plakophilin. Moreover, these transcription factors suppress the synthesis of basement membrane components.41 Overall, these changes in gene expression disrupt epithelial tissue architecture and barrier function, while impeding the formation of new junctions. An additional consequence of the loss of E-cadherin is the increased expression of N-cadherin, which is typically associated with mesenchymal cells. Further downstream signaling can occur via the crosstalk of TGF-β and matrix ligand adhesion, particularly PI3K-AKT, ERK MAPK, p38 and JNK pathways, which has recently been reviewed elsewhere.42
At the cellular level, EMT is associated with dramatic alterations in the cytoskeleton and morphology (Figure 2D).43 Initially epithelial cells lose apicobasal polarization and gain front-back polarization, driven in part by Rho GTPases that regulate actin dynamics.44 In particular, RAC1 and CDC42 activate actin polymerization and membrane protrusion formation. Simultaneously, RHOA displays a coordinated localization to the rear in order to facilitate actomyosin contractility for cell retraction. As a consequence, mesenchymal cell lines can display highly asymmetric tractions at the front and rear relative to epithelial cell lines.45,46 Moreover, EMT results in dramatic increases in actin stress fiber formation as well as actin-rich invadopodia that can direct local proteolytic cleavage of the matrix through the release of MMPs at the leading edge.32 Finally, the composition of intermediate filaments is also switched from cytokeratin to vimentin, which enhances cell deformability.47
Established Biomaterials for Tumor Invasion
Collagen I
Collagen I is a fibrillar biopolymer constituted from natural sources (typically rodent tail or bovine dermis) and was first utilized as a transparent hydrogel substrate by Ehrmann and Gey.48 Since collagen is maintained in its monomeric form in acidic solution and at low temperature,49 controlled collagen polymerization into a hydrogel can occur by increasing the pH and incubation in the presence of cells and culture media. Qualitatively, lower collagen concentrations result in lower fibril density and larger mesh size.50 Moreover, lower polymerization temperatures can result in larger mesh sizes and larger fibril diameters.51 Nevertheless, collagen hydrogels display a complicated dependence on mechanical stiffness and ligand density, making it difficult to decouple these mechanisms in epithelial cell processes. Typically, the elastic moduli for collagen gels can vary from 0.1 – 2 kPa for concentrations of rat-tail collagen from 2.0 – 4.0 mg/mL,52 although these values are quite sensitive to preparation conditions.53 Moreover, collagen hydrogels can be extensively degraded and remodeled, which can be advantageous for studies of directed cell migration but may also limit long term stability.54 Finally, collagen as a naturally-derived biomaterial can display some batch-to-batch variability. Overall, collagen I represents a reasonable mimic of the tumor ECM and has been utilized extensively to investigate EMT ever since the initial experiments by Greenburg and Hay.8
Reconstituted Basement Membrane (Matrigel)
Reconstituted Basement Membrane (rBM) was first extracted from mouse Engelbreth-Holm-Swarm (EHS) sarcoma tumors by Swarm and colleagues55, which is commercially available as Matrigel. Major components of rBM typically include laminin, collagen IV, entactin, as well as proteoglycans such as heparan sulfate. 56 In particular, the chief constituent of rBM, laminin, supports the differentiation and polarization of epithelial and endothelial cells and facilitates their attachment to the basement membrane. Furthermore, it is worth noting that there are 11 distinct chains that make up the laminin heterotrimer of α, β, and γ subunits, respectively. In particular, the specific isoform of laminin derived from EHS (laminin α1β1γ1) is critical in embryogenesis and development, however, adult epithelial tissues and carcinomas predominantly express laminin α3 and α5 isoforms.23,57,58 rBM is liquid near freezing temperatures, but solidifies into a hydrogel at physiological temperatures. Bissell and coworkers pioneered the use of rBM to investigate mammary epithelial morphogenesis.9,10 By embedding cells within rBM, or seeding cells on top of rBM with a dilute overlay, epithelial cells self-organized into spherical acini with apicobasal polarity, hollow lumens, and tightly controlled growth and proliferation.11,59 Weaver et al. subsequently used rBM to investigate the role of integrins β1 and β4 in driving a polarized 3D epithelial architecture, as well as reversion of a malignant phenotype.60,61 Thin rBM coatings on a porous plastic membrane have also been utilized in a classical invasion assay62 based on the Boyden chamber (Transwell assay)63 This membrane partitions two fluidic compartments, with migratory cells plated at the top and an optional chemoattractant solution underneath. Over time, migratory cells translocate across the membrane to the bottom, where they can be counted. Although established, this assay is limited in that it occurs as an endpoint and also biases towards individual cell migration, due to the relatively small pore size.
Transitions to invasion have been observed by combining rBM with fibrillar collagen I. For instance, Paszek et al. showed that mammary acini initially seeded on relatively soft rBM ( ~0.18 kPa) transitioned towards disorganized structures with destabilized cell-cell junctions when overlaid with collagen I with progressively increasing stiffness.64 Similarly, Guzman et al. have demonstrated that blending fibrillar collagen I into rBM can promote individual cell invasion.65 It should be noted that rBM displays significant batch-to-batch variability in biochemical composition and mechanical properties 56 and these biomaterial properties of rBM cannot be easily tuned for systematic studies. Nevertheless, rBM has been crucial in driving biological insights underlying epithelial morphogenesis and EMT, and remains widely used.
Polyacrylamide
Polyacrylamide (PA) is a versatile hydrogel with tunable mechanical and biochemical properties, first demonstrated as a biomaterial substrate by Pelham and Wang.66 PA is often polymerized as a thin film coating by the reaction of acrylamide monomer and bis-acrylamide crosslinker.67 The elastic modulus of this hydrogel can be systematically varied from 0.1–100 kPa by changing the relative concentrations of monomer and cross-linker, permitting well-controlled mechanical properties.68 It should be noted that PA lacks adhesion ligands and is largely bioinert, which deters cell attachment. To address this issue, ECM proteins such as collagen I, collagen IV, laminin, or fibronectin can be conjugated to PA through a bifunctional crosslinker such as sulfo-SANPAH.68 The surface ligand density can be controlled via the ligand concentration in solution, independently of mechanical properties. PA substrates have found great success in mechanobiology, particularly when fluorescent tracer particles are added for traction force microscopy.69 Nevertheless, PA is limited in that the hydrogel precursors are highly cytotoxic, so it cannot be used to encapsulate cells in a fully 3D microenvironment. Moreover, PA cannot be degraded by cells. As a consequence, PA is primarily used as a “soft” 2D biomaterial substrate. Hybrid approaches have also been demonstrated where cells adherent to PA are overlaid with a second hydrogel.
Polydimethylsiloxane
Polydimethylsiloxane (PDMS) is a moderately soft elastomer that can be micromolded with submicron fidelity, first demonstrated as a deformable cell substrate by Harris and coworkers.70 PDMS is relatively straightforward to prepare, and displays ideal material properties for soft lithography approaches based on replica molding against a microfabricated silicon master.71 In particular, PDMS is highly conformal and can adhere reversibly or irreversibly to other surfaces. PDMS can be used in microcontact printing as a topographically patterned “stamp” which can transfer molecular “ink” to a new surface.72 This gentle transfer is particularly useful for microscale patterning of soft substrates, such as the PA hydrogels discussed in the previous section.73 The irreversible bonding of PDMS microstructures to a glass substrate is also useful to prepare confined geometries for manipulation of fluid flows74 or cell migration.75 Oxygen plasma treatment of PDMS can render it (temporarily) hydrophilic, which permits adequate protein physisorption for cell adhesion.76 Although PDMS is advantageous for rapid prototyping, concerns have been raised over potentially adverse effects for long-term cell culture, including evaporation, adsorption of small hydrophobic molecules, and instability of surface treatments.77 It should be noted that the formulation of PDMS typically utilized for rapid prototyping (Sylgard 184 at 10:1 base:curing agent) is still significantly stiffer than ECM or tissue (~1 MPa). Instead, a higher ratio of base to curing agent (> 50:1) can result in softer PDMS (~ 5 kPa), although there is some discrepancy in the reported values, likely due to the significant viscoelastic effects.78,79 Palchesko has recently demonstrated that a blend of two different PDMS types (Sylgard 184 and Sylgard 527) can result in a softer substrate of 5 kPa, mechanically tunable up to 1 MPa.80 Overall, PDMS is highly effective for patterning microstructures with well controlled geometries, but may not mimic other physiochemical features of natural ECM.
On the Edge: 2D Cell Monolayers in Microfabricated Geometries
Epithelial cell monolayers cultured on top of glass or polystyrene surfaces are widely used to study cancer cell biology. These substrate materials are six orders of magnitude stiffer than native ECM (~GPa), presenting an asymmetrical mechanical cue that results in apicobasal polarization perpendicular to the surface. Interestingly, cells adjacent to empty space at the periphery of a monolayer can display a loss of apicobasal polarization that is analogous to EMT. This phenomenon is commonly used in “wound-healing” or scratch assays where an empty region is suddenly created by locally removing a part of the monolayer or some obstacle.81 “Leader” cells can then establish a front-back polarization oriented towards the empty region (parallel to the surface). The activation of RhoGTPases RAC1 and CDC42 can trigger lamellipodial and filopodial protrusions at the leading edge through actin polymerization.82 These leader cells typically retain some cell-cell junctions at the sides and rear, which can transmit mechanical signals that repolarize neighboring cells over longer length scales. Gilles et al. used a fluorescent reporter to show that mammary epithelial cells at the monolayer edge transiently express vimentin, consistent with an EMT.83 Subsequently, vimentin is downregulated once the cells fully occupy the empty region and re-establish a continuous monolayer. Thus, cells at the periphery of an epithelial monolayer recapitulate some biological behaviors observed at the periphery of a tumor or tissue.
Nelson et al. demonstrated the importance of the tissue periphery using a hybrid fabrication approach that utilized both microfabrication and soft hydrogels.84 Briefly, an elastomeric PDMS stamp was replicated from a silicon master and used to imprint microscale cavities of different shapes into a thin collagen hydrogel. Cells were seeded within the cavity, which was then capped with a second flat sheet of collagen. For instance, epithelial cells in a square geometry treated with TGF-β displayed vimentin and α-SMA only at the edges and corners, suggesting that geometric and mechanical cues mediated EMT induction.85 It was further shown that EMT induction was mediated by intercellular transmission of cytoskeletal tension. Subsequent work showed that a single tumor cell co-cultured with epithelial cells would display enhanced proliferation and invasion when located at the periphery relative to the interior.86 These regions were associated with a geometric enhancement of mechanical stress, which acts through RHOA and focal adhesion kinase (FAK) activity.
Surface topography can also play a critical role in modulating cell shape and directed migration, a phenomenon known as contact guidance.6 Curtis and Wilkinson demonstrated the early use of microfabrication techniques to pattern grooved substrates, along which fibroblasts would align and elongate.90 In particular, the width of these grooves can be comparable to the aligned collagen fibrils in desmoplasmic ECM. Analogously, PDMS channels have been used to confine cells within a tube-like environment,91 which mimics interstitial geometries or the aligned “microtracks” generated by invasive tumor cells. These approaches have primarily been used to investigate individual cell migration and have been recently reviewed elsewhere.75 Overall, microfabrication techniques can thus permit excellent geometric control over the monolayer periphery, as well as to confine individual cells along defined tracks.
Epithelial cell migration on microscale glass wires
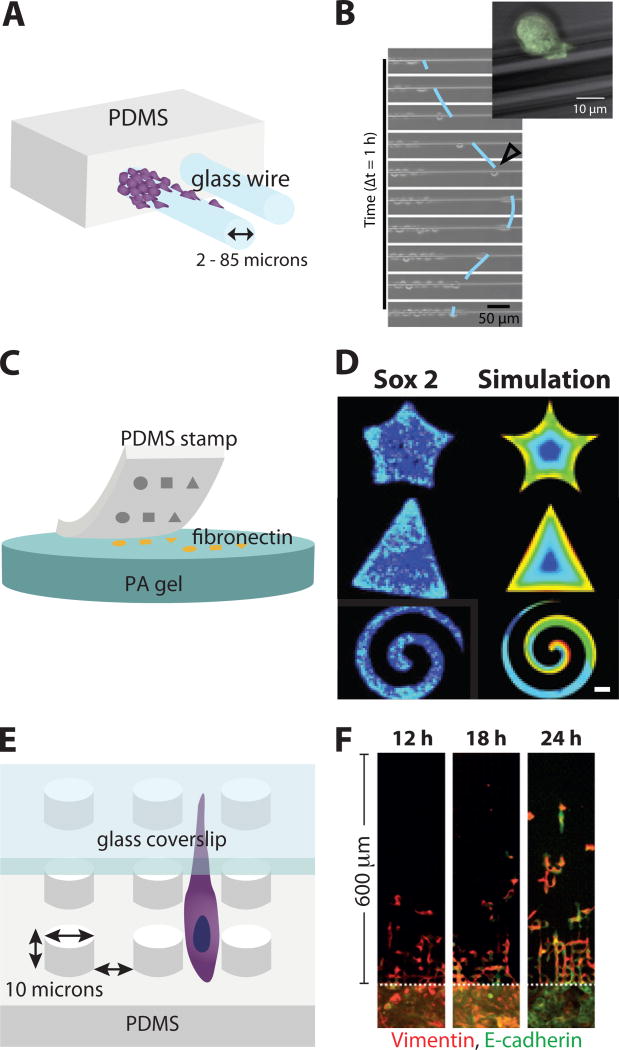
Yevick et al. characterized epithelial cell migration on microscale glass wires,87 comparable in size to bundled collagen fibers.6 Epithelial cells (MDCK) were seeded on a PDMS block, adjacent to protruding, fibronectin-coated glass wires with radii ranging from 2 µm to 85 µm (Figure 3A). Cells migrated collectively as a cohesive strand that encircled the wider glass wires (> 40µm). In contrast, single cells occasionally broke away individually on narrower wires (< 40µm). These individual cells displayed relatively rapid migration (up to 100 µm/h) with alternating elongated and rounded morphology, but frequently reversed course and rejoined the collectively migrating group (Figure 3B). Interestingly, for narrow wires, cells displayed actin stress fibers that were perpendicular to the wire axis and extended circumferentially around the wire, suggestive of mechanical connectivity. Moreover, the leading edge also exhibited a tensile actin cable, reminsicent of those utilized for closing epithelial wounds through a purse-string like mechanism.92 An interesting possibility would be to examine whether these actin cables enhanced migration on a more compliant substrate. Moreover, these actin cables may exert an effective surface tension that restricts the detachment of individual cells along the wire.
Fig. 3.
2D Cell Monolayers in Microfabricated Geometries. (A) Microscale glass wires embedded in PDMS, (B) Individual cells can detach and scatter along glass wires from multicellular groups. Reproduced from87 with permission from the National Academy of Sciences. (C) Microcontact printing using PDMS stamps to pattern fibronectin shapes on soft PA gels. (D) Cell monolayers display stem-like markers at the periphery and corners of these shapes. Reproduced from88 with permission from Nature Publishing Group. (E) Cells undergoing confined migration within arrays of microscale PDMS pillars, with a glass ceiling. (F) Cells transition from collective to individual migration after EMT. Reproduced from89 with permission from Nature Publishing Group.
Cancer stem-like cells and multicellular geometries
Lee et al. characterized the expression of stem-like biomarkers within multicellular islands with varying geometric shape.88 Soft lithography was used to transfer ECM proteins in specific geometric patterns onto hydrazine-modified PA hydrogels (Figure 3C). A murine melanoma cell line was cultured on these different shapes and immunostained for putative cancer stem-like cell markers (e.g. CD271, CD133, and ABCB5), as well as molecular markers of pluripotency and tumor initiation (e.g. Nestin, Nanog, Jarid1b, Oct4, Sox2, and Nanog). Strikingly, expression of these biomarkers occurred at the periphery, particularly at sharp corners and convex features with geometrically enhanced stress (Figure 3D). In order to maximize the number of cells located at an edge and minimize the number of cells at the interior, a narrow spiral shape was patterned. Gene expression profiling of cells on spirals relative to uniform PA gels revealed that stem-like phenotypes were activated by integrin α5β1, MAPK and STAT signaling.
A differentiated phenotype could be rescued by chemical inhibition of MAPK signaling (particularly through p38 and ERK), blocking integrin α5β1, as well as inhibition of JNK. These stem-like cells were also observed at the periphery when multicellular aggregates were cultured in three-dimensional geometries, including PA microwells, as well as encapsulation within PEG hydrogels. Subsequent wound-healing and Transwell assays confirmed the increased invasiveness of cells cultured on curved patterns. Moreover, stem-like cells from spirals displayed increased metastatic potential after tail vein injection into a murine model, with lung metastases and poor survival. Qualitatively similar expression of stem-like markers (e.g. CD133, CD144) was observed for various other murine and human cell lines. Overall, this work reveals that the geometry of a curved edge can apply mechanical stimuli that affect cell polarization and activate a stem-like phenotype, consistent with previous work on wound healing assays.83 It should be noted cells also experience asymmetric cell-cell interactions, which are strengthened towards the interior. Future work could examine the crosstalk between cell-cell and cell-matrix interactions in activating a stem-like phenotype and EMT.
Transitions between collective and individual migration in confined micropillar arrays
Wong et al. measured collective and individual migration phenotypes after EMT in confined micropillar arrays.89 PDMS devices were fabricated with soft lithography, bonded to glass cover slips and coated with fibronectin. The resulting confined geometries consisted of a square array of micropillars with 10 µm pillar-to-pillar separation, and 10 µm height, effectively restricting cells to single file migration with elongated phenotypes (Figure 3E). Remarkably, mammary epithelial cells that had undergone a controlled EMT through Snail induction migrated through the pillars led by individual cells, which broke away from a collectively advancing front (Figure 3F). Time-lapse fluorescence microscopy and automated tracking of cell nuclei revealed two distinct subpopulations with distinct migratory behaviors. In particular, individually migrating cells traveled with faster and straighter trajectories relative to collectively migrating cells, which can be classified using a Gaussian mixture model. This reversion of induced mesenchymal cells to an epithelial phenotype is likely to occur due to differences in cell-cell contact within the micropillar array relative to the flat loading region behind it. This scenario has a physical analogy with the solidification of an undercooled binary mixture,93 suggesting a competition of phenotypic inter-conversion and “sorting out” at the interfacial front.94 One limiting case of this model occurs when the two species are effectively insoluble, and sort out with minimal interconversion, which is well known from classical “differential adhesion” experiments by Steinberg and others.95 Indeed, differences in effective cell “surface tension” were proposed to arise from differences in cadherin expression, which is qualitatively consistent with EMT. More generally, these microfabrication techniques permit exquisite control of geometric features in order to define cell-cell and cell-matrix interactions. When combined with single cell tracking techniques, this approach enables the identification of rare cells and exceptional phenotypes that are associated with EMT.
Spreading Out: 3D Multicellular Spheroids on Planar Surfaces
Multicellular spheroids can be prepared through the aggregation of single cells under low matrix adhesion conditions.96 For instance, dispersed cells in solution can be continuously agitated,97 sedimented onto low adhesion surfaces,98 or confined within an air-liquid interface (hanging drops).99 Essentially, these various approaches enhance cell-cell adhesion while minimizing cell-matrix adhesion. The formation of multicellular spheroids by aggregation represents a facile approach for preparing tissue-scale constructs consisting of hundreds to thousands of cells. Moreover, multicellular spheroids recapitulate many of the hallmarks of cancer observed in vivo, including heterogeneity, necrotic microregions, and differential responses to therapeutic treatment.100
Remarkably, the aggregation of multiple dispersed cell types can display self-organizing behaviors analogous to tissue development.94 Early work by Steinberg showed that mixtures of embryonic cells appeared to “sort out” based on differences in cell-cell adhesion, which he termed the “differential adhesion hypothesis.”101 In particular, Steinberg proposed that cell sorting could be explained as the demixing of two liquids with different surface tensions. For instance, if two cell types had stronger homotypic adhesions than heterotypic adhesion, they were likely to actively self-segregate into subpopulations of like type. Instead, if these cell types had heterotypic adhesions that were comparable or stronger than their homotypic adhesions, they would remain randomly dispersed. Finally, if the first cell type displayed stronger homotypic adhesions than the second, the first cell type would be segregated into the interior while the second type would be segregated to the periphery. Interestingly, these adhesions are dependent (in part) on cadherin expression, so that cells with high cadherin expression would be expected to sorted to the interior while cells with low cadherin expression would be sorted to the exterior.102 This is qualitatively consistent with some tumor architectures, particularly since EMT at the periphery results in decreased cadherin expression.36 Nevertheless, Pawlizak et al. have reported that spheroids consisting of a mixture of epithelial and mesenchymal cell lines under low adhesion conditions do not self-sort according to cadherin expression.103 In contrast, Carey et al report that mixed epithelial and mesenchymal spheroids embedded in collagen I display outward invasion led by mesenchymal cells,104 suggesting that sorting emerges as a competition of cell-cell and cell-matrix interactions.105 These physical analogies are intriguing since they may facilitate quantitative predictions of tumor progression. Nevertheless, biological systems are thermodynamically far-from-equilibrium, which must be carefully addressed in these physical models. A natural extension of this physical analogy is to consider how droplets (tumor spheroids) interact with solid surfaces.
Disorganizing Spheroids as a Droplet Wetting Transition
Douezan et al. measured the disorganization of multicellular spheroids on flat substrates with varying stiffness.106 Murine sarcoma cells were prepared as 3D aggregates under agitation and low-adhesion conditions, then deposited onto fibronectin-coated substrates with varying stiffness. Quantitatively, a spreading parameter can be defined S = WCS – WCC, where WCS and WCC represent the cell-surface and cell-cell adhesion energies, respectively. If S < 0, then the cell-cell adhesion is stronger than the cell-surface adhesion (i.e. WCS < WCC), so that the cellular aggregate does not spread (e.g. partial wetting) (Figure 4Ai). Instead, when 0 < S, then the cell-surface adhesion is stronger than the cell-cell adhesion (i.e. WCC < WCS), and the aggregate spreads out as a monolayer, either cohesively as a “liquid” or dispersing as a “gas” (Figure 4Aii). In particular, on very soft substrates (< 10 kPa), aggregates remained intact without any cell dispersal (Figure 4Bi). For substrates of intermediate stiffness (~10 kPa), aggregates partially flattened into a spherical cap surrounded by dispersed single cells (Figure 4Bii). Finally, for relatively stiff glass or PDMS substrates (~ MPa-GPa), aggregates spread cohesively as a continuous monolayer (Figure 4Biii). This scenario has physical analogies with a liquid droplet on a solid surface, which will stabilize with a characteristic contact angle dependent on the relative interfacial energies.107
Fig. 4.
Multicellular spheroids spreading on planar surfaces. A. Partial or complete wetting is governed by the relative cell-surface and cell-cell adhesions. B. Substrate stiffness governs the solid, liquid, or gas-like dispersion of cells. Reproduced from106 with permission from the Royal Society of Chemistry.
The spreading dynamics of the aggregate could be explained by an effective “friction coefficient” for the “slippage” of the cell monolayer on the substrate.106 This friction coefficient was negligible for very soft substrates, where cells do not adhere. Instead, the friction coefficient was maximum for intermediate substrate stiffness (Ec ~ 8 kPa),. Finally, for stiffer substrates, the friction coefficient asymptotically saturated to smaler, constant value. This biphasic dependence of cell adhesion on substrate stiffness is qualitatively consistent with a motor-clutch model.108 Experimentally, as the substrate stiffness approaches EC, the cell motility is observed to increase, enhancing spreading. Moreover, the cell-cell adhesion also weakens below EC, resulting in individual cell dissemination. Further validation of this model occurred by varying substrate functionalization with fibronectin, effectively varying cell-substrate adhesion WCS.109 Moreover, mixtures of cells with varying E-cadherin expression were used to vary average cell-cell adhesion WCC, resulting in a transition from collective to individual migration.109 Overall, this proposed wetting model has analogies with the binary solidification model previously discussed,89 as well as jamming-unjamming transitions.110 The application of theoretical concepts inspired by soft matter physics may enable new fundamental insights into tumor invasion and EMT, but must be carefully applied in the context of biological signaling and adaptation.111
Multicellular Aggregates Interact through Collagen Fibers
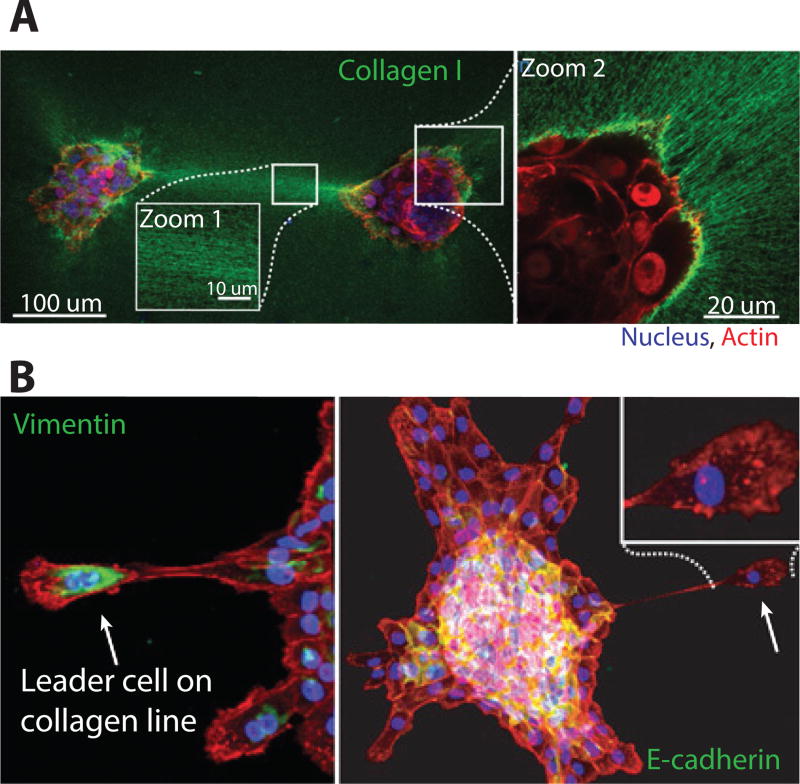
Shi et al. analyzed the disorganization and dissemination of mammary epithelial acini on collagen I surfaces (Figure 5A).112 Epithelial cells were cultured as acini in a rBM overlay assay for 8 days, then transferred onto a collagen I substrate. Over 40 h, the acini disorganized into collective and individually migrating cells, which traveled along the substrate. Motile cells at the periphery displayed increased vimentin, with decreased E-cadherin and β-cateinin, consistent with EMT (Figure 5B). Moreover, these cells displayed nuclear localization of the YAP/TAZ mechanotransducer, indicative of mechanical tension on their cytoskeleton. In contrast, cells that remained localized within the acini retained elevated E-cadherin and β-catenin, with minimal vimentin, indicative of an epithelial phenotype. These behaviors were attributed to the mechanical tension acting on the acini, so that tensile stress above some critical threshold resulted in an EMT-like transition to malignant phenotype. A non-invasive epithelial phenotype could be rescued by a rectangular cut around the acini that released the mechanical tension from its surroundings.
Fig. 5.
Multicellular aggregates spreading on collagen I substrates. A. Long-range mechanical interactions occur through collagen I bundling and alignment B. Dissemination of leader cells with vimentin expression and polarized morphologies. Reproduced from112 with permission from the National Academy of Sciences.
Shi et al. further utilized a photoactivatable collagen binding protein to visualize local matrix remodeling and large scale deformation. Remarkably, each acini displayed aligned collagen bundles which emanated radially outward. An analysis of acini pairs interacting along a collagen line revealed that they disorganized rapidly, sending cells in the direction of the other, over length scales of millimeters. Guo et al. observed qualitatively consistent interactions using a different biomaterial architecture based on a rBM substrate with a collagen I overlay.113 They showed that initial cell movements were necessary to locally bundle and align collagen fibers, which in turn directed cell migration along this path. Over longer time periods, multicellular strands migrated outwards along these collagen fibers before condensing into elongated tubules. In the work of both Shi et al. and Guo et al., the formation of aligned and bundled collagen permits relatively long range force transmission through the matrix (> 500 µm). Accounting for these nonlinear and discrete fiber effects in ECM will likely require revised constitutive equations coupled with careful experimental validation.114
Enter the Matrix: Epithelial Morphogenesis and Dissemination in 3D Biomaterials
Multicellular invasion is impeded within fully 3D biomaterial environments relative to 2D substrates and microfabricated geometries. In particular, the matrix architecture can present a wide variety of topographies, rigidity and adhesiveness, and can spatially confine cells.12 In response, tumor cells can undergo significant morphological changes as well as remodel the local matrix architecture.34 Although there is increasing use of natural hydrogels such as rBM and collagen I to investigate tumor invasion and EMT, there remain many fundamental aspects of cell-matrix interactions that remain poorly understood. The development of synthetic hydrogel materials with improved control over mechanical properties, biochemistry and degradation serves to complement these pioneering studies.
Transitions to Invasion in Collagen I Gels
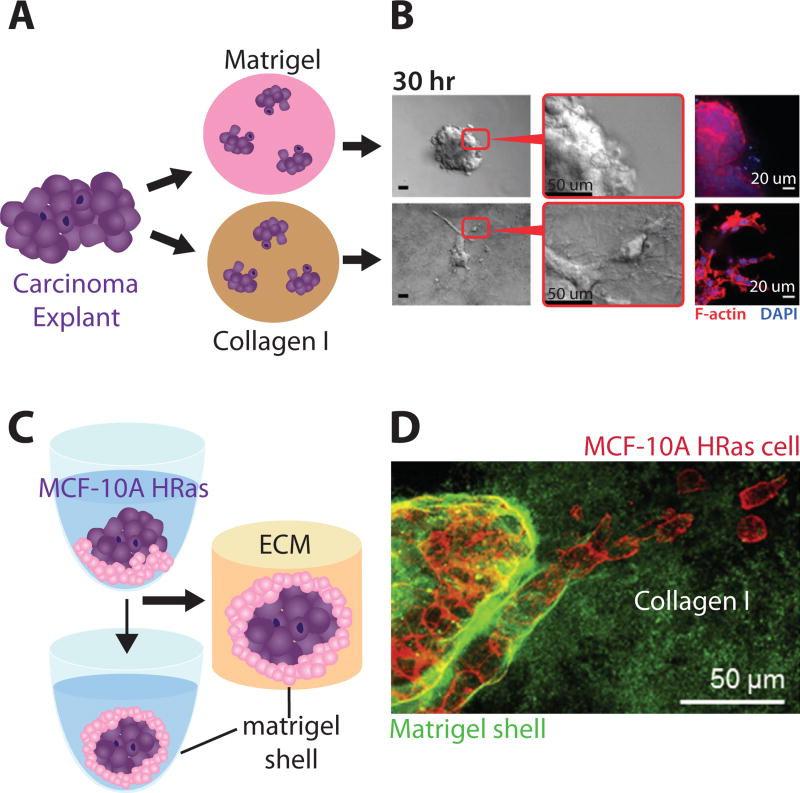
Nguyen-Ngoc et al. directly compared cell dissemination from epithelial tissue explants in rBM and collagen I (Figure 6A).115 Primary human or mouse breast tumors were mechanically dissociated into explants consisting of hundreds of cells, and then embedded in 3D matrix. Explants in rBM remained localized with a rounded tissue morphology (Figure 6B). Instead, explants in collagen I exhibited protrusions at the periphery with extensive local dissemination as mesenchymal, amoeboid and collective phenotypes (Figure 6B). Subsequently, explants were removed from 3D matrix and then re-embedded in either rBM or collagen I. Remarkably, the subsequent behavior of these explants was primarily determined by the new matrix conditions. For instance, rounded explants in rBM displayed local dissemination when transferred to collagen I, and vice versa. In contrast, explants from normal mammary tissue displayed transient dissemination in collagen I, reverting back to branching morphogenesis due to the deposition of basement membrane. Subsequent work from this group showed that these migration behaviors I depended sensitively on the pre-incubation time of collagen I at 4° C, which prepolymerized collagen fibrils and varied the matrix architecture.116 Overall, these experiments represent a highly promising approach to translate natural ECM as a preclinical assay to characterize cell motility from human patient samples.
Fig. 6.
Epithelial morphogenesis and invasion in natural 3D hydrogels. (A, B) Mammary epithelial explants remain localized in rBM but invade in collagen I. Reproduced from115 with permission from the National Academy of Sciences. (C) Multicellular spheroids can be encapsulated within a rBM shell, then embeded in collagen I. (D) Mammary epithelial cells display reduced invasion with a rBM shell. Reproduced from117 with permission from Elsevier.
Ahmadzadeh et al. computationally and experimentally investigated how spheroids assembled from a melanoma cell line invaded through collagen I matrix of varying initial concentration.118 Their mechanochemical model incorporated a feedback between cell contractility (i.e. through Rho/ROCK) and collagen fiber alignment. A major prediction of this model was that a critical stiffness existed, below which the spheroid was dominated by cell-cell adhesions and remained localized, and above which cell-matrix adhesion would dominate, permitting cells to break away from the spheroid. Furthermore, above a certain stiffness (effectively, collagen I concentration), cell invasion was impeded by fibril density, independent of MMP degradation. Their results show impressive agreement between theory and experiment, particularly based on a continuum model that did not explicitly account for cellular heterogeneity. An interesting future direction could be to incorporate these cellular and molecular details through multiscale modeling.
Carey et al. investigated the morphogeneis of single mammary epithelial cells embeded in mixtures of rBM and collagen.119 Multicellular clusters displayed were primarily acini-like in rBM, with increasing fractions of protrusions and invasive morphologies with increasing collagen I, consistent with Nguyen-Ngoc’s resuls.115 Moreover, gene expression profiling of cells in collagen I relative to rBM indicated a characteristic EMT signature, including loss of E-cadherin, with a gain of Snail, vimentin, fibronectin and MT-MMP1. An epithelial phenotype could be rescued in collagen I by chemical inhibition or knockdown of MT-MMP1, as well as inhibitors of PI3K and Rac1. A controlled stiffening of collagen I by non-enzymatic glycation revealed that increasing matrix stiffness also promoted an invasive phenotype.
Finally, Guzman et al. demonstrated an innovative technique to self-assemble tumor spheroids with a rBM shell in a collagen I matrix.117 First, dispersed mammary epithelial cells and diluted rBM were seeded in a concave, low-adhesion well and centrifuged, causing them to self-assemble into a tumor spheroid with a rBM coating after 24 h (Figure 6C). Tumor spheroids were then embedded into collagen I. The thickness of the rBM shell could be tuned by its diluted concentration in precursor solution, which was able to contain non-transformed mammary epithelial (MCF-10A) cells. Nevertheless, transformed MCF-10A Ras cells were capable of breaching the basement membrane shell using collective and individual invasion phenotypes. Moreover, MCF-10A Ras spheroids with a rBM shell displayed a significant reduction in invasion after to MMP inhibition, relative to shell-free spheroids embedded in collagen I (Figure 6D). However, some multicellular streaming was observed, in contrast to complete inhibition of migration for shell-free spheroids embedded in a mixture of rBM and collagen I. Thus, tumors with rBM shells display some features of tumor pathophysiology that may not be recapitulated by matrix consisting of mixed rBM and collagen I.
Epithelial Morphogenesis in PEG Hydrogels
Gill et al. investigated the formation of acini from lung epithelial cells cultured in poly(ethylene glycol) (PEG).120 PEG is a synthetic, hydrophilic polymer with non-fouling properties that renders it relatively “inert.”121 The matrix backbone incorporated a proteolytically degradable PQ peptide flanked by PEG chains on either side. In addition, PEG chains were modified with a fibronectin-derived RGDS peptide to allow cell adhesion. By varying the composition of PEG-PQ and PEG-RGDS, respectively, the stiffness and biochemistry of the hydrogel could be independently tuned. Murine lung adenocarcinoma cells were encapsulated within photopolymerized PEG hydrogels, eventually forming epithelial acini with hollow lumens over 2 weeks. As hydrogel stiffness or ligand density was increased, the average acini diameter decreased due to reduced proliferation. Instead, a higher percentage of acini formed lumens due to increased apoptosis. Lumenized acini were then treated with TGF-β to induce EMT, which resulted in a loss of polarized organization and lumens, with random proliferative and apoptotic events. Molecular analysis using RT-PCR revealed a consistent downregulation of miR-200, resulting in a decrease in classical epithelial gene expression (CDH1, CRB3), and an increase in mesenchymal genes (CDH2, VIM, ZEB1) for varying matrix stiffness and ligand density. It should be noted that TGF-β treatment did not result in MMP secretion, which may have impeded cell outgrowth and dissemination.
Enemchukwu also investigated the organization of epithelial acini in PEG hydrogels with tunable ligand density, mechanics and proteolytic degradation.122 Their approach utilizd a four-arm PEG macromer with maleimide groups at each terminus, which permitted better defined microstructure, ligand stoichiometry and cross-linking efficiency. Kidney epithelial cells (MDCK II) were encapsulated within PEG hydrogels of varying concentration, revealing that the formation of lumenized acini with apicobasal polarity occurred within a relatively narrow window of stiffness ( E ~ 4 kPa), RGD ligand density (~250 µM), and degradable crosslinker density. Acini typically displayed defects in polarization below this window and failed to form above this window. Overall, this modular and well-controlled hydrogel represents a minimal system that can recapitulate many features of epithelial morphogenesis.
EMT driven by Substrate Stiffness through TWIST1-G3BP2
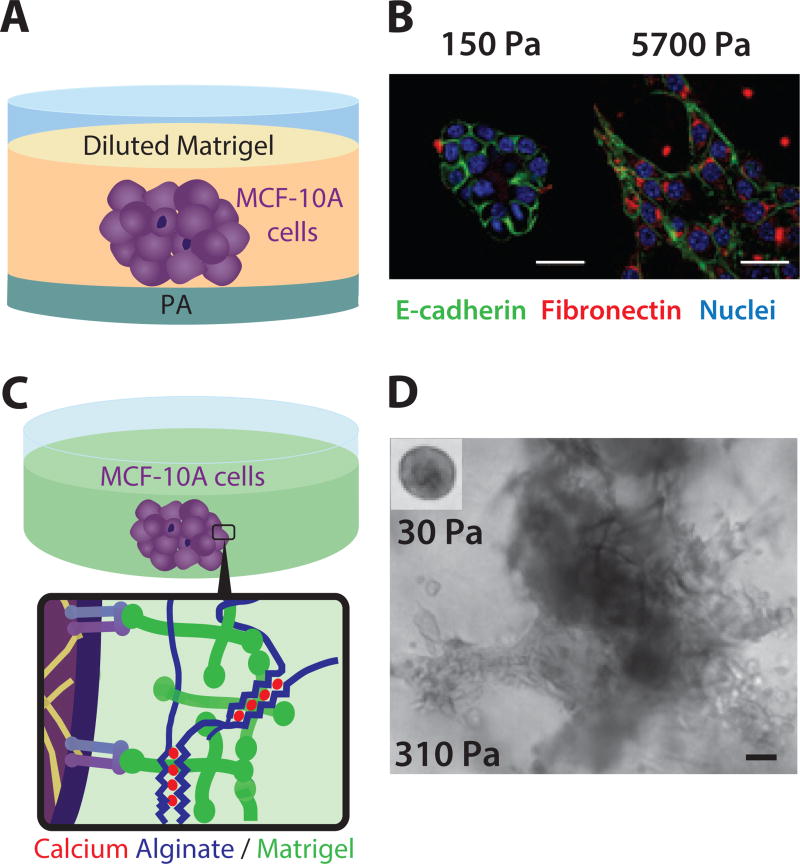
Wei et al. investigated the role of matrix stiffness on EMT, but utilized a collagen I-functionalized polyacrylamide (PA) gel with an overlay of diluted (2%) rBM.123 (Figure 7A) Non-transformed mammary epithelial cells was observed to form acini on soft PA gels (E ~ 0.15 kPa), while displaying invasive outgrowths and partial EMT on stiffer PA gels (E ~ 5.7 kPa) (Figure 7B). EMT induction by mechanical stiffness was found to require the nuclear localization of transcription factor TWIST1, which was found to be dependent on integrin β1. Further investigation revealed that on softer matrix, TWIST1 is retained in the cytoplasm through its interaction with Ras GTPase binding protein (G3BP2). Knockdown of G3BP2 resulted in a downregulation of E-cadherin and disruption of basement membrane, indicative of an EMT, as well as cell invasion. TWIST1-G3BP2 was further demonstrated to promote metastasis in a xenograft model, as well as poor prognosis (with increasing collagen organization) in breast cancer patients. Altogether, Wei et al. show that stiffer matrix causes TWIST1 to be released from its cytoplasmic anchor G3BP2, resulting in nuclear localization and EMT. Interestingly, cells appear to disseminate collectively on stiffer substrates, which is suggestive of a partial EMT phenotype. The lack of individual scattering may occur due to the uniform topography of the PA gel.
Fig. 7.
Epithelial morphogenesis and invasion in synthetic 3D hydrogels. (A) Multicellular clusters on PA substrates with variable stiffness, overlaid with rBM. (B) Stiffness-dependent epithelial disorganization and EMT. Reproduced from123 with permission from Nature Publishing Group. (C) Multicellular clusters embedded within tunable alginate-rBM networks. (D) Stiffness-dependent epithelial proliferation, disorganization and invasion. Reproduced from124 with permission from Nature Publishing Group.
Malignant Transitions driven by ECM through Integrin β4
Chaudhuri et al. utilized an interpenetrating network of alginate and reconstituted basement membrane to investigate the role of matrix stiffness in epithelial invasion.124 Alginate is a flexible polysaccharide derived from seaweed, which does not display mammalian cell adhesive ligands.125 Alginate can incorporate blocks of sequential guluronic acid resides (‘G blocks’) that can be crosslinked with divalent cations (Ca2+). Remarkably, these ‘G blocks’ form ‘egg box’ conformations that permit occupancy (binding) by varying numbers of divalent cations (Figure 7C). As a consequence, the crosslinker strength and mechanical stiffness of alginate hydrogels could be tuned by cation concentration, without affecting the corresponding pore size or hydrogel microstructure. Non-transformed mammary epithelial cells (MCF-10A) cultured in relatively soft matrix (E ~ 0.1 kPa) organized into growth-arrested acini, consistent with previously reported results with rBM.59 In contast, this same cell type in stiffer matrix (E ~ 1 kPa) formed larger multicellular clusters with invasive outgrowths (Figure 7D), even though ligand density and pore size were consistent with soft matrix. This mechanotransduction and malignant invasion in stiffer matrix was found to occur through signaling of integrin β4 through PI3K and Rac1 activation. Nevertheless, a growth-arrested acini phenotype could be rescued at stiff matrix by increasing basement membrane density. To explain this stiffness and composition dependent response, Chaudhuri et al. proposed that integrin β4 bound to laminin on soft ECM can undergo relatively large lateral fluctuations along the cell membrane, allowing them to form hemidesmosomes. However, when integrin β4 is bound to laminin at comparable density on stiff IPN, they are more constrained, reducing integrin β4 clustering and the formation of hemidesmosomes. As a consequence, unsequestered integrin β4 is free to drive downstream activation of PI3K and Rac1, yielding the malignant phenotype observed in stiff matrix. Finally, increasing the laminin density will increase the corresponding integrin β4 density on the cell membrane, making it more likely that they can associate and form hemidesmosomes. Overall, Chaudhuri et al. demonstrated an elegant biomaterial system for orthogonal control of ligand density, microstructure, and bulk rheology, which will likely have wide applications for mechanobiology. Moreover, this approach enables unique insights into the integrated transduction of chemical and mechanical signals by epithelial cells, particularly the role of integrin β4. It should be noted that the alginate used in this study displays limited biodegradation, which may hinder cell invasion. Future work could potentially address this through the addition of MMP-cleavable groups.126
Conspiring Across Borders: Co-Culture of Tumor and Stroma
The tumor microenvironment in vivo consists not only of aberrant ECM, but also a wide range of stromal cells.4 For instance, tumor cells can interact with cancer-associated fibroblasts, macrophages, and other immune cells which drive tumor-promoting inflammation.29 A first step towards recapitulating these heterotypic interactions is to co-culture tumor cells with other cell types. For instance, early work by Ronnov-Jessen et al. showed that co-culture of tumor cells with fibroblasts in 3D culture resulted in a transition towards an invasive phenotype.127 Modern biomaterials permit more compartmentalized tumor-like architectures that replicate human pathophysiology, as well as improved imaging of multicellular interactions.
Cancer Associated Fibroblasts lead Tumor Invasion
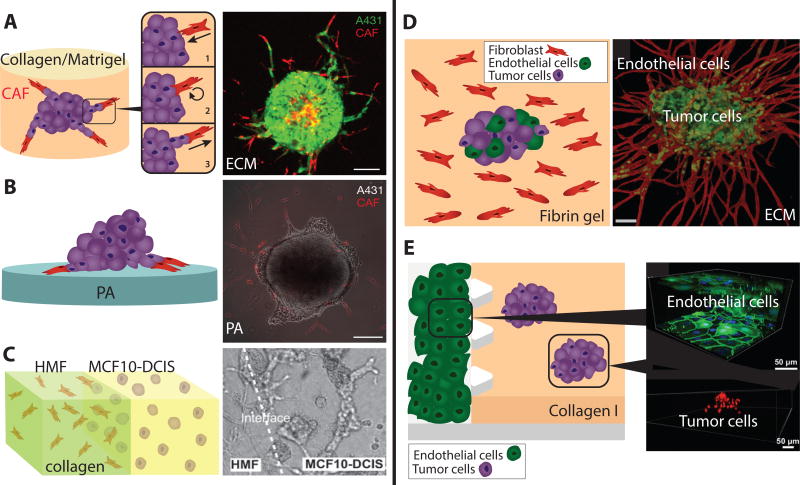
Labernadie et al. investigated the role of fibroblasts in tumor invasion based on spheroid dissemination onto both 3D matrix and 2D substrates.128 Multicellular spheroids consisting of vulval squamous cell carcinoma (SCC) cells (A431) were embedded in a 3D mixture of collagen and rBM, which also included cancer associated fibroblasts. Remarkably, fibroblasts were observed to approach tumors from afar and attach to tumor cells through heterophilic E-cadherin/N-cadherin junctions (Figure 8A). Subsequently, these fibroblasts reversed directions and led multicellular tumor invasion into the matrix. These leader and follower behaviors also occurred when the spheroids were plated on a soft 2D substrate and surrounded by fibroblasts (Figure 8B). In this latter system, traction force microscopy was used to show that fibroblast leaders exert pulling forces on cancer cell followers through these heterophilic cell-cell junctions. Further knockout of E-cadherin disrupted cell-cell junctions, preventing tumor cells from migrating collectively behind fibroblast leaders. It should be noted that these tumor cells were not defective at migration, but were simply unable to function as collective followers. Finally, these heterophilic junctions were observed for patient-derived cells cultured in fully 3D matrix. Overall, co-culture of tumor cells and fibroblasts revealed a cooperative mechanism whereby fibroblasts lead tumor cells into the matrix, while tumor cells sustain fibroblast polarization. It is also remarkable that heterophilic E-cadherin/N-cadherin junctions are mechanically active and comparable in activity to homophilic junctions. These new results must be considered in the physical theories that incorporate differential-adhesion like mechanisms.95
Fig. 8.
Co-culture models of tumor cells and stromal cells. (A). Fibroblasts are recruited to tumor spheroids and then lead collective invasion in 3D collagen-rBM hydrogels. (B) Fibroblasts display similar leader cell behavior on soft 2D PA substrates. Reproduced from128 with permission from Nature Publishing Group. (C) Microfluidic patterning of fibroblasts and tumor cells into adjacent compartments. Reproduced from129 with permission from the Royal Society of Chemistry. (D) Endothelial cells sprout into microvascular networks in fibrin gels, which permit intravasation-like behaviors by tumor cells. Reproduced from130 with permission from the Royal Society of Chemistry. (E) Targeted EMT inhibitors can be screened in microfluidic devices with tumor spheroids and endothelial barriers. Reproduced from131 with permission from the Royal Society of Chemistry.
Sung et al. utilized a microfluidic device to pattern tumor cells and fibroblasts in adjacent compartments, revealing that soluble signals could trigger morphological changes, but that cell-cell contact is necessary for invasion (Figure 8C).129,132 Singh et al. encapsulated melanoma cells within a PEG hydrogel core, which was then enclosed with primary human dermal fibroblasts in a collagen I hydrogel. They found that co-culture suppressed epithelial cluster growth, but subsequently activated a program of collective or individual invasion.133 Overall, the patterning of tumor and stromal cells into discrete compartments represents an important step towards reverse engineering the complex tumor architecture.
Tumor Intravasation and Extravasation in Endothelial Vessels
Ehsan et al. investigated the entry and migration of tumor cells within endothelial vessel networks in a 3D fibrin gel.130 Fibrin is a natural polymer that arises from a cascade of enzymatic reactions during blood clotting.134 Multicellular spheroids consisting of human tumor cells mixed with primary human endothelial cells were seeded in fibrin and surrounded by fibroblasts. Endothelial cells rapidly sprouted from these spheroids and formed highly branched networks within the gel over the course of a week (Figure 8D). Moreover, endothelial cells reorganized at the periphery of the spheroid, infiltrating into the spheroid in order to form an internally connected network. At the same time, individual tumor cells broke away from the spheroid and infiltrated the fibrin gel. Remarkably, a colon cancer cell line was observed to migrate along the lumens within the sprouting vascular network. This intravasation-like behavior was enhanced under hypoxic conditions, which was found to depend on the EMT transcription factor Slug. This complex intermixing of tumor cells and endothelial cells may have unexpected implications for abnormal angiogenesis in more advanced tumors in vivo.
Aref et al. perturbed multicellular invasion using targeted EMT inhibitors in a microfluidic 3D culture system,131 utilizing a compartmentalized microfluidic device where vertical endothelial monolayer could be cultured adjacent to a 3D hydrogel135 Aref et al. utilized lung adenocarcinoma (A549) cells with some propensity to revert from a partial mesenchymal phenotype to a more epithelial phenotype after inhibition of EMT-related pathways. Cells were self-assembled into 40–70 µm spheroids in a low-attachment dish, then seeded in a collagen matrix within the microfluidic device (Figure 8E). Interestingly, spheroids dispersed due to the presence of endothelial cells, but not in culture media supplemented with growth factors (FGF, EGF). Subsequent treatment with targeted inhibitors of EMT pathways (e.g. AkT, EGFR, IGF1, MEK, PDGFR< SRC and TGF-βR1) resulted in decreased proliferation and migration. Unexpectedly, the effective IC50 observed to inhibit migration in these 3D hydrogels was found to be roughly 5 to 10-fold lower than the corresponding IC50 needed to inhibit spreading of these spheroids onto a 2D substrate. The effective concentrations in 3D culture were reported to be comparable to plasma concentration that were effective in clinical trials. Overall, microfluidic devices may enable controlled application of molecular gradients, forces, and flows, which could recapitulate essential pathophysiological features.136 These so-called “organ-on-a-chip” microphysiological systems have great potential to complement existing animal models as predictive models of drug efficacy and toxicity.137 Nevertheless, careful consideration of scaling will be necessary to compare drug pharmacokinetics with animal models or human patients.138
Future Directions
What is the Matrix? Beyond Stiffness and Mesh Size
Synthetic and natural biomaterials have typically been characterized in terms of bulk mechanical properties, such as elastic modulus (E) or shear modulus (G).139 In the publications highlighted here, an overall trend is that epithelial cells cultured on stiffer biomaterials exhibit aberrant epithelial morphogenesis,120,122 enhanced invasiveness,124 and EMT induction.123 This transition to malignant behavior is qualitatively consistent with tumor progression in vivo, where ECM becomes dramatically stiffened with highly crosslinked and bundled collagen I.30 It should be noted that synthetic hydrogels such as PA or PEG can be understood as flexible polymer networks, which can be engineered with a controlled stiffness and well-defined linear response.140 Instead, natural hydrogels and physiological ECM exhibit additional physical complexity that is not captured solely by elastic modulus. In particular, natural hydrogels can be more fibrous with highly nonlinear rheology, requiring more sophisticated physical descriptions based on semiflexible or rigid polymers.141 Reconstituted collagen I or fibrin networks can display significant strain stiffening as a generic consequence of nonlinear force extension in semiflexible polymers.142 This nonlinear behavior has important biological implications, since cells or tissues have been observed to mechanically interact over much longer distances than they would in a linear elastic material.112,113 Moreover, physiological ECM can exhibit significant viscoelasticity. For instance, Chaudhuri et al. also showed that mesenchymal stem cells display faster cell spreading, proliferation, and osteogenic differentiation in hydrogels with faster stress relaxation times, even when the initial elastic modulus was held constant.143 Further work is needed to characterize and fundamentally understand the role of nonlinear ECM rheology in tumor invasion and EMT.
Cancer cell invasion is also restricted by the characteristic mesh size of the hydrogel, with a limit of ~7 µm2 imposed by nuclear deformability.50 For synthetic hydrogels, the mesh size of flexible polymers is typically quite small, on the order of 1–100 nm,139 requiring significant matrix degradation to facilitate cell migration. In contrast, natural hydrogels can exhibit larger mesh sizes on the order of microns, which may be more permissive for cell migration. Conceptually, natural hydrogels and pathological ECM may behave more as a discrete network of fibers rather than a continous porous architecture. Indeed, microstructural architectures may shape cells through contact-guidance like mechanisms, complementing the role of ligand density and bulk mechanical stiffness.6,12 Thus, some care is necessary when utilizing continuum metrics such as an elastic modulus, or average properties such as a mesh size. Instead, more systematic characterization of network geometry144 and pore size distribution145 may yield new physical insights. Indeed, an intriguing approach is to quantify the connectivity of a given matrix architecture (i.e. a percolation threshold for a nucleus or cell-sized object),146 and compare this with actual migration trajectories. These characterization tools may be further combined with recent advances in 3D traction force microscopy to visualize localized cell-generated forces.147–149 Overall, understanding the interplay between invasion, matrix architecture and mechanical properties will also be facilitated by improved materials characterization techniques at cellular and subcellular length scales.
Follow That Cell: Tracking Phenotypic Heterogeneity
Cells that disseminate from the tumor periphery represent uncommon phenotypes, which are likely to differ from the cells that remain within the tumor. One advantage of engineered biomaterial platforms is their experimental accessibility relative to animal models, which is particularly useful for optical microscopy. In the publications highlighted here, fluorescence microscopy was utilized to reveal cell migration at the periphery,87,89,109,115,117,118,150 distinct biomarker expression,88 disorganization of epithelial architecture,112,113,119,120,122–124 as well as coordination between tumor and stromal cells.128–131,133 However, many of these publications utilized manual analysis of selected cells or tissues, which samples a limited subset of the population with low throughput. Instead, computer vision and machine learning represent a powerful approach for phenotypic profiling.151 In general, fluorescently labeled cells must be individually identified and distinguished from the image background. Next, cell shape, texture, and biomarker expression can be analyzed as a set of quantitative features. Finally, the most biologically relevant parameters are determined based on feature selection or dimensionality reduction. Single cell features can then be profiled through unsupervised approaches that cluster distinct phenotypic subpopulations, or supervised approaches for classification based on linear or nonlinear decision boundaries. We have recently demonstrated automated profiling of EMT in 2D monolayer culture based on single cell morphology and biomarker expression,152 as well as cell morphology on nanotextured materials.153 It should be noted that even with automated image analysis, migratory cells may be detected in limited numbers, which are insufficient to define a phenotypic cluster. Unsupervised outlier detection may be useful to profile cells which are highly dissimilar from the others, but are biologically significant. Moreover, new algorithms (i.e. SPADE) can downsample abundant cells in high-dimensional feature space in order to facilitate the clustering of rarer cells.154
Continuous longitudinal tracking of single cell behaviors represents the next step beyond single cell profiling based on discrete “snapshots.”155 These high-resolution measurements are crucial to capture rare and transient events, such as the detachment of cells from the tumor periphery. Nevertheless, these experiments are associated with significant technical challenges, including maintaining consistent cell-friendly conditions (i.e. temperature, CO2, humidity, and medium osmolality), minimizing phototoxicity and photobleaching, as well as optimization of live cell fluorescent markers and reporters. Moreover, image analysis requires not only accurate cell segmentation (as previously described), but also linking cell positions into continuous trajectories across time-lapse images. Nevertheless, this approach permits unique insights into the cell dissemination. In particular, we have shown that distinct migration phenotypes can be classified based on quantitative metrics such as the speed and straightness of cell migration trajectories, as well as lifetime averaged nearest neighbors.89 Interestingly, the percentage of cells classified as individual was relatively small (~ 16%), and these cells could be further subdivided into subpopulations that were mostly collective with some individual migration, as well as mostly individual with some collective migration. An ongoing challenge is to keep track of cells as they proliferate, which results in increasing cell density with enhanced cell contact or overlap. We and others have tracked cell nuclei, since they remain relatively well dispersed with a relatively well defined shape. Our recent results have utilized these approaches to track the migration and self-organization of heterogeneous mixtures of epithelial and mesenchymal tumor cells.156
Increasing use of fully 3D hydrogel microenvironments will permit increased physiological relevance but will also result in new experimental challenges. First, three-dimensional fluorescence imaging requires the use of spinning disk confocal microscopes, multiphoton microscopy, or other advanced superresolution techniques.157 The imaging of cell and matrix volumes, rather than areas, will result in exponentially larger datasets, which must be stored, visualized and analyzed differently.158 In addition, immunofluorescent staining of cells will be complicated by the slow transport and non-specific adsorption of reagents through the hydrogel, requiring additional experimental optimization. Next, integration of single cell imaging with molecular analyses remains complicated. In particular, measurements of single cell gene expression, epigenetics and protein content are typically destructive and must be conducted at an endpoint. Moreover, cells must be isolated from the biomaterial, which is challenging to achieve with 100% extraction efficiency and loses spatial information. Overall, considerable challenges remain in elucidating multicellular invasion in biomaterials, particularly a mechanistic understanding of how microenvironmental cues are transduced to downstream molecular signaling, resulting in phenotypic decisions.
Scaling Up: Standardization using Multiwell Plates
One advantage of engineered biomaterials is that they can be utilized with multiwell plates, which enables higher throughput screens of experimental conditions in a massively parallel format. In practice, much of the research described here has been implemented in some type of multiwell plate in order to improve optical imaging conditions and limit aqueous solutions from leaking. For instance, rBM or collagen I can be manually dispensed,115 or PDMS devices can be bonded to glass bottom 24-well plates.89 It has also been shown that standardized PA gel substrates can be cast in a 96 well plate format.159 Nevertheless, the transition to increased laboratory automation, including plate handling robotics, high precision fluid dispensing, and automated high-content microscopy remains somewhat limited. Clevers and colleagues have demonstrated that tumor and normal organoids can be derived from colorectal carcinoma patients in a 384 well plate format and screened against a panel of 83 drugs.160 Lutolf and coworkers have developed modular approaches to screen synthetic PEG hydrogels with well-defined mechanical and biochemical properties. For instance, a robotic liquid-dispensing platform was demonstrated that enabled independent control of cell density, hydrogel stiffness, MMP sensitivity, extracellular matrix components, cell-cell interaction components, and soluble factors.161 This screen of 3D culture conditions revealed that physical confinement of induced pluripotent stem cells could enhance reprogramming through an accelerated mesenchymal-epithelial transition and epigenetic modifications.162 As the well size becomes progressively smaller, some consideration of scaling effects will likely be necessary, particularly the depletion of biochemical factors in solution. Overall, the use of standardized biomaterials in an automated format should enable preclinical therapeutic testing, rational drug design, as well as predictive and prognostic assays of human samples.
Biological Questions: EMT and Interpatient Heterogeneity?
Tumors display a variety of collective and individual invasion phenotypes, a phenomenon that remains poorly understood.1 In particular, the role of EMT remains controversial, given the reliance on 2D monolayer culture and animal models in the field.35 Recent reports by Fischer et al. and Zheng et al. using mouse models suggest that EMT is not required for invasion and metastasis, but play a role in therapeutic resistance.163,164 Interestingly, circulating tumor cells from human patients can exhibit a co-expression of epithelial and mesenchymal biomarkers in an intermediate or partial EMT,165 which is not observed for classical EMT in embryonic development.2 The interconversion between epithelial and mesenchymal phenotypes remains similarly controversial, due in part to the analysis at endpoints, as well as the lack of tumor-specific mesenchymal biomarkers. The technologies and future directions discussed here may enable improved experimental control of the ECM, as well as enhanced spatiotemporal resolution to address these ongoing questions. We envision that these approaches are more generally applicable beyond EMT to profile phenotypic heterogeneity and plasticity in tumors, which represents a subtle and complex problem.
Our review of multicellular behaviors in biomaterials is closely related to recent developments in organoid culture, which have been broadly defined. In particular, Shamir and Ewald suggest that: “…in the field of mammary gland biology, the term organoids refers to primary explants of epithelial ducts into 3D extracellular matrix (ECM) gels. Conversely, in studies of intestinal biology, organoids can refer to clonal derivatives of primary epithelial stem cells that are grown without mesenchyme or can refer to epithelial-mesenchymal co-cultures that are derived from embryonic stem cells or induced pluripotent stem cells”166,167 Recent successes have been based on extensive use of rBM or collagen I scaffolds with optimized media formulations, but Gjorevski et al. have demonstrated a tunable and modular PEG-based scaffold for organoid culture.168 Interestingly, normal intestinal stem cells required a soft matrix and laminin-based adhesion, and could be differentiated by gradual softening through hydrolytic degradation. In contrast, stiffer and MMP-degradable matrix resulted in tissue disorganization and the upregulation of stress and inflammatory gene expression, representing a pathological state. These approaches could be used to investigate how patient-derived cells respond to alterations in the microenvironment, as well as to maintain them more effectively within certain phenotypes. Overall, the potential derivation of organoids from human patients could potentially bridge the gap between transformed cancer cell lines and patient-derived xenograft models.13 An exciting prospect is to utilize organoids for biomarker and drug discovery, as well as preclinical screens for personalized therapies.
Conclusions
Natural and artificial biomaterials recapitulate important physical or biochemical features of the ECM, which can be used to elucidate tumor disorganization and dissemination. In this review, we highlight a selection of recent results that address multicellular invasion and EMT within biomimetic microenvironments. First, microfabrication techniques can precisely define interfacial geometries, revealing the detachment and directed migration of individual cells, as well as the emergence of stem-like phenotypes. Second, multicellular aggregates spread on planar surfaces as a balance of cell-cell and cell-matrix adhesions, which has physical analogies with droplet wetting. Moreover, when plated on fibrillar collagen I substrates, these aggregates mechanically interact and reorganize collagen into aligned bundles. Third, epithelial cells embedded in 3D hydrogels can organize into well-defined glandular architectures in reconstituted basement membrane, but undergo a transition to invasion in fibrillar collagen I or stiffer biomaterials. Fourth, co-culture of tumor cells with stromal cells such as fibroblasts or endothelial cells can drive heterotypic soluble or mechanical signaling. Interestingly, fibroblasts can act as leader cells to enhance multicellular invasion, while tumor cells can intravasate into endothelial networks. Finally, we consider future directions for the field, including improved physical understanding of ECM, profiling of single cell heterogeneity, standardization of biomaterials in a multiwell plate format, as well as the use of organoid culture. Altogether, we envision that the technologies reviewed here will facilitate fundamental insights into reciprocal interactions between tumor progression and the ECM, as well as enable patient-specific biomarker discovery and drug testing for precision medicine.
Acknowledgments
We apologize to colleagues whose work could not be included due to space constraints and thank anonymous reviewers for insightful comments. The authors gratefully acknowledge support from NIEHS through the T32 Training Grant in Environmental Pathology (5T32ES007272-25), NIGMS through the COBRE Center for Cancer Research Development at Rhode Island Hospital (1P30GM110759-01A1), a Rhode Island Foundation Medical Research Grant (20144137), as well as Start-Up Funds from Brown University.
Biographies
Susan E. Leggett is a Ph.D. candidate in the Pathobiology Graduate Program at Brown University. She is currently investigating EMT induction in engineered 2D and 3D biomimetic microenvironments. She is a recipient of the Barry Goldwater Scholarship, the Brown University Simper-Ronan Graduate Award in Cancer Research, the Frederic Poole Gorham Predoctoral Fellowship, and is a Predoctoral Trainee on an NIEHS T32 Training Grant in Environmental Pathology. She received her B.S. in Biochemistry (Cum Laude) with a minor in Mathematics from the University of Vermont in 2013.
Amanda S. Khoo is a Ph.D. candidate in the Biomedical Engineering Graduate Program at Brown University. She is currently investigating EMT in microfluidic models of the 3D tumor microenvironment. She received her B.S in Bioengineering from Santa Clara University (Cum Laude) in 2016.
Ian Y. Wong is an Assistant Professor of Engineering and of Medical Science at Brown University. He received an A.B. magna cum laude in Applied Mathematics from Harvard University and a Ph.D. in Materials Science & Engineering from Stanford University. He then completed postdoctoral research at the BioMEMS Resource Center of Massachusetts General Hospital and Harvard Medical School. His research group at Brown engineers interfacial biomaterials and microfluidics to elucidate tumor invasion, drug resistance, and heterogeneity. He also explores unconventional fabrication techniques for self-assembly and patterning of 2D nanomaterials. He has received the NSF Graduate Research Fellowship, the Damon Runyon Cancer Research Fellowship, and the Brown University Pierrepont Award for Outstanding Advising.
References
- 1.Friedl P, Alexander S. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Nieto MA, Huang RY-J, Jackson RA, Thiery JP. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Rørth P. Annu. Rev. Cell Dev. Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 4.Lu P, Weaver VM, Werb Z. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu L, Mooney DJ. Nat. Rev. Mater. 2016;16:56–66. doi: 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasiorowski JZ, Murphy CJ, Nealey PF. Annu. Rev. Biomed. Eng. 2013;15:155–176. doi: 10.1146/annurev-bioeng-071811-150021. [DOI] [PubMed] [Google Scholar]
- 7.Caliari SR, Burdick JA. Nat Meth. 2016;13:405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenburg G, Hay ED. J. Cell Biol. 1982;95:333–339. doi: 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debnath J, Brugge JS. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 12.Charras G, Sahai E. Nat Rev Mol Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 13.Sachs N, Clevers H. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Kleinsmith LJ. Principles of Cancer Biology, Pearson Education Limited. 2013 [Google Scholar]
- 15.Siegel RL, Miller KD, Jemal A. CA: Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 16.Kumar V, Abbas AK, Aster JC. Robbins & Cotran Pathologic Basis of Disease (Robbins Pathology) Saunders; 2014. [Google Scholar]
- 17.Nguyen DX, Bos PD, Massagué J. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 18.Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang K-J, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, Musah S, suke Torisawa Y, van der Meer AD, Villenave R, Yadid M, Parker KK, Ingber DE. Annu Rev Pathol. 2015;10:195–262. doi: 10.1146/annurev-pathol-012414-040418. [DOI] [PubMed] [Google Scholar]
- 19.Massagué J, Obenauf AC. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Høye AM, Erler JT. AJP: Cell Physiology. 2016;310:C955–67. doi: 10.1152/ajpcell.00326.2015. [DOI] [PubMed] [Google Scholar]
- 21.Barney LE, Jansen LE, Polio SR, Galarza S, Lynch ME, Peyton SR. Curr Opin Chem Eng. 2016;11:85–93. doi: 10.1016/j.coche.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeBleu VS, MacDonald B, Kalluri R. Exp. Biol. Med. 2007;232:1121–1129. doi: 10.3181/0703-MR-72. [DOI] [PubMed] [Google Scholar]
- 23.Beck K, Hunter I, Engel J. FASEB J. 1990;4:148–60. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 24.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouw JK, Ou G, Weaver VM. Nat Rev Mol Cell Biol. 2014;15:771–785. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. Mat Biol. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butcher DT, Alliston T, Weaver VM. Nat Rev Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik R, Lelkes PI, Cukierman E. Trends Biotechnol. 2015;33:230–236. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egeblad M, Rasch MG, Weaver VM. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnans C, Chou J, Werb Z. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hynes RO. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 34.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bill R, Christofori G. FEBS Lett. 2015;589:1577–1587. doi: 10.1016/j.febslet.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Brabletz T. Nat Rev Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 37.Müller P, Schier AF. Dev Cell. 2011;21:145–158. doi: 10.1016/j.devcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Lamouille S, Derynck R. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peinado H, Olmeda D, Cano A. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Mani SA, Weinberg RA. Cancer Res. 2006;66:4549–4552. doi: 10.1158/0008-5472.CAN-05-3850. [DOI] [PubMed] [Google Scholar]
- 41.Puisieux A, Brabletz T, Caramel J. Nature Cell Biology. 16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 42.Lamouille S, Xu J, Derynck R. Nature Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilmaz M, Christofori G. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 44.Hodge RG, Ridley AJ. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 45.Koch TM, Münster S, Bonakdar N, Butler JP, Fabry B. PLoS ONE. 2012;7:e33476. doi: 10.1371/journal.pone.0033476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraning-Rush CM, Califano JP, Reinhart-King CA. PLoS ONE. 2012;7:e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. J. Biol. Chem. 2015;290:17145–17153. doi: 10.1074/jbc.R115.640359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrmann RL, Gey GO. J. Natl. Cancer Inst. 1956;16:1375–1403. [PubMed] [Google Scholar]
- 49.Walters BD, Stegemann JP. Acta Biomater. 2014;10:1488–1501. doi: 10.1016/j.actbio.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y-l, Motte S, Kaufman LJ. Biomaterials. 2010;31:5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 52.li Yang Y, Leone LM, Kaufman LJ. Biophys J. 2009;97:2051–2060. doi: 10.1016/j.bpj.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL. Biopolymers. 2011;95:77–93. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grinnell F, Petroll WM. Annu Rev Cell Dev Biol. 2010;26:335–361. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- 55.Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. J Exp Med. 1977;145:204–220. doi: 10.1084/jem.145.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Adv. Drug Deliv. Rev. 2014;79–80:3–18. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Schéele S, Nyström A, Durbeej M, Talts JF, Ekblom M, Ekblom P. J Mol Med. 2007;85:825–836. doi: 10.1007/s00109-007-0182-5. [DOI] [PubMed] [Google Scholar]
- 58.Aumailley M. Cell Adh Migr. 2013;7:48–55. doi: 10.4161/cam.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson CM, Bissell MJ. Semin. Cancer Biol. 2005;15:342–352. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weaver VM, Lelièvre S, Lakins JN, Chrenek MA, Jones JCR, Giancotti F, Werb Z, Bissell MJ. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 63.Boyden S. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Guzman A, Ziperstein MJ, Kaufman LJ. Biomaterials. 2014;35:6954–6963. doi: 10.1016/j.biomaterials.2014.04.086. [DOI] [PubMed] [Google Scholar]
- 66.Pelham RJ, Wang Yl. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kandow CE, Georges PC, Janmey PA, Beningo KA. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 68.Tse JR, Engler AJ. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. Chapter 10, Unit 10.16. [DOI] [PubMed] [Google Scholar]
- 69.Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. Soft Matter. 2014;10:4047–4055. doi: 10.1039/c4sm00264d. [DOI] [PubMed] [Google Scholar]
- 70.Harris AK, Wild P, Stopak D. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 71.Folch A. Introduction to BioMEMS. CRC Press; 2016. [Google Scholar]
- 72.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 73.Wang N, Ostuni E, Whitesides GM, Ingber DE. Cell Motil. Cytoskeleton. 2002;52:97–106. doi: 10.1002/cm.10037. [DOI] [PubMed] [Google Scholar]
- 74.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 75.Paul CD, Hung W-C, Wirtz D, Konstantopoulos K. Annu. Rev. Biomed. Eng. 2016;18:159–180. doi: 10.1146/annurev-bioeng-071114-040654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JN, Jiang X, Ryan D, Whitesides GM. Langmuir. 2004;20:11684–11691. doi: 10.1021/la048562+. [DOI] [PubMed] [Google Scholar]
- 77.Berthier E, Young EWK, Beebe D. Lab on a Chip. 2012;12:1224–1237. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 78.Brown XQ, Ookawa K, Wong JY. Biomaterials. 2005;26:3123–3129. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, Smith ML. Lab Chip. 2007;7:1074–1077. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
- 80.Palchesko RN, Zhang L, Sun Y, Feinberg AW. PLOS ONE. 2012;7:e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashby WJ, Zijlstra A. Integr Biol (Camb) 2012;4:1338–1350. doi: 10.1039/c2ib20154b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayor R, Etienne-Manneville S. Nat Rev Mol Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 83.Gilles C, Polette M, Zahm JM, Tournier JM, Volders L, Foidart JM, Birembaut P. J Cell Sci. 1999;112(Pt 24):4615–4625. doi: 10.1242/jcs.112.24.4615. [DOI] [PubMed] [Google Scholar]
- 84.Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gomez EW, Chen QK, Gjorevski N, Nelson CM. J. Cell. Biochem. 2010;110:44–51. doi: 10.1002/jcb.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boghaert E, Gleghorn JP, Lee K, Gjorevski N, Radisky DC, Nelson CM. Proc Natl Acad Sci USA. 2012;109:19632–19637. doi: 10.1073/pnas.1118872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yevick HG, Duclos G, Bonnet I, Silberzan P. Proc Natl Acad Sci USA. 2015;112:5944–5949. doi: 10.1073/pnas.1418857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J, Abdeen AA, Wycislo KL, Fan TM, Kilian KA. Nature Mat. 2016;15:856–862. doi: 10.1038/nmat4610. [DOI] [PubMed] [Google Scholar]
- 89.Wong IY, Javaid S, Wong EA, Perk S, Haber DA, Toner M, Irimia D. Nature Mat. 2014;13:1063–1071. doi: 10.1038/nmat4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clark P, Connolly P, Curtis AS, Dow JA, Wilkinson CD. Development. 1990;108:635–644. doi: 10.1242/dev.108.4.635. [DOI] [PubMed] [Google Scholar]
- 91.Irimia D, Toner M. Integr Biol (Camb) 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ravasio A, Cheddadi I, Chen T, Pereira T, Ong HT, Bertocchi C, Brugues A, Jacinto A, Kabla AJ, Toyama Y, Trepat X, Gov N, Neves de Almeida L, Ladoux B. Nat Commun. 2015;6:7683. doi: 10.1038/ncomms8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis SH. Theory of Solidification. Cambridge University Press; 2001. [Google Scholar]
- 94.Lancaster MA, Knoblich JA. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 95.Steinberg MS. Curr Opin Genet Dev. 2007;17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 96.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. J. Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 97.Sutherland RM, McCredie JA, Inch WR. J. Natl. Cancer Inst. 1971;46:113–120. [PubMed] [Google Scholar]
- 98.McAllister RM, Reed G, Huebner RJ. J. Natl. Cancer Inst. 1967;39:43–53. [PubMed] [Google Scholar]
- 99.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Biotechnol Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 100.Sutherland RM. Science. 1988;240:177–184. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 101.Steinberg MS. Science. 1963;141:401–408. doi: 10.1126/science.141.3579.401. [DOI] [PubMed] [Google Scholar]
- 102.Steinberg MS, Takeichi M. Proc Natl Acad Sci USA. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pawlizak S, Fritsch AW, Grosser S, Ahrens D, Thalheim T, Riedel S, Kieβling TR, Oswald L, Zink M, Manning ML, Käs JA. New J Phys. 2015;17:1–17. [Google Scholar]
- 104.Carey SP, Starchenko A, McGregor AL, Reinhart-King CA. Clin Exp Metastasis. 2013;30:615–630. doi: 10.1007/s10585-013-9565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cerchiari AE, Garbe JC, Jee NY, Todhunter ME, Broaders KE, Peehl DM, Desai TA, LaBarge MA, Thomson M, Gartner ZJ. Proc Natl Acad Sci USA. 2015;112:2287–2292. doi: 10.1073/pnas.1410776112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Douezan S, Dumond J, Brochard-Wyart F. Soft Matter. 2012;8:4578–4583. [Google Scholar]
- 107.de Gennes PG. Rev. Mod. Phys. 1985;57:827–863. [Google Scholar]
- 108.Bangasser BL, Rosenfeld SS, Odde DJ. Biophys J. 2013;105:581–592. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Douezan S, Guevorkian K, Naouar R, Dufour S, Cuvelier D, Brochard-Wyart F. Proc Natl Acad Sci USA. 2011;108:7315–7320. doi: 10.1073/pnas.1018057108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park J-A, Atia L, Mitchel JA, Fredberg JJ, Butler JP. J Cell Sci. 2016;129:3375–3383. doi: 10.1242/jcs.187922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gonzalez-Rodriguez D, Guevorkian K, Douezan S, Brochard-Wyart F. Science. 2012;338:910–917. doi: 10.1126/science.1226418. [DOI] [PubMed] [Google Scholar]
- 112.Shi Q, Ghosh RP, Engelke H, Rycroft CH, Cassereau L, Sethian JA, Weaver VM, Liphardt JT. Proc Natl Acad Sci USA. 2014;111:658–663. doi: 10.1073/pnas.1311312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo C-L, Ouyang M, Yu J-Y, Maslov J, Price A, Shen C-Y. Proc Natl Acad Sci USA. 2012;109:5576–5582. doi: 10.1073/pnas.1114781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang H, Abhilash AS, Chen CS, Wells RG, Shenoy VB. Biophys J. 2014;107:2592–2603. doi: 10.1016/j.bpj.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nguyen-Ngoc K-V, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, Ewald AJ. Proc Natl Acad Sci USA. 2012;109:E2595–604. doi: 10.1073/pnas.1212834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nguyen-Ngoc KV, Ewald AJ. Journal of Microscopy. 2013;251:212–223. doi: 10.1111/jmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guzman A, Sánchez Alemany V, Nguyen Y, Zhang CR, Kaufman LJ. Biomaterials. 2017;115:19–29. doi: 10.1016/j.biomaterials.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 118.Ahmadzadeh H, Webster MR, Behera R, Jimenez Valencia AM, Wirtz D, Weeraratna AT, Shenoy VB. Proc Natl Acad Sci USA. 2017:201617037. doi: 10.1073/pnas.1617037114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carey SP, Martin KE, Reinhart-King CA. Sci Rep. 2017;7:42088. doi: 10.1038/srep42088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gill BJ, Gibbons DL, Roudsari LC, Saik JE, Rizvi ZH, Roybal JD, Kurie JM, West JL. Cancer Res. 2012;72:6013–6023. doi: 10.1158/0008-5472.CAN-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin C-C, Anseth KS. Pharm Res. 2009;26:631–643. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Enemchukwu NO, Cruz-Acuña R, Bongiorno T, Johnson CT, García JR, Sulchek T, García AJ. J Cell Biol. 2016;212:113–124. doi: 10.1083/jcb.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J. Nat Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ. Nature Mat. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 125.Augst AD, Kong HJ, Mooney DJ. Macromol. Biosci. 2006;6:623–633. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 126.Fonseca KB, Bidarra SJ, Oliveira MJ, Granja PL, Barrias CC. Acta Biomater. 2011;7:1674–1682. doi: 10.1016/j.actbio.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 127.Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. J. Clin. Invest. 1995;95:859–873. doi: 10.1172/JCI117736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, González-Tarragó V, Elosegui-Artola A, Albertazzi L, Alcaraz J, Roca-Cusachs P, Sahai E, Trepat X. Nat Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Integr Biol (Camb) 2011;3:439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ehsan SM, Welch-Reardon KM, Waterman ML, Hughes CCW, George SC. Integr Biol (Camb) 2014;6:603–610. doi: 10.1039/c3ib40170g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aref AR, Huang RY-J, Yu W, Chua K-N, Sun W, Tu T-Y, Bai J, Sim W-J, Zervantonakis IK, Thiery JP, Kamm RD. Integrative Biology. 2013;5:381–389. doi: 10.1039/c2ib20209c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sung KE, Su X, Berthier E, Pehlke C, Friedl A, Beebe DJ. PLoS ONE. 2013;8:e76373. doi: 10.1371/journal.pone.0076373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Singh SP, Schwartz MP, Tokuda EY, Luo Y, Rogers RE, Fujita M, Ahn NG, Anseth KS. Sci Rep. 2015;5:17814. doi: 10.1038/srep17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weisel JW, Litvinov RI. Blood. 2013;121:1712–1719. doi: 10.1182/blood-2012-09-306639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chung S, Sudo R, Mack PJ, Wan C-R, Vickerman V, Kamm RD. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 136.Griffith LG, Swartz MA. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 137.Bhatia SN, Ingber DE. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 138.Abaci HE, Shuler ML. Integr Biol (Camb) 2015;7:383–391. doi: 10.1039/c4ib00292j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oyen ML. Int Mater Rev. 2013;59:44–59. [Google Scholar]
- 140.van Oosten AS, Galie PA, Janmey PA. Gels Handbook: Fundamentals, Properties and Applications, World Scientific. 2016:67–79. ch. 4. [Google Scholar]
- 141.Broedersz CP, MacKintosh FC. Rev Mod Phys. 2014;86:995–1036. [Google Scholar]
- 142.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 143.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H-p, Lippens E, Duda GN, Mooney DJ. Nature Mat. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stein AM, Vader DA, Jawerth LM, Weitz DA, Sander LM. J Microsc. 2008;232:463–475. doi: 10.1111/j.1365-2818.2008.02141.x. [DOI] [PubMed] [Google Scholar]
- 145.Mickel W, Münster S, Jawerth LM, Vader DA, Weitz DA, Sheppard AP, Mecke K, Fabry B, Schröder-Turk GE. Biophys J. 2008;95:6072–6080. doi: 10.1529/biophysj.108.135939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ashworth JC, Mehr M, Buxton PG, Best SM, Cameron RE. Adv Healthc Mater. 2015;4:1317–1321. doi: 10.1002/adhm.201500197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Münster S, Aifantis KE, Fabry B. Nat Meth. 2015:1–9. doi: 10.1038/nmeth.3685. [DOI] [PubMed] [Google Scholar]
- 148.Stout DA, Bar-Kochba E, Estrada JB, Toyjanova J, Kesari H, Reichner JS, Franck C. Proc Natl Acad Sci USA. 2016;113:2898–2903. doi: 10.1073/pnas.1510935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hall MS, Alisafaei F, Ban E, Feng X, Hui C-Y, Shenoy VB, Wu M. Proc Natl Acad Sci USA. 2016;113:14043–14048. doi: 10.1073/pnas.1613058113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Beaune G, Stirbat TV, Khalifat N, Cochet-Escartin O, Garcia S, Gurchenkov VV, Murrell MP, Dufour S, Cuvelier D, Brochard-Wyart F. Proc Natl Acad Sci USA. 2014;111:8055–8060. doi: 10.1073/pnas.1323788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grys BT, Lo DS, Sahin N, Kraus OZ, Morris Q, Boone C, Andrews BJ. J. Cell Biol. 2017;216:65–71. doi: 10.1083/jcb.201610026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Leggett SE, Sim JY, Rubins JE, Neronha ZJ, Williams EK, Wong IY. Integr Biol (Camb) 2016;8:1133–1144. doi: 10.1039/c6ib00139d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Z, Tonderys D, Leggett SE, Williams EK, Kiani MT, Steinberg RS, Qiu Y, Wong IY, Hurt RH. Carbon. 2016;97:14–24. doi: 10.1016/j.carbon.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Skylaki S, Hilsenbeck O, Schroeder T. Nat Biotechnol. 2016;34:1137–1144. doi: 10.1038/nbt.3713. [DOI] [PubMed] [Google Scholar]
- 156.Gamboa Castro M, Leggett SE, Wong IY. Soft Matter. 2016;12:8327–8337. doi: 10.1039/c6sm01287f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pampaloni F, Reynaud EG, Stelzer EHK. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 158.Driscoll MK, Danuser G. Trends Cell Biol. 2015;25:749–759. doi: 10.1016/j.tcb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mih JD, Sharif AS, Liu F, Marinković A, Symer MM, Tschumperlin DJ. PLoS ONE. 2011;6:e19929. doi: 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VSW, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RGJ, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP. Proc Natl Acad Sci USA. 2016;113:E6831–E6839. doi: 10.1073/pnas.1603529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Caiazzo M, Okawa Y, Ranga A, Piersigilli A, Tabata Y, Lutolf MP. Nature Mat. 2016;15:344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 163.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong STC, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu C-C, LeBleu VS, Kalluri R. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shamir ER, Ewald AJ. Nat Rev Mol Cell Biol. 2014;15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Simian M, Bissell MJ. J Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]