Abstract
The lack of mechanistic explanations for many genotype-phenotype associations identified by GWAS precludes thorough assessment of their impact on human health. Here, we conducted an expression quantitative trait locus (eQTL) mapping analysis in erythroblasts and found erythroid-specific eQTLs for ATP2B4, the main calcium ATPase of red blood cells (rbc). The same SNPs were previously associated with mean corpuscular hemoglobin concentration (MCHC) and susceptibility to severe malaria infection. We showed that Atp2b4–/– mice demonstrate increased MCHC, confirming ATP2B4 as the causal gene at this GWAS locus. Using CRISPR-Cas9, we fine mapped the genetic signal to an erythroid-specific enhancer of ATP2B4. Erythroid cells with a deletion of the ATP2B4 enhancer had abnormally high intracellular calcium levels. These results illustrate the power of combined transcriptomic, epigenomic, and genome-editing approaches in characterizing noncoding regulatory elements in phenotype-relevant cells. Our study supports ATP2B4 as a potential target for modulating rbc hydration in erythroid disorders and malaria infection.
Keywords: Genetics, Hematology
Introduction
GWAS have identified hundreds of loci associated with common human diseases and other clinically relevant traits. Most of these DNA-sequence variants map to noncoding regions of the human genome. The functional characterization of genotype-phenotype associations implicating noncoding variants remains a major bottleneck. Some noncoding variants influence phenotypic variation by modulating the activity of cell- or tissue-specific gene regulatory elements (1–3). The statistical enrichment of GWAS-implicated SNPs in regulatory sequences predicted from epigenomic profiling suggests a promising strategy for fine mapping (4, 5). However, relatively few examples of regulatory mechanisms at individual loci have been described in detail, limiting the ability to design informative high-throughput experiments to characterize causal variants and genes.
Erythropoiesis — the differentiation of hematopoietic stem cells into mature enucleated red blood cells (rbc) — is an auspicious system for dissecting how noncoding genetic variants influence phenotypes. The process is largely cell autonomous and driven by a small set of master transcription factors. Well-established cell-culture protocols exist to monitor proliferation and differentiation. Furthermore, GWAS have already revealed more than 100 loci associated with the number, size, or hemoglobin content of rbc (6, 7). Fine mapping these genetic associations with rbc traits promises not only to provide new illustrations of how noncoding variants influence complex phenotypes through effects on gene expression, but to also reveal genes that control rbc biology in health and disease.
Results
eQTL mapping in erythroblasts identifies cell-specific associations with gene expression.
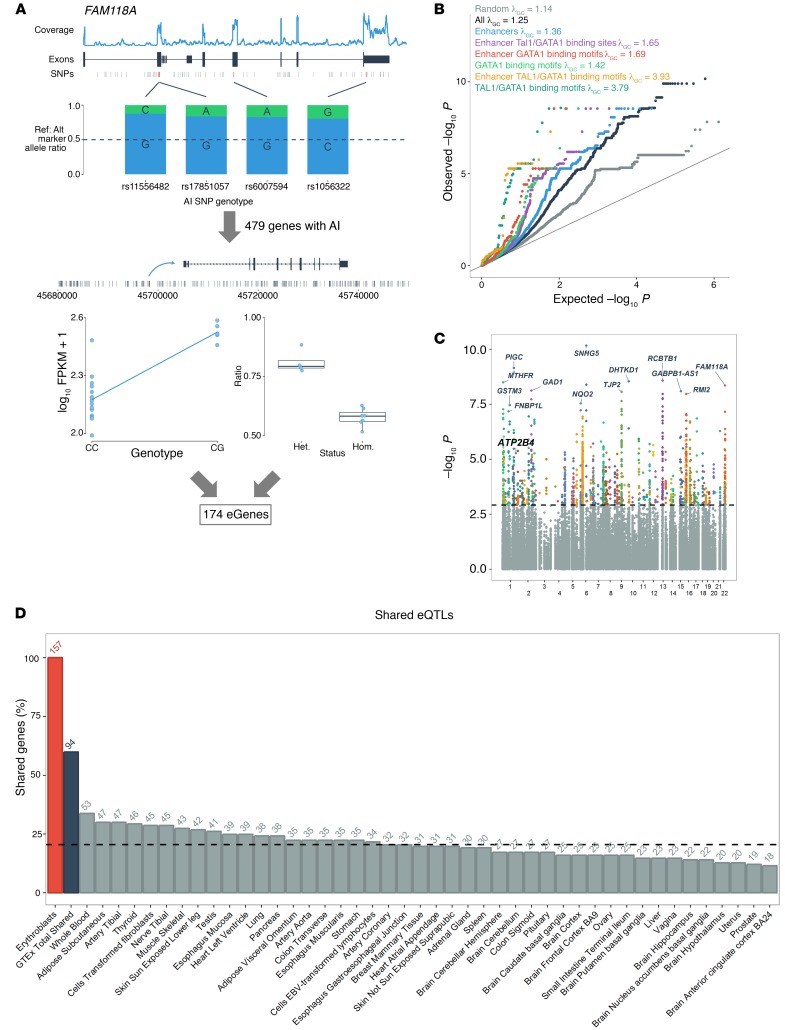
We mapped expression quantitative trait loci (eQTLs) in ex vivo–differentiated human erythroblasts, the nucleated precursors of mature rbc (8). To increase statistical power, we focused the eQTL search on 479 genes that display allelic imbalance (AI) (P < 2 × 10–5) (Methods and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI94378DS1). For each of these 479 genes, we tested to determine whether nearby SNPs (within 100 kb) were associated with their expression levels and had genotypes consistent with the observed AI effect (Figure 1A and Methods). We observed a strong enrichment of eQTLs among variants located near AI genes (Figure 1B). In total, we identified 6,325 significant eQTLs associated with the expression of 174 different genes at a false discovery rate (FDR) of less than 0.05 (Figure 1C and Supplemental Table 2). We observed further enrichment of erythroblast eQTLs within erythroid enhancers identified by DNAse I hypersensitive site (DHS) and histone tail modification analyses and ChIP-sequencing (ChIP-seq) binding sites for the erythroid master transcriptional regulators GATA1 and TAL1 as well as the short binding motifs (12–18 bp) for GATA1 and GATA1::TAL1 (Figure 1C). We noted that the cooccurring GATA1::TAL1 motifs showed the greatest inflation among these annotations. Thus, epigenome features prioritize variants that control gene expression in human erythroblasts. Variants associated with rbc traits by GWAS were also overrepresented among significant erythroblast eQTLs (Supplemental Figure 1 and Supplemental Table 2) (7).
Figure 1. eQTL mapping in erythroblasts.
(A) To identify eQTLs in erythroblasts (n = 24), we first focused on genes that show AI in at least 1 sample (n = 479 AI genes). Then we tested to determine whether SNPs located within 100 kb of these AI genes were associated with their expression level (left panel) and whether their genotypes were consistent with the expected AI ratio of reference allele/alternate allele (right panel). In this example, we highlight the candidate eQTL variant rs7287869 that is associated with the expression of the AI gene FAM118A. (B) Quantile-quantile plot of eQTL P values for variants located within 100 kb of 479 AI genes in human erythroblasts (black). Given that this analysis is limited to AI genes, we expected to observe a strong inflation of the eQTL test statistics (λGC = 1.25). In comparison, the inflation is reduced (λGC = 1.14) when analyzing variants located near 479 randomly selected non-AI genes (gray). This residual inflation could be explained if some of these genes have real eQTLs in the absence of AI or if they have AI effects that merely miss statistical significance. We generated subsets of SNPs overlapping erythroid enhancers (blue), GATA1 and TAL1 ChIP-seq peaks inside erythroid enhancers (purple), GATA1- or GATA1-TAL1–binding motifs inside erythroid enhancers (red and yellow, respectively), or all GATA1- or GATA1-TAL1–binding motifs (light and dark green, respectively). These subsets of variants show substantial enrichment (as summarized by the λGC statistic) when compared with all SNPs (black). (C) Manhattan plot of eQTL P values. The dashed line corresponds to FDR q value = 0.05. (D) Number of genes that share at least 1 eQTL between erythroblasts and the GTEx tissues (at P < 0.001). The dashed line corresponds to the mean percentage of shared eGenes (mean = 20.8%).
We compared our eQTL results with the GTEx data set (9). Although GTEx does not include erythroblasts, it is a powerful resource for confirming eQTL effects that are shared across cell types. Of the 5,924 erythroid eQTLs for which results were available in GTEx, 4,502 (76%) were replicated at P < 0.001 in at least 1 tissue. On average, human erythroblasts and individual GTEx tissue share 1,755 eQTLs that control the expression of 32 genes (Figure 1D and Supplemental Figures 2–4). We found 63 genes with candidate erythroblast-specific eQTLs (Supplemental Table 3). Overall, genes with eQTLs in erythroblasts were enriched for genes implicated in heme biosynthesis (P < 6.6 × 10–7) and mouse rbc phenotypes (P < 8.9 × 10–7) (Supplemental Table 4).
ATP2B4 eQTLs and rbc traits.
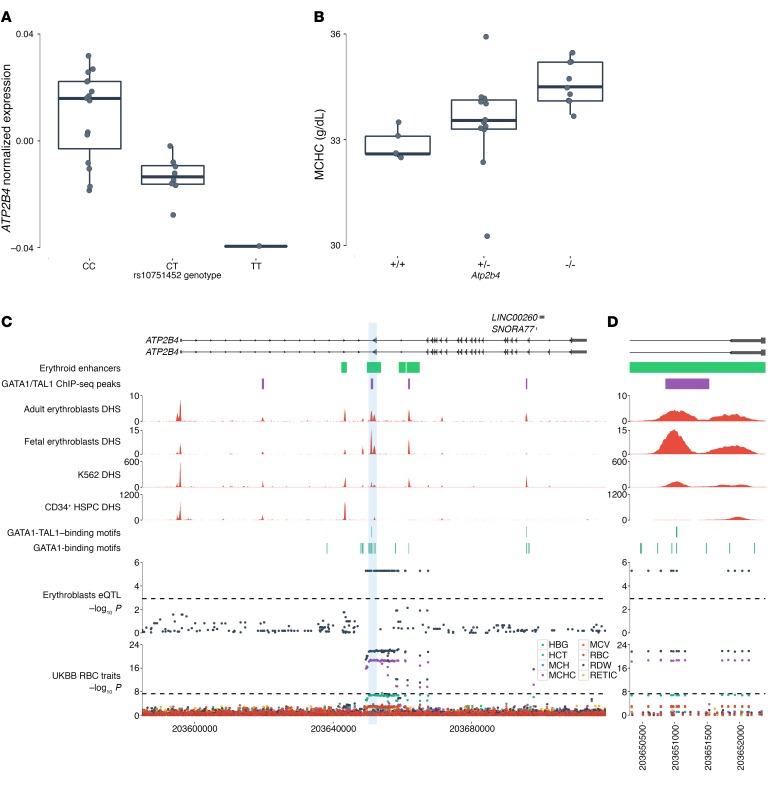
Our eQTL search highlighted many genes without known functions in erythropoiesis (e.g., MTHFR, GSTM3, RCBTB1) (Figure 1B and Supplemental Tables 1 and 2). Because many erythroid eQTLs are also associated with rbc traits, our results are useful for prioritizing candidate causal genes at these GWAS loci even if these genes have no known roles in rbc biology (e.g., SUCO, PTTG1IP, CDH1) (Supplemental Table 2). Among the top genes with erythroblast-specific eQTLs, we were particularly interested in ATP2B4 (also known as PMCA4) because it encodes the main calcium ATPase of rbc (Figure 2A and Supplemental Figure 5) and because the ATP2B4 locus is characterized by an interesting erythroid-specific chromatin landscape (see below). GTEx has identified eQTLs for ATP2B4, but these variants are in weak linkage disequilibrium (LD) with the erythroblast-specific eQTLs (r2 < 0.09 in the 1000 Genomes Project, http://www.internationalgenome.org/) and are not associated with ATP2B4 expression levels in erythroblasts (P > 0.05 after correction for multiple testing) (Supplemental Figure 6). We noted that the same SNPs associated with the expression of ATP2B4 in human erythroblasts had previously been associated with mean corpuscular hemoglobin concentration (MCHC, a measure of rbc hydration) and susceptibility to severe malaria infection by GWAS (10–13), implicating ATP2B4 as the likely causal gene for these rbc-related phenotypes. Additional support for our results comes from a recent report that showed that the same SNPs are associated with ATP2B4 protein levels in human rbc (14).
Figure 2. ATP2B4 eQTLs overlap an erythroid-specific regulatory region and are associated with rbc traits.
(A) Association of the rs10751452 C allele with decreased ATP2B4 expression in human erythroblasts (n = 24, P = 5.3 × 106). LD between rs10751452 and the ATP2B4 sentinel GWAS SNP, rs7551442, is r2 = 1.0. Normalized expression corresponds to residuals of log10(FPKM) after correcting for cell developmental stage in a linear regression model. Box plots represent the median (central line), the first and third quartiles (hinges), and the lowest and highest values inside 1.5 times the interquartile range from the hinges (whiskers). (B) Knockout of Atp2b4 in mice induces a dose-dependent increase in MCHC (n = 26, P = 0.0027). (C) ATP2B4 eQTLs overlap erythroid enhancers and cluster around an erythroid-specific DHS bound by GATA1 and TAL1. This site is not present in undifferentiated CD34+ HSPCs. Several GATA1-binding motifs are clustered inside this regulatory region. eQTLs for ATP2B4 are associated with MCHC and RDW in the UKBB. (D) Zoom-in of the erythroid-specific regulatory region (left DHS peak) in intron 1 of ATP2B4. We note the near-perfect concordance between the UKBB association results and the eQTL results in erythroblasts as well as the overlap of these SNPs with an erythroid-specific enhancer and GATA1/TAL1 sites. All statistical tests were performed using linear regression.
To test the role of Atp2b4 in rbc phenotypes, we analyzed blood from mice with a targeted deletion of this gene. Atp2b4-knockout mice are viable, but characterized by male infertility and protection against pathological cardiac hypertrophy (15, 16). We found that MCHC was elevated in these Atp2b4–/– mice (Figure 2B), consistent with the observation that the allele associated with low ATP2B4 expression in erythroblasts is associated with higher MCHC in humans (12). These results corroborate that Atp2b4 plays a causal role in maintaining MCHC in vivo.
To extend the characterization of variants at the ATP2B4 locus and their effects on rbc phenotypes, we used the first release of the UK Biobank (UKBB) (http://www.ukbiobank.ac.uk/) to test the association between ATP2B4 erythroblast-specific eQTLs and 8 rbc traits (Supplemental Table 5). We observed a strong association between the A allele of rs7551442 and increased MCHC, replicating the signal from previous GWAS (P = 2.6 × 10–19, Figure 2, C and D, and Supplemental Figure 7) and consistent with a recent report (7). We also detected an association of this allele with decreased rbc distribution width (RDW) (P = 1.2 × 10–22) and increased hemoglobin levels (P = 2.1 × 10–7) (Figure 2, C and D). The ATP2B4 genetic association signals with rbc traits in the UKBB were essentially identical to the ATP2B4 erythroid eQTL association signals in human erythroblasts (Figure 2, C and D). This concordance supports the hypothesis that the variants act on rbc traits and malaria susceptibility through an effect on the expression of ATP2B4 in erythroid cells.
An erythroid-specific regulatory element is required for ATP2B4 expression.
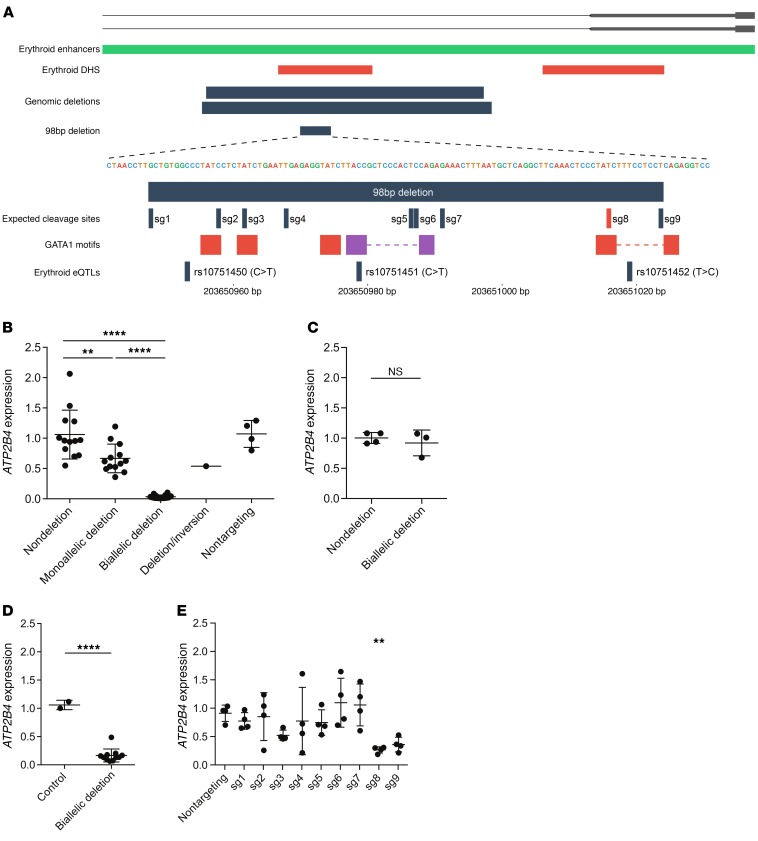
Characterization of DHSs at the ATP2B4 locus revealed an intronic DHS peak that was present in both adult and fetal erythroblasts (Figure 2, C and D) (17). This DHS peak was also present in the K562 erythroleukemic cell line, but absent from 229 other cell types and tissues, including CD34+ hematopoietic stem/progenitor cells (HSPCs) (Supplemental Figure 8) (18, 19). Further annotation of this DHS revealed that it overlapped with an erythroid enhancer chromatin signature as defined in primary human erythroid precursors (17) and with several GATA1-binding motifs, and harbored multiple erythroblast-specific eQTLs in strong LD that were associated with ATP2B4 expression and rbc phenotypes (Figure 2D and Figure 3A). Supporting the regulatory potential of this DHS in erythroid cells, a recent analysis of ENCODE data found that it showed allele-specific transcription factor binding only in K562 cells (20).
Figure 3. Genome editing at the ATP2B4 erythroid enhancer.
(A) Upper panel shows deletions engineered around the ATP2B4 regulatory region. Lower panel shows sgRNA cleavage sites inside the 98-bp core enhancer. The alternative allele of rs10751451 creates a GATA1 motif (purple box). Boxes separated by dashed lines correspond to GATA1-TAL1 motifs. (B) ATP2B4 expression measured in HUDEP-2 cells. Combined analysis of 2 sgRNA pairs that result in overlapping 927-bp and 889-bp deletions. Nondeletion (n = 13), monoallelic deletion (n = 13), and biallelic deletion (n = 14) are clones with, respectively, 0, 1, or 2 ATP2B4 enhancer alleles deleted. None of these clones has an inversion allele. One clone was identified with 1 deletion allele and 1 inversion allele. Four clones exposed to nontargeting sgRNAs are shown for comparison. Gene expression is normalized to mean of nondeletion clones for the same sgRNA pair. Bars and whiskers show mean and SD. The difference in ATP2B4 expression levels between nondeletion and nontargeting clones is not significant. (C) ATP2B4 expression in 293T cell clones exposed to enhancer targeting sgRNA pairs, but without deletion (n = 4) or with biallelic deletion (n = 3). (D) ATP2B4 expression in HUDEP-2 cells with 98-bp core enhancer deletion. Control (n = 2) refers to 1 nontargeting control clone and unedited parental cells as compared with biallelic deletion clones (n = 11). (E) ATP2B4 expression in HUDEP-2 cells with individual sgRNAs specifying cleavages within the 98-bp core enhancer. Each dot indicates an edited bulk population that is an independent transduction of cells. Mean and SD are plotted for each of 4 biological replicates. Gene expression is normalized to unedited cells. In B and E, we used 1-way ANOVA with Bonferroni’s correction for the number of comparisons tested. In C and D, we used Welch’s t test. All P values are 2 tailed. **P < 0.01; ****P < 0.0001.
We posited that this region may harbor the causal variant or variants influencing ATP2B4 expression, rbc traits, and malaria susceptibility. To test this hypothesis, we deleted this region in a human erythroid precursor cell line (HUDEP-2) using the CRISPR-Cas9 system (21). We introduced 2 independent pairs of guide RNAs, one of which results in a 927-bp deletion and the other in a 889-bp deletion (Supplemental Table 6). We observed a dose-dependent reduction in ATP2B4 expression upon enhancer deletion, with biallelic deleted clones displaying only 3% residual ATP2B4 expression (Figure 3B). There was an intermediate phenotype in monoallelic enhancer deleted cells (Figure 3B). A clone in which one copy of the enhancer was deleted and the other copy was inverted showed an expression pattern similar to that of the monoallelic enhancer deleted clones, suggesting that the enhancer can function independently of orientation in situ (Figure 3B).
To test the requirement of the enhancer element in nonerythroid cells, we generated 293T cells (human embryonic kidney derived) with the same 927-bp deletions. In contrast with HUDEP-2 cells, no change in ATP2B4 expression was noted upon enhancer deletion in the 293T cells (Figure 3C). These results suggest that the ATP2B4 intronic element is required for ATP2B4 expression in erythroid cells, but dispensable in nonerythroid cells.
We identified core sequences of the element that included 3 SNPs (rs10751450, rs10751451, rs10751452) associated with ATP2B4 expression by eQTL analysis and with rbc traits and malaria susceptibility by GWAS. These core sequences included 5 GATA1 or composite half-E-box/GATA1–binding motifs, binding sites for the transcription factors GATA1 and TAL1 (Figure 3A). We introduced into HUDEP-2 cells a pair of guide RNAs to generate a 98-bp deletion that removed the 3 SNP positions and these GATA1 motifs (Figure 3A and Supplemental Table 6). We observed that clones with biallelic deletion of this 98-bp segment had 83% reduction in expression of ATP2B4 when compared with ATP2B4 expression in WT cells (Figure 3D). To fine map the gene-expression regulatory element, we introduced 9 individual guide RNAs to produce small insertions-deletions (indels) at their cleavage sites within the 98-bp enhancer core (Figure 3A and Supplemental Table 6). We observed a significant reduction of ATP2B4 expression with one of the 9 guides (sg8 in Figure 3E). This guide RNA cleaves the enhancer directly over a GATA1-binding motif. Overall, these results demonstrate a hierarchical requirement for trait-associated sequences at the erythroid-specific enhancer of ATP2B4.
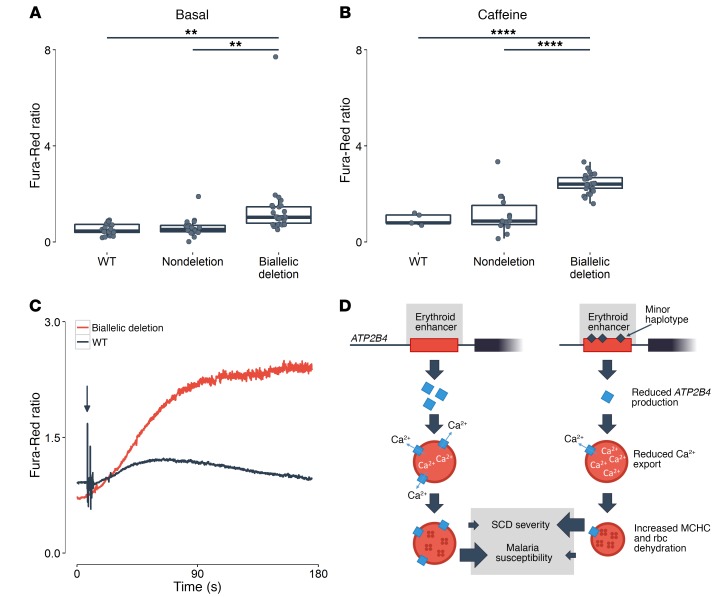
Finally, given ATP2B4’s role in rbc calcium homeostasis, we measured intracellular calcium concentration by ratiometric imaging in unedited HUDEP-2 cells as well as cells with a deletion of the ATP2B4 enhancer element. At baseline or upon stimulation (see Methods), we found higher intracellular calcium levels in ATP2B4-edited HUDEP-2 cells, indicating that cells that do not express ATP2B4 cannot efficiently pump calcium outside of the cytoplasm (Figure 4, A and B). In response to endoplasmic reticulum calcium release by caffeine stimulation, ATP2B4-deficient cells demonstrate exaggerated cytoplasmic calcium accumulation and persistence (Figure 4C). These results provide a physiological link between common regulatory SNPs at ATP2B4, a gene that encodes a major calcium pump, an ion homeostatic defect in erythroid cells, and human complex phenotypes, such as rbc hydration and susceptibility to severe malaria infection.
Figure 4. ATP2B4 activity and calcium homeostasis in erythroid cells.
Ratiometric Fura-Red fluorescence (A) prior to and (B) following application of caffeine (10 μM) in HUDEP-2 WT (basal, n = 21; caffeine treated, n = 5), nondeleted (nondeletion: basal, n = 25; caffeine treated, n = 14), and ATP2B4 enhancer–deleted cells (biallelic deletion: basal, n = 30; caffeine treated, n = 27). Box plots represent the median (central line), the first and third quartiles (hinges), and the lowest and highest values inside 1.5 times the interquartile range from the hinges (whiskers). Statistical analyses used Welch’s t test. Two-tailed P values are reported. **P < 0.01; ****P < 0.0001. (C) Representative time course of the fluorescence Fura-Red ratio following exposure to a single bolus of caffeine (black arrow) in a control cell (black, WT) and in a cell with biallelic deletion of the ATP2B4 enhancer (red; biallelic deletion). Intracellular calcium store release induced by caffeine exposure substantially increased cytoplasmic levels in both cells, but elevated calcium concentrations persisted only in cells with the ATP2B4 enhancer deleted. (D) Model of genetic variation at the ATP2B4 erythroid enhancer influencing rbc traits and malaria susceptibility. Erythroid cells carrying the minor haplotype at the erythroid-specific enhancer express lower levels of ATP2B4. The rbc with reduced ATP2B4 accumulate cytoplasmic calcium, resulting in dehydration and elevated MCHC. Although relatively resistant to infection by the malaria parasite P. falciparum, dehydrated rbc may increase the severity of erythroid disorders such as sickle cell disease (SCD).
Discussion
Few GWAS discoveries have been investigated at the molecular and cellular levels. To explore the genetic architecture of regulatory variants that control rbc traits in humans, we undertook an eQTL search in human in vitro–differentiated erythroblasts. Although we identified more than 4,500 eQTLs that replicated in the GTEx data set, we acknowledge that some of our findings might be false-positive associations; independent replication studies in the same cell type are needed. Furthermore, because we limited our eQTL analyses to genes with AI and variants located within 100 kb of these genes in order to increase statistical power, we would have missed genes without exonic variants (necessary to monitor AI) or that are controlled by long-range regulatory variants. Despite these limitations, we found strong eQTLs for ATP2B4, validating our experimental design. Our own functional results and the recent report that the same SNPs are associated with ATP2B4 protein levels in human rbc (14) strongly argue that this is a true association signal.
Using human erythroblasts, knockout mice, and erythroid cells amenable to genome editing, we undertook the detailed characterization of the ATP2B4 locus and its roles in rbc biology. This comprehensive approach allowed us to identify the causal regulatory variants within an erythroid-specific enhancer, to confirm ATP2B4 as the causal gene, and to highlight the calcium homeostasis defect as one possible effector pathway responsible for the association with MCHC and malaria susceptibility (Figure 4D). Excess intracellular calcium activates a calcium-activated potassium channel (the Gardos channel), resulting in potassium efflux, rbc volume loss, and elevated MCHC. Hydration of rbc has been linked with clinical severity in the hemoglobin disorder sickle cell disease (22) and with infectivity by the malaria agent Plasmodium falciparum (23). Supported by our genetic and mechanistic results, the development of therapies that specifically modulate ATP2B4 activity could have a broad impact on rbc diseases that affect millions of individuals worldwide.
Methods
Cell culture, RNA sequencing, and DNA genotyping.
The cell culture protocol to proliferate and differentiate human CD34+ HSPCs into erythroblasts has been described before (8, 24). We purchased human fetal (fetal liver, n = 12) and adult (bone marrow, n = 12) CD34+ HSCs from DV Biologics and Lonza, respectively. This sample size was selected, as it provides 90% power to detect a 3 SD difference in gene-expression levels between fetal and adult erythroblasts at α = 1 × 10–5. We have also described elsewhere the protocol for RNA extraction and RNA sequencing (RNA-seq) (8). Briefly, we performed RNA-seq with an Illumina HiSeq2000 sequencer using stranded cDNA library and a paired-ends 100-bp protocol. We mapped reads to the genome (hg19) using Tophat2 (v.2.0.9, with options –library-type fr-firststrand –microexon-search –coverage-search) and estimated transcript abundance with Cufflinks (v.2.2.1, with options –library-type fr-firststrand –max-bundle-frags 50000000) (25, 26). All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE90878). Genomic DNA extraction, genotyping on the Illumina HumanOmniExpress-12 v1.1 BeadChip array, and quality control were performed as previously described (8). We imputed genotypes using the Michigan Imputation Server with the Haplotype Reference Consortium (HRC) panel (v. r1.1) (27).
AI and eQTL mapping.
We measured AI at each heterozygous genotype covered by RNA-seq in the 24 human erythroblast samples. We only considered SNPs directly genotyped or with high imputation quality (R2 > 0.6). We removed duplicated reads using the Picard MarkDuplicates tool (v. 1.96). We counted each read using the samtools (v 1.1) mpileup software and genome build hg19 and kept uniquely mapping reads using the –q 50 argument (mapping quality > 50) and sites with base quality greater than 10. We further restricted the analysis to uniquely mapping sites as per the ENCODE 50-mer mappability track (score = 1) and removed sites showing mapping bias in simulations (28). We excluded sites with less than 30 overlapping reads. For a given heterozygous SNP, we determined the statistical significance of AI, that, is the difference between the observed and expected ratio of reference allele/alternate allele, with a binomial test. To account for read-mapping bias, we summed all reads overlapping all heterozygous SNPs in the RNA-seq data set and calculated the expected ratio for each combination of alleles in each sample independently. For SNPs with high sequencing coverage, we downsampled the number of reads that fell in the top 25th coverage percentile so that the most covered sites did not bias the expected ratio (29). We used Bonferroni’s correction to account for the number of tests performed: the significance threshold for this AI experiment was α = 2 × 10–5.
Next, we mapped the regulatory variants responsible for differential gene-expression phenotypes. Given our limited sample size, we focused on genes that showed AI, reasoning that the likelihood of finding significant eQTL was higher in this subset of genes. We developed a method that combines statistical evidence of AI and eQTL effects. First, we tested by linear regression the association between SNP genotypes (additive model) and gene-expression levels (expressed as log10[FPKM + 1], where FPKM indicates fragments per kilobase of transcript per million reads), adjusting for cell type (fetal or adult). For these analyses, we only considered SNPs located within 100 kb of the AI genes. Second, we hypothesized that samples that are homozygous for the tested SNP (either reference/reference or alternate/alternate) should not show AI, whereas heterozygote samples should have AI. In other words, the reference allele:alternate allele ratio in heterozygote samples should be further from the expected 50:50 ratio than the ratio observed in homozygote samples. We tested this hypothesis using a 1-sided t test. Because the linear regression and t test P values were not correlated, we metaanalyzed these statistics using Fisher’s method to obtain a final P value. In situations of perfect LD between the potential regulatory and exonic variants, that is, when there are no homozygous samples at the exonic variants that are heterozygous at the regulatory variants, we cannot perform the concordance t test and simply report the linear regression results. We used a FDR methodology to correct for multiple testing, considering SNPs with a q value less than 0.05 as significant eQTLs.
Replication of eQTLs in GTEx.
We used the GTEx database to replicate the eQTLs that we identified in human erythroblasts (9). GTEx does not include erythroblasts, but we reasoned that it would still represent a valid source of replication for non–tissue-specific eQTLs. We downloaded from the GTEx portal (version 6) all SNP–gene association results across all available tissues (http://www.gtexportal.org/home/) (9). We considered as replicated erythroid eQTLs with a P < 0.001 in any other samples from the GTEx data set. More stringent thresholds gave consistent results.
eQTL enrichment analyses.
We used 3 sources of information to test the enrichment of erythroid eQTL within specific genomic annotations. First, we obtained the coordinates of erythroid-specific enhancers defined using DHSs and histone tail modifications (17). From the same study, we also obtained genomic coordinates of GATA1 and TAL1 peaks determined by ChIP-seq (17). Finally, we used Homer software to identify binding motifs for GATA1 (MA0035.2) and GATA1::TAL1 (cooccurring GATA1 and half-E box motifs, MA0140.2) across the human genome, or specifically within erythroid enhancer regions (30, 31). We carried out gene ontology and pathway enrichment analyses using the ToppGene suite (toppgene.cchmc.org) (32).
Analyses of rbc traits in Atp2b4–/– mice.
Mice (male only, n = 26) with complete inactivation of Atp2b4 have been generated and described elsewhere (15, 16). The mice are in mixed background of 129/sv × C57BL/6. All mice used were males between 9 and 13 weeks of age. Mice were anesthetized with isoflurane (2.5%), and blood was collected from the jugular vein by venipuncture. The samples were measured within 6 hours of collection at room temperature in the biological research unit at Cancer Research UK Manchester Institute. Evaluation of hematological parameters was carried out in 2 batches on a Sysmex XT-2000iV (Sysmex) automated hematology analyzer using a mouse profile. Quality control was carried out before running each batch of samples. No randomization was used, and experimenters who did the complete blood count analyses were blinded to the animals’ Atp2b4 genotypes.
Replication of the association between ATP2B4 and rbc phenotypes in the UKBB.
We tested the association between genotypes at the ATP2B4 locus (2 Mb) and rbc traits in the July 2015 release of the UKBB data set. We excluded participants with blood cancer, leukemia, lymphoma, bone marrow transplant, congenital or hereditary anemia, HIV, end-stage kidney disease, dialysis, splenectomy, or cirrhosis and those with extreme rbc trait measurements (>8 SD from the mean). We limited our analysis to participants of British ancestry with imputed genotype data available. In total, we tested the association between 8 rbc traits (hemoglobin, hematocrit, rbc count, mean corpuscular volume, mean corpuscular hemoglobin, MCHC, RDW, and reticulocyte count) and genotypes (additive model) in 136,727 participants with PLINK1.9 (https://www.cog-genomics.org/plink2). This sample size provides more than 99% power to replicate the association between ATP2B4 SNPs and MCHC at α = 0.05. After applying exclusion criteria, we corrected the rbc traits for age, sex, recruitment center, and cell counter and then normalized the residuals using inverse normal transformation. As covariates, we included in the association tests the 10 first principal components calculated using FlashPCA (33).
Generating ATP2B4 deletions in cell lines.
HUDEP-2 cells and 293T cells with stable expression of Cas9 were generated by lentiviral transduction (lentiCas9-Blast, Addgene plasmid ID 52962) and blasticidin selection as previously described (34). We chose to perform genome-editing experiments in HUDEP-2 and not K562 cells for several reasons. Unlike K562 cells that were originally isolated from a naturally occurring human malignancy (chronic myeloid leukemia), HUDEP-2 cells were prospectively isolated from primary hematopoietic stem and progenitor cells transduced by an inducible viral oncogene and continuously cultured under conditions permissive to expansion of erythroid precursors. HUDEP-2 cells are an in vitro model of human erythropoiesis that mimics erythroid development from the proerythroblast to reticulocyte stages (21, 35). We have previously established robust methods to perform CRISPR-Cas9 genome editing in HUDEP-2 cells (34, 36). Also HUDEP-2 cells are disomic for chromosome 1 (unlike K562 cells), which simplifies the analysis of genome-editing outcomes at ATP2B4. Tandem sgRNA lentiviruses were produced based on a modification of lentiGuide-Puro (Addgene plasmid ID 52963) to carry 2 U6 promoter-guide RNA cassettes per construct as previously described (34). Tandem sgRNA lentiviruses were transduced into HUDEP-2 cells or 293T cells with stable Cas9 expression. The tandem sgRNA constructs expressed 2 sgRNAs (Supplemental Table 6) and thus were designed to produce interstitial deletions. Cells were also transduced with a pool of lentiviruses containing 10 unique nontargeting sequences (Supplemental Table 6). After transduction, bulk cultures were incubated for 7 to 10 days with 10 μg ml–1 blasticidin and 1 μg ml–1 puromycin selection to select for cells with edited alleles. Those bulk cultures transduced with tandem sgRNA lentiviruses were plated clonally at limiting dilution. 96-well plates with greater than 30 clones per plate were excluded to avoid mixed clones. After approximately 14 days of clonal expansion, genomic DNA was extracted using 50 μl QuickExtract DNA Extraction Solution per well (Epicentre). Clones were screened for deletion by conventional PCR, with 1 PCR reaction using primers internal to a segment to be deleted (nondeletion amplicon) and 1 gap-PCR reaction using primers across the deletion junction (deletion amplicon) that would produce a characteristic short amplicon in the presence of deletion (Supplemental Table 7). Clones bearing inversion alleles were also identified with one primer outside the segment to be deleted and the other primer inside the segment to be deleted, both in the same orientation with respect to the reference genome, as previously described. PCR was performed using the QIAGEN HotStarTaq 2× Master Mix and the following cycling conditions: 95°C for 15 minutes, 45 cycles of 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 1 minute, 72°C for 10 minutes. Biallelic deletion clones were identified based on the presence of a deletion PCR band with absence of a nondeletion PCR band. Inversion clones were also identified as previously described. Compound deletion-inversion clones had 1 deleted allele and 1 inverted allele without the presence of nondeletion alleles. For disruption of individual GATA1 motifs, stable Cas9-expressing HUDEP-2 cells were transduced with lentiviruses carrying individual guide RNAs (lentiGuide-Puro) (Figure 3A and Supplemental Table 6). Edited populations of cells were selected with puromycin and blasticidin and RNA was isolated 7 to 10 days following transduction. Genome editing with indel rates exceeding 75% was confirmed by isolating gDNA from each of these bulk populations of cells, performing a PCR reaction with primers flanking the edited region (Supplemental Table 6), and Sanger sequencing the amplicon with analysis according to a publicly available sequence deconvolution algorithm (37).
Reverse transcription–quantitative PCR.
RNA was extracted for each selected clone using a kit (QIAGEN). 1 μg of RNA per clone was converted to cDNA using the iScript cDNA kit. Real-time reverse-transcription–quantitative PCR (RT-qPCR) was subsequently performed using SYBR Select Master Mix (Thermo Fisher Scientific). Primers were designed to span exon 5 and exon 6 of the ATP2B4 gene and were empirically validated for efficiency by serial dilution analysis (Supplemental Table 7). Gene expression was normalized to that of GAPDH. All gene expression data reported represent the mean of at least 3 technical replicates.
Intracellular calcium monitoring.
Intracellular calcium levels in HUDEP-2 cell lines were monitored using Fura-Red, a ratiometric fluorescent calcium indicator, with a laser-scanning confocal microscope. Cells were first seeded (1 hour) in a coverslip-bottom chamber coated with Cell-Tak (Corning). Cells were then washed with HEPES-PSS and incubated with Fura-Red (10 μM) for 45 minutes at 37°C. Intracellular calcium levels were recorded on an LSM-Duo confocal microscope (Zeiss) with a 40× objective (Plan APO Oil DIC, 1.3 NA). Single emission fluorescence (LP575) was collected (10 FPS) upon alternate excitation (405 and 489 nm, solid state lasers) on a 512 × 256 field of view. Stacks of interleaved images (16 bits) were then analyzed using FIJI (ImageJ, NIH). Upon background subtraction, 20 × 20 pixel circular regions of interest (ROIs) were manually positioned on cells and individual ROI mean fluorescence intensity was measured. Variations in intracellular calcium levels were expressed as the mean ratio of the fluorescence from calcium-bound (excitation wavelength 405 nm) and calcium-free (excitation wavelength 489 nm) Fura-Red of each ROI from 10 consecutive images.
Statistics.
Statistical tests were performed using Welch’s t test, binomial test, linear regression, or 1-way ANOVA when appropriate (see specific Methods subsections and figure legends). Multiple testing correction was performed using Bonferroni’s method or an FDR procedure. Adjusted P values of less than 0.05 or q values of less than 0.05 were considered significant.
Study approval.
Mouse experiments were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act 1986 and were approved by the University of Manchester Ethics Committee. Human genetic analyses were approved by the Montreal Heart Institute Ethics Committee, and informed consent was obtained from all participants.
Author contributions
SL, ESG, DEB, and GL conceived and designed the experiments. SL, ESG, MB, PGS, FS, AA, SP, and JL performed the experiments. SL, ESG, JL, DEB, and GL analyzed the data. RK, YN, EB, and DO contributed reagents and materials. SL, ESG, DEB, and GL wrote the manuscript with contributions from all authors.
Supplementary Material
Acknowledgments
Part of this research has been conducted using the UKBB resource under application number 11707. The authors wish to thank Gerald Lilly and Louis R. Villeneuve for assistance and Seth L. Alper, Carlo Brugnara, and Elizabeth Egan for comments. SL holds fellowships from the Canadian Institute of Health Research (CIHR) and the “Fondation Pierre Lavoie.” ESG was the recipient of an American Society of Hematology HONORS award. AA holds a Cancer Research UK Clinical Training Award (C147/A19496). EB is supported by a Cancer Research UK start-up grant (C5759/A20971). DO was supported by a British Heart Foundation Intermediate Fellowship (FS/09/046/28043) and British Heart Foundation project grants (PG/13/12/30017 and PG/16/77/32400). DEB was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (K08DK093705, R03DK109232), the National Heart, Lung, and Blood Institute (DP2OD022716), the Burroughs Wellcome Fund, the American Society of Hematology, and the Doris Duke Charitable, Charles H. Hood, and Cooley’s Anemia foundations. GL was supported by grants from the CIHR (no. 123382), the Doris Duke Charitable Foundation, the National Sciences and Engineering Research Council of Canada (RGPIN-2016-04597), the Montreal Heart Institute Foundation, and the Canada Research Chair Program.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(8):3065–3074.https://doi.org/10.1172/JCI94378.
Contributor Information
Samuel Lessard, Email: lessards@mhi-humangenetics.org.
Mélissa Beaudoin, Email: melissa.beaudoin@mhi-humangenetics.org.
Patrick G. Schupp, Email: patrickgschupp@gmail.com.
Falak Sher, Email: Falak.Sher@childrens.harvard.edu.
Sukhpal Prehar, Email: sukhpal.prehar@manchester.ac.uk.
Ryo Kurita, Email: r-kurita@jrc.or.jp.
Yukio Nakamura, Email: yukio.nakamura@riken.jp.
Esther Baena, Email: Esther.Baena@cruk.manchester.ac.uk.
Jonathan Ledoux, Email: jonathan.ledoux@umontreal.ca.
Delvac Oceandy, Email: delvac.oceandy@manchester.ac.uk.
Guillaume Lettre, Email: guillaume.lettre@mhi-humangenetics.org.
References
- 1.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chami N, et al. Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am J Hum Genet. 2016;99(1):8–21. doi: 10.1016/j.ajhg.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167(5):1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessard S, Beaudoin M, Benkirane K, Lettre G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med. 2015;7(1):1. doi: 10.1186/s13073-014-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malaria Genomic Epidemiology Network, Malaria Genomic Epidemiology Network Reappraisal of known malaria resistance loci in a large multicenter study. Nat Genet. 2014;46(11):1197–1204. doi: 10.1038/ng.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timmann C, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489(7416):443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- 12.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum Mol Genet. 2013;22(7):1457–1464. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zambo B,et al. Decreased calcium pump expression in human erythrocytes is connected to a minor haplotype in the ATP2B4 gene. Cell Calcium. doi: 10.1016/j.ceca.2017.02. [published online ahead of print February 3, 2017]. https://doi.org/10.1016/j.ceca.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Schuh K, et al. Plasma membrane Ca2+ ATPase 4 is required for sperm motility and male fertility. J Biol Chem. 2004;279(27):28220–28226. doi: 10.1074/jbc.M312599200. [DOI] [PubMed] [Google Scholar]
- 16.Mohamed TM, et al. The plasma membrane calcium ATPase 4 signalling in cardiac fibroblasts mediates cardiomyocyte hypertrophy. Nat Commun. 2016;7:11074. doi: 10.1038/ncomms11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23(4):796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli M, et al. Allele-specific transcription factor binding to common and rare variants associated with disease and gene expression. Hum Genet. 2016;135(5):485–497. doi: 10.1007/s00439-016-1654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurita R, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartolucci P, et al. Erythrocyte density in sickle cell syndromes is associated with specific clinical manifestations and hemolysis. Blood. 2012;120(15):3136–3141. doi: 10.1182/blood-2012-04-424184. [DOI] [PubMed] [Google Scholar]
- 23.Tiffert T, Lew VL, Ginsburg H, Krugliak M, Croisille L, Mohandas N. The hydration state of human red blood cells and their susceptibility to invasion by Plasmodium falciparum. Blood. 2005;105(12):4853–4860. doi: 10.1182/blood-2004-12-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panousis NI, Gutierrez-Arcelus M, Dermitzakis ET, Lappalainen T. Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. Genome Biol. 2014;15(9):467. doi: 10.1186/s13059-014-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS ONE. 2014;9(4):e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda T, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351(6270):285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canver MC, et al. Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat Genet. 2017;49(4):625–634. doi: 10.1038/ng.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.