Abstract
Defective protein quality control (PQC) systems are implicated in multiple diseases. Molecular chaperones and co-chaperones play a central role in functioning PQC. Constant mechanical and metabolic stress in cardiomyocytes places great demand on the PQC system. Mutation and downregulation of the co-chaperone protein BCL-2–associated athanogene 3 (BAG3) are associated with cardiac myopathy and heart failure, and a BAG3 E455K mutation leads to dilated cardiomyopathy (DCM). However, the role of BAG3 in the heart and the mechanisms by which the E455K mutation leads to DCM remain obscure. Here, we found that cardiac-specific Bag3-KO and E455K-knockin mice developed DCM. Comparable phenotypes in the 2 mutants demonstrated that the E455K mutation resulted in loss of function. Further experiments revealed that the E455K mutation disrupted the interaction between BAG3 and HSP70. In both mutants, decreased levels of small heat shock proteins (sHSPs) were observed, and a subset of proteins required for cardiomyocyte function was enriched in the insoluble fraction. Together, these observations suggest that interaction between BAG3 and HSP70 is essential for BAG3 to stabilize sHSPs and maintain cardiomyocyte protein homeostasis. Our results provide insight into heart failure caused by defects in BAG3 pathways and suggest that increasing BAG3 protein levels may be of therapeutic benefit in heart failure.
Keywords: Cardiology
Introduction
Proteinopathy, the toxic aggregation of misfolded or damaged proteins, is involved in the pathogenesis of multiple diseases, including neurodegenerative disorders, skeletal myopathies, and heart failure (1–3). A common feature of these diseases is a deterioration of the protein quality control (PQC) system, a collection of pathways devoted to the repair or clearance of faulty proteins crucial for normal cell and organ function (4). In the heart, defective PQC pathways and toxic protein accumulation have been associated with hypertrophic and dilated cardiomyopathies, as well as idiopathic and ischemic heart disease (5–8). Central players of the PQC system are molecular chaperones and co-chaperones that mediate and stabilize the correct conformation of nascent proteins, refold aberrant proteins to their functionally active native state, or target unfolded/misfolded or aggregated substrates for degradation (9–11).
Mutations in the co-chaperone protein BCL-2–associated athanogene 3 (BAG3) are associated with cardiac myopathies (12–17). A GWAS, involving multiple families with heart failure due to dilated cardiomyopathy (DCM), identified associated mutations in BAG3 (17). In the same study, analysis of a large, 4-generation family identified a single amino acid substitution of glutamic acid 455 with lysine (E455K) in BAG3 as the unequivocal cause of DCM (17). The E455 residue lies within a highly evolutionarily conserved region within the BAG domain of BAG3 (17). BAG3 interacts with the nucleotide-binding domain (NBD) of the molecular chaperone HSP70/HSC70 via its BAG domain, thereby modulating chaperone activity and regulating PQC (18). A recent study showed that mutation of this residue perturbed the interaction of BAG3 with HSC70 in vitro (19).
Decreased levels of BAG3 have also been described in a family with idiopathic DCM and in unrelated patients with end-stage heart failure (14). BAG3 is a member of an evolutionarily conserved family of molecular co-chaperones (20) that interact with chaperones of the HSP family, including HSP70, to modulate their function (18, 21–23). BAG3 is prominently expressed in cardiac and skeletal muscle tissues (24). However, little is known regarding the specific requirements for BAG3 in cardiomyocytes or the mechanisms by which the BAG3 E455K missense mutation leads to the progression of cardiomyopathy.
Results
Loss of BAG3 in the heart leads to DCM, heart failure, and premature death.
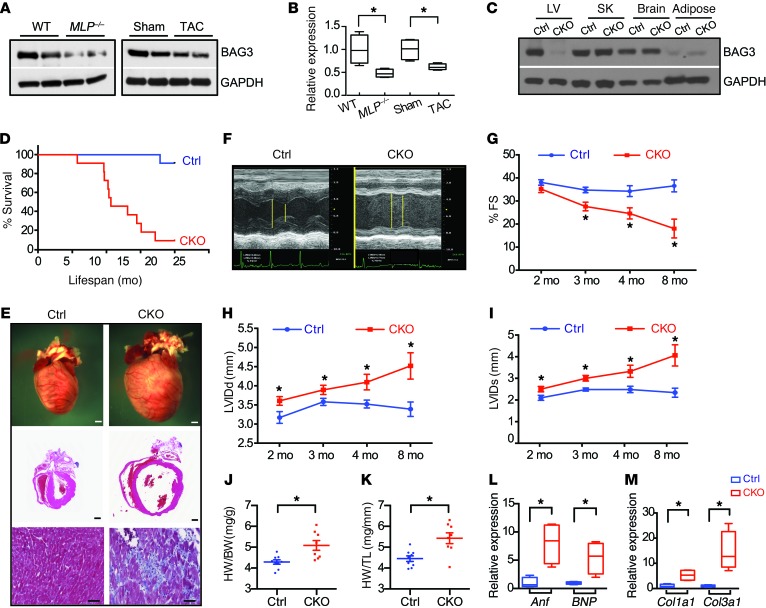
Decreased levels of BAG3 protein are found in failing human hearts (14). To ascertain whether failing mouse hearts, like their human counterparts, exhibit decreased BAG3, we performed Western blot analysis on cardiac tissue from 2 mouse models of cardiomyopathy. Muscle LIM protein–KO (MLP-KO) mice are a model of DCM and show many signs seen clinically in heart failure (25). Mice submitted to chronic transverse aortic constriction (TAC) develop cardiac hypertrophy, which progresses to dilation and heart failure, again modeling human disease (26). As with human failing hearts, we observed decreased BAG3 protein levels in both dilated MLP-KO mouse hearts and hypertrophic hearts isolated 4 weeks after TAC (Figure 1, A and B), supporting the idea of a crucial role of BAG3 in cardiac function.
Figure 1. BAG3 deficiency leads to DCM and premature death.
(A and B) Representative Western blot (A) and corresponding quantitative densitometric analysis (B) of BAG3 in WT and MLP-KO (MLP–/–) mouse hearts and hearts isolated from mice that had undergone TAC for 4 weeks. GAPDH served as a loading control. n = 4 mice per group. (C) Western blot analysis of BAG3 in different tissues from Bag3fl/fl αMHC-Cre–positive (CKO) and control (Ctrl) mice. GAPDH served as a loading control. n = 3 mice per group. SK, skeletal muscle. (D) Kaplan-Meier survival curves for CKO (n = 11) and control (n = 11) mice. (E) Representative microscopic views of whole mouse hearts (top, scale bars: 1 mm) and cross-sectional views of H&E-stained (middle, scale bars: 1 mm) and Masson’s trichrome–stained (bottom, scale bars: 50 μm) hearts isolated from control and CKO mice at 6 months of age. (F) Representative echocardiographic images of control and CKO mice at 4 months of age. (G–I) Echocardiographic measurements for control and CKO mice (n = 10–11 mice at 2, 3, 4, and 8 months of age) of (G) LV FS (% FS), (H) LVIDd, and (I) LVIDs. (J and K) HW to BW ratio (J) and HW to TL ratio (K) for control (n = 9) versus CKO (n = 8) mice at 4 months of age. (L–M) qRT-PCR analysis of cardiac fetal gene markers (L) and profibrotic genes (M) in control (n = 9) and CKO (n = 8) mouse hearts at 4 months of age. Data were normalized to corresponding 18S levels, and CKO is expressed as the fold-change versus control. Data are represented as the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test.
To investigate the in vivo role of BAG3 in cardiac function, we generated BAG3-KO mice by crossing targeted mice containing Bag3-floxed alleles (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI94310DS1) with global deleter Sox2-Cre mice (27). Global deficiency of BAG3 resulted in impaired postnatal growth, with Bag3–/– mice exhibiting decreased body weight (BW) and early lethality at 3 to 4 weeks of age (Supplemental Figure 1, B–D), consistent with previous reports (24, 28).
Early lethality of BAG3-KO mice precluded the study of the role of BAG3 in adult heart function. To investigate the specific role of BAG3 in cardiomyocytes, we crossed Bag3-floxed (Bag3fl/fl) mice with cardiomyocyte-specific α-myosin heavy chain Cre-transgenic mice (αMHC-Cre mice) (29). This αMHC-Cre line was specifically shown to have normal cardiac function (30). Consistent with published data, our echocardiography studies also showed that this αMHC-Cre mouse line in the same genetic background (C57/B6) as that of our mouse models has normal cardiac function (Supplemental Figure 2). Western blot analysis confirmed specific and effective deletion of Bag3 in Bag3fl/fl α-MHC-Cre–positive (BAG CKO) hearts compared with those of control Cre-negative Bag3fl/fl hearts (Figure 1C). Loss of BAG3 did not result in increased expression of other BAG family members (Supplemental Figure 1E). BAG3-CKO mice were viable at birth; however, they had a striking susceptibility to premature death (Figure 1D) consequent to DCM and heart failure (Figure 1, E–M). Only 9% of the BAG3-CKO mice survived past 20 months of age compared with 91% of the control littermates (Figure 1D). Morphological and histological analysis of BAG3-CKO hearts revealed marked cardiac enlargement with severe fibrosis in surviving 6-month-old CKO mice (Figure 1E). Echocardiography revealed an age-dependent decrease in left ventricular (LV) systolic function (percentage of fractional shortening [FS]) in mice deficient for BAG3 (Figure 1, F and G). Consistent with histological observations, loss of BAG3 resulted in LV chamber dilation, as evidenced by a significant increase in end-diastolic LV internal diameter (LVIDd) and end-systolic LV internal diameter (LVIDs) (Figure 1, H and I). Heart weight to BW (HW/BW) and HW to tibia length (HW/TL) ratios were also significantly increased in CKO mice compared with controls at 4 months of age (Figure 1, J and K), with no observed difference in BW (data not shown).
Consistent with molecular evidence of cardiac remodeling (31), cardiac fetal gene markers, atrial natriuretic factor (Anf), and B-type natriuretic peptide (BNP) were significantly increased in the hearts of CKO mice at 4 months of age (Figure 1L), as were the profibrotic genes collagen α1 types I (Col1a1) and III (Col3a1) (Figure 1M). In addition, hemodynamic analyses showed that both the peak rate of pressure rise (dP/dtmax) and peak rate of pressure decline (dP/dtmin) were significantly reduced in CKO mice at as early as 10 weeks of age, indicating early impairment of cardiac contractile function in BAG3-deficient mice (Supplemental Figure 1, F and G). Consistent with hemodynamic studies, single myocyte contraction induced by field stimulation was also significantly reduced in BAG3-CKO cardiomyocytes when compared with control cardiomyocytes (Supplemental Figure 1, H and I), although the profiles of induced Ca2+ transients, including amplitude and tau, were not significantly affected (Supplemental Figure 1, J and K). Taken together, these data demonstrated that loss of BAG3 impaired the cardiomyocyte contractile apparatus and that BAG3 was essential for maintaining normal cardiac structure and function.
BAG3 deficiency leads to the downregulation of protein levels of small HSPs and impaired autophagic flux.
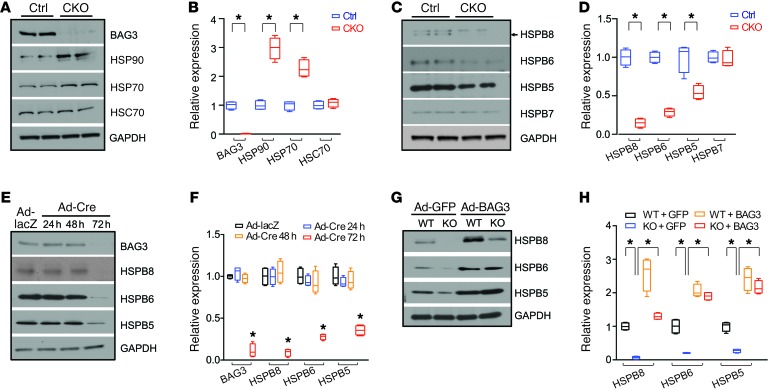
We next explored the molecular mechanisms by which BAG3 deficiency caused DCM. As a co-chaperone protein, BAG3 interacts with both ATP-dependent high-molecular-weight HSPs and ATP-independent small HSPs (sHSPs) in large, functionally distinct multi-chaperone complexes (18, 32–34). BAG3 interacts with sHSPs through its 2 IPV motifs (32, 33, 35). To determine the effects of Bag3 deletion on components of the chaperone complex, we evaluated protein samples from isolated adult cardiomyocytes of 2-month-old CKO mice and control mice. Western blot analysis revealed that the levels of stress-inducible HSPs, such as HSP90 and HSP70, were significantly upregulated in BAG3-CKO cardiomyocytes, whereas levels of the constitutively expressed HSC70 were unchanged (Figure 2, A and B). These data suggested increased protein stress in cardiomyocytes deficient in BAG3. Interestingly, protein levels of sHSPs, such as HSPB8 (also known as HSP22), HSPB6 (also known as HSP20), and HSPB5 (also known as αB-crystallin) were significantly decreased in BAG3-CKO cardiomyocytes (Figure 2, C and D). It is noteworthy that the levels of HSPB7, a sHSP that does not interact with BAG3 (36), were not changed in CKO-mutant mouse hearts. To determine whether this reduction was a primary or secondary effect, we next isolated neonatal cardiomyocytes from Bag3fl/fl mice and deleted Bag3 using Cre adenovirus (Ad-Cre). Acute loss of BAG3 in cardiomyocytes led to the downregulation of sHSPs in a time-dependent manner (Figure 2, E and F). Furthermore, reexpression of BAG3 in BAG3-KO cells was able to rescue the expression of sHSPs (Figure 2, G and H). Next, we sought to determine why sHSP levels were diminished in the absence of BAG3. No changes in the mRNA levels of sHSPs were detected in WT or BAG3-KO hearts (Supplemental Figure 3A). We then performed a cycloheximide (CHX) chase experiment to determine HSPB8 protein stability. Unfortunately, as shown in Supplemental Figure 3B, HSPB8 was undetectable, even at basal conditions without CHX treatment in BAG3-KO cardiomyocytes. To overcome this problem, we then infected neonatal cardiomyocytes isolated from BAG3-KO and WT control mice with adenovirus to overexpress tagged HSPB8 (Dendra2-HSPB8). As shown in Supplemental Figure 3C, the levels of overexpressed HSPB8 were significantly diminished 18 hours after CHX treatment and were undetectable 24 hours after CHX treatment in BAG3-KO neonatal cardiomyocytes. In contrast, in control WT neonatal cardiomyocytes, approximately 50% of the overexpressed HSPB8 was still present 24 hours after CHX treatment. Taken together, these data indicated that upon loss of BAG3, the levels of affected sHSPs were diminished as a result of protein instability.
Figure 2. BAG3 deficiency leads to the downregulation of protein levels of sHSPs.
(A–D) Representative immunoblots (A and C) and quantification analysis (B and D) of BAG3, HSP90, HSP70, HSC70 (A and B), and HSPB5, -6, -7, and -8 (C and D) in adult cardiomyocytes isolated from control and CKO mice. n = 4 mice per group. (E and F) Neonatal cardiomyocytes were isolated from Bag3fl/fl mice and infected with Ad-Cre (20 MOI) for 24, 48, and 72 hours. Ad-lacZ was used as a control. Representative immunoblots (E) and quantification analysis (F) of BAG3 and HSPB5, -6, and -8. n = 4. GAPDH served as a loading control. (G and H) Neonatal cardiomyocytes were isolated from Bag3 global-KO mice or WT controls and infected with BAG3-overexpressing adenovirus (Ad-BAG3) (20 MOI) for 36 hours. GFP adenovirus (Ad-GFP) was used as a control. Representative immunoblots (G) and quantification analysis (H) of HSPB5, -6, and -8. n = 4. GAPDH served as a loading control. Data are represented as the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test or 2-way ANOVA.
As it has also been reported that BAG3 is involved in autophagy (37), we next investigated how the loss of BAG3 might affect autophagic flux by examining the levels of the autophagosome-associated protein light chain 3B (LC3B) (38), which is present in cytosolic form (LC3B-I) until it becomes targeted to nascent autophagosomes (LC3B-II form) (39, 40). As expected, Western blotting showed a significant decrease in LC3B-II levels in BAG3-CKO hearts compared with levels in control littermate hearts (Supplemental Figure 4, A and B). The selective autophagy receptor p62 was also increased in BAG3-CKO hearts (Supplemental Figure 4, C and D). Moreover, in cultured neonatal cardiomyocytes, starvation-induced autophagic LC3B-II levels were further decreased in BAG3-KO cells when compared with levels in WT cells (Supplemental Figure 4, E and F). To monitor autophagic flux in vivo by fluorescence microscopy, we next crossed BAG3-CKO mice with GFP-LC3–transgenic mice (41). After 18 hours of starvation, we observed a reduction in LC3-GFP accumulation in BAG3-CKOLC3-GFP myocardium compared with controlLC3-GFP mouse myocardium, indicating that autophagic flux was suppressed in BAG3-deficient hearts (Supplemental Figure 4, G and H). Autophagic flux is a dynamic process that starts with the formation of autophagosomes and ends with their degradation after fusion with lysosomes (42). Owing to the difficulty of monitoring this process in vivo, we analyzed BAG3-CKO mouse neonatal cardiomyocytes cultured in the absence or presence of bafilomycin-A1, a v-ATPase inhibitor (43), or chloroquine, an inhibitor of autophagosome-lysosome fusion and autophagosome degradation (39), to assess whether BAG3 regulates the formation and/or clearance of autophagosomes. Following bafilomycin-A1 and chloroquine treatment, LC3B-II remained at reduced levels in BAG3-CKO cardiomyocytes compared with levels in control cells (Supplemental Figure 4, I and J), indicating that BAG3 regulated autophagosome formation. Taken together, these results indicated that BAG3 deficiency results in the suppression of autophagosome formation.
Impaired autophagy suggested that BAG3 deficiency might result in misfolded protein aggregates. In the chaperone network, sHSPs function as “holdases,” a term that refers to their ability to bind and stabilize denatured or non-native proteins against aggregation, subsequently bringing them to be refolded by HSP70 (44, 45). Thus, dramatic downregulation of HSPB5, -6, and -8 in BAG3 mutants would also be expected to lead to misfolded protein aggregates. To assess the presence of misfolded protein aggregates in BAG3 mutants, we first evaluated p62, a common component of cytoplasmic inclusions in protein aggregates (46), by immunofluorescence. Although p62 levels were increased in BAG3-CKO hearts (Supplemental Figure 5A), we did not detect aberrant aggregation of p62 or the aggregation-prone proteins desmin and αB-crystallin (CryAB) (Supplemental Figure 5, B and C). Moreover, electron-dense perinuclear protein aggregates, characteristic of aggresomes (7, 47), were not visible by transmission electron microscopic analysis of BAG3-CKO myocardium (data not shown). We also investigated whether the expression levels of some critical cardiac-specific proteins were changed with loss of BAG3 (Figure 3B). Quantitative proteomic analysis of total proteins from BAG3-CKO and control hearts revealed no significant differences (Supplemental Figure 6).
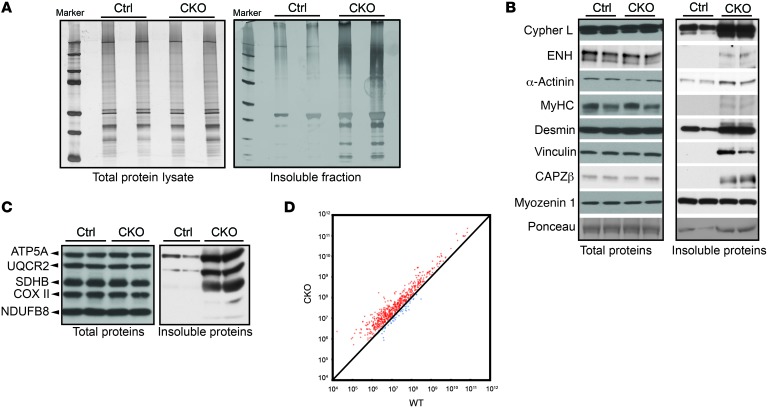
Figure 3. BAG3 deficiency leads to increased levels of a subset of insoluble proteins.
(A) Silver staining of total and insoluble proteins in control and CKO mice. n = 4. (B) Western blot analysis of cypher L, ENH, α-actinin, myosin heavy chain (MyHC), desmin, vinculin, CAPZβ, and myozenin 1 in total and insoluble protein fractions of control and CKO mouse hearts. n = 4 mice per group. The blots shown were derived from replicate samples run on parallel gels. (C) Western blot analysis of the oxidative phosphorylation complexes in total and insoluble protein fractions from control and CKO mouse hearts. n = 4. (D) Scatter plot comparing the intensity of proteins from MS analysis of the insoluble fraction from CKO and control mouse hearts.
BAG3 deficiency leads to increased levels of a subset of insoluble proteins.
Recent studies have found that, despite a paucity of age-dependent differences in the total cardiac proteome, significant age-dependent differences are observed among proteins isolated from detergent-insoluble protein aggregates (48, 49). Therefore, since we observed no significant changes in the total proteome of cardiac tissue from BAG3-deficient mice (Supplemental Figure 6), we next analyzed insoluble proteins by isolating detergent-insoluble protein fractions from BAG3-CKO and control hearts. Strikingly, upon equal loading of total protein lysates (Figure 3A, left), the sarcosyl-insoluble protein fraction from the aggregated protein components relative to those from control littermate hearts (Figure 3A, right). To identify constituent proteins of the insoluble fraction, we used a candidate-based approach to evaluate the effects of BAG3 deficiency on the levels of a number of filament proteins critical for cardiac function (cypher L [LDB3], ENH, α-actinin, myosin heavy chain, desmin, vinculin, and CAPZβ). The results demonstrated a marked accumulation of these proteins in the insoluble protein fraction of BAG3-CKO hearts (Figure 3B). Conversely, these proteins were unchanged in total lysate samples (Figure 3B). We also detected an accumulation of components of the mitochondrial oxidative phosphorylation (OXPHOS) pathway in both total and insoluble protein fractions using an OXPHOS antibody cocktail (Figure 3C).
To profile the constituents of the insoluble protein fraction in BAG3-deficient hearts, we used label-free mass spectrometry (MS) to quantify protein profiles from control and BAG3-CKO mouse hearts. We identified approximately 700 proteins within insoluble fractions of either WT or CKO samples. Scatter-plot analysis revealed that most of these proteins were more abundant in the insoluble protein fraction of BAG3-CKO hearts relative to that seen in controls (Figure 3D), with the levels of 101 proteins increased by more than 5-fold. We next validated the MS results by Western blot analysis and found that, in accordance with our proteomics data, a number of PQC pathway proteins (HSP70, HSP90, HSPB5, and p62), filament proteins (FLNC and tropomyosin), and mitochondrial proteins (FIS1, VDAC2, and MTCO2) were increased in the insoluble protein fraction of BAG3-CKO hearts. Myozenin and HADHB were unchanged, while UQCRB was decreased (Supplemental Figure 7A). Gene Ontology Enrichment Analysis (GOEA) with Cytoscape modeling to provide a visual biomolecular interaction network (50) revealed that proteins with increased levels in the insoluble fraction of BAG3-CKO hearts are involved in multiple essential pathways for maintaining normal cardiac function, including muscle contraction, actin filament capping, cytoskeleton regulation, translation initiation, protein folding, ATP synthesis–coupled electron transport, the acetyl-CoA biosynthetic process, mitochondrial organization, response to ROS, cell junction assembly, and the proteasome core complex (Supplemental Figure 7B). These results indicated that BAG3 regulated several specific targets related to metabolic and contractile pathways. It should also be pointed out that BAG3 could be detected in multiple cellular compartments, including mitochondrial and cytoplasmic fractions, and could also be found colocalizing with α-actinin at the Z-disc (Supplemental Figure 8). Altogether, these findings suggested that perturbed PQC and insoluble protein accumulation of a subset of functionally important cardiomyocyte proteins were responsible for the development of heart failure in BAG3-CKO mice.
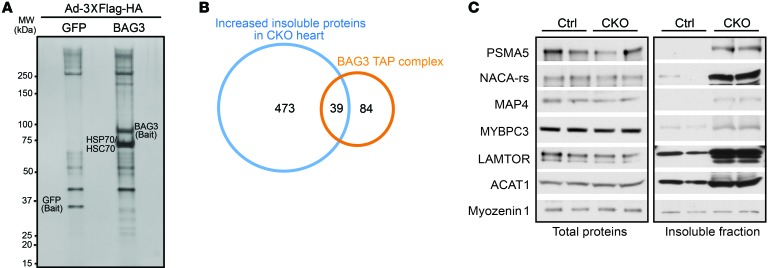
To further investigate specific substrates of the BAG3-chaperone complex in cardiomyocytes, we performed tandem affinity purification (TAP) in combination with MS analysis. We delivered an adenovirus expressing an N-terminal 3XFlag-HA tandem affinity–tagged full-length BAG3 (3XFlag-HA-BAG3) as a bait protein into neonatal cardiomyocytes to affinity-purify endogenous proteins that interacted with BAG3. Compared with the 3XFlag-HA-GFP control affinity purification, multiple bands were present in the immune-precipitated BAG3-containing complex (Figure 4A). Using gel digestion MS, we identified the most intense band at 70 kDa as HSP70 and HSC70, which are known interactors of BAG3 (35). To comprehensively profile the BAG3 interactome, we performed on-bead digestion following TAP and subjected the immunoprecipitated products to label-free proteomics (51). A total of 1,490 proteins were affinity purified from cardiomyocytes expressing 3XFlag-HA-BAG3 compared with 983 proteins pulled down from 3XFlag-HA-GFP controls. To identify high-confidence BAG3 interactors, proteins were filtered to have 2 or more matching queries and an enrichment of at least 20-fold over the GFP control group. This stringent cutoff yielded 123 potential binding partners. Interestingly, several ATPase-independent sHSP family members (HSPB5, -6, and -8), and ATPase-dependent HSP70 family members (HSP70 and HSC70), previously identified as part of the BAG3 complex (35), were among the 123 potential binding partners identified in this manner. Notably, of the 123 potential binding partners, 39 were overlapping with proteins identified as being increased within the insoluble protein fraction from BAG3-CKO hearts (Figure 4, B and C, Figure 3, B and E, and Supplemental Table 1). A 2-tailed Fisher’s exact test indicated that the overlap was highly significant (P < 2.2 × 1016). These results indicated that BAG3 directly interacted with ATPase-independent and -dependent HSPs to ensure protein quality control of specific protein substrates, several of which also directly interact with the BAG3-chaperone complex.
Figure 4. Accumulation of BAG3-binding partners in the insoluble protein fraction from BAG3-CKO heart.
(A) Silver staining of the TAP BAG3-interacting complex. MW, molecular weight. (B) Venn diagram showing overlapping proteins between the BAG3 TAP complex and increased insoluble proteins in CKO mouse hearts. (C) Western blot validation of proteins in the BAG3 interactome from total and insoluble protein fractions from control and CKO mouse hearts. The blots shown were derived from replicate samples run on parallel gels. n = 4 mice per group.
Interaction between BAG3 and HSP70 is critical for sHSP stability.
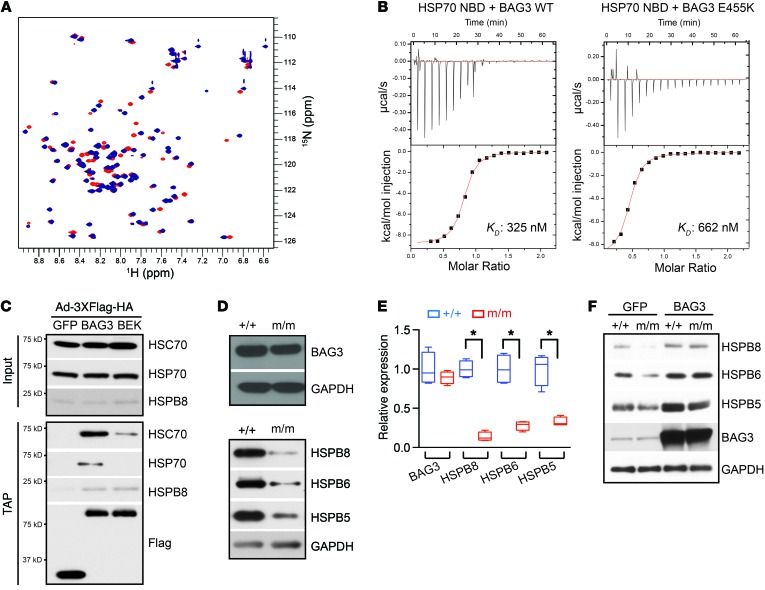
A human genetic linkage study involving a large, 4-generation family identified a single amino acid substitution of glutamic acid 455 with lysine (E455K) in BAG3 as the unequivocal cause of DCM (17). However, the pathogenic effects of this mutation are not known. BAG3 interacts with the NBD domain of HSP70/HSC70 via its BAG domain. We first determined whether the mutation affects BAG domain folding by performing NMR spectra analyses. 1H15N HSQC spectra of both the WT and mutant BAG domain showed sharp, well-dispersed peaks characteristic of folded proteins, revealing that the mutation is compatible with folding (Figure 5A). We then determined the binding affinity between the BAG domain of BAG3 and the NBD (ATPase domain) of HSP70 by isothermal titration calorimetry (ITC). Strikingly, the E455K mutation weakened the interaction by almost 50%, yielding comparable heat-release values of 8.8 and 8.9 kcal/mol, but differing KD affinities of 325 and 662 nM for WT and BAG3 E455K mutants, respectively (Figure 5B). Furthermore, TAP analysis using GFP control, WT BAG3, and E455K-mutant BAG3 proteins (BEK) overexpressed in neonatal cardiomyocytes revealed that E455 was critical for the interaction between BAG3 and HSP70 (Figure 5C). Interestingly, the E455 mutation did not affect BAG3 binding to HSPB8 (Figure 5C).
Figure 5. The interaction between BAG3 and HSP70 is critical for sHSP stability.
(A) Overlay of WT (blue) and E455K-mutant (orange) BAG3 domain 1H15N heteronuclear single-quantum correlation (HSQC) spectra. (B) ITC of the interaction of the HSP70 NBD with the WT (left) and E455K-mutant (right) BAG domain of BAG3. Upper panels represent the measure of enthalpy during titration. Lower panels represent the integrated binding isotherm of the titration and experimental fit to a single site model. (C) Representative Western blot analysis of HSC70, HSP70, and HSPB8 in input and the TAP BAG3-interacting complex. n = 3 (D and E) Representative Western blot (D) and corresponding quantitative analysis (E) of BAG3 and HSPB5, -6, and -8 in WT (+/+) and BAG3 E455K-mutant (m/m) mouse hearts. GAPDH served as a loading control. n = 4 mice per group. Data are represented as the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test. (F) Western blot analysis of HSPB5, -6, and -8 and BAG3 in neonatal cardiomyocytes isolated from WT (+/+) and BAG3 E455K-mutant (m/m) hearts infected with WT Ad-BAG3 (20 MOI) for 36 hours. Ad-GFP was used as a control, and GAPDH served as a loading control. n = 4.
To elucidate the biological and physiological consequences of the BAG3 E455K mutation, we generated BAG3 E455K–knockin (BEK-knockin) mice, in which the endogenous Bag3 gene was replaced with mutant BAG3 containing the E460K mutation (equivalent to the human E455K mutation; Supplemental Figure 9, A–C). Western blot analysis showed that BAG3 levels in the hearts of BAG3 homozygous E455K mice were comparable to BAG3 protein levels in WT controls (Figure 5, D and E), suggesting that the E455K mutation does not impair the expression or stability of BAG3. Conversely, the protein levels of sHSPs (HSPB5, HSPB6, and HSPB8) were downregulated in E455K-mutant hearts (Figure 5, D and E), and overexpression of WT BAG3 protein rescued sHSP levels in BAG3 E455K-mutant cardiomyocytes (Figure 5F). Notably, the BAG3 E455K mutation did not affect BAG3 binding to sHSPs (Figure 5C) (35). Taken together, we concluded that the ability of BAG3 to interact with HSP70 was essential to the stability of sHSPs. These results suggested that the BAG3 E455K mutant would result in dysfunction of the chaperone complex, thus leading to cardiomyopathy.
The BAG3 E455K mutation impairs BAG3 function and leads to DCM.
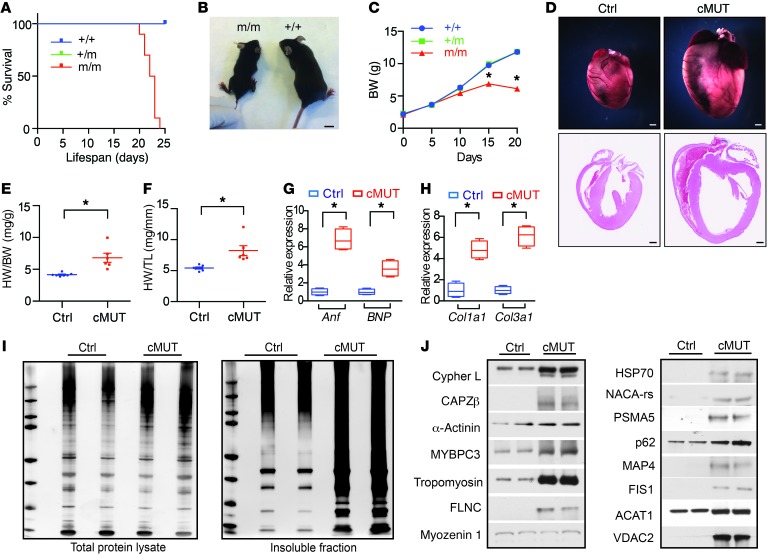
Indeed, homozygous global BAG3 E455K mutants exhibited impaired postnatal growth and premature lethality at 4 weeks of age (Figure 6, A–C), reminiscent of BAG3 homozygous global null mutants. Given that BAG3 protein levels were unchanged (Figure 5, D and E), our results suggested that the E455K mutation impaired the essential functions of BAG3. As early lethality of homozygous BAG3 E455K mutants precluded the study of BAG3 E455K mutation effects on adult heart function, we generated cardiac-specific BAG3 E455K-mutant (cMUT) mice using the αMHC-Cre–transgenic mouse line (29). Immunostaining analyses confirmed that there were no alterations in BAG3 protein levels or in BAG3 localization in cMUT hearts compared with control hearts (Supplemental Figure 9D). As observed with CKO animals, the hearts of cMUT animals had increased levels of stress-inducible HSPs and impaired autophagic flux (Supplemental Figure 9, E and F). In addition, cMUT mice had phenotypes comparable to those of BAG3-CKO mice, with marked cardiac enlargement, diminished systolic function, and activation of a fetal cardiac gene program (Figure 6, D–H, and Supplemental Figure 10).
Figure 6. Amino acid E455 is critical for BAG3 function.
(A) Kaplan-Meier survival curves for BAG3 E455K-mutant homozygous (m/m), heterozygous (m/+), and WT control (+/+) mice. n = 10 mice per group. (B) Representative image illustrating the significant growth retardation in BAG3 E455K-mutant homozygous mice (m/m) compared with littermate control (+/+) mice at 20 days of age. Scale bar: 1 cm. (C) Growth curves of BAG3 E455K-mutant homozygous (m/m), heterozygous (m/+), and control (+/+) mice. n = 10 mice per group. (D) Representative microscopic views of whole mouse hearts and cross-sectional views of H&E-stained hearts isolated from cardiac-specific cMUT and control mice at 6 months of age. n = 3 mice per group. Scale bars: 1 mm. (E and F) HW to BW ratio (E) and HW to TL ratio (F) for control (n = 7) versus cMUT (n = 6) mice at 5 months of age. (G and H) qRT-PCR analysis of cardiac fetal gene markers (G) and profibrotic genes (H) in control (n = 4) and cMUT (n = 4) mouse hearts at 5 months of age. Data were normalized to corresponding 18S levels, and cMUT is expressed as the fold-change versus control. (I) Silver staining of total and insoluble proteins in control and cMUT mice. n = 4. (J) Western blot analysis of proteins in the insoluble fraction of control and cMUT mouse hearts. n = 4 mice per group. Data are represented as the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test.
As found in BAG3-CKO mice, the abundance of insoluble proteins (Figure 6I) was dramatically increased in cMUT hearts. To determine whether the same proteins identified in the insoluble fraction of BAG3-CKO mouse hearts were also in the insoluble fraction of BAG3 cMUT mouse hearts, we performed Western blots analysis with antibodies against 14 proteins that had been found to be upregulated in the insoluble fraction of BAG3-CKO mouse hearts. As shown in Figure 6J, all 14 of the investigated proteins were upregulated in the insoluble fraction from cMUT hearts. Taken together, our results demonstrate that the BAG3 E455K mutation is a loss-of-function mutation and imply that interaction with HSP70 is essential for normal BAG3 function in cardiomyocyte protein homeostasis and cardiac function.
Discussion
While it is clear that mutations in BAG3 result in human cardiac myopathies (12–17), little is known about the role of BAG3 in cardiomyocytes or the adult heart, or about the mechanisms by which the BAG3 E455K missense mutation leads to the progression of cardiomyopathy. The present study demonstrated an essential role of BAG3 in cardiomyocytes for the maintenance of normal heart function, with loss of BAG3 leading to DCM. Specifically, BAG3 maintained normal heart function by exercising quality control over a specific subset of proteins required for the metabolic and contractile function of cardiomyocytes. We also found that cardiac-specific BAG3 E455K-mutant mice had DCM comparable to that of cardiomyocyte-specific BAG3-KO mice, demonstrating that E455K is a loss-of-function mutation and that this residue is critical for essential functions of BAG3.
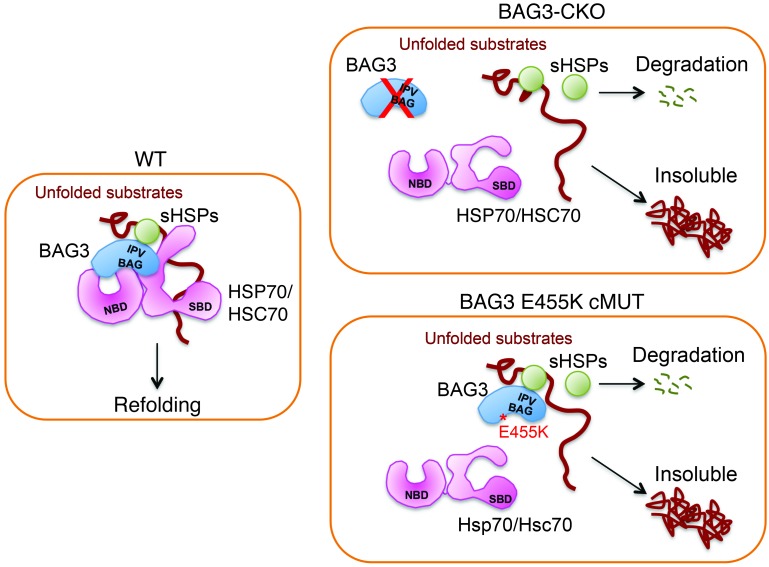
The ATP-dependent HSP70 and the ATP-independent small sHSP/HSPB family, which includes HSPB8, HSPB6, and HSPB5, constitute an ancient system for protecting proteins under conditions of proteotoxic stress (52). One major gap in the understanding of the chaperone network is how sHSPs link to other network components. This question is important, because sHSPs themselves lack the ATPase activity that is required for active refolding. The BAG domain of BAG3 binds HSP70 with high affinity (53), suggesting that BAG3 is a functionally important partner of HSP70. BAG3 also interacts with HSPB8 and other members of the sHSP/HSPB family through its 2 IPV motifs (32, 54), thus allowing BAG3 to assemble large multi-chaperone complexes. Our study demonstrated that the human BAG3 E455K mutation diminished the interaction of BAG3 with HSP70. Although the E455K mutation did not affect the interaction of BAG3 with sHSPs, it markedly affected the stability of sHSPs (Figure 7).
Figure 7. Model for mechanisms underlying cardiomyopathy in BAG3 mutants.
WT BAG3 plays a critical role in the formation of multi-chaperone complexes and the stability of sHSPs, maintaining the PQC of BAG3 complex substrates and normal heart function. Cardiomyocyte-specific KO of BAG3 (CKO) leads to dysfunction of the chaperone complex and destabilization of sHSPs, resulting in an increased insolubility of BAG3 complex substrates and cardiomyopathy. The BAG3 E455K mutation leads to dysfunction of the chaperone complex and destabilization of sHSPs by disrupting the interaction between BAG3 and HSP70, resulting in an increased insolubility of BAG3 complex substrates and cardiomyopathy. SBD, substrate-binding domain.
We were not able to detect large aggregates in BAG3-CKO or cMUT mice by microscopic methods. Aggregates are operationally defined by poor solubility in aqueous or detergent solvents, aberrant subcellular or extracellular localization, and a non-native secondary structure (55, 56). By using biochemical and proteomic methods, we found evidence for increased insoluble aggregates in BAG3-CKO and cMUT mice. Accumulating evidence suggests that proteotoxicity resulting from small aggregates of proteins can contribute to heart failure and hereditary heart diseases (57, 58). Small protein aggregates may interfere with critical cellular functions by binding and obstructing key signaling or trafficking molecules, potentially leading to cell death (57, 58). In this regard, proteotoxicity resulting from small protein aggregates could be an important factor leading to the observed DCM phenotype in CKO and cMut animals. We found increased amounts of specific proteins in the insoluble fraction from BAG3-CKO and cMUT mice compared with that detected in controls. Notably, our pulldown studies with BAG3 demonstrated a significant overlap between proteins that were complexed with BAG3 and those that were increased in the insoluble fraction of mutants.
Altogether, our observations suggested that interaction between BAG3 and HSP70 is essential for the execution of BAG3 function in stabilizing sHSPs and maintaining cardiomyocyte protein homeostasis (Figure 7). These findings indicate that the downregulation of BAG3 protein levels in the failing human heart (14) is likely to be of functional significance and to contribute to human heart failure, regardless of etiology. Our results suggest that increasing the protein levels of BAG3 may be of therapeutic benefit in heart failure.
Methods
Mouse models.
C57BL/6 mice (strain code: 027) were purchased from Charles River Laboratories. FLPase (FLP) mice [B6.SJL-Tg(ACTFLPe)9205Dym/J, stock no. 005703] and Sox2-Cre mice [Tg(Sox2-cre)1Amc/J, stock no. 004783] were purchased from The Jackson Laboratory. Mice with Bag3 conditional alleles and lacZ and neo cassettes (B6N-Bag3tm1a, MGI: 1352493) (Supplemental Figure 1) were obtained from the International Knockout Mouse Consortium. Bag3-floxed (Bag3fl/fl) mice were generated by crossing B6N-Bag3tm1a mice with FLP deleter mice (59) to excise the lacZ and neo cassettes. BAG3 global-KO mice were subsequently generated by crossing B6N-Bag3tm1a mice with Sox2-Cre mice (27). To generate BAG3 cardiomyocyte–specific KO (CKO) mice, Bag3fl/fl mice were crossed with α-myosin heavy chain–transgenic (αMHC-Cre) mice (29). The corresponding littermate controls were Bag3fl/fl αMHC-Cre–negative mice. The GFP-LC3–transgenic mouse line (41) was from Noboru Mizushima’s laboratory at The University of Tokyo and was used to generate BAG3-CKOLC3-GFP mice.
BAG3 E455K (equivalent to the mouse E460K) knockin mice were generated by standard techniques using a targeting construct (60), in which codon 455 GAA (encoding glutamic acid) in exon 4 of Bag3 was changed to AAA (encoding lysine) and which contained a neomycin selection cassette flanked by FRT sites (59). Following electroporation of the targeting vector into R1 embryonic stem (ES) cells, G418-resistant ES cells were screened for homologous recombination by Southern blot analysis as described below. One heterozygous recombinant ES clone was identified and microinjected into blastocysts from C57BL/6J mice to generate male chimeras. Male chimeras were bred with female C57BL/6J mice to generate germline-transmitted BAG3 E455K mice (Bag3m-neo/+). Bag3m-neo/+ mice were then crossed with FLP deleter mice to remove the neomycin gene. Bag3m/+ heterozygous mutant mice were backcrossed onto a C57BL/6 background for at least 10 generations. Bag3m/+ heterozygous mutant mice were subsequently intercrossed to generate homozygous Bag3m/m mice (BEK mice).
BAG3-CKO male mice (Bag3fl/fl αMHC-Cre–positive) were crossed with Bag3m/+ heterozygous mutant female mice to generate BAG3 cardiomyocyte–specific cMUT mice (Bag3fl/m αMHC-Cre–positive). BAG3 cMUT mice were bred with Bag3fl/fl mice to generate BAG3 cMUT (Bag3fl/m αMHC-Cre–positive) and littermate control (Bag3fl/fl αMHC-Cre–negative) mice. Thus, the cMUT mice had a –/m genotype in cardiomyocytes and a fl/m genotype in all other cells of the body.
Animal procedures, echocardiography, and hemodynamics.
Echocardiography was performed as previously described (60, 61). Briefly, mice were anesthetized with 1% isoflurane and underwent echocardiography using a FUJIFILM VisualSonics SonoSite Vevo 2100 ultrasound system with a 32- to 55-MHz linear transducer. The percentage of FS was used as an indicator of systolic cardiac function. Measurements of heart rate (HR), LVIDs, and LVIDd were determined from the LV M-mode tracing. For hemodynamics, 10-week-old adult mice were anesthetized using 100 mg/kg ketamine and 10 mg/kg xylazine and subjected to hemodynamic measurements as described previously (60).
Cells, plasmids, adenoviral vectors, and reagents.
Cardiomyocytes from adult mouse hearts were isolated using enzymatic digestion as described previously (62) and directly lysed for Western blot analysis. Neonatal mouse cardiomyocytes (NMCMs) were prepared as previously described (63) and maintained in DMEM supplemented with 10% horse serum, 5% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin for 24 hours before treatment. GFP or mouse Bag3 cDNA (Thermo Fisher Scientific) was cloned and inserted into a 3XFlag-HA vector (gift of Rhonda Bassel-Duby, UT Southwestern Medical Center, Dallas, Texas, USA) to generate 3XFlag-HA–tagged GFP control (3XFlag-HA-GFP) and BAG3-expressing (3XFlag-HA-BAG3) vectors, respectively. 3XFlag-HA harboring the E455K-mutant BAG3 vector (3XFlag-HA-BEK) was generated using the method of site-directed mutagenesis as previously described (64). Mouse HSPB8 (OriGene) fusion with photoconvertible Dendra2 fluorescent protein (64) was cloned into the pCDNA3.1 vector. 3XFlag-HA-GFP, 3XFlag-HA-BAG3, 3XFlag-HA-BEK, and Dendra2-HSPB8 adenoviruses were constructed by Welgen Inc. as previously described (63). Adenoviruses expressing Cre and lacZ (Ad-Cre and Ad-lacZ) were obtained from the UCSD Viral Vector Core. The other reagents used were from Sigma-Aldrich unless otherwise specified.
Protein isolation and Western blot analysis.
Total protein extracts were prepared by suspending ground heart tissue or isolated cardiomyocytes in urea lysis buffer (8 M urea, 2 M thiourea, 3% SDS, 75 mM DTT, 0.03% bromophenol blue, 0.05 M Tris-HCl, pH 6.8) (65) or RIPA (50 mM Tris, 10 mM EDTA, 150 mM NaCl, 0.25% deoxycholic acid, 0.1% SDS, 2% NP-40 substitute, 0.01% sodium azide), respectively. Protein lysates were separated on 4% to 12% SDS-PAGE gels (Life Technologies, Thermo Fisher Scientific) and transferred overnight at 4°C onto PVDF membranes (Bio-Rad). After blocking for 1 hour in TBS containing 0.1% Tween-20 (TBST) and 5% dry milk, the membranes were incubated overnight at 4°C with the indicated primary antibody (listed in Supplemental Table 2) in blocking buffer containing 2% dry milk. Blots were washed and incubated with HRP-conjugated secondary antibody generated in rabbit (1:5,000) or mouse (1:2,000) (Dako) for 1 hour at room temperature (66). Immunoreactive protein bands were visualized using ECL reagent (Thermo Fisher Scientific).
Quantitative RT-PCR.
Total RNA was extracted from mouse LVs or isolated cardiomyocytes using TRIzol reagent (Life Technologies, Thermo Fisher Scientific) according to the manufacturer’s recommendations. cDNA was synthesized using MMLV Reverse Transcriptase (Bio-Rad) (67). Primer sequences for quantitative RT-PCR (qRT-PCR) are listed in Supplemental Table 3. RT-PCR reactions were performed using SsoFast EvaGreen Real-Time PCR Master Mix (Bio-Rad) in 96-well, low-profile PCR plates in a Bio-Rad CFX96 Thermocycler.
Histology.
Hearts were isolated from age- and sex-matched littermates, washed in PBS, and fixed overnight in 4% paraformaldehyde. Hearts were subsequently dehydrated in 70% ethanol, embedded in paraffin, and coronally sectioned (10-μm thickness). Sections were stained with Masson’s trichrome or H&E as previously described (65, 66) and then mounted and imaged using a Hamamatsu NanoZoomer 2.0HT Slide Scanning System.
NMR studies.
WT and mutant BAG domains were produced in synthetic M9 medium with 15NH4Cl as a source of nitrogen. NMR spectra were recorded at the NMR facility of the UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences. Two-dimensional 1H15N HSQC spectra on 0.25-mM samples were acquired at 22°C on a Bruker BMA600 equipped with cryogenic triple-resonance probes and processed with TopSpin software (Bruker).
ITC.
The NBD of murine HSP70 (UniProtKB/Swiss-Prot entry Q61696, residues 1–388) and the BAG domain of BAG3 (WT or E455K-mutant) (UniProtKB/Swiss-Prot entry Q9JLV1.2, residues 419–509) were produced in E. coli. The constructs were cloned into PETM-11 vectors as N-terminal fusions with a His-tag and a TEV protease cleavage site and purified as described previously (68). The purified samples were dialyzed overnight into ITC buffer (25 mM HEPES, pH 7.5, 150 mM KCl, 5 mM MgCl2, 0.25 mM TCEP). ITC measurements were done at the Protein Analysis Core of the Sanford Burnham Prebys Medical Discovery Institute using a MicroCal ITC200 System (Malvern). The measurements were collected by injecting BAG3 WT or E455K-mutant protein into the cell filled with HSP70-NBD solution. Control experiments were performed by titrating BAG3 WT or E455K-mutant protein into the cell filled with ITC buffer alone. The data were processed using MicroCal Origin Software (Malvern).
Additional details on the methods used in this study are provided in the Supplemental Methods.
Statistics.
Data are presented as the mean ± SEM unless indicated otherwise. Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software), with a 2-tailed Student’s t test or 2-way ANOVA for comparisons among groups. Survival data were calculated using Kaplan-Meier survival analysis with a log-rank statistical method. P values of less than 0.05 were considered statistically significant.
Study approval.
The UCSD animal care personnel maintained all animals, and the IACUC of UCSD approved all experimental procedures. UCSD has an Animal Welfare Assurance document (A3033-01) on file with the Office of Laboratory Animal Welfare and is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International.
Author contributions
XF, JB, WZ, TW, CL, KO, XM, ZZ, YM, MJS, DHL, NDD, YG, CW, MW, YL, SL, and KLP performed the research; XF, JB, TW, and JC designed the research; and XF, JV, MJS, SME, and JC wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Hongyu Qiu at Loma Linda University (Loma Linda, California, USA) for providing HSPB8 antibodies. We thank Rhonda Bassel-Duby at UT Southwestern Medical Center for providing the 3XFlag-HA vector. We thank Yishi Jin at UCSD for providing the Dendra2 taq plasmid. We thank Sherin Hashem, Hemal Patel, and Jan Shilling at UCSD (La Jolla, California, USA) for mitochondrial samples. We thank Alexandre Rosa Campos at the Sanford Burnham Prebys Medical Discovery Institute’s Proteomics Shared Resource (La Jolla, California, USA) for help with the proteomics study. JC is funded by grants (HL130295, HL123626, and HL066100) from the NIH and the Foundation Leducq (TNE-13CVD04) and holds an American Heart Association Endowed Chair in Cardiovascular Research. XF is supported by an American Heart Association postdoctoral fellowship (16POST30960067). JB is supported by the European Commission’s Marie Sklodowska-Curie Individual Fellowship (Titin Signals, 656636). SME is supported by grants from the NIH (HL123747 and HL119967). MJS is supported by an American Heart Association postdoctoral fellowship (13POST17060120). OK is supported by the National Science Foundation of China (31370823 and 91439130); the Shenzhen Basic Research Foundation (JCYJ20140509093817680 and JCYJ20160428154108239); and the Guangdong Province Basic Research Foundation (2016A020216003). SL is supported by NIH grant HL128457.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(8):3189–3200.https://doi.org/10.1172/JCI94310.
See the related Commentary at BAG3 plays a central role in proteostasis in the heart.
Contributor Information
Xi Fang, Email: xifang@ucsd.edu.
Julius Bogomolovas, Email: jbogomolovas@ucsd.edu.
Tongbin Wu, Email: tow003@ucsd.edu.
Wei Zhang, Email: wzhang1984@gmail.com.
Canzhao Liu, Email: cal126@ucsd.edu.
Jennifer Veevers, Email: jelowe@ucsd.edu.
Zhiyuan Zhang, Email: zzhang@ucsd.edu.
Xiaolong Ma, Email: xim034@ucsd.edu.
Yongxin Mu, Email: yomu@ucsd.edu.
Yusu Gu, Email: yogu@ucsd.edu.
Celine Wang, Email: cew001@ucsd.edu.
Michael Wang, Email: miw066@ucsd.edu.
Yan Liang, Email: yal225@ucsd.edu.
Stephan Lange, Email: slange@ucsd.edu.
Kunfu Ouyang, Email: ouyangkunfu@pkusz.edu.cn.
Ju Chen, Email: juchen@ucsd.edu.
References
- 1.Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65(2):184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296(5575):1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 4.Moreno-Gonzalez I, Soto C. Misfolded protein aggregates: mechanisms, structures and potential for disease transmission. Semin Cell Dev Biol. 2011;22(5):482–487. doi: 10.1016/j.semcdb.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heling A, et al. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86(8):846–853. doi: 10.1161/01.RES.86.8.846. [DOI] [PubMed] [Google Scholar]
- 6.Kostin S, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92(7):715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 7.Sanbe A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101(27):10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3(2):208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 9.Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583(16):2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 11.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5(10):781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 12.Arimura T, Ishikawa T, Nunoda S, Kawai S, Kimura A. Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat. 2011;32(12):1481–1491. doi: 10.1002/humu.21603. [DOI] [PubMed] [Google Scholar]
- 13.Chami N, et al. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can J Cardiol. 2014;30(12):1655–1661. doi: 10.1016/j.cjca.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Feldman AM, et al. Decreased levels of BAG3 in a family with a rare variant and in idiopathic dilated cardiomyopathy. J Cell Physiol. 2014;229(11):1697–1702. doi: 10.1002/jcp.24615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norton N, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selcen D, et al. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65(1):83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villard E, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3(10):E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 19.Rauch JN, Zuiderweg ER, Gestwicki JE. Non-canonical Interactions between Heat Shock Cognate Protein 70 (Hsc70) and Bcl2-associated Anthanogene (BAG) Co-Chaperones Are Important for Client Release. J Biol Chem. 2016;291(38):19848–19857. doi: 10.1074/jbc.M116.742502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doong H, Vrailas A, Kohn EC. What’s in the ‘BAG’?--A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188(1-2):25–32. doi: 10.1016/S0304-3835(02)00456-1. [DOI] [PubMed] [Google Scholar]
- 21.Shiber A, Ravid T. Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co-chaperones in targeting misfolded proteins to the proteasome. Biomolecules. 2014;4(3):704–724. doi: 10.3390/biom4030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J Biol Chem. 2014;289(3):1402–1414. doi: 10.1074/jbc.M113.521997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su F, et al. Bcl-2-associated athanogene 3 protects the heart from ischemia/reperfusion injury. JCI Insight. 2016;1(19):e90931. doi: 10.1172/jci.insight.90931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169(3):761–773. doi: 10.2353/ajpath.2006.060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arber S, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/S0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 26.Rockman HA, et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr Patterns. 2002;2(1-2):93–97. doi: 10.1016/S0925-4773(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 28.Youn DY, et al. Bis deficiency results in early lethality with metabolic deterioration and involution of spleen and thymus. Am J Physiol Endocrinol Metab. 2008;295(6):E1349–E1357. doi: 10.1152/ajpendo.90704.2008. [DOI] [PubMed] [Google Scholar]
- 29.Abel ED, et al. Cardiac hypertrophy with preserved contractile function after selective deletion of GLUT4 from the heart. J Clin Invest. 1999;104(12):1703–1714. doi: 10.1172/JCI7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buerger A, et al. Dilated cardiomyopathy resulting from high-level myocardial expression of Cre-recombinase. J Card Fail. 2006;12(5):392–398. doi: 10.1016/j.cardfail.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 32.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283(3):1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs M, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2009;425(1):245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 34.Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauch JN, et al. BAG3 is a modular, scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins. J Mol Biol. 2017;429(1):128–141. doi: 10.1016/j.jmb.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vos MJ, et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet. 2010;19(23):4677–4693. doi: 10.1093/hmg/ddq398. [DOI] [PubMed] [Google Scholar]
- 37.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sciarretta S, et al. Activation of NADPH oxidase 4 in the endoplasmic reticulum promotes cardiomyocyte autophagy and survival during energy stress through the protein kinase RNA-activated-like endoplasmic reticulum kinase/eukaryotic initiation factor 2α/activating transcription factor 4 pathway. Circ Res. 2013;113(11):1253–1264. doi: 10.1161/CIRCRESAHA.113.301787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salama F, Ready MA, Sharawy M, Hanes CM. Successive internal resorption. J Pedod. 1990;14(3):165–169. [PubMed] [Google Scholar]
- 43.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266(26):17707–17712. [PubMed] [Google Scholar]
- 44.Basha E, et al. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem. 2004;279(9):7566–7575. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- 45.Haslbeck M, Vierling E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol. 2015;427(7):1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zatloukal K, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160(1):255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walther DM, Mann M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol Cell Proteomics. 2011;10(2):M110.004523. doi: 10.1074/mcp.M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayyadevara S, et al. Age- and hypertension-associated protein aggregates in mouse heart have similar proteomic profiles. Hypertension. 2016;67(5):1006–1013. doi: 10.1161/HYPERTENSIONAHA.115.06849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1-2):228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 52.Lanneau D, Wettstein G, Bonniaud P, Garrido C. Heat shock proteins: cell protection through protein triage. ScientificWorldJournal. 2010;10:1543–1552. doi: 10.1100/tsw.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274(2):781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 54.Fuchs M, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2009;425(1):245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 55.Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3(1):R9–23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 56.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/S0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 57.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med. 2013;368(5):455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 58.McLendon PM, Robbins J. Proteotoxicity and cardiac dysfunction. Circ Res. 2015;116(11):1863–1882. doi: 10.1161/CIRCRESAHA.116.305372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25(2):139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, et al. Normalization of Naxos plakoglobin levels restores cardiac function in mice. J Clin Invest. 2015;125(4):1708–1712. doi: 10.1172/JCI80335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, et al. Postnatal loss of kindlin-2 leads to progressive heart failure. Circ Heart Fail. 2016;9(8):e003129. doi: 10.1161/CIRCHEARTFAILURE.116.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheikh F, et al. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J Clin Invest. 2012;122(4):1209–1221. doi: 10.1172/JCI61134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang X, et al. Adipocyte-specific loss of PPARγ attenuates cardiac hypertrophy. JCI Insight. 2016;1(16):e89908. doi: 10.1172/jci.insight.89908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang X, Fang L, Liu A, Wang X, Zhao B, Wang N. Activation of PPAR-δ induces microRNA-100 and decreases the uptake of very low-density lipoprotein in endothelial cells. Br J Pharmacol. 2015;172(15):3728–3736. doi: 10.1111/bph.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Q, et al. IP3 receptors regulate vascular smooth muscle contractility and hypertension. JCI Insight. 2016;1(17):e89402. doi: 10.1172/jci.insight.89402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirai M, Arita Y, McGlade CJ, Lee KF, Chen J, Evans SM. Adaptor proteins NUMB and NUMBL promote cell cycle withdrawal by targeting ERBB2 for degradation. J Clin Invest. 2017;127(2):569–582. doi: 10.1172/JCI91081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boogerd CJ, et al. Probing chromatin landscape reveals roles of endocardial TBX20 in septation. J Clin Invest. 2016;126(8):3023–3035. doi: 10.1172/JCI85350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bogomolovas J, Simon B, Sattler M, Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr Purif. 2009;64(1):16–23. doi: 10.1016/j.pep.2008.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.