Abstract
Of the two 18F-labeled PET ligands currently available to image metabotropic glutamate receptor 5 (mGluR5), [18F]FPEB is reportedly superior because [18F]SP203 undergoes glutathionlyation, generating [18F]-fluoride ion that accumulates in brain and skull. To allow multiple PET studies on the same day with lower radiation exposure, we prepared [11C]FPEB and [11C]SP203 from [11C]hydrogen cyanide and compared their abilities to accurately quantify mGluR5 in human brain, especially as regards radiometabolite accumulation. Genomic plot was used to estimate the ratio of specific-to-nondisplaceable uptake (BPND) without using a receptor blocking drug. Both tracers quantified mGluR5; however [11C]SP203, like [18F]SP203, had radiometabolite accumulation in brain, as evidenced by increased distribution volume (VT) over the scan period. Absolute VT values were ∼30% lower for 11C-labeled compared with 18F-labeled radioligands, likely caused by the lower specific activities (and high receptor occupancies) of the 11C radioligands. The genomic plot indicated ∼60% specific binding in cerebellum, which makes it inappropriate as a reference region. Whole-body scans performed in healthy subjects demonstrated a low radiation burden typical for 11C-ligands. Thus, the evidence suggests that [11C]FPEB is superior to [11C]SP203. If prepared in higher specific activity, [11C]FPEB would presumably be as effective as [18F]FPEB for quantifying mGluR5 in human brain.
Keywords: FPEB, genomic plot, mGluR5, SP203, PET imaging
Introduction
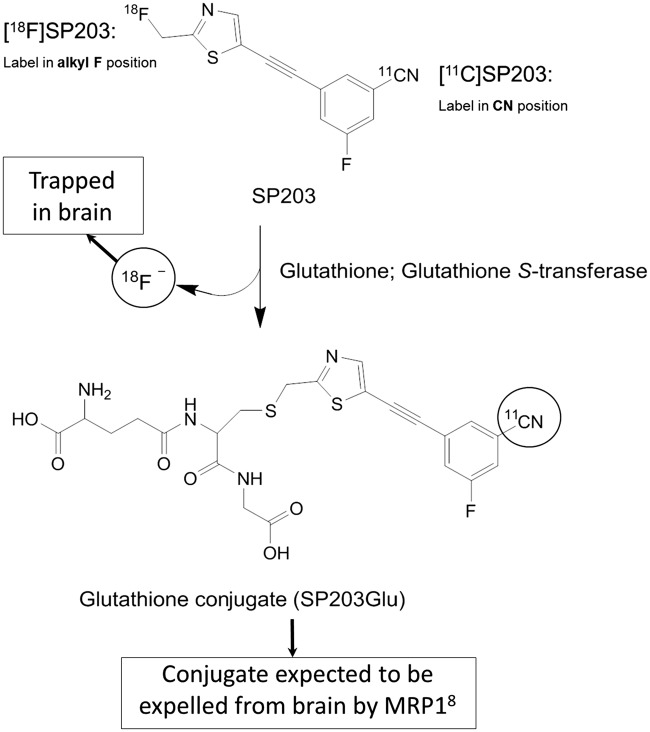
Two 18F-labeled radioligands, FPEB and SP203, have been reported as high performing radioligands to measure metabotropic glutamate receptor 5 (mGluR5) in human brain; of these, [18F]FPEB1,2 is clearly superior to [18F]SP203.3,4 Although both radioligands have high brain uptake and relatively fast washout that allows quantitation of mGluR5s with compartmental modeling, [18F]SP203 has two limitations compared with [18F]FPEB. First, [18F]SP203 is metabolized in vivo with subsequent uptake of the radiometabolites into bone, including skull, which spills over and contaminates adjacent neocortex.2,5 Second, the apparent distribution volume (VT) of [18F]SP203 gradually increases during the scan, consistent with the accumulation of radiometabolites. Previous studies from our laboratory found that [18F]SP203 is metabolized in rat brain by glutathionylation, which generates [18F]fluoride ion that becomes trapped within brain due to poor ability to cross the blood–brain barrier (Figure 1).3,6 The apparent VT of [18F]SP203 increases about 10% per hour in human subjects. [18F]FPEB—currently the best 18F-labeled radioligand for measuring mGluR5s—provides unusually stable measures of VT from 90 to 360 min.1
Figure 1.
The structure of SP203 and its radiometabolites showing different labeling positions for 18F and 11C, and the different metabolism by glutathione S-transferase.
Although the longer half-life of fluorine-18 allows distribution of radioligands to more distant PET centers, the shorter half-life and lower radiation exposure of carbon-11 allows multiple scans to be safely performed in the same subject on the same day. Such repeated administrations are particularly useful for studies that involve some intervention—e.g. measuring receptor occupancy before and after administration of a novel therapeutic. Here, we labeled FPEB and SP203 with carbon-11, each in the nitrile position. Thus, for each radioligand, the 11C- and 18F-labeled versions have the same chemical structure, but the radionuclide is on opposite sides of the molecule (Figure 1).7 Part of the rationale for preparing [11C]SP203 was that placing the radionuclide on the opposite end of the molecule from [18F]SP203 might avoid radiometabolite accumulation in brain. For example, if glutathionylation of [11C]SP203 occurs in human brain, the glutathionylated conjugate would likely be expelled from brain by multidrug resistance-associated protein 1 (MRP1) leaving only unchanged [11C]SP203 in brain.8
The primary goals of this study were three-fold. First, we sought to compare [11C]FPEB and [11C]SP203 with regard to their ability to quantify mGluR5 in human brain by using radioligands with the same nuclidic label (i.e. 11C) and imaged for the same length of time (i.e. 120 min) in order to reduce as many inherent differences as possible. Second, we measured time stability of VT to determine whether [11C]FPEB or [11C]SP203 generates radiometabolites that accumulate in brain, as indirectly shown by an apparent increase in VT during the course of the scan. Third, we sought to compare the two radioligands with regard to the ratio of specific to nondisplaceable uptake (i.e. BPND) as determined by genomic plot.9,10 This plot is an adaptation of the Lassen/occupancy plot and correlates regional PET radioligand binding with the density of the cognate mRNA gene transcript. Similar to the Lassen/occupancy plot, the genomic plot allows extrapolation to an imaginary region lacking mRNA and receptors, which is the nondisplaceable uptake (VND) in brain. Unlike the Lassen/occupancy plot, the genomic plot does not use a receptor blocking drug. The secondary goal of this study was to estimate, based on whole body imaging, the radiation dosimetry of the two radioligands, given that they might be used in future human studies.
Materials and methods
Chemistry
Materials and methods for chemistry, and description of the synthesis of precursor for labeling FPEB are contained in Supporting Information.
Synthesis of [11C]FPEB
Radiosynthesis was performed within a modified11 remotely controlled Synthia apparatus12 in a lead-shielded hot-cell for radiation protection (Supplementary Fig. 1).
NCA [11C]hydrogen cyanide (∼14 GBq) in nitrogen carrier gas was produced as described in Supporting Information and passed into a solution of KH2PO4 (2.5 mg) and kryptofix 2.2.2 (4 mg) in DMSO (0.30 mL) at ∼300 mL/min, until the collected radioactivity reached a maximum. The reaction mixture was then heated to 80℃ and purged with nitrogen at 200 mL/min for 1 min. Labeling precursor (0.6 mg) and tetra-kis(triphenylphosphine)palladium(0) (0.5 mg) in DMSO (0.2 mL) were added to the reaction mixture, which was then sealed and heated at 80℃ for 4 min. The reaction mixture was then injected onto a Luna C18 column (5 µm, 10 mm × 250 mm, Phenomenex) eluted with a mixture (61: 39 v/v) of MeCN and aqueous TFA (0.1% w/v) at 6 mL/min, with eluate monitored for radioactivity and for absorbance at 254 nm. The fraction containing [11C]FPEB (tR = 13.2 min) was collected. The incorporation of initial [11C]cyanide ion into [11C]FPEB was calculated from the radiochromatogram. The collected [11C]FPEB fraction was diluted in water (30 mL) and passed through a C18 cartridge (SepPak Plus; Waters Corp, Milford, MA). The cartridge was washed with water (10 mL) and eluted with dehydrated alcohol injection (USP, 2 mL) followed by 0.9% sodium chloride injection (USP, 2 mL). The combined eluent was passed through a sterile filter (Millex-MP, 0.22 µm, Millipore, Taunton, MA) into a sterile dose vial containing 0.9% sodium chloride injection (USP, 16 mL) and measured for radioactivity.
Analysis of [11C]FPEB
The radiochemical purity, chemical purity, and specific activity of each batch of formulated [11C]FPEB were determined with HPLC on a Luna C18 column (10 µm, 4.6 mm × 250 mm; Phenomenex) eluted at 3 mL/min with MeCN-10 mM HCOONH4 (45: 55 v/v) with column eluate monitored for radioactivity and absorbance at 254 nm ([11C]FPEB, tR = 5.6 min). The analytical method was calibrated for absorbance response (peak area) versus mass of injected FPEB to allow the amount of carrier FPEB in a known amount of [11C]FPEB to be measured, thereby giving the specific activity of the radioligand at the time of analysis. This value was corrected for radioactive decay to the end of synthesis. The identity of [11C]FPEB was also confirmed by LC-MS of the associated carrier. The radiochemical stability of formulated [11C]FPEB was checked by repeat HPLC analysis after the batch had been kept at room temperature for 60 min.
Synthesis of [11C]SP203
[11C]SP203 was labeled using an iodo precursor and [11C]cyanide ion as the labeling agent, as previously described.7
Radioligand preparation
The radioligands were prepared according to our Investigational New Drug Application ([11C]FPEB: 118,353, [11C]SP203: 105,222), which was submitted to the U.S. Food and Drug Administration; a copy is available at http://pdsp.med.unc.edu/snidd/. [11C]FPEB was obtained with high radiochemical purity (>99%) and specific activity of 22 ± 7 GBq/µmol at the time of injection (n = 14 batches). [11C]SP203 was obtained with high radiochemical purity (>99%) and specific activity of 33 ± 11 GBq/µmol at the time of injection (n = 14 batches).
Subjects
Approval for this study was obtained from the Institutional Review Board of the National Institute of Mental Health (NIMH), Ethics Committee and the Radiation Safety Committee of the National Institutes of Health (NIH). All the studies were conducted according to the Declaration of Helsinki. All subjects gave written informed consent. For [11C]FPEB, eight separate healthy volunteers participated in the brain PET scans (four males, four females; 28 ± 8 years of age). Six separate healthy volunteers participated in the whole-body PET (n = 4) or PET/CT (n = 2) scans (three males, three females; 33 ± 10 years of age). For [11C]SP203, six healthy volunteers participated in the brain positron emission tomography (PET) scans (three males, three females; 30 ± 12 years of age). Eight separate healthy volunteers participated in the whole-body PET/computed tomography (CT) scans (five males, three females; 32 ± 9 years of age). All subjects were free of current medical or psychiatric illnesses as determined by medical history, physical examination, electrocardiogram (EKG), urinalysis, and laboratory blood tests (complete blood count, serum chemistries, and thyroid function test). The subject’s vital signs were recorded before radioligand injection and at 15, 30, 90, and 120 min after injection. Repeat urinalysis and blood tests were conducted within 2 h of completing the PET scan.
Brain PET
Scan procedure
Brain PET scans were performed on a GE Advance tomograph (GE Medical Systems, Waukesha, WI). [11C]FPEB (700 ± 24 MBq, n = 8) or [11C]SP203 (649 ± 63 MBq; n = 6) was intravenously injected over 1 min and dynamic three-dimensional emission scans were acquired for 120 min in 33 frames of increasing duration from 30 s to 5 min. One brain transmission scan using 68Ge rods was acquired before the injection of the radioligand. The position of the transmission scan was corrected for motion before applying attenuation correction.
Measurement of [11C]FPEB and [11C]SP203 in plasma
To determine arterial input function for brain PET scans, blood samples (1.5 mL each) were drawn from the radial artery at 15-s intervals until 150 s, followed by 3 mL samples at 3, 4, 6, 8, 10, 15, 20, 30, 40, and 50 min, and 5 mL at 60, 75, 90, and 120 min. The concentration of parent radioligand was measured using HPLC as previously described.13 For [11C]FPEB, the separation was done on preparatory X-Terra C18 column (10 µm, 7.8 × 300 mm2; Waters Corp.) and a mobile phase of MeOH:H2O:Et3N (70:30:0.1, by volume). For [11C]SP203, the HPLC column used was Novapak C18 (100 × 8 mm2; Waters Corp) with radial compression module RCM-100 and a mobile phase of MeOH:H2O:Et3N (65:35:0.1, by volume). In addition, the plasma free fraction (fp) was measured by ultrafiltration, as previously described.14
Blood data processing
The time-activity curves of parent [11C]FPEB or [11C]SP203 concentrations were calculated by multiplying total plasma activity with fractions of [11C]FPEB or [11C]SP203 measured in plasma. Tri-exponential fitting was performed to the measured parent concentrations by weighting data according to errors in the measurement of each sample based on Poisson distribution. Whole blood radioactivity was used to correct radioactivity in brain that represents the vascular compartment (∼5% of tissue volume).
Image data processing
Head movements were corrected after the scan by realigning all frames from each subject using Statistical Parametric Mapping, SPM (Version 8 for Windows, Wellcome Department of Cognitive Neurology, UK). Brain regional data were obtained after coregistering magnetic resonance (MR) to PET images and spatial normalization to Montreal Neurologic Institute (MNI) space as previously described.15 The data were obtained in the following 10 volumes of interest (VOIs) in the MNI space: frontal (432 cm3), parietal (247 cm3), occipital (172 cm3), temporal (251 cm3), medial temporal (36 cm3), and cingulate (28 cm3) cortices; caudate (16 cm3); putamen (17 cm3); thalamus (17 cm3); and cerebellum (195 cm3). Image and kinetic analyses were performed using pixelwise modeling software (PMOD3.17, PMOD Technologies Ltd, http://www.pmod.com/).
Calculating distribution volume (VT)
The equilibrium value of VT was calculated three different ways to assess whether the methods applicable to voxel data yielded VT values similar to those obtained from VOI data using the gold standard method of compartmental modeling.
Compartmental modeling
Brain time-activity data were analyzed with compartmental modeling. Rate constants (K1, k2, k3, and k4) in standard one- and two-tissue compartment models were estimated with the weighted least-squares method and the Levenberg–Marquardt algorithm. Brain data for each frame were weighted by assuming that the standard deviation of the data was proportional to the inverse square root of noise equivalent counts. To correct brain data for the vascular component, radioactivity in serial whole blood was measured and subtracted from the PET measurements, assuming that cerebral blood volume is 5% of total brain volume. The delay between the arrival of radioligand in the radial artery and brain was estimated by fitting the whole brain, excluding white matter.
Linear and multilinear graphical analyses
The graphical approach of Logan16 was used to calculate VT. With the maximum deviation between the regression and all measurements set to 20%, the equilibration start time (t*) and the slope of the linear portion of the Logan plot (VT) were fitted for each brain region that included data from multiple voxels within a VOI (LoganVOI). To decrease the bias induced by noise in the measurements, VT was also calculated by Ichise’s bilinear MA1 analysis,17 using the same t* obtained from the Logan analysis for each brain region (MA1VOI).
Parametric modeling
To determine whether VT could be determined from the voxel data, parametric images of VT (Loganvoxel and MA1voxel) were generated using data from each voxel of dynamic PET images. For both Logan and MA1 methods, PET frames used for the regression were selected based on t* obtained from the time-activity curve of whole brain excluding white matter and by setting the maximum deviation between the regression and all measurements to 20%. The same plasma input function as that used for compartmental modeling was used for the parametric modeling. After spatial normalization of the parametric images to the MNI space, VT values were obtained from the 10 regions described above to compare the results from kinetic analyses using the VOI data.
Time-stability analysis
To determine the minimum scan length needed for reliable measurement, as well as to indirectly assess whether radiometabolites enter brain, the time stability of VT was examined by increasingly truncating the scan duration from 0–120 min to 0–30 min in 10-min increments.
Statistical analysis
The optimal compartment model (i.e. one- vs. two-tissue compartment) was chosen based on the Akaike information criterion (AIC),18 model selection criterion (MSC) proposed by Micromath®, Saint Louis, MO, http://www.micromath.com/products.php?p=scientist&m=statistical_analysis), and F-test.19 The most appropriate model would be the one with the smallest AIC and the largest MSC value. F-statistics were used to compare goodness-of-fit by one- and two-tissue compartment models. A value of p < 0.05 was considered significant. The identifiability (%) of rate constants was expressed as a percentage and equaled the ratio of the standard error (SE) of the kinetic variables divided by the value of the kinetic variables themselves. Identifiability (%) of VT was calculated from the covariance matrix using the generalized form of error propagation equation, where correlations among parameters (K1 and k2, or K1, k2, k3, and k4) were taken into account. A lower percentage indicates better identifiability.
For each subject, we used simple Logan and MA1 regression methods to compare regional brain VT values obtained from VOI and voxel data with VT values obtained from the two-tissue compartment model applied to VOI data. Differences in VT between methods were examined with repeated measures analyses of variance with region as the repeated factor. All statistical analyses were performed with SPSS (Version 17 for Windows, SPSS Inc., Chicago, IL). Group data are expressed as mean ± SD. The mean and SD for parametric images were calculated in each voxel across subjects. Values from all voxels were then averaged within each VOI.
Genomic plot
The genomic plot9 was used to compare [11C]FPEB and [11C]SP203 with regard to the ratio of specific to nondisplaceable uptake (i.e. BPND) without the use of a receptor blocking drug. The method is an adaption of the Lassen (or occupancy) plot20 and allows estimation of VND using the brain maps of mRNA transcripts of the target receptor as a surrogate measure of tracer-specific binding.
The genomic plot was implemented as previously described.9,10 Briefly, individual mRNA expression maps for mGluR5, as derived from the Allen Human Brain atlas (http://www.brain-map.org),21 were spatially normalized to the standard stereotaxic space (MNI/ICBM152). Based on their original anatomical labeling, the mRNA samples belonging to the same regions of interest (ROIs) were averaged across different donors.
The regional mRNA measures for mGluR5 gene expression were then converted from log2 intensity into linear scale and linearly regressed with the population-average VT estimates for the corresponding ROIs for both PET tracers. The ROIs considered were: frontal, parietal, temporal and occipital lobes, cingulate gyrus, striatum, thalamus, and cerebellum. These regions were chosen to match the gene analysis with PET data quantification and to guarantee a sufficiently large number of samples per region (>10 per region and per donor).
The squared Pearson’s correlation coefficient (R2) was used to quantify whether the relative gene transcript was proportional to VT. Following the assumption that mRNA expression can be used as a surrogate of tracer specific binding, VND was estimated as the x-intercept of the plot of mGluR5 transcript density vs. VT across brain regions. When specific binding is absent (x-intercept), the value of VT equals VND. For both [11C]FPEB and [11C]SP203, the VND estimates were then used to calculate the tracer binding potentials (BPND = (VT/VND − 1)).
The processing of the mRNA data was done with MENGA (Multimodal Environment for Neuroimaging and Genomic Analysis),22 while genomic plot implementation was performed using Matlab®2012b (The MathWorks, Inc., Natick, MA) on a Windows 7 computer.
Whole body PET or PET/CT
Scan procedure
Whole-body PET scans were performed on an Advance tomograph (GE Medical Systems, Waukesha, WI) and whole-body PET/CT scans were performed on a Siemens Biograph™ mCT (Siemens Healthcare, Malvern, PA). [11C]FPEB (547 ±210 MBq) or [11C]SP203 (633 ± 125 MBq) was intravenously injected over 20 s, and dynamic three-dimensional emission scans were acquired for ∼120 min in 13 frames of increasing duration from 15 s to 4 min by serial imaging of the body from head to middle thigh in eight contiguous segments. One whole body 68Ge or CT transmission scan was used for attenuation correction.
Dosimetry analysis
Source organs were identified on the individual coronal slices of whole body PET/CT images. The frames with the highest organ uptake were averaged and smoothed by a Gaussian filter to increase the contrast between the organ and surrounding tissue. Brain, lungs, heart, liver, kidneys, gallbladder, urinary bladder, small intestine, lumbar vertebrae, and thyroid ([11C]FPEB only) were identified as the source organs.
Uptake in each source organ was corrected for recovery of measured activity, which was calculated by averaging the activity of the second through sixth frames where the image quality was good with a large ROI placed over the entire body visible by PET/CT.
At each time point, the activity not corrected for decay of the source organ was expressed as the fraction of the injected activity (IA). The area under the time-activity curve of each organ was calculated by the trapezoidal method until scan acquisition ended. The area after the last image to infinity was calculated by assuming that the subsequent decline of radioactivity occurred only via physical decay, without biological clearance. The area under the curve of percent IA from time zero to infinity equals the time-integrated activity coefficient, formerly known as “residence time” as per the 1991 Medical Internal Radiation Dose (MIRD) Primer of the organ (MIRD Pamphlet 21).23 The time-integrated activity coefficient of all red marrow in the body was estimated from that of the lumbar vertebrae as previously described.5 To calculate the time-integrated activity coefficient for the remainder of the body, the time-integrated activity coefficients for all of the source organs were summed and subtracted from the fixed theoretical value of t1/2/ln2 = 0.49 h.
Radiation absorbed doses were calculated according to the MIRD scheme using OLINDA/EXM 1.1 (http://www.doseinfo-radar.com/OLINDA.html) and the model for a 70-kg adult male.
Results
Chemistry
The labeling of FPEB with carbon-11 was based on transition metal mediated aryl 11C-cyanation,24 and has been applied to the preparation of other mGluR receptor PET radioligands, such as [11C]SP2037 and [11C]AZD9272.25 Incorporation yields of [11C]FPEB determined with HPLC were 47 ± 7% (n = 14). [11C]FPEB was obtained ready for intravenous administration in useful radiochemical yields of 2.5 ± 0.7 GBq (n = 14). Single pass reverse phase HPLC afforded [11C]FPEB in high radiochemical purity (>99%), and in the absence of any significant chemical impurities. Formulated [11C]FPEB was stable when kept at room temperature for 60 min.
Pharmacological effects
The injected mass dose of [11C]FPEB (468 ± 187 pmol/kg (n = 14)) or [11C]SP203 (296 ± 108 pmol/kg (n = 14)) caused no pharmacological or subjective effects in any subject during the 2-h PET scan. Specifically, it did not change blood pressure, pulse, respiratory rate, EKG, or pulse oximetry. Similarly, no significant effects were noted in any of the blood or urine tests acquired at the end of the scan.
Plasma analysis
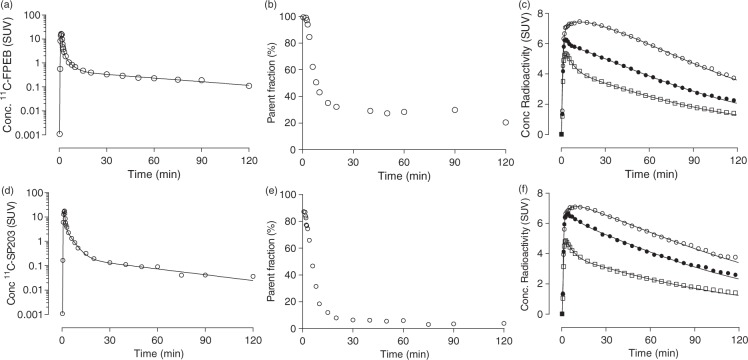
[11C]FPEB concentrations in arterial plasma peaked to 21 ± 3 SUV at 1.3 min after injection and then rapidly declined, followed by a slow terminal clearance phase. The fitting of plasma parent curves converged by tri-exponential function in all subjects (Figure 2(a)). The fraction of [11C]FPEB, expressed as a percentage of total plasma radioactivity, declined rapidly and reached 31 ± 6% at 15 min, followed by a gradual decline (Figure 2(b)). Radiometabolites appeared quickly in plasma and, after 5 min, became the predominant component of plasma radioactivity. Radiometabolites A–D eluted before the more lipophilic parent and were not well resolved among themselves by reverse-phase HPLC (Supplementary Fig. 2A). The fp of [11C]FPEB was 6.3 ± 0.8% (n = 8).
Figure 2.
Radioactivity concentrations, parent radioligand fraction in plasma, and brain time activity curves from a representative subject injected with [11C]FPEB (a–c) or [11C]SP203 (d–f). The first panels (a and d) for each radioligand plot the concentration of parent radioligand, separated from radiometabolites as a function of time after injection. The middle panels (b and e) show the percentage of total radioactivity in plasma that represents parent radioligand. The last panels (c and f) plot the concentrations of radioactivity in three brain regions as a function of time after injection. The lines represent the unconstrained two-tissue compartment model that well fit all data points. (o) putamen, (•) thalamus, and (□) cerebellum.
[11C]SP203 concentrations in arterial plasma peaked to 15 ± 3 SUV at 1.5 min after injection, and then rapidly declined, followed by a slow terminal clearance phase. The fitting of plasma parent curves converged by tri-exponential function in all subjects (Figure 2(d)). The fraction of [11C]SP203, expressed as a percentage of total plasma radioactivity, declined rapidly and reached 12 ± 5% at 15 min, followed by a gradual decline (Figure 2(e)). Radiometabolites appeared quickly in plasma and, after 5 min, became the predominant component of plasma radioactivity. Radiometabolites A–D eluted before the more lipophilic parent and were not well resolved among themselves by reverse-phase HPLC (Supplementary Fig. 2B). A small peak, radiometabolite E, was seen just after the parent. The fp of [11C]SP203 was 2.8 ± 0.1% (n = 6).
Brain radioactivity and kinetic analysis
After [11C]FPEB or [11C]SP203 injections, all subjects showed high concentrations of radioactivity followed by quick washout in all regions. [11C]FPEB radioactivity in brain peaked at 6.4 ± 0.7 SUV (n = 8) within 10 min and decreased to 52% of the peak by 120 min (Figure 2(c)). [11C]SP203 radioactivity in brain peaked at 5.5 ± 1.0 SUV (n = 6) within 10 min and decreased to 60% of the peak by 120 min (Figure 2(f)). The regional distribution of radioactivity was consistent with the known distribution of mGluR5 in the human brain, with high levels in neocortex and striatum and lowest levels in cerebellum.26
Kinetic analysis of brain and plasma data for [11C]FPEB and [11C]SP203 had three major findings: (a) regional brain data were well fit by a two-tissue compartment model; (b) voxel-level data analyzed with a bilinear method (Ichise MA1) gave accurate values compared with those from regional analyses; and (c) the time stability of VT for [11C]FPEB was better than that for [11C]SP203.
With regard to the first result, unconstrained two-tissue compartmental fitting converged in all regions and in all scans for both [11C]FPEB and [11C]SP203 (Figure 2(c) and (f)). An F-test showed that the two-tissue compartment model was superior to the one-tissue compartment model in all 10 regions of all subjects for both radiotracers, consistent with the presence of significant amounts of both specific and nonspecific binding in human brain. In addition, the two-tissue compartment model showed lower mean AIC and higher mean MSC scores than the one-tissue compartment model. The unconstrained two-tissue compartment model identified VT with an average SE across brain regions of 0.6% for [11C]FPEB and 2.1% for [11C]SP203. Regional values of two-tissue mean VT (mLċcm−3) for [11C]SP203 ranged from 26 in temporal cortex to 9.6 in cerebellum (Supplementary Table 1). Regional values of two-tissue mean VT (mLċcm−3) for [11C]FPEB ranged from 17 in temporal cortex to 6.7 in cerebellum (Supplementary Table 2). All the rate constants (K1, k2, k3, and k4) were relatively well-identified with an average SE of less than ∼20% across all regions (Supplementary Tables 3 and 4).
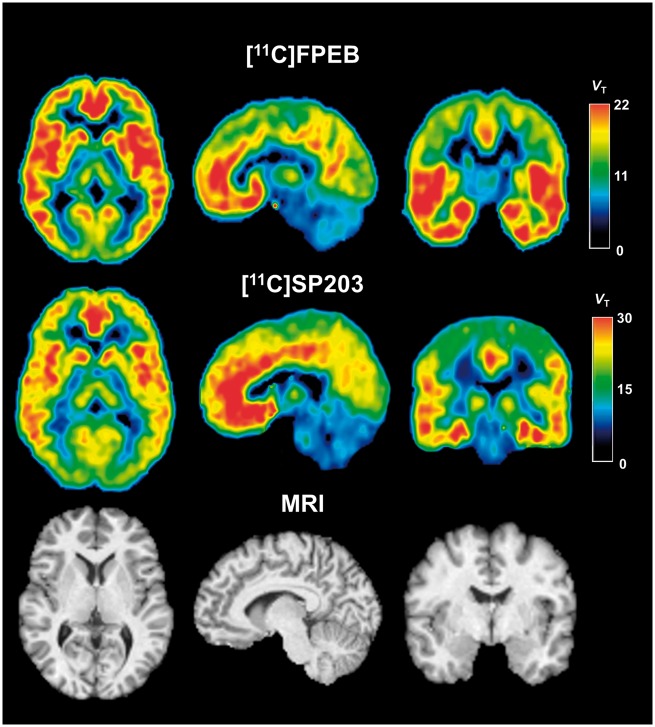
With regard to the second result, we first used large regional values to calculate VT with the Logan and MA1 methods and confirmed that these two mathematically simple methods gave VT values almost identical to those obtained by more complex compartmental modeling. We then applied Logan and MA1 methods to voxel data. The voxel-based Logan model (Loganvoxel) significantly underestimated VT compared with the two-tissue compartment model with a mean difference of 10% for [11C]FPEB and 15% for [11C]SP203. Voxel-based MA1 gave VT values almost identical to those obtained via two-tissue compartment modeling for VOI data with a mean difference of 6% for [11C]FPEB and 4% for [11C]SP203 (Supplementary Tables 1 and 2). Further, MA1voxel VT and two-tissue VT showed excellent correlation (p < 0.0001) across brain regions from all subjects for both radiotracers, with a regression slope close to one and a small intercept (Supplementary Fig. 3A and B). In addition, the parametric images were of high quality, with excellent contrast between gray and white matter; few voxels had unrealistically high or low VT values (Figure 3). Therefore, voxel-based MA1 appeared to be a valid method for measuring mGluR5s at the voxel level.
Figure 3.
PET and MR images in axial (left), sagittal (middle), and coronal (right) views. Top row: Parametric MA1 PET images of a 25-year-old healthy male injected with 730 MBq of [11C]FPEB and 2 h scan duration. Middle row: Parametric MA1 PET images of a 23-year-old healthy female injected with 670 MBq [11C]SP203 and 2 h scan duration. Bottom row: MR images of the same subject scanned with [11C]FPEB. Each voxel in the PET images represents distribution volume (VT), with its value encoded in the color scale on right.
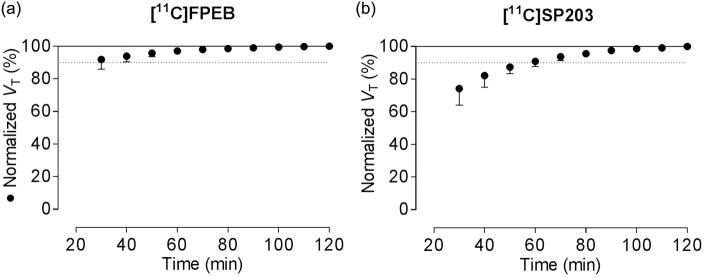
As regards the third result, and to determine the minimum scanning time needed to accurately measure VT, we increasingly truncated the entire scan in 10-min increments from 0–120 to 0–30 min. For [11C]FPEB, the increase in VT was less than 5% during the last hour of the 2-h scan, with stable values as early as 80 min (Figure 4(a)); this indicates negligible accumulation of radiometabolites and suggests that 80 min of scan data are adequate to calculate VT. In contrast, VT values for [11C]SP203 gradually increased by about 10% during the last hour of the 2-h scan and showed a rising trend without reaching a stable value (Figure 4(b)). We had previously found that VT values for [18F]SP203 also increased by about 10% during the same period.3 Similar increments with [11C]SP203 indicate that the radiometabolite (possibly glutathionylated conjugate) could be accumulating in the brain.
Figure 4.
Distribution volume (VT) as a function of duration of image acquisition for [11C]FPEB (a) and [11C]SP203 (b). VT was calculated for cerebellum (•) using an unconstrained two-tissue compartment model with increasingly truncated acquisition times. Values are normalized as percentage of terminal value attained from 120 min of imaging. Data represent mean ± SD of eight subjects for [11C]FPEB and six subjects for [11C]SP203.
Genomic plot
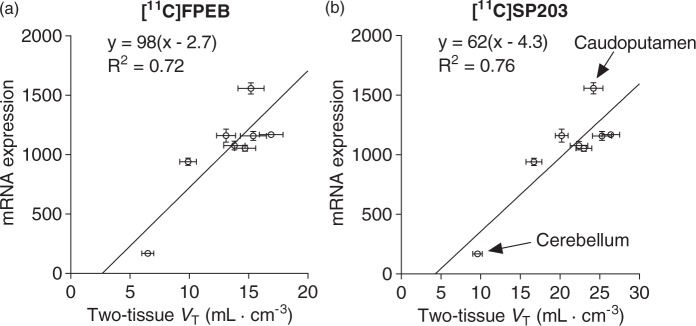
Regional values for the mGluR5 gene transcript were highly correlated with those of PET VT for both tracers (for [11C]FPEB, Pearson’s R2 = 0.72, p < 0.01, two-tailed; for [11C]SP203 Pearson’s R2 = 0.76, p < 0.01, two-tailed), supporting the use of the genomic plot for VND quantification. This high correlation was predominantly driven by the cerebellum, which is characterized by both low mRNA expression and low PET binding. Notably, brain mRNA expression for mGluR5 was highly consistent across all donors of the Allen Human Brain atlas (Pearson’s R2 = 0.925 ±0.033).
Following genomic plot application (Figure 5), the estimated VND was 2.7 mL·cm−3 for [11C]FPEB and 4.3 mL·cm−3 for [11C]SP203. The resulting binding potential estimates were highly comparable (Pearson’s R2 = 0.99, p < 0.001). The highest binding was found in the temporal cortex ([11C]FPEB BPND = 5.3; [11C]SP203 BPND = 5.1), followed by cingulate cortex ([11C]FPEB BPND = 4.7; [11C]SP203 BPND = 4.9). For both tracers, a significant amount of specific binding was found in the cerebellum, as BPND was 1.4 for [11C]FPEB and 1.2 for [11C]SP203. Because BPND is the ratio at equilibrium of specific to nondisplaceable uptake, these values imply that about 60% of total cerebellar uptake was specifically bound to mGluR5.
Figure 5.
Genomic plots of eight brain regions for [11C]FPEB (a) and [11C]SP203 (b). The y-axis is the gene expression for human mGluR5 from the Allen Brain Atlas and its density is expressed relative to expression of housekeeping gene. Data points (•) represent mean values, with standard error bars shown for both x and y values. The linear regressions shows an x-intercept ( = VND) of 2.7 mL·cm−3 for [11C]FPEB and 4.3 mL·cm−3 for [11C]SP203.
Whole-body biodistribution and dosimetry estimates
Whole-body images were notable for early distribution of radioactivity in the blood pool, accumulation in the target organ (i.e. brain), and excretion via hepatobiliary and urinary routes (Supplementary Figs. 4 and 5). At early time points, radioactivity was prominent in organs with high blood volume, including liver, kidneys, and brain. Consistent with its high densities of mGluR5, high radioactivity uptake was observed in brain, with a peak of 11–12% of injected activity 5–10 min after [11C]FPEB or [11C]SP203 injection. [11C]SP203 radioactivity was excreted via both hepatobiliary and urinary routes, with 9% of the injected activity in gallbladder, 8% in small intestine, and 19% in urinary bladder by the end of the scan. [11C]FPEB radioactivity was also excreted via both hepatobiliary and urinary routes, with 4.5% of the injected activity in liver, 8% in gallbladder, and 11% in urinary bladder by the end of the scan (Supplementary Fig. 6). Lumbar vertebrae were visible in the scans.
For [11C]FPEB, the three source organs with the highest time-integrated activity coefficients were liver, brain, and urinary bladder; for [11C]SP203, the organs were liver, brain, and red marrow (Supplementary Table 5). The three organs with the highest radiation-absorbed-doses (µSv/MBq) for [11C]FPEB were gallbladder wall (41.2 ± 20.5), urinary bladder wall (14.3 ± 3.8), and brain (12.2 ± 2.0); for [11C]SP203, the organs were gallbladder wall (38.1 ± 27.7), urinary bladder wall (17.1 ± 5.4), and kidneys (12.0 ± 3.0) (Supplementary Table 6). The effective dose, a weighted sum of overall exposure to the body, was 3.7 ± 0.2 µSv/MBq for [11C]FPEB and 4.4 ± 0.4 µSv/MBq for [11C]SP203.
Discussion
This study sought to compare the ability of two 11C-labeled radioligands to quantify mGluR5 in human brain, to determine whether radiometabolites accumulate in brain, and to determine whether cerebellum has specific binding to mGluR5. We found that both [11C]FPEB and [11C]SP203 effectively quantified regional mGluR5 distribution in human brain. However, [11C]FPEB was the superior radioligand because [11C]SP203, like its 18F version, generated radiometabolites that accumulate in brain, as evidenced by increasing values of apparent VT over the course of the scan. The genomic plot suggested that about 60% of cerebellar uptake for both radioligands reflected specific binding to mGluR5. As a secondary aim of this study, we also calculated the radiation dosimetry of [11C]FPEB and [11C]SP203 and found that their effective doses (ED) were low (∼4 µSv/MBq) and similar to other 11C-labeled PET radioligands.27
Using region and voxel-wise kinetic modeling approaches, mGluR5 was quantified in terms of VT for both [11C]FPEB and [11C]SP203. Both tracers yielded well identifiable VT values that were consistent with regional mGluR5 distribution in human brain (Supplementary Tables 1 and 2). However, the apparent VT of [11C]FPEB was more stable over time than that of [11C]SP203 (Figure 4), consistent with the ability of [11C]FPEB to generate few radiometabolites that accumulate in brain (Figure 4). With regard to [11C]SP203, which is known to undergo glutathionylation, the expectation that any glutathionylated metabolite generated will be expelled from brain did not appear to occur. Thus, it is unlikely that SP203 will be useful. Instead, we recommend FPEB for effective quantification of mGluR5.
Low specific activity of [11C]FPEB and [11C]SP203
The specific activity of the two radioligands was relatively low at the time of injection: ∼20 GBq/µmol for [11C]FPEB and ∼40 GBq/µmol for [11C]SP203 (Table 1). The carrier in these low specific activity radioligands likely led to significant receptor occupancy, thereby causing our VT values to be lower than those using the 18F-labeled versions, whose specific activity was ∼150–300 GBq/µmol (Table 1). We estimated receptor occupancies at the time of peak specific brain uptake, based on published values of receptor density (Bmax)26 (Table 2). The peak concentration of specific binding was determined from two-compartment modeling, which separately estimates concentration in the specific and nondisplaceable compartments. Occupancy of the receptor by carrier (i.e. nonradioactive parent radioligand) ranged from 4% for [11C]SP203 in frontal cortex to 27% for [11C]FPEB in cerebellum. The estimates for almost all regions was >5%, which is often regarded as a limit for a true tracer dose of radioligand. The resulting self-blockade would decrease our measured values of VT, which was apparent relative to the 18F-labeled radioligands that have higher specific activity (Table 1). A major limitation of our estimates of receptor occupancy is that we do not know the density of mGluR5s in vivo that are available to bind radioligand—i.e. Bavail. Instead, we used the in vitro measurement of receptor density (Bmax), for which we found only one published value in human brain.26 However, only a subset of receptors measured in vitro in homogenized tissue may be in the proper location or conjoined with other proteins to be able to bind radiolabeled drug in vivo. True measurements of in vivo receptor occupancy require baseline and partially blocked scans, which can then be analyzed with the so-called Lassen/occupancy plot.28
Table 1.
Comparison of distribution volume (VT), plasma free fraction (fP), and related parameters among mGluR5 radioligands used in human brain.
| PET ligand | [11C]FPEB | [18F]FPEB | [18F]FPEB | [18F]SP203 | [11C]SP203 |
|---|---|---|---|---|---|
| Reference | (Current study) | 1 | 2 | 3 | (Current study) |
| VT valuesa | 10–17 | 17–32 | 14–26 | 15–26 | 17–26 |
| VT (cerebellum) | 6.7 | 10 | – | 14 | 10 |
| Test–retest variability | – | – | ∼20% (VT) | – | – |
| ∼10% (BPND) | |||||
| Plasma free fraction (fP) | 6.3 ± 0.8% | 4.7 ± 0.6% | 8.1 ± 0.7% | 5.2 ± 0.2% | 2.8 ± 0.1% |
| 4.3 ± 0.7% | |||||
| VT/fP (Thalamus) | 163 | 351 | 173 | – | 596 |
| Specific activity at TOI (GBq/µmol) | 22 ± 6 | 144 ± 44 | 330 ± 210 | 156 ± 54 | 40 ± 6 |
| Injected mass dose (µg) | 7.4 ± 1.5 | 0.3 ± 0.1 | 0.10 ± 0.06 | 0.6 ± 0.2 | 4.3 ± 0.9 |
Regional VT values except for cerebellum.
TOI: time of injection. The VT/fP values were calculated by dividing average VT for thalamus with average fp values for each PET ligand from the current and reported studies.
Table 2.
Receptor occupancies achieved by [11C]FPEB and [11C]SP203 at time of peak specific binding in three brain regions.
| Brain region | mGluR5 Bmax (nM)a | Peak brain concentration (nM) |
% Occupancy at peak brain concentration |
Peak specific brain concentration (nM)b |
% Occupancy at peak specific brain concentrationc |
||||
|---|---|---|---|---|---|---|---|---|---|
| [11C]FPEB | [11C]SP203 | [11C]FPEB | [11C]SP203 | [11C]FPEB | [11C]SP203 | [11C]FPEB | [11C]SP203 | ||
| Frontal cortex | 25 ± 5.6 | 3.4 ± 1.0 | 1.2 ± 0.3 | 13 ± 4.1 | 4.9 ± 1.4 | 3.0 ± 1.0 | 1.1 ± 0.4 | 12 ± 3.9 | 4.2 ± 1.4 |
| Caudate | 17 ± 2.3 | 3.1 ± 1.0 | 1.2 ± 0.3 | 18 ± 5.6 | 7.0 ± 2.0 | 2.6 ± 1.0 | 1.0 ± 0.3 | 15 ± 5.7 | 5.8 ± 2.0 |
| Cerebellum | 5.1 ± 2.0 | 2.8 ± 0.9 | 1.0 ± 0.3 | 55 ± 17 | 19 ± 5.9 | 1.4 ± 0.5 | 0.5 ± 0.2 | 27 ± 10 | 10 ± 3.0 |
Data are mean ± SD from eight subjects for [11C]FPEB and six subjects for [11C]SP203.
Bmax values are from Patel et al.26 and assume that 10 mg brain contains 1 mg protein.
The concentration of specific binding came from the two-tissue compartment model, which separates radioligand into specific and nondisplaceable compartments.
Occupancy for each region was calculated by dividing peak specific concentration by its Bmax.
The cause of the low specific activity of the radioligands was the low specific activity of the [11C]hydrogen cyanide used as the labeling agent. In this study, [11C]hydrogen cyanide was prepared from cyclotron-produced [11C]carbon dioxide.25 Improved specific activity may be obtainable by using [11C]hydrogen cyanide produced from cyclotron-produced [11C]methane29 rather than from [11C]carbon dioxide.
Genomic plot
Here, we used the genomic variant of the Lassen plot to estimate the specific and nondisplaceable components of both radioligands, a task that would otherwise require administration of blocking doses of a non-radioactive drug. The method is generally applicable to any protein PET target that shows good correlation of regional densities of gene transcript from a genomic atlas with the density of expressed protein measured with PET.9 In fact, both [11C]FPEB and [11C]SP203 showed high correlations among brain regions between mGluR5 mRNA expression and the VT estimates (R2 = 0.72 and 0.76, respectively) (Figure 5). A similar correlation can be obtained using the [18F]FPEB VT regional estimates published by Sullivan and colleagues (R2 = 0.67).1
The genomic plot for mGluR5 had two major results. First, both [11C]FPEB and [11C]SP203 provided a high percentage of specific binding in human brain. For example, in the highest density region—the temporal cortex—specific binding (VS) of both radioligands was 84% of total uptake VT (Supplementary Tables 1 and 2), assuming VND is 2.7 mL·cm−3 for [11C]FPEB and 4.3 mL·cm−3 for [11C]SP203. Second, even the lowest density region, the cerebellum, had a high percentage (∼60%) of specific binding. This finding is consistent with that for another mGluR5 radioligand, [11C]AZD9272, but that study required administration of an mGluR5 blocking drug.30 Taken together, the data indicate that the cerebellum is not a suitable reference region—i.e. one that lacks specific binding. Furthermore, if the cerebellum were going to be used as a pseudo-reference region (i.e. one with low specific binding), it must be shown to be similar between patient and control groups.31,32
Conclusion
Results from both prior studies using the 18F-labeled versions and the current study, which used 11C-labeled versions, indicate that FPEB is superior to SP203 as a ligand for quantifying mGluR5. The major advantage of FPEB is that its VT, a measure of receptor density, is quite stable after 60 to 90 min of imaging, consistent with it generating minimal radiometabolites that accumulate in brain. Because stable values are achieved within the practical imaging time for 11C (T1/2 = 20 min), [11C]FPEB could be used for multiple studies in a single subject within 1 day; this would, however, require higher specific activity than was achieved in this study.
Supplementary Material
Acknowledgments
We thank Denise Rallis-Frutos, Desiree Ferraris Araneta, Emily Page, David Clark, Kimberly Jenko, Jeih-San Liow, and the staff of the PET Department for [11C]hydrogen cyanide production and help in completing the studies, PMOD Technologies (Zurich, Switzerland) for providing its image analysis and modeling software, and Ioline Henter for editorial assistance. This work was performed during Dr. Lohith’s and Dr. Tsujikawa’s postdoctoral fellowships at the NIH.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Intramural Research Program of the National Institute of Mental Health, NIH (project numbers ZIAMH002852 and ZIAMH002793), under clinical protocols NCT00538798 (07 -M-0082) and NCT01896843 (13 -M-0166), and Society of Nuclear Medicine Wagner-Torizuka Fellowship (2011–2013) to Tetsuya Tsujikawa.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
TGL and TT contributed significantly to study design, data collection, analysis, interpretation, drafting and critically revising manuscript. FGS and VWP contributed to study design, data analysis, interpretation, drafting and critically revising manuscript. MV contributed to data analysis, interpretation, drafting and critically revising manuscript. SSZ contributed to study design, data collection, analysis, interpretation, and critically revising manuscript. CHL and YK contributed to study design, data analysis, interpretation, and critically revising manuscript. CLM contributed to data collection, analysis, and critically revising manuscript. MF and RBI contributed to study design, data interpretation, drafting and critically revising manuscript.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data
References
- 1.Sullivan JM, Lim K, Labaree D, et al. Kinetic analysis of the metabotropic glutamate subtype 5 tracer [(18)F]FPEB in bolus and bolus-plus-constant-infusion studies in humans. J Cereb Blood Flow Metab 2013; 33: 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong DF, Waterhouse R, Kuwabara H, et al. 18F-FPEB, a PET radiopharmaceutical for quantifying metabotropic glutamate 5 receptors: A first-in-human study of radiochemical safety, biokinetics, and radiation dosimetry. J Nucl Med 2013; 54: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AK, Kimura Y, Zoghbi SS, et al. Metabotropic glutamate subtype 5 receptors are quantified in the human brain with a novel radioligand for PET. J Nucl Med 2008; 49: 2042–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura Y, Simeon FG, Zoghbi SS, et al. Quantification of metabotropic glutamate subtype 5 receptors in the brain by an equilibrium method using 18F-SP203. Neuroimage 2012; 59: 2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura Y, Simeon FG, Hatazawa J, et al. Biodistribution and radiation dosimetry of a positron emission tomographic ligand, 18F-SP203, to image metabotropic glutamate subtype 5 receptors in humans. Eur J Nucl Med Mol Imaging 2010; 37: 1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shetty HU, Zoghbi SS, Simeon FG, et al. Radiodefluorination of 3-fluoro-5-(2-(2-[18F](fluoromethyl)-thiazol-4-yl)ethynyl)benzonitrile ([18F]SP203), a radioligand for imaging brain metabotropic glutamate subtype-5 receptors with positron emission tomography, occurs by glutathionylation in rat brain. J Pharmacol Exp Ther 2008; 327: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simeon FG, Liow JS, Zhang Y, et al. Synthesis and characterization in monkey of [11C]SP203 as a radioligand for imaging brain metabotropic glutamate 5 receptors. Eur J Nucl Med Mol Imaging 2012; 39: 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura T, Kikuchi T, Okada M, et al. Noninvasive and quantitative assessment of the function of multidrug resistance-associated protein 1 in the living brain. J Cereb Blood Flow Metab 2009; 29: 504–511. [DOI] [PubMed] [Google Scholar]
- 9.Veronese M, Zanotti-Fregonara P, Rizzo G, et al. Measuring specific receptor binding of a PET radioligand in human brain without pharmacological blockade: The genomic plot. Neuroimage 2016; 130: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanotti-Fregonara P, Xu R, Zoghbi SS, et al. The PET radioligand 18F-FIMX images and quantifies metabotropic glutamate receptor 1 in proportion to the regional density of its gene transcript in human brain. J Nucl Med 2015; 57: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith D. Synthia gets extreme makeover courtesy of National Instruments, ftp://ftpnicom/pub/branches//NIDaysBookletpdf (2008, accessed 29 Ausgust 2016), pp.24–25.
- 12.Bjurling P, Reineck R, Westerburg G, et al. Synthia, a compact radiochemistry system for automated production of radiopharmaceuticals. In: Link JM and Ruth TJ (eds) Proceedings of sixth workshop on targetry and target chemistry. Vancouver: TRIUMF, 1995, pp.282–284.
- 13.Zoghbi SS, Shetty HU, Ichise M, et al. PET imaging of the dopamine transporter with 18F-FECNT: a polar radiometabolite confounds brain radioligand measurements. J Nucl Med 2006; 47: 520–527. [PubMed] [Google Scholar]
- 14.Gandelman MS, Baldwin RM, Zoghbi SS, et al. Evaluation of ultrafiltration for the free-fraction determination of single photon emission computed tomography (SPECT) radiotracers: beta-CIT, IBF, and iomazenil. J Pharm Sci 1994; 83: 1014–1019. [DOI] [PubMed] [Google Scholar]
- 15.Lohith TG, Zoghbi SS, Morse CL, et al. Brain and whole-body imaging of nociceptin/orphanin FQ peptide receptor in humans using the PET ligand 11C-NOP-1A. J Nucl Med 2012; 53: 385–392. [DOI] [PMC free article] [PubMed]
- 16.Logan J, Fowler JS, Volkow ND, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab 1990; 10: 740–747. [DOI] [PubMed] [Google Scholar]
- 17.Ichise M, Toyama H, Innis RB, et al. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. J Cereb Blood Flow Metab 2002; 22: 1271–1281. [DOI] [PubMed] [Google Scholar]
- 18.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control 1974; 19: 716–723. [Google Scholar]
- 19.Hawkins RA, Phelps ME, Huang SC. Effects of temporal sampling, glucose metabolic rates, and disruptions of the blood-brain barrier on the FDG model with and without a vascular compartment: studies in human brain tumors with PET. J Cereb Blood Flow Metab 1986; 6: 170–183. [DOI] [PubMed] [Google Scholar]
- 20.Lassen NA, Bartenstein PA, Lammertsma AA, et al. Benzodiazepine receptor quantification in vivo in humans using [11C]flumazenil and PET: application of the steady-state principle. J Cereb Blood Flow Metab 1995; 15: 152–165. [DOI] [PubMed] [Google Scholar]
- 21.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzo G, Veronese M, Expert P, et al. MENGA: A new comprehensive tool for the integration of neuroimaging data and the allen human brain transcriptome atlas. PloS One 2016; 11: e0148744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolch WE, Eckerman KF, Sgouros G, et al. MIRD pamphlet No. 21: a generalized schema for radiopharmaceutical dosimetry–standardization of nomenclature. J Nucl Med 2009; 50: 477–484. [DOI] [PubMed] [Google Scholar]
- 24.Andersson Y, Långström B. Transition metal-mediated reactions using [11C]cyanide in synthesis of 11C-labelled aromatic compounds. J Chem Soc Perkin Trans I 1994, pp. 1395–1400. [Google Scholar]
- 25.Andersson JD, Seneca N, Truong P, et al. Palladium mediated (1)(1)C-cyanation and characterization in the non-human primate brain of the novel mGluR5 radioligand [(1)(1)C]AZD9272. Nucl Med Biol 2013; 40: 547–553. [DOI] [PubMed] [Google Scholar]
- 26.Patel S, Hamill TG, Connolly B, et al. Species differences in mGluR5 binding sites in mammalian central nervous system determined using in vitro binding with [18F]F-PEB. Nucl Med Biol 2007; 34: 1009–1017. [DOI] [PubMed] [Google Scholar]
- 27.Zanotti-Fregonara P, Innis RB. Suggested pathway to assess radiation safety of 11C-labeled PET tracers for first-in-human studies. Eur J Nucl Med Mol Imaging 2012; 39: 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham VJ, Rabiner EA, Slifstein M, et al. Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. J Cereb Blood Flow Metab 2010; 30: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Airaksinen AJ, Andersson J, Truong P, et al. Radiosynthesis of [11C]ximelagatran via palladium catalyzed [11C]cyanation. J Labell Comp Radiopharm 2008; 51: 1–5. [Google Scholar]
- 30.Kagedal M, Cselenyi Z, Nyberg S, et al. Non-linear mixed effects modelling of positron emission tomography data for simultaneous estimation of radioligand kinetics and occupancy in healthy volunteers. Neuroimage 2012; 61: 849–856. [DOI] [PubMed] [Google Scholar]
- 31.Lyoo CH, Ikawa M, Liow JS, et al. Cerebellum can serve as a pseudo-reference region in Alzheimer disease to detect neuroinflammation measured with PET radioligand binding to translocator protein. J Nucl Med 2015; 56: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turkheimer FE, Selvaraj S, Hinz R, et al. Quantification of ligand PET studies using a reference region with a displaceable fraction: application to occupancy studies with [(11)C]-DASB as an example. J Cereb Blood Flow Metab 2012; 32: 70–80. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.