Abstract
During nervous system development, neurons extend axons along well-defined pathways. The current understanding of axon pathfinding is based mainly on chemical signalling. However, growing neurons interact not only chemically but also mechanically with their environment. Here we identify mechanical signals as important regulators of axon pathfinding. In vitro, substrate stiffness determined growth patterns of Xenopus retinal ganglion cell (RGC) axons. In vivo atomic force microscopy revealed striking stiffness gradient patterns in the embryonic brain. RGC axons grew towards the tissue’s softer side, which was reproduced in vitro in the absence of chemical gradients. To test the importance of mechanical signals for axon growth in vivo, we altered brain stiffness, blocked mechanotransduction pharmacologically, and knocked down the mechanosensitive ion channel Piezo1. All treatments resulted in aberrant axonal growth and pathfinding errors, suggesting that local tissue stiffness–read out by mechanosensitive ion channels–is critically involved in instructing neuronal growth in vivo.
Keywords: mechanosensitivity, durotaxis, AFM, axon guidance, biomechanics, stretch-activated ion channels, brain stiffness, stiffness gradient
During the development of the central nervous system (CNS), each neuron extends an axon, which is the dominant cell process, and a number of finer, branched dendrites. Before connecting with their targets, axons grow along well-defined pathways in a stereotypic manner. Since the introduction of Sperry’s chemoaffinity hypothesis more than 50 years ago1, it has generally been accepted that axon guidance is regulated primarily by chemical signals2,3.
In the developing Xenopus laevis optic pathway, which is one of the best understood model systems of axon pathfinding, retinal ganglion cell (RGC) axons leave the retina via the optic nerve, cross the midline at the optic chiasm, grow along the contralateral brain surface in the optic tract (OT), and terminate in the optic tectum. Axon guidance along the OT, and particularly the caudal turn of axons in the mid-diencephalon, is thought to be mainly controlled by the repellent chemical cues Slit1, Slit2 and Semaphorin3A (Sema3A)4–6, which are expressed in the telencephalon and diencephalon.
Growth implies motion, however, and motion is driven by forces. Growing axons must exert forces on their environment and interact with it not only chemically but also mechanically7–10. Neuronal growth may thus be influenced by the mechanical properties of the environment, as originally shown in vitro11. Recent work has revealed that, in vitro, neuronal growth patterns, neurite extension, branching patterns, and neuronal traction forces all change with substrate stiffness10,12–16. Since CNS tissue is mechanically heterogeneous17–23, growing axons are likely to encounter tissue regions with different mechanical properties. While local tissue mechanics might thus provide important signals to growing axons, the mechanical properties of developing CNS tissue in vivo are currently unknown, and the potential neuronal response to mechanical signals in vivo is poorly understood.
Here, we used in vitro mechanosensitivity assays, in vivo atomic force microscopy (AFM), and interfered with brain tissue stiffness and neuronal mechanosensitivity, to investigate how mechanical signals affect neuronal growth. We found that axonal growth patterns depend strongly on the local mechanical properties of the surrounding tissue, suggesting that, in vivo, growing neurons respond not only to chemical but also to mechanical signals.
Results
RGC axons are mechanosensitive
To investigate Xenopus RGC axon mechanosensitivity, we first cultured eye primordia, which contain intact retinae from which RGCs extend their axons, on polyacrylamide substrates of controlled stiffness24. We used two different substrates to probe axonal mechanosensitivity: ‘stiff’ substrates with a shear modulus of 1 kPa and ‘soft’ substrates with a shear modulus of 0.1 kPa, which correspond approximately to the upper and lower bounds of brain tissue stiffness, respectively25. The substrates were coated with laminin, the density of which was independent of substrate stiffness (Supplementary Fig. 1c). Thus, cells cultured on soft and stiff substrates were only exposed to different mechanical signals, while their chemical environments were similar.
After 24 hours, axons grown on stiff substrates were significantly longer than those grown on soft substrates, as assessed by Sholl analysis (P < 10-6, One-Way-ANOVA followed by Bonferroni post-hoc test) (Fig. 1a,b,d,e). RGCs cultured on substrates coated with fibronectin, which engages different integrins from laminin26, also grew significantly longer axons on stiff substrates (P < 10-5, two-tailed t-test) (Supplementary Fig. 2a-d), suggesting that neurons were mechanosensitive irrespective of the type of integrins involved in cell adhesion.
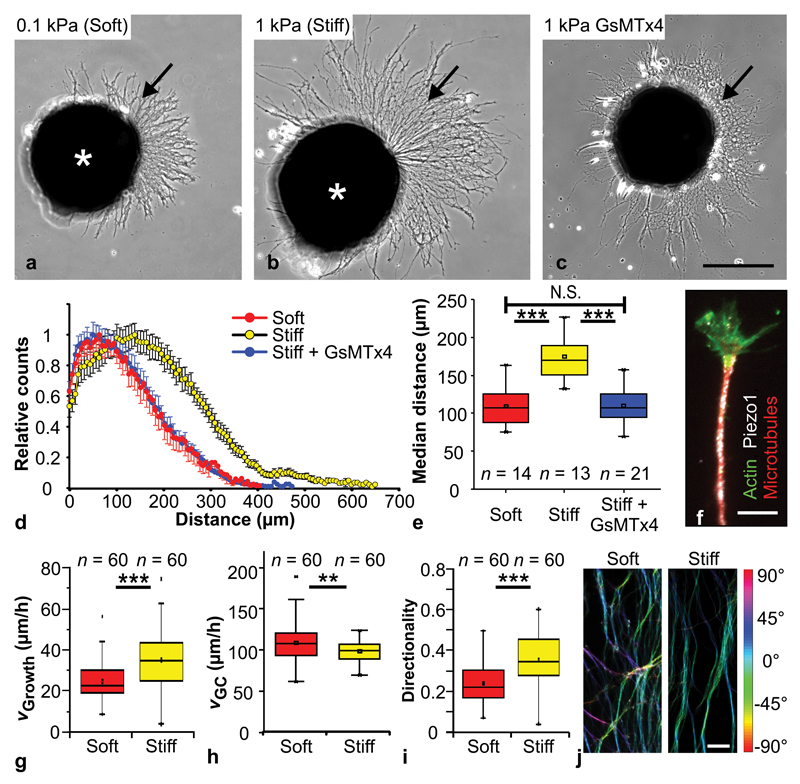
Figure 1. Mechanosensitivity of RGC axons in vitro.
(a, b) Cultures of Xenopus eye primordia (asterisks) on (a) ‘soft’ (0.1 kPa) and (b) ‘stiff’ (1 kPa) substrates. Arrows indicate axons. (c) Eye primordium grown on a stiff substrate and treated with GsMTx4, a blocker of mechanosensitive ion channels. Scale bar: 200 µm. (d) Sholl analysis of axon lengths after 24 hours (normalized counts as mean ± S.E.M.). (e) Median distances shown in (d). Axons were significantly longer on stiffer substrates than either on soft ones (One-Way-ANOVA followed by Bonferroni post-hoc test; P = 2.79 × 10-7, t = 6.354,) or after GsMTx4 treatment (P = 5.01 × 10-8, t = -6.855). Neurons grown on stiff substrates and treated with GsMTx4 resembled neurons grown on soft substrates (P = 1.00, t = 0.082). n = number of eye primordia from three biological replicates. (f) Immunocytochemistry showing f-actin (green), beta-tubulin (red) and the mechanosensitive ion channel Piezo1 (white). Scale bar: 10 µm. (g) The extension velocity of axons was higher on stiff substrates (Mann-Whitney-Test; P = 9.32 × 10-6, Z = 4.432). (h) On soft substrates, growth cones explored their environment more and migrated significantly faster than on stiff ones (two-tailed t-test; P = 0.00867, t = 2.669). (i) On stiff substrates, axon growth was more directed (i.e., straight) than on soft substrates (Mann-Whitney-Test; P = 1.10 × 10-6, Z = 4.873). n = number of axons from three biological replicates. (j) Processed fluorescence images of beta-tubulin-labelled RGC axons; colour represents local angular orientation of axonal segments. On soft substrates, axons grew less directionally persistent (from bottom to top; cf. Supplementary Fig. 2e-g). Scale bar: 15 µm. All experiments were repeated three times, and representative images are shown. Boxes show the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data.
In agreement with the Sholl analysis, time-lapse movies revealed that the average extension velocity of axons, i.e., the average distance between the proximal and distal ends of the axon per time interval, was significantly higher on stiff than on soft substrates (median velocities of 35.0 µm/h vs. 22.6 µm/h; P < 10-5, Mann-Whitney-Test) (Fig. 1g, Supplementary movies 1-2). However, when we assessed the absolute distance growth cones, which are the tips of advancing axons, moved in a given time, we found that they actually moved faster and explored their environment more on soft as compared to stiff substrates (109 ± 3 µm/h vs. 99 ± 2 µm/h; P<0.01, two-tailed t-test) (Fig. 1h). Thus, the directionality of axonal growth, which is defined as the ratio between axon extension and the length of the path covered by the growth cone, was significantly reduced on soft substrates (P < 10-6; Fig. 1i), indicating that axon growth is more directionally persistent on stiffer substrates.
The more explorative motion of growth cones on soft substrates was reflected in the appearance of the explant cultures. While on stiff substrates, axons grew rather straight and parallel to each other (i.e., formed bundles), on soft substrates axons grew less coherently, crossed each other more frequently, and appeared to splay apart (Fig. 1j, Supplementary Fig. 2e-g). Together, these experiments showed that, in vitro, growing Xenopus RGC axons respond to mechanical signals.
Mechanosensing is mediated by stretch-activated ion channels
Previous in vitro experiments have suggested an involvement of stretch-activated ion channels in neuronal mechanosensitivity27–30. As the opening probability of mechanosensitive ion channels increases on stiffer substrates27–29,31, impeding the channels’ activity should prevent neurons from detecting ‘stiff’ and result in the equivalent of a ‘soft’ phenotype.
Piezo1, which forms a mechanically activated cation channel32,33, has been identified as a key player in several mechanotransduction cascades in developing systems31,34,35. Using immunocytochemistry, we found a positive signal for Piezo1 distributed in punctate patterns all along axons and in growth cones of Xenopus RGCs (Fig. 1f). We then applied the spider venom peptide GsMTx4, which blocks the activity of mechanosensitive ion channels including Piezo136, to eye primordia cultures. As predicted, axons grown on stiff substrates and treated with GsMTx4 were significantly shorter than axons in the control group (P < 10-7, One-Way-ANOVA followed by Bonferroni post-hoc test) and strongly resembled axons grown on soft substrates (P = 1.00) (Fig. 1c-e), indicating that mechanosensing of Xenopus RGCs is mediated by mechanosensitive ion channels.
Stiffness gradients in developing brain tissue
In order to investigate the mechanical environment RGC axons encounter in vivo, we developed an atomic force microscopy (AFM)-based approach to map the local tissue stiffness of the exposed intact developing brain in vivo at different developmental stages by force-indentation measurements (Fig. 2a; for details see Methods). Tissue stiffness was quantified by the reduced apparent elastic modulus K; the larger K the stiffer the tissue. Stiffness maps had a spatial resolution of 20 - 25 µm, a length scale relevant to individual neuronal growth cones16. RGC axons were visualized using epifluorescence by introducing an Ath5/GFP reporter.
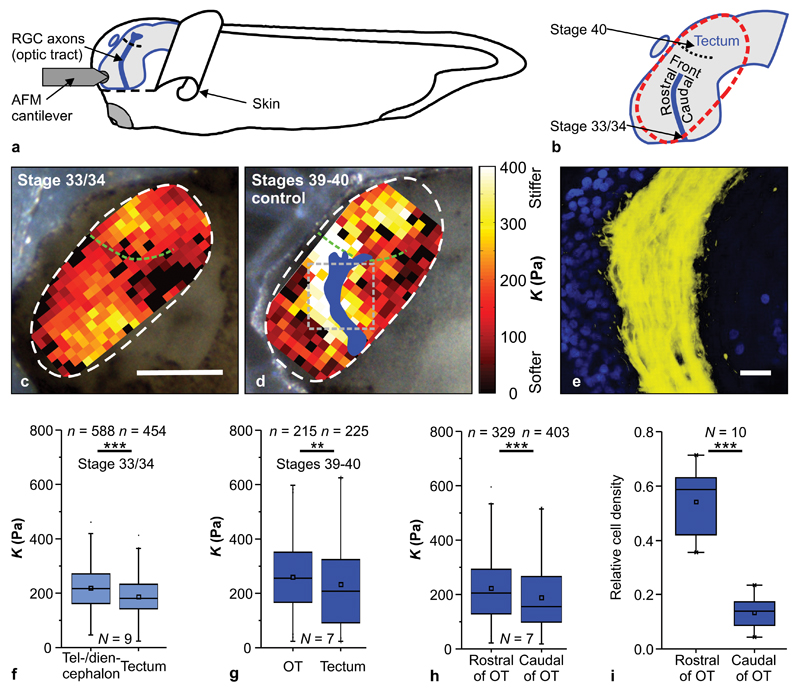
Figure 2. In vivo brain mechanics.
(a) Schematic of the experimental setup. (b) Xenopus brain. The dashed red line indicates the stiffness map area. (c, d) Images of Xenopus embryos with overlaid AFM-based stiffness maps of exposed in vivo brain tissue. Colour encodes the apparent elastic modulus K assessed at an indentation force of 7 nN. Blue shape in (d) shows the OT location (based on fluorescence images, Supplementary Fig. 3). Scale bar: 200 µm. At both stage 33/34 (c) and stages 39-40 (d), brain tissue was mechanically heterogeneous and displayed clearly visible stiffness gradients. Green dashed lines indicate tectum boundaries. The grey dashed square in (d) indicates a region as shown in (e). (e) Immunohistochemistry demonstrated a significantly higher density of cell nuclei (blue) rostral to the OT (yellow) than caudal to it. Scale bar: 20 µm. (f) The tectum was softer than the tel-/diencephalon at stage 33/34 (Mann-Whitney-Test; P = 2.26 × 10-9, Z = 5.978) and (g) than the OT at stages 39-40 (P = 0.0033, Z = 2.933). (h) At stages 39-40, tissue rostral of the OT was significantly stiffer than caudal of it (P = 2.97 × 10-5, Z = 4.163). (i) Quantification of cell density on both sides of the OT; cell density was significantly higher rostral to the OT (paired two-tailed t-test; P = 3.96 × 10-6, t = 9.879). n = number of measurements, N = animal numbers. All representative images and stiffness maps shown are from three biological replicates. Boxes show the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data.
AFM measurements were carried out at two different developmental time points: when the first axons had left the optic chiasm and entered the ventral OT (developmental stage 33/34) and again when the first axons had reached the optic tectum (stages 39-40) (Fig. 2b). At both time points, tissue stiffness was heterogeneously distributed (Fig. 2c, d; Supplementary Fig. 3a, b). An analysis of the average stiffness of distinct tissue regions revealed that, at stage 33/34, the tel-/diencephalon, and, at stages 39-40, the OT, were significantly stiffer than the optic tectum (P33/34 < 10-8, P39-40 < 10-2) (Fig. 2f, g). Furthermore, at stages 39 - 40, tissue rostral to the OT was significantly stiffer than that caudal to it (P < 10-4) (Fig. 2h), resulting in a stiffness gradient perpendicular to the growth direction of RGC axons.
In order to determine the structural origin of this stiffness gradient in the brain, we first tested if RGC axons themselves were involved. We ablated eye primordia in early Xenopus embryos. Brains of those embryos developed normally, but no RGC axons entered the diencephalon. We found similar stiffness distributions in the tel-/diencephalon of these brains as in control brains (Supplementary Fig. 3c), suggesting that RGC axons themselves did not cause the observed stiffness gradients.
While the extracellular matrix (ECM) contributes to overall CNS tissue stiffness22,37, mechanical heterogeneities appear to be largely established by the tissue’s cellular constituents19,22,38. In other CNS tissues stiffness scales with cell body density22; we therefore used immunohistochemistry to investigate the distribution of cell nuclei in the vicinity of the OT. Nuclei in whole mount brain preparations were labelled using DAPI and the area they occupied in a given region was determined. Cell body densities rostral to the OT were significantly higher than those caudal to it (P < 10-5, paired two-tailed t-test) (Fig. 2e,i), suggesting a functional connection of the local cell body distribution to the presence and direction of the observed stiffness gradient.
Axons grow towards soft tissue
We found that RGC axons grow perpendicular to a local stiffness gradient in the brain (Fig. 2d, h). To assess the impact of this stiffness gradient on axon growth, we quantified the local curvature C = 1/R of the OT approximately every 40 µm along its length by fitting a circle of radius R to the OT as indicated in Figure 3a. We then calculated the local stiffness gradient M perpendicular to the OT at each position by subtracting the apparent elastic modulus K averaged over an area of (100 × 100) µm2 caudal of the OT from the average K rostral of it. This difference in K was then divided by 100 µm, resulting in a stiffness gradient M expressed in Pa/µm (Fig. 3a).
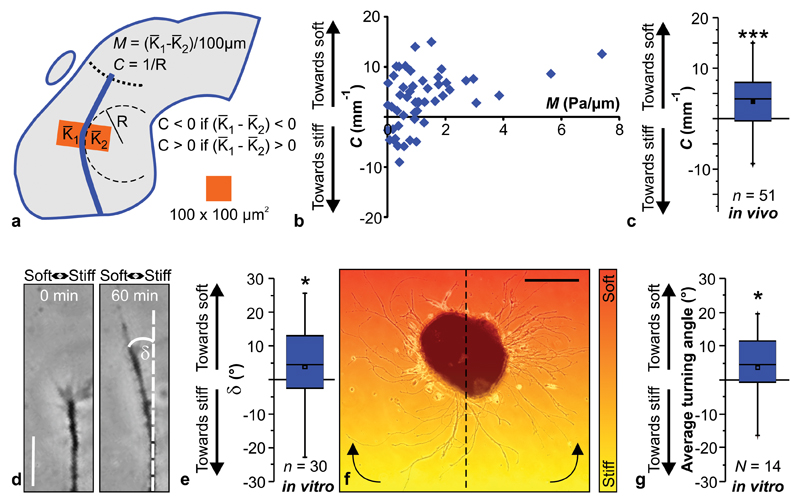
Figure 3. Neurons grow towards soft tissue.
(a) Schematic showing how local gradients in brain tissue stiffness perpendicular to the RGC axon growth direction M and the local OT curvature C were determined. (b) Relationship between M and C. (c) Same data as in (b), pooled. Axons in vivo preferentially turned towards the softer side of the tissue (one-sample Wilcoxon Signed Rank Test; P = 1.44 × 10-4,Z = 3.801). n = number of measurement points from 7 animals. (d, e) Time-lapse imaging of individual axon bundles growing on a stiffness gradient matching that found in vivo (Mmax ∼ 2 Pa/µm; cf. Supplementary Fig. 1) revealed that also in vitro, in the absence of chemical gradients, RGC axons preferentially turned towards the softer side of the substrate (one-sample Wilcoxon Signed Rank Test; P = 0.0549; Z = -1.913). Scale bar: 20 µm. Representative images from 11 biological replicates are shown. (f) Eye primordium cultured on a similar stiffness gradient (indicated by colour). Axons growing more clockwise in the left half and more counter-clockwise in the right half of the image turned towards the soft side of the substrate. Scale bar: 200 µm. The experiment was repeated three times, and a representative image is shown. (g) Quantification of individual axon segment orientations similarly revealed preferential turning towards the soft side of the substrate (one-sample Wilcoxon Signed Rank Test; P = 0.0264, Z = 2.197). Boxes show the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data.
The local curvature C of axons at any position along the OT strongly correlated with the tissue’s local stiffness gradient M at that position. RGC axons encountering stiffness gradients in vivo clearly turned away from stiffer and grew towards softer tissue (P < 10-3, one-sample Wilcoxon Signed Rank Test) (Fig. 3b,c). Interestingly, the curvature of the OT was independent of the absolute tissue stiffness (Supplementary Fig. 4c).
To test whether mechanical signals imposed by stiffness gradients are instructive rather than just permissive to axon growth, we developed cell culture substrates incorporating stiffness gradients similar to those found in vivo (Supplementary Fig. 1, for details see Methods), which allowed us to reduce the complexity of the neuronal environment and to study the effect of stiffness gradients on axon growth in the absence of chemical guidance cues. To test the correlation between stiffness gradients and axon turning in these in vitro assays, eye primordia were cultured on stiffness gradients, imaged, and the orientation of axons analysed using two complementary approaches.
We first used time-lapse microscopy to quantify the dynamic turning of individual axon bundles growing perpendicular to the stiffness gradient (with respect to their initial growth direction; Fig. 3d, e). We verified these results by analyzing the orientation of all axons of an explant (∼2800 segments per eye primordium on average) relative to their original orientation at the end of an experiment (Fig. 3f, g, see Online Methods). Both approaches showed that axon bundles preferentially turned towards soft also in vitro (P ≤ 0.05) – in the absence of chemical gradients.
Softening of brain tissue leads to aberrant axon growth
As mentioned above, the ECM contributes to overall CNS tissue stiffness (although less to mechanical heterogeneities). To test if brain tissue stiffness provides an instructive signal for RGC axon growth in vivo, we first perturbed the mechanical properties of developing Xenopus brains by manipulating their ECM composition. Chondroitin sulphate (CS) proteoglycans are a main ECM component in the developing brain39. After applying CS to exposed brains in vivo, we still found similar stiffness gradients as in control brains; however, the overall tissue stiffness was significantly decreased (P < 10-9, Mann-Whitney-Test) (Fig. 4a-c, Supplementary Fig. 3d), particularly in the region in front of the OT (P < 10-10) (Fig. 4c).
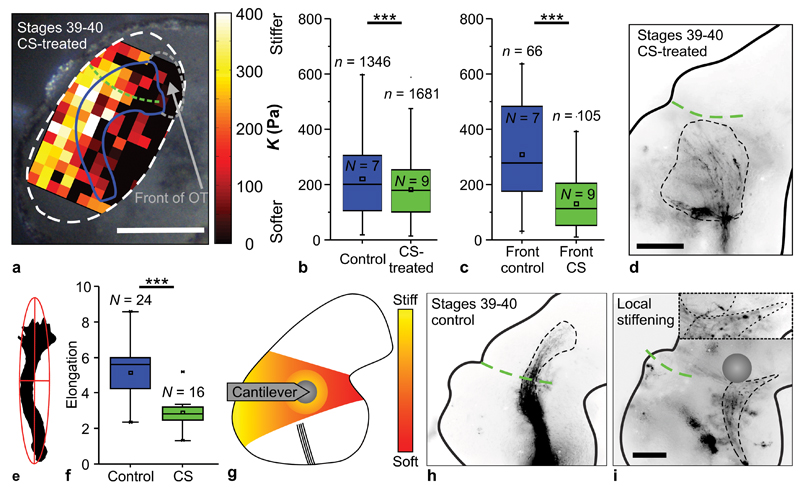
Figure 4. Changing brain stiffness leads to axonal pathfinding errors.
(a) Xenopus brain treated with 15 mg/ml CS, overlaid with an AFM-based stiffness map. Blue curve indicates OT location (based on fluorescence images, Supplementary Fig. 3), green dashed curve tectum boundary, grey dashed line the region in front of the OT. Scale bar: 200 µm. (b) Tissue was significantly stiffer in controls (blue) compared to CS-treated brains (green) (Mann-Whitney-Test; P = 6.62 × 10-10, Z = 6.175), particularly in front of the OT (c) (P = 8.57 × 10-11, Z = 6.490). n = number of measurements, N = animal numbers. (d) Image of CS-treated, softened brain. The dashed black curve indicates the outline of the OT. RGC axons dispersed widely from their normal trajectory. Scale bar: 100 µm. (e) Example of a fit of an ellipse around the outline of an OT; the ratio of long and short axis determines the elongation. (f) CS treatment significantly decreased the elongation of the OT (two-tailed t-test; P = 3.12 × 10-6, t = 5.462). n = number of animals. (g) Schematic illustration of local mechanical brain manipulation. Soft tissue caudal to the presumptive caudal turn of the OT was locally indented with an AFM cantilever for ∼6 hours. (h) In control brains, the OT grew normally. (i) When a force was locally exerted on the tissue (position of the probe is indicated), axons grew away from the cantilever probe, thus deviating from their normal pathway. Inset: magnification of the distal part of the OT. Scale bar: 100 µm. All experiments were repeated three times, and representative images are shown. Boxes show the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data.
Because the turning of RGC axons towards soft tissue did not depend on absolute brain stiffness but rather on the strength of the local stiffness gradient (Fig. 3b, Supplementary Fig. 4c), axons still preferentially turned towards softer tissue in CS treated brains (Supplementary Fig. 4). However, in agreement with our in vitro experiments and a previous study39, RGC axons in these softened brains dispersed widely from their normal trajectory, with reduced directionality and fasciculation (Fig. 4d), thus resembling axon behaviour on soft substrates in vitro (Fig. 1). To quantify OT phenotypes, we fitted ellipses around OTs and compared the ratios between the long and short axes of the ellipses, i.e., their elongation24 (Fig. 4e). The elongation of the OT in CS-treated brains was significantly decreased compared to controls (P < 10-5, two-tailed t-test) (Fig. 4f), suggesting that different mechanical instructions provided by CS-treated, softened brain tissue likely contributed to RGC axon defasciculation and aberrant growth.
Local mechanical manipulation of CNS tissue redirects axons
As CS proteoglycans are likely involved in binding and presenting various trophic and tropic factors to axons39, the OT phenotype observed after increasing the brain’s CS content might be due not only to mechanical but also to chemical changes in the tissue. To perturb brain mechanics without altering the chemical environment, we used AFM to apply a sustained compressive force F = 30 nN to exposed embryonic brains in vivo (illustrated in Fig. 4g) with a spheroidal probe of 87 µm diameter. The resulting mechanical stress σ = F / A ∼ 30 pN / µm2 = 30 Pa, where A corresponds to the contact area, led to a maximum tissue strain ε = σ / Kmax ∼ 7.5 % (corresponding to a maximum indentation δmax = (3.6 ± 0.2) µm).
The experiment commenced at stage 35/36, when the fluorescently labelled OT was already visible but had not started to turn caudally in the mid-diencephalon (Supplementary Fig. 5a), and finished ∼6 hours later, when embryos had reached stage 39. The compressive force was applied to the region just caudal to the anticipated OT turn, where the tissue would normally be soft (cf. Fig. 2d). Importantly, brain tissue is nonlinearly elastic and stiffens under compression18,21,40 (P < 0.01, Kruskal-Wallis ANOVA) (Supplementary Fig. 5h).
In all experiments, axons avoided growing underneath the centre of the probe, where the compressive strain was largest. While in time-matched control embryos the OT grew normally and frequently formed a caudal bend in the mid-diencephalon (Fig. 4h), in 2 of 5 experiments axons grew straight, passed the site of indentation laterally, and the OT did not substantially turn (Supplementary Fig. 5f, g). In the other 3 cases, RGC axons deviated from their normal path to grow either away from or around the compressed tissue (Fig. 4i, Supplementary Fig. 5b-e). These experiments showed that mechanically interfering with brain tissue alone is sufficient to alter axon growth patterns.
Mechanosensing through Piezo1 is critical for axon growth
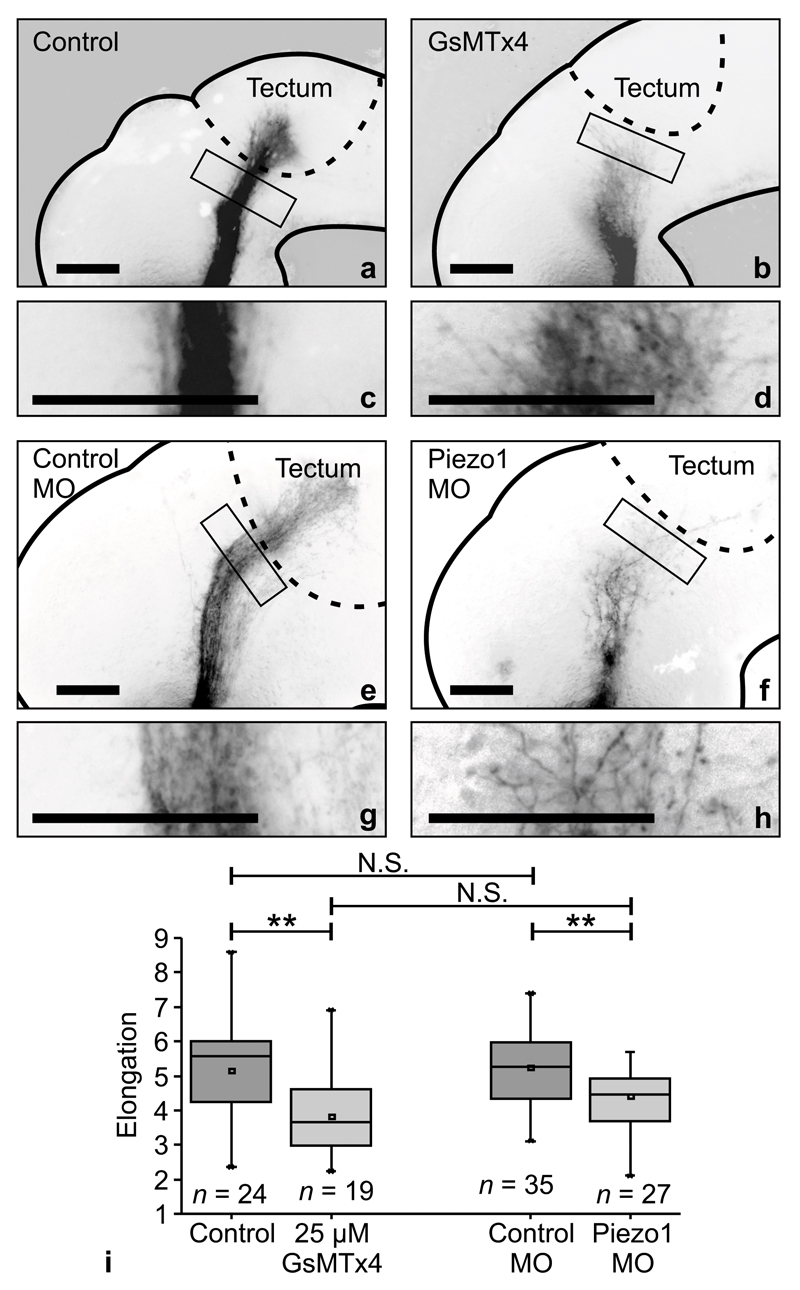
The previous set of experiments indicated that RGC axons respond to mechanical signals in vivo. To test if their mechanosensitive behaviour was mediated by mechanosensitive ion channels, as in our in vitro experiments (Fig. 1), we either applied GsMTx4 to exposed brains of stage 33/34 embryos (cf. Fig. 2b), or down-regulated Piezo1 expression by ∼42% using morpholino knockdown in vivo (cf. Supplementary Fig. 6d,e). Following both manipulations, at stages 39-40 RGC axons dispersed widely from their normal trajectory and assumed a phenotype similar to axons grown on softer substrates in vitro (cf. Fig. 1), with directional incoherence, decreased length, and overall reduced elongation (P < 0.01, two-tailed t-test) (Fig. 5).
Figure 5. In vivo manipulation of mechanosensitive ion channels disrupts axon pathfinding.
(a-d) The application of 25 µM GsMTx4 disrupted axon pathfinding in vivo. Axons were shorter and spread more, resembling axons cultured on soft substrates in vitro. (c-d) Enlarged boxes shown in (a) and (b). (e-h) Similarly, MO knockdown of Piezo1 led to aberrant axon growth in vivo. Scale bars: 100 µm. Experiments were repeated three (a-d) or four times (e-h), and representative images are shown. (i) Quantification of OT morphology. Interfering with mechanotransduction led to a significantly decreased elongation of the OT. (two-tailed t-test; PGsMTx4 = 0.00243, t = 3.231; PPiezo1-MO = 0.00476, t = 2.932). Phenotypes of controls were similar (P = 0.782, t = 0.278), as well as those of GsMTx4-treated and Piezo1 knockdown animals (P = 0.0976, t = 1.692). n = number of animals. Boxes show the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data.
This phenotype was maintained in the Piezo1 morpholino-treated animals at stage 42, at which point OT growth and tectal innervation are normally complete (∼1 day older than the ones shown in Fig. 5; Supplementary Fig. 6a-c), indicating that Piezo1 knockdown led to pathfinding abnormalities rather than merely to slowed axon growth. Hence, our experiments suggested that RGC mechanosensitivity, which is involved in controlling axon growth, is mediated by Piezo1 in vivo.
Discussion
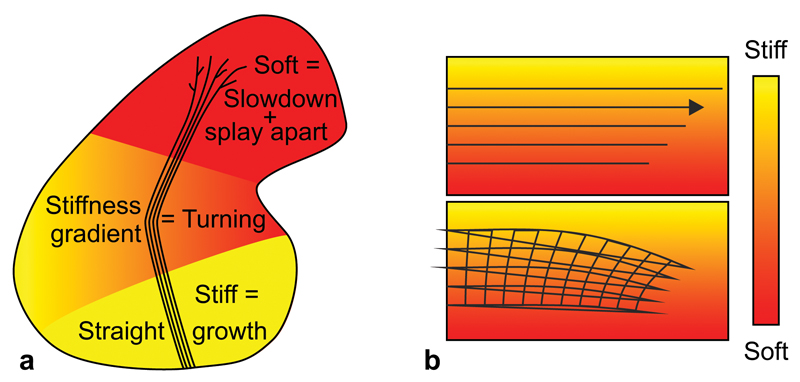
We have shown that, in vitro as well as in vivo, Xenopus RGC axons respond to mechanical signals in their environment (summarised in Fig. 6a). Tissue stiffness regulates the length of axons and their degree of spreading: axons grow faster, straighter and more parallel on stiffer substrates (Fig. 1). A higher stiffness, causing persistent growth and facilitating fasciculation, thus seems favourable for tissues through which axons have to grow. A lower stiffness, on the other hand, promoting slowed exploratory growth and splaying of axons, seems beneficial for regions where axons have to search for their targets and form synapses. Accordingly, tissue in the vicinity of the OT, where axons are tightly bundled, is stiffer than the rostral part of the tectum (Fig. 2g), where axons splay apart, branch, and form synapses. In line with this observation, spinal cord neurons in vitro branch more on softer substrates12.
Figure 6. Schematics of the mechanical control of axon growth.
(a) Schematic summary of this study. Shown is the outline of a Xenopus brain and the OT. Mechanical signals contribute to neuronal growth in the developing CNS. RGC axons grow faster, straighter, and more parallel on stiffer than on softer substrates (Fig. 1). Accordingly, the part of the brain these axons have to pass is stiffer than the tectum (Fig. 2), where axon growth slows down and eventually stops. The softness of the tectum then facilitates unbundling and branching. On their way, axons encounter an area with a stiffness gradient, which contributes to the turning of the OT towards the softer side of the brain (Fig. 3). (b) Schematic illustration of a mechanism by which axon bundles encountering a perpendicular stiffness gradient might turn towards the softer side of the substrate. The velocity of axons is larger on stiffer substrates (upper panel). As axons in the OT fasciculate, they are mechanically coupled, so the faster axons growing on the stiffer side may be ‘pulled’ towards the slower axons on the softer side, leading to a reorientation of the axon bundle and overall growth towards the softer side.
Furthermore, local stiffness gradients guide RGC axon growth. Non-neuronal cell types responding to stiffness gradients in vitro usually migrate towards the stiffer side of their substrate in a process termed durotaxis41. Durotaxis only depends on the strength of the gradient and is independent of the absolute substrate stiffness42, similar to what we observed in our in vivo experiments (Supplementary Fig. 4c, d). However, in contrast to other cell types, RGC axon bundles turned towards the softer side of their growth substrate, both in vitro and in vivo. While individual growth cones might possess neuron-specific mechanotransduction mechanisms causing this response27,28, growth of axon bundles towards soft areas could also at least partly be a collective effect (Fig. 6b). Growth velocities of axons are higher on stiffer substrates (Fig. 1g). When axon bundles grow perpendicular to a stiffness gradient, the faster axons on the stiffer side will be ‘pulled’ towards the tightly coupled slower axons on the softer side, and consequently turn towards them (similar to phototropism in plants, where cells on the side of the stem farthest from the light extend more than cells closer to the light).
The mechanical properties of embryonic brain tissue have been studied in two different systems before20,43. While Xu and colleagues suggested that embryonic chick brain is mechanically rather homogeneous and does not change its elastic stiffness over time43, in the mouse embryonic cerebral cortex, tissue stiffness is region- and developmental stage-dependent20. Both studies were done ex vivo. Using an in vivo AFM approach, we obtained stiffness maps of the intact Xenopus brain with a spatial resolution on the order of the size of neuronal growth cones. We found that the stereotypic caudal bend of the OT in the mid-diencephalon coincides with a strong stiffness gradient in the tissue (Fig. 2, Supplementary Fig. 3). Axons in that region turned away from stiffer tissue as in our in vitro experiments, and the curvature of the OT correlated strongly with the strength of the gradient but not with the absolute tissue stiffness (Fig. 3, Supplementary Fig. 4).
The stiffness gradient likely originates mostly from the change in cell body density from rostral to caudal of the OT. Similarly, in the mouse spinal cord higher cell densities are associated with larger tissue stiffness22. While RGC axons respond to the mechanical stiffness of their environment (cf. Fig. 1), the space available for axons to grow through might contribute an additional mechanical signal controlling axon growth in vivo. Higher cell body densities likely also result in a decrease in available space and thus increased steric hindrance, which could add to the mechanotactic growth of RGC axons in the mid-diencephalon towards softer and less dense regions of the brain (cf. Fig. 2e).
Similarly, the change in RGC growth direction following the local application of a sustained compressive force to Xenopus brains (Fig. 4) is likely the result of a combination of different mechanical signals directly impacting neuronal growth. (1) As CNS tissue stiffens under compression18,21,40, the stiffness of the tissue underneath the AFM probe was increased (Fig 4g, Supplementary Fig. 5h). Such a change in stiffness can be read out by RGC axons via Piezo1, leading to slowed growth and decreased directionality (Figs. 1, 5). Also, compression of the tissue via a bead should lead to a local gradient in stiffness, with a maximum stiffening occurring at the centre of the probe where the strain is highest. This induced stiffness gradient might repel axon bundles as described above. (2) Local tissue compression could additionally lead to a local decrease in the available space, causing RGC axons to avoid and grow around the denser region. (3) Finally, pushing on the tissue might also directly trigger an immediate mechanosensitive response of neurons27, which at least at short time scales might contribute to changing growth directions. Moreover, all three mechanisms may potentially impact chemical signaling pathways, thus also indirectly contributing to changing RGC axon growth directions (see below).
Mechanosensitive ion channels open more frequently in neurons grown in stiffer environments27–29,31. Blocking these channels thus prevents neurons from detecting ‘stiff’ (Fig. 1c-e), suggesting a plausible mechanism by which RGC axons were shorter, grew with reduced directionality and fanned out from the OT in brains treated with GsMTx4 or Piezo1 morpholinos. It is likely that there are other mechanisms involved in neuronal mechanotransduction25,44; however, interfering with these ion channels alone was sufficient to significantly impact axon growth in vitro as well as in vivo.
While we cannot rule out that our treatments also affected other cells in the tissue35, they indicate that changing either tissue mechanics or the cellular susceptibility for mechanical signals impact RGC axon growth (Figs. 4, 5). The mechanism by which CS-treatment disrupts axon pathfinding in the OT is still poorly understood. It has been speculated that CS might modulate axon pathfinding via CS-binding molecules39. We here provide an alternative or additional mechanism. The less directionally persistent axonal growth and the breakdown of fasciculation could at least partly be attributed to a softening of the tissue, likely due to increased hydration through the introduction of additional sulphate groups45.
It is possible that interfering with mechanotransduction not only directly but also indirectly altered RGC axon growth. Cells are likely to integrate all signals they can detect, chemical and mechanical ones alike, resulting in a response that is the consequence of all available information. As one signal may modulate the response of a cell to another signal46, perturbing mechanotransduction will likely alter the way in which neurons respond to chemical signals in the environment and vice versa. Furthermore, other cells in the vicinity of the OT, which, for example, secrete signalling molecules such as Sema3A or Slits, might also be mechanosensitive. Manipulating mechanotransduction in these cells could then change chemical signals that contribute to controlling RGC growth. This intimate cross-talk between chemical and mechanical signalling supports the view that mechanosensing is just as critical as biochemical signalling for axon growth in the developing brain.
Axon pathfinding is a highly complex process; several chemical guidance cues have been shown to be important for instructing axon growth2–6. We have identified local tissue stiffness as another critical signal in a developing organism, which together with membrane-bound and diffusible chemical cues controls cell growth and tissue organisation. Similar mechanical cell-tissue interactions are likely to be important for the development of the CNS in general44 and in other organ systems across species. It is also likely to be critical for regenerative processes in which cells have to migrate or re-grow through damaged tissue with altered mechanical properties.
Online Methods
All chemicals were obtained from Sigma-Aldrich and all antibodies were obtained from Abcam if not stated differently.
Preparation of polyacrylamide hydrogels
Plain substrates
Compliant polyacrylamide (PAA) hydrogel culture substrates were prepared as described previously23. Briefly, 21 mm glass-bottom Petri dishes (μ-Dish35mm, high; IBIDI, Germany) were dabbed with a 0.1 N NaOH solution using a cotton bud and air-dried. The glass was then treated with 200 μl (3-aminopropyl)trimethoxysilane (APTMS) for 3 min and washed with distilled water. 400 μl of 0.5% glutaraldehyde solution was then added for 30 min. Petri dishes were again washed and air-dried. 19 mm diameter glass coverslips were cleaned with 70% ethanol and distilled water and then treated with Rain-X solution (Shell Car Care International Ltd, UK) for 10 min, resulting in a non-adhesive coating. Rain-X solution was removed and the coverslips dried using lint-free wipes.
The shear storage modulus G’ of the PAA gel was adjusted using pre-defined ratios of 60% phosphate buffered saline (PBS; Fisher Scientific, UK), 40% (w/v) acrylamide (AA) solution (Sigma), and 2% bis-acrylamide (Bis-AA) solution (Fisher Scientific, UK), as described previously23. Gel pre-mix solutions (500 µl) corresponding to ∼0.1 kPa (5% AA & 0.04% Bis-AA) and 1 kPa (7.5% AA & 0.06% Bis-AA) were desiccated for 10 min. Polymerization was initiated by addition of 5 µL freshly prepared 10% ammonium persulphate (APS) and 1.5 µL N,N,N’,N’-tetramethylethylenediamine (TEMED) to the PAA pre-mixes. 40 µl of this solution was pipetted onto the treated glass-bottom Petri dish and then covered with the Rain-X-coated coverslip. After 15 min, coverslips were submerged in PBS for a further 20 min before the top coverslip was removed. Gels were then washed in filter-sterilised 60% PBS three times and functionalized with 5 - 10 µg/ml laminin (from Engelbreth-Holm-Swarm murine sarcoma basement membrane) for 2 h before culturing, or 1 µg/ml fibronectin (from bovine plasma) for 3 h prior to culturing. To allow laminin or fibronectin to adhere to the substrates, gels were either prepared by incubating them in hydrazine hydrate for 4 h, 5% acetic acid solution (Fisher Scientific, UK) for 1 h, sterile 60% PBS (3 times thorough washing), 10 or 100 µg/ml poly-D-lysine solution (PDL; MW 70,000-150,000) overnight at 4 °C, and sterile 60% Leibovitz L15 medium; three times washing23, or by incubating them in 40 µg/ml Cell-Tak™ (BD Biosciences, UK) for 2 h at room temperature and subsequent washing in sterile 60% PBS10.
Gradient substrates
Substrates incorporating stiffness gradients with a similar strength to those we found in vivo were produced by filling custom-designed chambers first with PAA gel premixes for ∼10 kPa (12% AA & 0.2% Bis-AA) immediately after polymerization was initiated, and then with 0.1 kPa gel premixes (see above). In the 10 kPa premix, 5 µl PBS were substituted by 5 µl of 1% (w/v) fluorescein O,O’-dimethacrylate diluted in DMSO. Diffusion led to a linear gradient in stiffness and fluorescence signal (Supplementary Fig. 1).
Gel chambers were assembled from two Parafilm-covered microscope slides, enclosing a glutaraldehyde-treated coverslip (22 × 22) mm2 and a RainX-treated coverslip (22 × 40) mm2, which were placed with the treated sides facing each other but separated by a U-shaped Parafilm spacer. Chambers were held together with bulldog clips and stood vertically, with the gap between the two coverslips at the top, into which the gels premixes were pipetted (Supplementary Fig. 1).
Substrate coating measurements
PAA gels were coated with 10 µg/ml laminin as described above. Substrates were then incubated overnight (18 h) at room temperature with 300 µL of a polyclonal rabbit anti-laminin antibody (ab11575) diluted 1:200 with PBS and 1% bovine serum albumin (BSA). Gels were then washed (4 × 10 min) with PBS and subsequently incubated for 4 h in secondary antibody solution (donkey anti-rabbit Alexa Fluor® 488; Invitrogen, A-21206, 1:1000 dilution) containing 1% BSA in PBS. Gels were washed again (5 × 10 min) with PBS and imaged immediately using a Nikon Eclipse Ni-E microscope (60x W; NA = 1.0). 24 images were captured from each gel (three gels per group). For each image, six distinct regions of interest were randomly chosen and the mean fluorescence intensity calculated using ImageJ software (NIH, MD, USA).
Animal model
All animal experiments were approved by the Ethical Review Committee of the University of Cambridge and complied with Home Office guidelines. Xenopus laevis embryos of both sexes were obtained by in vitro fertilization, raised in 0.1x Modified Barth's Saline (MBS) at 14 – 18 °C, and staged according to the tables of Nieuwkoop and Faber47. For AFM experiments, embryos were injected in one blastomere at the 4 cell stage with an Ath5-GFP construct (50 pg/ 5 nl) in order to label RGC axons in one retina. Fluorescently labelled RGC axons crossed the optic chiasm into the unlabelled brain, allowing visualization of the OT for AFM experiments.
For ablated eye primordia experiments, in which brains not containing RGC axons were measured by AFM at stages 39-40, stage 31-32 embryos were transferred to MR solution48 (composition: 1x MBS with 0.04% (w/v) MS222 anaesthetic (3-aminobenzoic acid ethyl ester methanesulfonate) and 1x Penicillin / Streptomycin / Fungizone (P/S/F; Life Technologies, UK), pH 7.4) and both eye primordia carefully removed with 0.1 mm minutien pins. Embryos were allowed to recover at 18°C in 0.5x MBS + 0.02% (w/v) MS222 for 1 hour, followed by 0.25x MBS + 0.01% (w/v) MS222 overnight. Embryos were then transferred to 0.1x MBS and left at 18°C to reach stages 39-40.
For ex vivo eye primordia cell culture experiments, stage 33/34 or 35/36 embryos were transferred to a 35 mm Petri dish coated with SYLGARD® 184 and anesthetized with 0.04% (w/v) MS222 solution (24.75 ml 10x MBS, 250 µl 100X P/S/F, and 100 mg MS222 dissolved in 225 ml ddH2O, adjusted to pH 7.7, and filter-sterilized). Whole eye primordia were dissected, placed onto PAA hydrogels, and cultured at 20 °C for 24–36 h in culture medium. Xenopus culture medium was composed of 60% L15 medium, 100 U/ml penicillin, 100 µg/ml streptomycin and 2.5 µg/mL amphotericin B (Life Technologies, UK), pH 7.7.
For GsMTx4 treatment experiments, 1-5 µM GsMTx4 (from Fred Sachs and Abcam, UK, ab141871) was added to eye primordia two hours after they had been placed on gels. After 22 additional hours of incubation, explants were fixed in 2% PFA + 7.5% sucrose for 25 minutes and washed several times with 100% PBS prior to imaging.
Exposed Brain Preparations
Exposed brain experiments were carried out in MR solution48. Stage 33/34 embryos were transferred to a 35 mm Petri dish coated with SYLGARD® 184 and immobilized with bent 0.2 mm minutien pins with the side of the body facing up. Skin, dura and eye were carefully dissected out using fine forceps and a 0.1 mm minutien pin to expose the brain from the dorsal to ventral midline and from the hindbrain, just anterior of the otic vesicle, to the telencephalon. The embryos were then transferred to a 4 well plate and submerged in solution containing either MR alone (control), or MR + 25 µM GsMTx4, or MR + 15 mg/ml chondroitin sulphate, and kept in the incubator at 14 or 18 °C until reaching stages 39-40.
Morpholino injections
Fluorescein-tagged translation blocking morpholino oligonucleotides (MOs) against Piezo-1 RNA (5’- CACAGAGGACTTGCAGTTCCATCCC-3’) were designed and synthetized by GeneTools (GeneTools, OR, USA). 15 ng of MO or control scrambled MO (GeneTools, OR, USA; 5’-CCTCTTACCTCAGTTACAATTTATA-3’) were injected into each dorsal blastomere of four cell-stage embryos, as described previously49. The embryos were transferred to a 14 °C incubator until stage 40, when they were imaged.
Western blots
Heads of stage 39-41 Xenopus embryos were dissected from control animals, control (scrambled) MO-injected animals and those injected with Piezo-1 MO as described above. While for the characterization of axon growth only the healthiest embryos, which looked closest in phenotype to the controls, were taken, for Western blot analysis all embryos containing the morpholino were used. Heads were homogenized in a protease inhibitor cocktail diluted in lysis buffer (NaCl 150mM , TritonX-100 1%, Sodium deoxycholate 0.5%, Sodium dodecyl sulphate 4 mg/ml Tris buffer 50mM, pH8), with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, UK), and protein samples prepared for Western blot, as described previously50. The Bradford (Bio-Rad Protein Assay Dye Reagent Concentrate, #5000006; Bio-Rad Laboratories, UK) calorimetric assay was used to calculate total protein concentration, and the loading volume adjusted accordingly. Samples were run on 4-15% SDS-PAGE gradient gels (Bio-Rad Laboratories, UK) and transferred to a nitrocellulose membrane. Membranes were blocked for 1 h in a blocking solution of 5% skim milk powder diluted in TBS-T (pH 7.4), and incubated overnight at 4°C with a polyclonal rabbit anti-Piezo1 (anti-FAM38A) primary antibody (ab82336; 1:500 dilution), and monoclonal mouse anti-α-tubulin (ab7291; 1:8 000 dilution) as a loading control. Excess primary antibodies were then washed off and the nitrocellulose membrane incubated for 1 h at room temperature in polyclonal goat anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (ab97069; 1:2000 dilution) for Piezo1, and a polyclonal goat anti-mouse antibody conjugated to HRP (ab6789; 1:15 000 dilution) secondary for α-tubulin. Western Blots were developed using Novex ECL HRP Chemiluminescent Substrate Reagent kit (Invitrogen, CA, USA) and X-ray developer. Densitometry was performed on 8-bit greyscale images imported into Fiji software (NIH, MD, USA). The Gel analysis tool was used to correct for background and to measure relative band intensities. The ratio of relative intensities of Piezo 1 to α-tubulin was used to compare different groups.
Imaging of axons in situ
For morphological characterization, eye primordia were imaged after 24 hours in culture by phase contrast microscopy on a Nikon Eclipse TE2000-U or a Zeiss AxioObserver A1 microscope (10x Ph1, NA = 0.3). For time-lapse imaging of axon growth, eye primordia were imaged after 18–24 h in culture using a Nikon Eclipse TE3000 microscope (10x Ph1 objective, NA = 0.25). Images were captured every 30 seconds for ∼3 h using a CCD camera (AxioCam ERc 5s ; Zeiss, UK) and ZEN 2011 software. For turning assays (see below), axons were imaged after 12-24 hours in culture using a Nikon Eclipse TE3000 (20x Ph1, NA = 0.45). Images were captured every 30 seconds using a Hamamatsu c4742-95 camera (Hamamatsu Photonics, Japan) and OpenLab software.
Immunofluorescence
To visualize axons for orientation analysis (see below), eye primordia were fixed after 24 h in 2% PFA + 7.5% sucrose and stained for β-tubulin (primary: ab6046, 1:1000; secondary: ab175470, 1:1000). Actin was visualized with phalloidin (A12379; Life Technologies, UK). Images were taken of axons close to eye primordia using a Zeiss AxioObserver.A1 (40x water immersion objective, NA = 1.1) and an sCMOS camera (Zyla 4.2, Andor). To study the Piezo1 distribution, eye primordia were cultured on glass-bottom microwell dishes (MatTek corporation, MA, USA) fixed as above, stained with β-tubulin (primary: ab6046, 1:1000; secondary: ab150075, 1:1000) and Piezo1 (primary: sc164319, 1:200; secondary: ab175704, 1:1000); actin was visualized with phalloidin (A12379; Life Technologies, UK). Images of axons and growth cones were taken using a Nikon Eclipse TE2000-U inverted fluorescence microscope (60x oil immersion, NA = 1.4).
OT imaging in intact brains
To visualize the OT and cell nuclei for cell density measurements, the lens primordia of stage 40 embryos were removed, and a plug of semidried HRP (30% HRP in 1% lysolecithin) was placed in the lens cavity. Embryos were fixed 30 min later in 4% paraformaldehyde for 1h at room temperature. After fixation, brains were dissected and reacted with diaminobenzidine (DAB; 1 tablet DAB into 15ml 0.1M Tris buffer and 12µl 30% H2O2). Nuclei were labelled using 4,6-diamidino-2-phenylindole (DAPI, 1 μg ml−1). Brains were mounted in Fluoromount-G (eBioscience, UK), and the lateral view of the OT was imaged using a confocal microscope (SP8, Leica Microsystems, UK; 20x objective, N.A. = 0.75).
To visualize the OT in mechanically manipulated brains, embryos were fixed in 4% Paraformaldehyde overnight at 4°C. DiI crystals were diluted in ethanol and injected at the boundary between lens and the retina as previously described51. After 24h of incubation at room temperature, brains were dissected out, mounted in PBS, and the lateral view of the OT was imaged using a Nikon Eclipse 80i microscope (10x, NA = 0.3; and 20x, NA = 0.75).
In vivo AFM experiments
Tipless silicon cantilevers (Arrow-TL1; NanoWorld, Switzerland) were mounted on a JPK Nanowizard Cellhesion 200 (JPK Instruments AG, Germany), which was set up on a x/y-motorized stage of an inverted optical microscope (Axio ObserverA1, Zeiss, UK). Cantilever spring constants were determined via the thermal noise method52 and cantilevers with spring constants between 0.01 and 0.03 N/m selected. Monodisperse polystyrene beads (diameter: (37.28 ±0.34) µm; microParticles GmbH, Germany) were glued to the cantilevers as probes53,54.
Xenopus embryos were anesthetized and one hemisphere of the intact brain exposed by removing skin and dura as described in Chien et al.48 (Fig. 2a). Embryos were then transferred to a Petri dish onto the motorized stage, and immobilized using a harp slice grid (ALA Scientific, NY, USA). Epifluorescence and bright field images were taken to identify the OT. On the exposed brains, the region containing the OT was selected. Images of the upper right and lower left corners of the selected region were taken with a CCD camera (Imaging Source, UK) mounted on a TopViewOptics™ upright imaging system (JPK Instruments AG, Germany), to identify the region of the brain mapped by the AFM after data analysis. Force-distance curves (maximum indentation force: 7 nN, approach speed: 10 µm/s, Data rate: 1000 Hz) were taken every 20 or 25 µm apart in a raster scan using a custom-written script.
For local brain stiffening experiments, anaesthetised stage 35/36 Xenopus embryos with one brain hemisphere exposed as described above were transferred to 1.3x MR solution (composition: 1.3x MBS with 0.04% (w/v) MS222 and 1X P/S/F (pH 7.4); the higher osmolarity retards skin regrowth for the duration of the experiment.). Epifluorescence and bright field images were collected using a modified AxioZoom V.16 system (Zeiss, UK) connected to an Andor Zyla 4.2 CMOS camera to identify the position of the OT. To induce local strain stiffening at the mid-diencephalon, tipless silicon cantilevers (Shocon-TL; AppNano, CA, USA) with attached polystyrene beads of 89.3 µm diameter (microParticles GmbH, Germany) were used to apply a constant force of 30 nN to a region towards the front of the advancing OT. The force was applied for ∼6 hours at 25°C until embryos had reached stage ∼39. Controls were treated in the same way except for the AFM application. After removal of the cantilever, manipulated and control embryos were fixed in 4% PFA, and the optic tract labelled with DiI as described above for analysis.
Data analysis
Image pre-processing of in vitro experiments
Images of Xenopus eye primordia were imported into ImageJ. Some images were corrected for uneven background illumination using a Fourier bandpass filter. Large features in the background were manually removed and images binarised. The resulting binary images were used as input for two independent analyses, Sholl analysis and turning angle analysis (see below). For the turning angle analysis, the region corresponding to the eye primordium was manually adjusted by an ellipse in each image. All the pixels inside this ellipse were set to zero (background), as well as those pixels belonging to connected components of size smaller than 45 pixels (18.8 µm2). Each eye primordium’s centre of mass was set as the origin of a 2D coordinate system with the gradient orientated along the y-axis.
Sholl analysis55
Images were analyzed using the Sholl Analysis plugin in ImageJ. An ellipse was fit to the eye primordium and the initial radius set to where A is the area of the ellipse. The outer radius was set beyond the extent of the longest axon bundle, and the radius step size (spacing between consecutive circles) was set to 6.25 µm (10px).
In vitro time-lapse experiments
8-bit greyscale images were imported into Fiji software. Twenty axons per eyeball were chosen at random and the path each growth cone traversed over a ∼3 h period was manually tracked using the ‘Tracking’ function of Fiji. After tracking growth cones of 60 axons on both stiff and soft substrates, we analysed the trajectories using a custom written MATLAB script (Mathworks, MA, USA). The growth velocity for each axon vGrowth was calculated by
with the vector to the start position and end position of the growth cone and the time t. The speed for each growth cone vGC was calculated by
with the position of the growth cone on the ith frame and the total frame number N. The directionality of the growth cones’ path D was calculated by
D = 1 corresponds to straight growth, while D close to 0 corresponds to a random walk.
The direction of growth cone migration φi for the position i ∈ {x | x∈ℕ; x > 64; x ≤ N} was calculated as follows.
For the third component of
And for the third component of
Analysis of axon orientation on plain substrates
Images of axons stained for β-tubulin were imported into Fiji and cropped to ∼(140 × 45) µm2 regions. These were analyzed using the OrientationJ plugin. Orientation values were weighted by coherency (a parameter between 0 and 1, which indicates how locally co-aligned image features are). The distribution of angles for each image was adjusted so that the median angle was 0°.
AFM data
To quantify data from in vivo AFM indentation experiments, we used a custom-written automated routine based in MATLAB to analyse force-distance curves18. This routine is based on the Hertz model56:
with applied force F, Young’s modulus E, Poisson’s ratio v, indenter radius r, indentation depth δ, and apparent reduced elastic modulus . Curves were analyzed for defined indentation depths (Supplementary Fig. 3) or the maximum applied force F = 7nN (Figs. 2, 4). K values were then colour-coded and mapped onto the image of the Xenopus brain using a custom-written MATLAB script (hereafter termed ‘stiffness map’).
For the quantification of the mechanical properties of different regions of interest (ROIs), we defined the OT by manually drawing an outline as indicated in Supplementary Figs. 3b and d. Rostral and caudal of the OT (Fig. 2h) was defined as an approximately 50µm wide area rostral and caudal to the tract, respectively, while for the quantification of local gradients parallel to the OT (Fig. 3b, c) ROIs were defined as described in the text and shown in Fig. 3a. The front of the OT was defined as an approximately 50µm wide area in front of the tract, which was ∼25 µm wider than the OT on each side as indicated in Fig. 4a. The tectum was defined based on its anatomic location (cf. Figs. 2, 4). ROIs were selected 3 times by hand using a custom-written MATLAB script; a pixel was chosen to be part of the ROI if it was selected at least 2 out of 3 times. Afterwards, the image of the Xenopus brain was scaled down to match the resolution of the stiffness map. Element-wise multiplication of the stiffness map matrix with the downscaled selection matrix resulted in the selection of the measurements in the selected area. Subsequently, all selected measurements of all experiments were pooled and then further analyzed as described in the statistics section.
Cell density measurements
Image stacks were imported into Fiji. For each brain, the image where the OT was in focus at the caudal bend was determined, and a maximum projection of that image and the image before and after it made (2µm z-stack height). A Gaussian blur filter (sigma = 2.0) was used to remove noise. The resulting image was thresholded; thresholds were manually adjusted to ensure that most of the nuclei were captured. Regions of interest were selected manually at the rostral and caudal sides of the OT. The image was binarized and the ‘Analyze Particle’ function (size: 2-∞; circularity: 0.5-1.00) was used to acquire the area of nuclei in each ROI. The relative cell density was calculated by dividing the area with nuclei by the total area of the ROI.
OT curvature
To extract the relationship between the local stiffness gradient M perpendicular to the OT and the curvature of the OT at each position we used the following algorithm. The path of the centre of the OT was drawn by hand. Every 5th point of this line was selected and data was smoothed to reduce irregularities in the outline (points were approximately 40µm apart). The curvature C at the ith selected point was calculated by:
with the radius R (which can be negative or positive, depending on A) of the circumscribed circle of the triangle between the (i-1)th, ith and (i+1)th point, the ‘area’ A being the 3rd component of the three-dimensional (3D) vector which points from the (i-1)th to the ith point (3rd component of being 0), the 3D vector which points from the (i-1)th to the (i+1)th point (3rd component of being 0), the length the length b of the line between the ith and (i+1)th point and the length To calculate the corresponding local stiffness gradient perpendicular to the tract M we first fitted a line through 5 points to obtain the general direction around the middle point, for which we already calculated the curvature. Thereafter, we calculated the average of the apparent reduced elastic modulus of a (100 × 100) µm2 area left, K1, and right, K2, of this line (Fig. 3a). The gradient was then calculated by:
For better visualization of axon turning towards softer vs. stiffer tissue, the left side of the coordinate system showing local curvature C as a function of local stiffness gradient M < 0 was rotated by 180° (Fig. 3b).
In vitro turning angle analysis
To detect how many segments of axons in each half of the coordinate system (x < 0 and x > 0; see Image pre-processing of in vitro experiments) were aligned along the direction defined by the turning angle τ, binary images were rotated by this angle and searched for peaks higher than 20 pixels (∼13 µm) in the histogram of the rotated x-coordinate. Because the pixels that contribute to the same bin of the histogram may not belong to the same segment of axon in the binary image, we performed an additional connectivity analysis to identify how many segments contribute to each bin. This way, on average ∼2800 axon segments were analysed per eye primordium. To account for the clockwise growth of axons on 2D substrates57, turning angles were normalized by subtracting the median angle of the total explant from the median angle of the individual halves. Plotted is the distribution of median turning angles for both image halves; for better visualization of turning towards softer vs. stiffer substrates in Fig. 3e, the left side of the coordinate system (x < 0) was rotated by 180° and data points were pooled.
In a second approach, we utilized time lapse imaging of individual axons or tight axon bundles grown on substrates incorporating a linear stiffness gradient. The initial direction of the axon was determined by the most recent ∼50 µm of axon growth, and only straight axons growing approximately perpendicular to the stiffness gradient were selected. Axons were left to grow for one hour, and the new growth direction determined by determining the angle between the initial growth direction and the new axon segment (Fig. 3f). To correct for the clockwise growth of axons in 2D cultures57, equal numbers of axons were selected from each side of the stiffness gradient.
Shape characterization of the OT
Outlines of the OT distal of the optic chiasm were drawn manually in Corel Draw X5. The elongation of the OT was calculated by the major to minor axis ratio using an automated algorithm in MATLAB as previously described23. Briefly, the axes were determined by fitting ellipses – with the same normalized second central moment as the OT area – around OTs.
Statistics and visualization
Data were collected from at least 3 independent experiments (N ≥ 3). Sample sizes were chosen using a Power & Sample Size Calculator (http://www.statisticalsolutions.net/pss_calc.php). The order of data collection was randomized, no blinding was done, and no data were excluded from the analysis. Normality was tested for all data sets using the Shapiro-Wilk test; variances were compared using a χ2 test. In case of normal distribution, statistical comparison between two groups was done using two-tailed unpaired or paired t-tests, and between more than two groups using ANOVA. If data did not follow a Gaussian distribution, Kruskal-Wallis ANOVA, Mann-Whitney, and one-sample Wilcoxon Signed Rank Tests were used to statistically compare more than two groups, two groups, and data within one group, respectively. Data were plotted as box plots, with boxes showing the 25th, 50th (the median), and 75th percentiles, whiskers the spread of the data (excluding outliers), and an ‘O’ the mean score for a group. Asterisks in figures indicate significance levels: *=(P<0.05), **=(P<0.01), and ***=(P<0.001). A supplementary methods checklist is available.
Supplementary Material
Acknowledgements
We would like to thank D. Bray, A. Reichenbach, B. Simons, and S. Wolff for discussions about neuronal mechanics, K. Chalut and W. Harris for feedback on the manuscript, A. Christ for an AFM data analysis code, P. Moshayedi for help with establishing PAA gels, F. Sachs (University at Buffalo, USA) for providing GsMTx4, H. Wong for extensive help in the lab, A. Winkel and R. Field (JPK) for technical help, and the German National Academic Foundation (Scholarship to D.E.K.), Wellcome Trust and Cambridge Trusts (Scholarships to A.J.T.), Winston Churchill Foundation of the United States (Scholarship to S.K.F.), Herchel Smith Foundation (Research Studentship to S.K.F.), CNPq (307333/2013-2 to L.d.F.C.), NAP-PRP-USP, and FAPESP (11/50761-2 to L.d.F.C.), UK BBSRC (BT grant to J.G. and BB/M021394/1 to K.F.), Wellcome Trust (WT085314) and ERC (322817) grants (CEH); Alexander von Humboldt Foundation (Feodor Lynen Fellowship to K.F.), the Human Frontier Science Program (Young Investigator Grant to K.F.), the UK Medical Research Council (Career Development Award to K.F.), and the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number R21HD080585 (to K.F.) for funding support. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions: K.F. conceived the project; J.G., C.E.H., and K.F. designed the research; D.E.K., A.J.T., S.K.F, A.D., G.K.S., E.P., H.S., and K.F. performed the experiments; D.E.K., A.J.T., S.K.F, G.K.S., E.P, H.S., M.V., L.F.C., and K.F. analysed the data; all authors discussed the data; K.F. wrote the manuscript with contributions from all authors.
Competing financial interests: The authors declare no competing financial interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Sperry RW. Chemoaffinity in the Orderly Growth of Nerve Fiber Patterns and Connections. Proc Natl Acad Sci U S A. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 3.Erskine L, Herrera E. The retinal ganglion cell axon's journey: insights into molecular mechanisms of axon guidance. Dev Biol. 2007;308:1–14. doi: 10.1016/j.ydbio.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Campbell DS, et al. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J Neurosci. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piper M, et al. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson-Leadbeater K, et al. Dynamic expression of axon guidance cues required for optic tract development is controlled by fibroblast growth factor signaling. J Neurosci. 2010;30:685–693. doi: 10.1523/JNEUROSCI.4165-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 8.Moore SW, Biais N, Sheetz MP. Traction on immobilized netrin-1 is sufficient to reorient axons. Science. 2009;325:166. doi: 10.1126/science.1173851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betz T, Koch D, Lu YB, Franze K, Kas JA. Growth cones as soft and weak force generators. Proc Natl Acad Sci U S A. 2011;108:13420–13425. doi: 10.1073/pnas.1106145108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch D, Rosoff WJ, Jiang J, Geller HM, Urbach JS. Strength in the periphery: growth cone biomechanics and substrate rigidity response in peripheral and central nervous system neurons. Biophys J. 2012;102:452–460. doi: 10.1016/j.bpj.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss P. In vitro experiments on the factors determining the course of the outgrowing nerve fiber. Journal of Experimental Zoology. 1934;68:393–448. [Google Scholar]
- 12.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;90:3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostic A, Sap J, Sheetz MP. RPTPalpha is required for rigidity-dependent inhibition of extension and differentiation of hippocampal neurons. J Cell Sci. 2007;120:3895–3904. doi: 10.1242/jcs.009852. [DOI] [PubMed] [Google Scholar]
- 15.Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. Effect of dynamic stiffness of the substrates on neurite outgrowth by using a DNA-crosslinked hydrogel. Tissue Eng Part A. 2010;16:1873–1889. doi: 10.1089/ten.TEA.2009.0574. [DOI] [PubMed] [Google Scholar]
- 16.Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Agarose gel stiffness determines rate of DRG neurite extension in 3D cultures. Biomaterials. 2001;22:1077–1084. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- 17.Elkin BS, Azeloglu EU, Costa KD, Morrison B., 3rd Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J Neurotrauma. 2007;24:812–822. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
- 18.Christ AF, et al. Mechanical difference between white and gray matter in the rat cerebellum measured by scanning force microscopy. J Biomech. 2010;43:2986–2992. doi: 10.1016/j.jbiomech.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Franze K, et al. Spatial mapping of the mechanical properties of the living retina using scanning force microscopy. Soft Matter. 2011;7:3147–3154. [Google Scholar]
- 20.Iwashita M, Kataoka N, Toida K, Kosodo Y. Systematic profiling of spatiotemporal tissue and cellular stiffness in the developing brain. Development. 2014;141:3793–3798. doi: 10.1242/dev.109637. [DOI] [PubMed] [Google Scholar]
- 21.Elkin BS, Ilankovan A, Morrison B., 3rd Age-dependent regional mechanical properties of the rat hippocampus and cortex. J Biomech Eng. 2010;132:011010. doi: 10.1115/1.4000164. [DOI] [PubMed] [Google Scholar]
- 22.Koser DE, Moeendarbary E, Hanne J, Kuerten S, Franze K. CNS cell distribution and axon orientation determine local spinal cord mechanical properties. Biophys J. 2015;108:2137–2147. doi: 10.1016/j.bpj.2015.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weickenmeier J, et al. Brain stiffness increases with myelin content. Acta Biomater. 2016 doi: 10.1016/j.actbio.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 24.Moshayedi P, et al. Mechanosensitivity of astrocytes on optimized polyacrylamide gels analyzed by quantitative morphometry. J Phys Condens Matter. 2010;22 doi: 10.1088/0953-8984/1022/1019/194114. [DOI] [PubMed] [Google Scholar]
- 25.Franze K, Janmey PA, Guck J. Mechanics in neuronal development and repair. Annu Rev Biomed Eng. 2013;15:227–251. doi: 10.1146/annurev-bioeng-071811-150045. [DOI] [PubMed] [Google Scholar]
- 26.Guan W, Puthenveedu MA, Condic ML. Sensory neuron subtypes have unique substratum preference and receptor expression before target innervation. J Neurosci. 2003;23:1781–1791. doi: 10.1523/JNEUROSCI.23-05-01781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franze K, et al. Neurite branch retraction is caused by a threshold-dependent mechanical impact. Biophys J. 2009;97:1883–1890. doi: 10.1016/j.bpj.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerstein PC, et al. Mechanosensitive TRPC1 channels promote calpain proteolysis of talin to regulate spinal axon outgrowth. J Neurosci. 2013;33:273–285. doi: 10.1523/JNEUROSCI.2142-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang QY, et al. Stiff substrates enhance cultured neuronal network activity. Sci Rep. 2014;4:6215. doi: 10.1038/srep06215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibasaki K, Murayama N, Ono K, Ishizaki Y, Tominaga M. TRPV2 enhances axon outgrowth through its activation by membrane stretch in developing sensory and motor neurons. J Neurosci. 2010;30:4601–4612. doi: 10.1523/JNEUROSCI.5830-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak MM, et al. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc Natl Acad Sci U S A. 2014;111:16148–16153. doi: 10.1073/pnas.1409802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coste B, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515:279–282. doi: 10.1038/nature13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottlieb PA, Sachs F. Piezo1: properties of a cation selective mechanical channel. Channels (Austin) 2012;6:214–219. doi: 10.4161/chan.21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majkut S, et al. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol. 2013;23:2434–2439. doi: 10.1016/j.cub.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walz A, Anderson RB, Irie A, Chien CB, Holt CE. Chondroitin sulfate disrupts axon pathfinding in the optic tract and alters growth cone dynamics. J Neurobiol. 2002;53:330–342. doi: 10.1002/neu.10113. [DOI] [PubMed] [Google Scholar]
- 40.Pogoda K, et al. Compression stiffening of brain and its effect on mechanosensing by glioma cells. New J Phys. 2014;16:075002. doi: 10.1088/1367-2630/16/7/075002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isenberg BC, Dimilla PA, Walker M, Kim S, Wong JY. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu G, et al. Opening angles and material properties of the early embryonic chick brain. J Biomech Eng. 2010;132:011005. doi: 10.1115/1.4000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franze K. The mechanical control of nervous system development. Development. 2013;140:3069–3077. doi: 10.1242/dev.079145. [DOI] [PubMed] [Google Scholar]
- 45.Singh T, Meena R, Kumar A. Effect of sodium sulfate on the gelling behavior of agarose and water structure inside gel networks. J Phys Chem B. 2009;113:2519–2525. doi: 10.1021/jp809294p. [DOI] [PubMed] [Google Scholar]
- 46.Hopker VH, Shewan D, Tessier-Lavigne M, Poo M, Holt C. Growth-cone attraction to netrin-1 is converted to repulsion by laminin-1. Nature. 1999;401:69–73. doi: 10.1038/43441. [DOI] [PubMed] [Google Scholar]
- 47.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. North-Holland Pub. Co.; 1967. [Google Scholar]
- 48.Chien CB, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- 49.Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalous A, Stake JI, Yisraeli JK, Holt CE. RNA-binding protein Vg1RBP regulates terminal arbor formation but not long-range axon navigation in the developing visual system. Dev Neurobiol. 2014;74:303–318. doi: 10.1002/dneu.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wizenmann A, et al. Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron. 2009;64:355–366. doi: 10.1016/j.neuron.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutter JL, Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Review of Scientific Instruments. 1993;64:1868–1873. [Google Scholar]
- 53.Mahaffy RE, Shih CK, MacKintosh FC, Kas J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys Rev Lett. 2000;85:880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- 54.Franze K. Atomic force microscopy and its contribution to understanding the development of the nervous system. Curr Opin Genet Dev. 2011;21:530–537. doi: 10.1016/j.gde.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- 56.Hertz H. Über die Berührung fester elastischer Körper. Journal für die reine und angewandte Mathematik. 1881;92:156–171. [Google Scholar]
- 57.Tamada A, Kawase S, Murakami F, Kamiguchi H. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J Cell Biol. 2010;188:429–441. doi: 10.1083/jcb.200906043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.