Abstract
Spinal Muscular Atrophy (SMA) is an autosomal recessive motor neuron disease that results in loss of spinal motor neurons, muscular weakness and, in severe cases, respiratory failure and death. SMA is caused by a deletion or mutation of the SMN1 gene and retention of the SMN2 gene that leads to low SMN expression levels. The measurement of SMN mRNA levels in peripheral blood samples has been used in SMA clinical studies as a pharmacodynamic biomarker for response to therapies designed to increase SMN levels. We recently developed a postnatal porcine model of SMA by the viral delivery of a short-hairpin RNA (shRNA) targeting porcine pSMN. scAAV9-mediated knockdown of pSMN mRNA at postnatal day 5 reliably resulted in denervation, weakness and motor neuron and ventral root axon loss that began 3–4 weeks after viral delivery, and this phenotype could be ameliorated by subsequent viral delivery of human SMN (hSMN). To determine if the effect of modulating SMN levels using gene therapy can be measured in blood, we measured expression of pSMN mRNA and hSMN mRNA by quantitative droplet digital PCR (ddPCR). We found that the endogenous expression of pSMN mRNA in blood increases in the first month of life. However, there were no significant differences in blood levels of pSMN mRNA after knock-down or of human SMN mRNA after gene therapy. Our results, obtained in a large animal model of SMA that is similar in size and anatomy to human infants, suggest that measurement of SMN mRNA levels in blood may not be informative in SMA clinical trials involving intrathecal delivery of SMN-modulating therapies.
Keywords: ddPCR, gene-therapy, scAAV9-shRNA, scAAV9-SMN, SMA, SMN
Introduction
Spinal muscular atrophy (SMA) is the leading genetic cause of death in infants, exhibits a wide range of clinical severity and has an incidence of 1 in 11,000 live births (Pearn 1973, Sugarman, Nagan et al. 2012). SMA is an autosomal recessive disease caused by deletion or mutation in the SMN1 (survival motor neuron 1) gene, and the retention of the nearly identical SMN2 (survival motor neuron 2) which leads to reduction of SMN protein levels (Lefebvre, Burglen et al. 1995, Coovert, Le et al. 1997, Lefebvre, Burlet et al. 1997). SMN1 functionally differs from SMN2 by a single nucleotide that results in the exclusion of exon 7 in approximately 90% of SMN transcripts insert (Lorson, Hahnen et al. 1999, Monani, Lorson et al. 1999). The majority of SMN mRNA from SMN2 lacks exon 7, (SMNΔ7) that produces a truncated protein that does not oligomerize efficiently and is rapidly degraded. There is a well-established positive correlation between SMN2 copy number and disease severity (McAndrew, Parsons et al. 1997, Mailman, Heinz et al. 2002, Prior, Krainer et al. 2009).
Over the last 5 years, promising results in preclinical models of SMA have led to a surge of clinical trials using small molecule, oligonucleotide-based therapies and viral-mediated gene therapies (Kolb and Kissel 2011, Arnold and Burghes 2013). Oral, intravenous and intrathecal routes of delivery have been explored in animal studies. The first FDA-approved therapy for SMA, nusinersen, involves delivery of an oligonucleotide via an intrathecal injection (Finkel, Chiriboga et al. 2017). An additional promising strategy has been to harness the ability of the self-complementary adeno-associated virus, serotype 9 (scAAV9) to transduce motor neurons with a copy of the human SMN1 cDNA under the chicken β-actin promoter when delivered intravascularly or intrathecally (Foust, Wang et al. 2010, Bevan, Duque et al. 2011, Duque, Arnold et al. 2015). These therapies demonstrated survival extension of mouse models of SMA from ~14 days to over 250 days and correction of neuromuscular junction transmission when initiated early in the course of disease (Foust, Wang et al. 2010). A Phase 1 clinical trial in infants with SMA using scAAV9-SMN delivered intravenously is nearing completion (Clinical Trials Identifier NCT02122952).
Recently, we reported the creation of a porcine model of SMA using intrathecal (IT) delivery of an scAAV9 containing a short hairpin RNA (shRNA) directed at the porcine SMN (pSMN) gene (Duque, Arnold et al. 2015). scAAV9-mediated knockdown of pSMN mRNA at postnatal day 5 reliably resulted in denervation, weakness, motor neuron and ventral root axon loss that began 3–4 weeks after viral delivery and a >70% reduction of SMN expression in motor neurons (Duque, Arnold et al. 2015). These piglets developed progressive weakness over the next 3–4 weeks and motor neuron degeneration was seen pathologically (Duque, Arnold et al. 2015). Subsequent IT delivery of the human SMN cDNA via scAAV9 dramatically prevented the progression of weakness in this model when delivered soon after pSMN knock-down (on PND6) (Duque, Arnold et al. 2015). Late delivery of SMN at the onset of symptoms (PND33–36) halted the progression of disease in the piglets (Duque, Arnold et al. 2015). Peripheral blood SMN levels serve as a potential pharmacodynamic biomarker and may be useful in the interpretation of clinical outcomes in the setting of SMA clinical trials (Brichta, Holker et al. 2006, Tiziano, Lomastro et al. 2013, Renusch, Harshman et al. 2015). In this study, we sought to evaluate SMN mRNA levels in blood as a potential pharmacodynamic biomarker upon pSMN knockdown and subsequent therapy in piglets.
Materials and Methods
Vectors
Small hairpin RNA (shRNA) sequence targeting pig SMN (or scrambled shRNA) under control of the H1 promoter was cloned into a self-complementary AAV9-based backbone plasmid along with reporter gene GFP under the chicken β-actin (CBA) promoter (Duque, Arnold et al. 2015). Human SMN cDNA under the CBA promoter was cloned into the same backbone plasmid as previously described (Foust, Wang et al. 2010). The scAAV9-shRNA against pSMN and scAAV9-SMN vectors were produced by tri-transfection of HEK293 cells as previously described (Foust, Wang et al. 2010). Vector titers were determined using Real-Time PCR and expressed as viral genome (vg) per milliliter.
Animal care and use
All animals procedures performed were in strict accordance to The Ohio State University Institutional Animal Care and Use Committees (IACUC).
Study Design
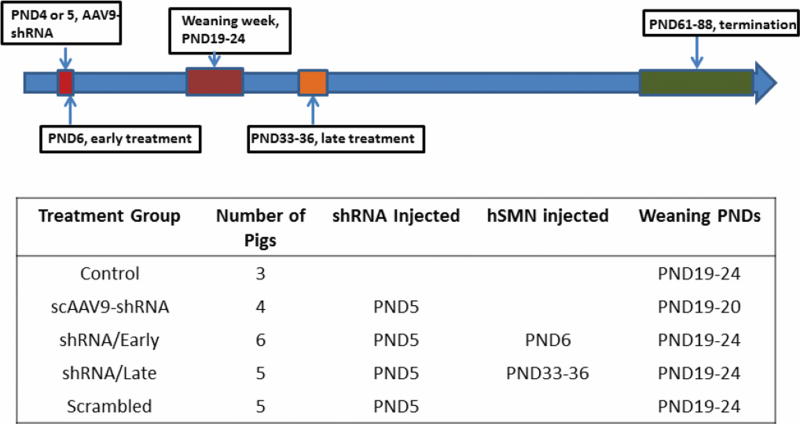
The sows (Sus scrofa domestica) were obtained from a regional farm (Hartley Farm, OH). The sow delivered piglets who had full access to her for nursing and comfort. Five-day old (postnatal day 5, PND5) piglets were induced and maintained under anesthesia by mask inhalation of 5% isoflurane in oxygen. Body temperature, electrocardiogram and respiratory rate were monitored throughout the procedure. Piglets were injected with scAAV9-shSMN, scAAV9-SMN, or scrambled shRNA in the cisterna magna as previously described (Bevan, Duque et al. 2011, Federici, Taub et al. 2012, Duque, Arnold et al. 2014). Piglets received scAAV9-shRNA at PND 4 or 5 (Figure 1). Fifteen piglets received scAAV9-shRNA knock-down virus against pSMN; 6 of these received a second intrathecal injection 24 hours later with the rescue vector scAAV9-SMN as an early treatment group (shRNA/Early group, n=6); 5 of scAAV9-shRNA-injected animals received a second intrathecal injection during PND33–36 at the onset of weakness, as a late treatment group (shRNA/Late group, n=5). Three piglets were in the control, non-injected group (Control group, n=3), while 5 piglets received a scrambled shRNA construct as a control, injected group (Scrambled group, n=5) (Figure 1).
Figure 1.
Illustration of experimental groups and longitudinal timelines in the study. The scAAV9-shRNA against pSMN or scrambled-shRNA was injected into five-day old piglets to generate a large-animal SMA model. The early-treatment group (shRNA/Early) consists of knockdown piglets receiving a second intrathecal injection 24 hours later with the rescue vector scAAV9-SMN. In the late-treatment group (shRNA/Late), SMA animals received the second intrathecal injection of scAAV9-SMN at the onset of symptoms at around postnatal day (PND) 33–36. Blood was withdrawn pre-weaning, post-weaning and every week thereafter till the end-point of the study.
Pig blood was collected from femoral vein into the PAXgene Blood RNA Tubes (PreAnalytiX, Franklin Lakes, New Jersey) at PND4 or 5 for all cohorts and again after weaning, which occurred over the course of 3–4 weeks and ranged from PND19–24. Therefore, the period of PND4–5 was defined as pre-wean and the first blood collection day after weaning was defined as post-wean PNDs (PND24–40). Blood was then collected every week (±2 days) for the remainder of the study. When the timing of scAAV9 delivery coincided with collection of a blood sample, as with the late treatment cohort (PND33–36), blood was drawn prior to administration of scAAV9-SMN.
Laser-capture Microdissection of spinal cord motor neurons (LCM) and cDNA synthesis of LCM-material
The lumbar spinal cord was flash frozen in liquid nitrogen-cooled isopentane and cryosectioned (IEC Minotome Plus). 14µm sections of lumbar spinal cord were collected onto PEN membrane slides (Zeiss), fixed in methanol (1 min), stained with 1% cresyl violet in methanol (1.5 min), quickly rinsed in methanol and immediately stored at −80°C for no more than a week with dessicant. Laser capture microdissection (LCM) was performed using the Palm Microbeam IV (Carl Zeiss MicroImaging) under 10× magnification. Motor neurons were identified based on their localization in the ventral horn and morphology (large cell body). Approximately 2,000,000 µm2 of motor neuron tissue was collected per sample. In addition, 2,000,000 µm2 of dorsal horn tissue was collected from the same sections. RNA was isolated using the RNaqueous Micro Kit (Ambion) according to the manufacturer’s instructions.
cDNA synthesis
Following collection in PAXgene Blood RNA Tubes (PreAnalytiX, Franklin Lakes, New Jersey), blood was allowed to stand at room temperature for up to 2 hours. RNA isolation was conducted using the PAXgene Blood RNA Kit according to the manufacturer’s instructions. RNA samples were then treated with TURBO DNase (Ambion) to remove contaminating genomic DNA. An aliquot of 1µg RNA was used to generate cDNA by two-step reactions using AMV-RT (Invitrogen) and random hexamer primers.
SMN mRNA Quantification by ddPCR
SMN mRNA levels were determined using digital droplet PCR (ddPCR, Biorad). For pig and human SMN mRNA analyses, the following primers and probes were used: Pig SMN (pSMN) Fd 5’-ggcggcagcggtgtt-3’, pSMN Rv 5’-gaatcatcactctggcctgca-3’, pSMN probe 5’FAM-ctgaggcggaggact-MGB; Human SMN (hSMN) Fd 5’-caaaaagaaggaaggtgctca-3’, hSMN Rv 5’-tccagatctgtctgatcgttt-3’, hSMN probe 5’FAM-ttaaggagaaatgctggcatagagcagcac-MGB. GAPDH quantification was used to normalize the SMN levels. The sequence of the GAPDH primers and probe are as follows: Pig GAPDH (pGAPDH) Fd 5’-ccccaacgtgtcggttgt-3’, pGAPDH Rv 5’-cctgcttcaccaccttcttga-3’, pGAPDH probe 5’VIC-agaaacctgcaaaata-MGB. The primers and probes for SMN and GAPDH were obtained from Applied Biosystems.
Technical replicates were performed in multiplex reactions with 1 µl of cDNA, 1.8 µl each of human SMN or pig SMN forward and reverse primers (900 nM final), 1.8 µl each of GAPDH forward and reverse primers, 0.5 µl of each probe (250nM final) and 10 µl of 2× ddPCR supermix (BioRad) in a 20 µl final volume. This reaction was transferred to an 8-row droplet generator cartridge (BioRad) wherein 70 µl of droplet generation oil was added. The cartridge was then placed in the QX100 droplet generator (BioRad). The machine generates ~17,000–20,000 droplets per sample and thus the PCR occurs as individual PCR reactions in droplets. The droplets were then transferred to a 96-well plate and placed in a conventional PCR machine and cycled under standard conditions: 95°C for 10min followed by 40 cycles of 94°C for 30sec, 60°C for 1min and a final step of 98°C for 10min. After PCR amplification, the fluorescence was read by the QX200 droplet reader (BioRad). Thus, ddPCR is an end-point quantification. Results were analyzed using the QuantaSoft analysis software, based on Poisson statistics (BioRad). Data is expressed as relative fluorescence units (RFU) of pSMN or hSMN normalized by GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Statistical analyses
Descriptive statistics were used to summarize all baseline values. Continuous values are expressed using means, standard deviations and other appropriate measures of spread. Categorical values are expressed using frequencies and percentages. Data management and T-tests were performed using Sigmaplot v12.0 (Systat Software Inc, CA, USA). Mixed models were used to estimate the slope of the ratio (SMN/GAPDH) overtime, for each treatment group (Figure 3). Time (as measured by PND) and the time & treatment interaction term were included in the models and litter was included as a random effect. Litter was not significant in the model (p=0.062). In order to compare treatment effects, the differences in slopes were estimated between pairwise groups. All reported p-values and confidence intervals are two sided and unadjusted for multiple comparisons. Statistical analyses were performed in SAS software 9.4 (SAS Institute Inc., Cary, NC, USA).
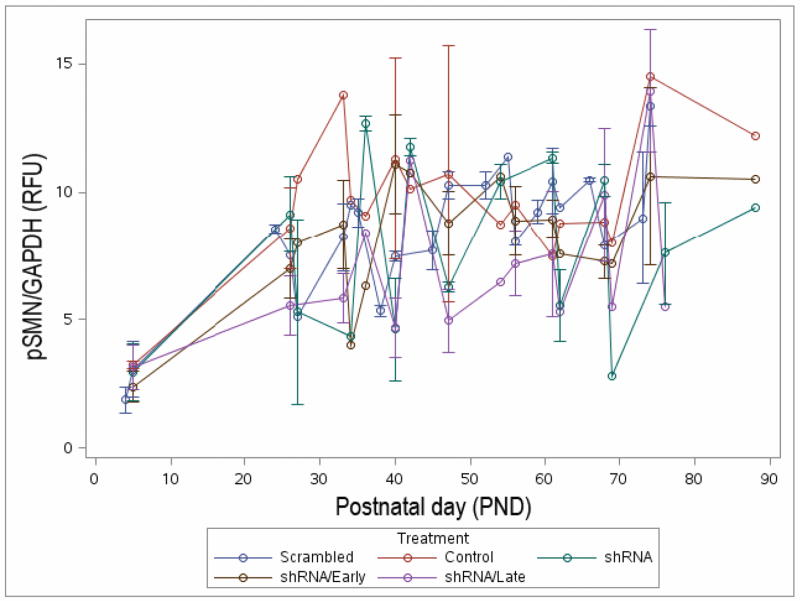
Figure 3.
There is no significant difference in longitudinal pSMN expression in blood. pSMN in blood was analyzed by ddPCR every week (±2 days) post-weaning up to end-point of the study (PND61–88).
Results
Porcine blood SMN mRNA expression increases in the first two months of life
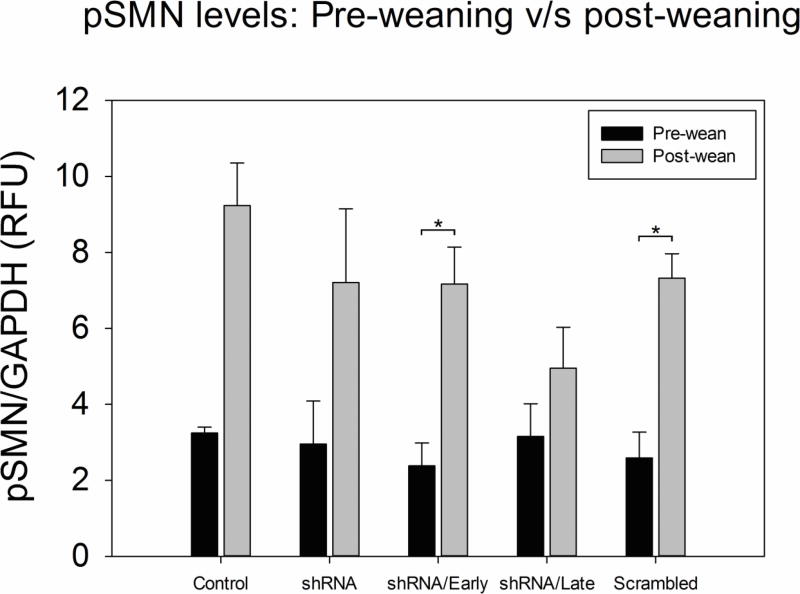
There were a total of 5 study groups as outlined in Figure 1. Pre-weaning blood samples were collected at PND4 or PND5. Weaning occurred approximately 3 weeks after birth (range: PND19 - 24), at which time blood was collected again. The pSMN mRNA levels in blood increased between pre-wean and post-wean time points for each group (Figure 2). Table 1 shows the mean data with standard deviation expressed as relative fluorescence units (RFU) of pSMN normalized to GAPDH. pSMN levels in the control (n=3) and scramble (n=5) groups increased by 2.8 fold upon weaning (control: 3.24 ± 0.15 RFU v/s 9.23 ± 1.12 RFU; scramble: 2.59 ± 0.67 RFU v/s 7.32 ± 0.64 RFU, p≤0.05). The shRNA (n=4) and shRNA/Late (n=5) groups showed a 2.4 and 1.5 fold increase in pSMN respectively post-weaning, while the shRNA/Early group showed a fold increase of 3 in pSMN levels post-weaning (2.38 ± 0.59 RFU v/s 7.17 ± 0.97 RFU, p≤0.05). Thus all groups, regardless of treatment, showed a 1.5 −3 fold increase in pSMN mRNA levels between PND3–4 and PND19–24.
Figure 2.
Pig Survival Motor Neuron (pSMN) expression in blood increased after weaning in all groups. The pSMN mRNA expression in blood was determined by droplet digital PCR (ddPCR) at two time points – before weaning (PND4–5) and at weaning (PND19–24) and normalized to GAPDH. The control (n=3) and scramble groups showed a 2.8 fold increase in pSMN post-weaning. The shRNA group (n=4) showed a 2.4 fold increase in pSMN post-weaning, while the shRNA/Early (n=6,) and shRNA/Late (n=5) showed a 3× and 1.5× increase respectively, in blood pSMN upon weaning. (*p≤0.05, t-test, error bars = sem)
Table 1.
pSMN mRNA levels in porcine blood before and after weaning
| Group | |||||
|---|---|---|---|---|---|
| Control (n = 3) |
Scramble (n =5) |
shRNA (n = 4) |
shRNA/Early (n = 6) |
shRNA/Late (n =5) |
|
| Pre-wean pSMN mRNA (RFU) | 3.2 ± 0.0.1 | 2.5 ± 0.6 | 2.9 ± 1.1 | 2.3 ± 0.5 | 3.1 ± 0.8 |
| Post-wean pSMN mRNA (RFU) | 9.2 ± 1.1 | 7.3 ± 0.6 | 7.2 ± 1.9 | 7.1 ± 0.9 | 4.9 ± 1.0 |
| Fold Change | 2.8 | 2.8 | 2.4 | 3 | 1.5 |
Porcine blood SMN levels were not significantly altered by intrathecal delivery of sacAAV9-SMN
Blood samples were collected from all cohorts every week ± 2 days for pSMN analysis after weaning. All animals were sacrificed between PND61 and PND88. None of the groups show clear trends in SMN levels in blood over time (Figure 3). There was large variability observed at all time-points across groups.
In order to reveal the trend changes in each group over time, we compared the difference in slopes between pair-wise groups. The slopes for the various treatments were estimated and then pairwise compared (Table 2). In general, there is insufficient evidence to suggest differences in the trajectories between groups (Table 2). Note that the difference in the estimated slopes for the shRNA/Late and shRNA/Early cohorts was only 0.020 (p=0.157). Additionally neither the shRNA/Early or shRNA/Late groups were significantly different from the scramble cohort with estimated slope differences of −0.032 (p=0.285) and −0.012 (p=0.710), respectively. Also, we attempted to measure blood levels of hSMN in the scAAV9-SMN early and late treatment groups. However, we were unable to detect any hSMN in the blood.
Table 2.
Comparison of Slopes between experimental groups
| Comparison | Estimate | 95% CI | p-value |
|---|---|---|---|
| scAAV9-shRNA v/s Control | −0.015 | (−0.089, 0.059) | 0.692 |
| shRNA/Early v/s Scrambled | −0.032 | (−0.090, 0.027) | 0.285 |
| shRNA/Late v/s Scrambled | −0.012 | (−0.072, 0.049) | 0.710 |
| shRNA/Late v/s shRNA/Early | 0.020 | (−0.038, 0.079) | 0.496 |
| scAAV9-shRNA v/s Scrambled | −0.045 | (−0.108, 0.018) | 0.157 |
Relationship between pSMN expression in blood and in motor neuron tissue
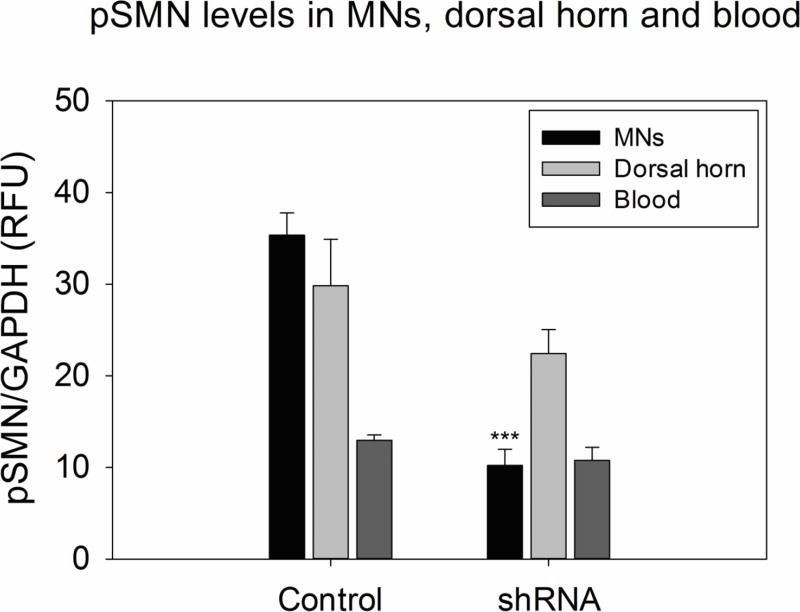
In our previous study, we reported the reduction of pSMN mRNA levels in ventral root motor neurons (MNs) and dorsal root ganglion cells in response to scAAV9-shRNA against pSMN at the end point of the study (PND61–88), (Duque, Arnold et al. 2015). Spinal cord motor neurons in the dorsal and ventral horn were assayed for pSMN and compared to pSMN expression in blood at that time point. Figure 4 shows the normalized pSMN levels in the MNs, dorsal horn and the blood between the control non-injected group and the shRNA-treated groups. The pSMN levels in MNs of the shRNA-knockdown groups show an effective 71.1% knockdown of pSMN in the motor neurons compared to samples from control littermates, thus indicating successful reduction of pSMN (p≤0.001). There was no significant reduction of pSMN in the dorsal horn and no significant reduction in pSMN detected in blood. (Table 3 and Figure 4).
Figure 4.
ddPCR quantification of pSMN mRNA in LCM-collected motor neurons in the dorsal and ventral horn, compared to pSMN levels in blood at the last time point. Ventral horn motor neurons showed a significant decrease in pSMN in the shRNA-knockdown group (71.7%) as compared to the non-injected control group. The dorsal horn and blood do not show any decrease in pSMN. (***p≤0.001, t-test, error bars = sem)
Table 3.
Comparison of pSMN mRNA levels in blood and spinal cord
| Tissue | Groups | |
|---|---|---|
|
| ||
| Control (n = 3) | shRNA (n = 10) | |
|
| ||
| Ventral Horn Motor Neuron SMN level (RFU) | 35.3 ± 2.4 | 10.2 ± 1.7 |
| Mean ± SE | ||
|
| ||
| Dorsal Horn SMN level (RFU) | 29.8 ± 5.0 | 22.4 ± 2.6 |
| Mean ± SE | ||
|
| ||
| Blood SMN level (RFU) | 12.9 ± 0.5 | 10.7 ± 1.4 |
| Mean ± SE | ||
Discussion
The increase in peripheral blood pSMN mRNA levels in the first two months of life suggests a developmental pattern of SMN expression that may not become stable until after birth. It is not yet known whether there is also an increase (or decrease) in pSMN levels in porcine motor neurons, or any other relevant cell type, in this time period. There is potential significance to this observation if a similar increase occurs in human infants in this time period, because it is in the first three months of life that infants with type 1 SMA usually present clinically with progressive weakness. Blood levels of SMN mRNA transcripts and protein appear to be stable in humans over periods of weeks (Sumner, Kolb et al. 2006, Zaworski, von Herrmann et al. 2016) and months (Renusch, Harshman et al. 2015), however these studies involved children and adults. The earliest measures of SMN mRNA in blood have been reported in infants with type 1 SMA and healthy infants ranging from 1 week old to 6 months of age (Kolb, Coffey et al. 2016). In that cross sectional report, there was no correlation between age and SMN mRNA levels in either cohort (Kolb, Coffey et al. 2016). Thus, it is possible that the increase observed here in piglets does not occur in human infants. Further study of developmental changes in SMN levels, including in the developing fetal pig, may allow for a more complete understanding of SMN gene regulation in this critical period.
Throughout this study, we observed large variability in pSMN levels in all cohorts across time. This variability made it difficult to determine differences between groups. It is unlikely that the ddPCR measurements contributed largely to this error, because all samples were measured in parallel. Potential confounders could be time of day or piglet stress levels. It will be important in future work to define the confounders of the measurement of SMN levels in piglet blood tissues as it is possible that the baseline variability seen here is biological and may more accurately reflect a day to day variability in human infant blood samples than in transgenic murine models.
Our results also demonstrate that intrathecal delivery of scAAV9-shRNA in piglets does not alter blood pSMN mRNA levels. Significant peripheral exposure to scAAV9 does occur after intrathecal delivery (Meyer, Ferraiuolo et al. 2015), however, expression in rapidly dividing myeloid progenitor cells is likely short-lived reducing the utility of blood cells for biomarker studies in this context. Thus, the measurement of SMN mRNA levels in blood is an attractive candidate for a pharmacodynamic biomarker in SMA clinical trials, however it may not have utility in clinical trials involving intrathecal delivery of therapeutics in general and of scAAV9-mediated therapeutics specifically.
Acknowledgments
We thank Dr. D. J. Coble, L. Mattox, and C.L. Sims from the ULAR Wiseman Hall Surgery Services at Ohio State University for excellent technical and supportive help with the piglets. This work is supported by NINDS R21NS083804 (to AHMB), U01NS079163 (to SJK), and NICHD 5K12HD001097-17 (to WDA).
Abbreviations
- CNS
central nervous system
- ddPCR
droplet digital polymerase chain reaction
- IT
intrathecal
- hSMN
human survival motor neuron
- MNs
motor neurons
- mRNA
messenger ribonucleic acid
- PND
postnatal day
- pSMN
porcine survival motor neuron
- scAAV9
self-complimentary adeno-associated virus serotype-9
- shRNA
short hairpin ribonucleic acid
- SMA
spinal muscular atrophy
References
- Arnold WD, Burghes AH. Spinal muscular atrophy: development and implementation of potential treatments. Ann Neurol. 2013;74(3):348–362. doi: 10.1002/ana.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan AK, Duque S, Foust KD, Morales PR, Braun L, Schmelzer L, Chan CM, McCrate M, Chicoine LG, Coley BD, Porensky PN, Kolb SJ, Mendell JR, Burghes AH, Kaspar BK. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol Ther. 2011;19(11):1971–1980. doi: 10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol. 2006;59(6):970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6(8):1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Duque SI, Arnold WD, Odermatt P, Li X, Porensky PN, Schmelzer L, Meyer K, Kolb SJ, Schumperli D, Kaspar BK, Burghes AH. A large animal model of Spinal Muscular Atrophy and correction of phenotype. Ann Neurol. 2014 doi: 10.1002/ana.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque SI, Arnold WD, Odermatt P, Li X, Porensky PN, Schmelzer L, Meyer K, Kolb SJ, Schumperli D, Kaspar BK, Burghes AH. A large animal model of spinal muscular atrophy and correction of phenotype. Ann Neurol. 2015;77(3):399–414. doi: 10.1002/ana.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici T, Taub JS, Baum GR, Gray SJ, Grieger JC, Matthews KA, Handy CR, Passini MA, Samulski RJ, Boulis NM. Robust spinal motor neuron transduction following intrathecal delivery of AAV9 in pigs. Gene Ther. 2012;19(8):852–859. doi: 10.1038/gt.2011.130. [DOI] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2017;388(10063):3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28(3):271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010 doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kolb SJ, Coffey CS, Yankey JW, Krosschell K, Arnold WD, Rutkove SB, Swoboda KJ, Reyna SP, Sakonju A, Darras BT, Shell R, Kuntz N, Castro D, Iannaccone ST, Parsons J, Connolly AM, Chiriboga CA, McDonald C, Burnette WB, Werner K, Thangarajh M, Shieh PB, Finanger E, Cudkowicz ME, McGovern MM, McNeil DE, Finkel R, Kaye E, Kingsley A, Renusch SR, McGovern VL, Wang X, Zaworski PG, Prior TW, Burghes AH, Bartlett A, Kissel JT, Neuro NCTN N. NS. M. A. B. I. on behalf of the. Baseline results of the NeuroNEXT spinal muscular atrophy infant biomarker study. Ann Clin Transl Neurol. 2016;3(2):132–145. doi: 10.1002/acn3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb SJ, Kissel JT. Spinal muscular atrophy: a timely review. Arch Neurol. 2011;68(8):979–984. doi: 10.1001/archneurol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, Burghes AH, Prior TW. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4(1):20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60(6):1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Ferraiuolo L, Schmelzer L, Braun L, McGovern V, Likhite S, Michels O, Govoni A, Fitzgerald J, Morales P, Foust KD, Mendell JR, Burghes AH, Kaspar BK. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol Ther. 2015;23(3):477–487. doi: 10.1038/mt.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Pearn JH. The gene frequency of acute Werdnig-Hoffmann disease (SMA type 1). A total population survey in North-East England. J Med Genet. 1973;10(3):260–265. doi: 10.1136/jmg.10.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, Burghes AH, Kissel JT. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85(3):408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renusch SR, Harshman SW, Hongyang P, Workman E, Wehr A, Li X, Swoboda KJ, Simard LR, Kissel JT, Battle DJ, Parthun MR, Freitas MA, Kolb SJ. Spinal Muscular Atrophy biomarker measurements from blood samples in a clinical trial of valproic acid in ambulatory adults. Journal of Neuromuscular Diseases. 2015;2:119–130. doi: 10.3233/JND-150081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, Flynn K, Hendrickson BC, Scholl T, Sirko-Osadsa DA, Allitto BA. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012;20(1):27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner CJ, Kolb SJ, Harmison GG, Jeffries NO, Schadt K, Finkel RS, Dreyfuss G, Fischbeck KH. SMN mRNA and protein levels in peripheral blood: biomarkers for SMA clinical trials. Neurology. 2006;66(7):1067–1073. doi: 10.1212/01.wnl.0000201929.56928.13. [DOI] [PubMed] [Google Scholar]
- Tiziano FD, Lomastro R, Di Pietro L, Barbara Pasanisi M, Fiori S, Angelozzi C, Abiusi E, Angelini C, Soraru G, Gaiani A, Mongini T, Vercelli L, Vasco G, Vita G, Luca Vita G, Messina S, Politano L, Passamano L, Di Gregorio G, Montomoli C, Orsi C, Campanella A, Mantegazza R, Morandi L. Clinical and molecular cross-sectional study of a cohort of adult type III spinal muscular atrophy patients: clues from a biomarker study. Eur J Hum Genet. 2013;21(6):630–636. doi: 10.1038/ejhg.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaworski P, von Herrmann KM, Taylor S, Sunshine SS, McCarthy K, Risher N, Newcomb T, Weetall M, Prior TW, Swoboda KJ, Chen KS, Paushkin S. SMN Protein Can Be Reliably Measured in Whole Blood with an Electrochemiluminescence (ECL) Immunoassay: Implications for Clinical Trials. PLoS One. 2016;11(3):e0150640. doi: 10.1371/journal.pone.0150640. [DOI] [PMC free article] [PubMed] [Google Scholar]