Abstract
Angioimmunoblastic T cell lymphoma (AITL) represents a distinct, aggressive form of peripheral T cell lymphoma with a dismal prognosis. Recent exome sequencing in patients with AITL has revealed the frequent coexistence of somatic mutations in the Rho GTPase RhoA (RhoAG17V) and loss-of-function mutations in the 5-methylcytosine oxidase TET2. Here, we have demonstrated that TET2 loss and RhoAG17V expression in mature murine T cells cooperatively cause abnormal CD4+ T cell proliferation and differentiation by perturbing FoxO1 gene expression, phosphorylation, and subcellular localization, an abnormality that is also detected in human primary AITL tumor samples. Reexpression of FoxO1 attenuated aberrant immune responses induced in mouse models adoptively transferred with T cells and bearing genetic lesions in both TET2 and RhoA. Our findings suggest a mutational cooperativity between epigenetic factors and GTPases in adult CD4+ T cells that may account for immunoinflammatory responses associated with AITL patients.
Keywords: Hematology, Immunology
Introduction
Peripheral T cell lymphomas (PTCLs) are a group of aggressive lymphomas derived from mature T cells or NK cells and have a dismal prognosis (1–4). Current treatments for PTCLs are ineffective, with high relapse rates that are largely due to the lack of targeted therapeutics and a thorough mechanistic understanding of the molecular pathogenesis (1, 3, 5). Recent exome sequencing has unveiled the coexistence of recurrent somatic mutations in the small GTPase RhoA and the 5-methylcytosine (5mC) oxidase TET2 in PTCLs, but not in other types of hematological malignancies (6–8). Most notably, mutations in both TET2 and RhoA are frequently observed (~60%–70%) in angioimmunoblastic T cell lymphoma (AITL) (6–8), one of the most common subtypes of PTCLs, with a median overall survival of 18 months (9). Patients with AITL often have autoimmune manifestations (e.g., hemolytic anemia, rheumatoid arthritis, or autoimmune thyroiditis) (1, 2, 10, 11), but the mechanistic underpinnings remain poorly understood.
TET2 belongs to the ten-eleven translocation (TET) family of 2-oxoglutarate–dependent (2OG-dependent) dioxygenases, a class of DNA-modifying enzymes that successively oxidize 5mC to 5-hydroxymethylcytosine (5hmC, the major catalytic product), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in the mammalian genome (12–16). These oxidative products can be ultimately excised and replaced by unmodified cytosine to achieve active DNA demethylation. Although TET2 loss-of-function mutations are observed in a large fraction of patients with lymphoid or myeloid malignancies (16–22), they are also present in healthy elderly individuals with clonal hematopoiesis (23). Genetic depletion of TET2 in mice causes increased self-renewal of hematopoietic stem and progenitor cells (HSPCs) and biased HSPC differentiation toward the myeloid lineage, but it seldom leads to overt development of hematological malignancies (20, 24–26). Together, these findings suggest that TET2 inactivation contributes to the initiation of lymphoma or leukemia (27), but that additional mutations are required for full-blown malignant transformation.
Ras homolog family member A (RhoA) is a small GTPase that cycles between an active GTP-bound state and an inactive GDP-bound state, the balance of which is controlled by the coordinated actions of GAPs and GEFs (28). Inhibition of Rho signaling in thymus by overexpressing the bacterial toxin C3 transferase resulted in aggressive thymic lymphoma of T cell origin (29), indicating that RhoA-mediated signaling is critical for the maintenance of T cell function (30). Recent exome sequencing has unveiled a spectrum of RhoA mutations in patients with leukemia or lymphoma (1, 3, 5, 31), with both loss- and gain-of-function mutations implicated in the development of malignant T cell phenotypes (31). Among those mutations, RhoAG17V, together with TET2 loss-of-function mutations, is most frequently detected in AITL, indicating a mutational cooperativity that might contribute to the disruption of proper CD4+ T cell function in this aggressive subtype of lymphoma. Here, we investigated the loss of TET2 together with an AITL-associated RhoA mutation (RhoAG17V) in mature T cells and mechanistically dissected how they cooperatively impinge on transcriptional and posttranslational outputs to induce aberrant immunoinflammatory responses that exacerbate disease progression.
Results
TET2 loss, together with RhoAG17V expression, induces abnormal immune responses in vivo.
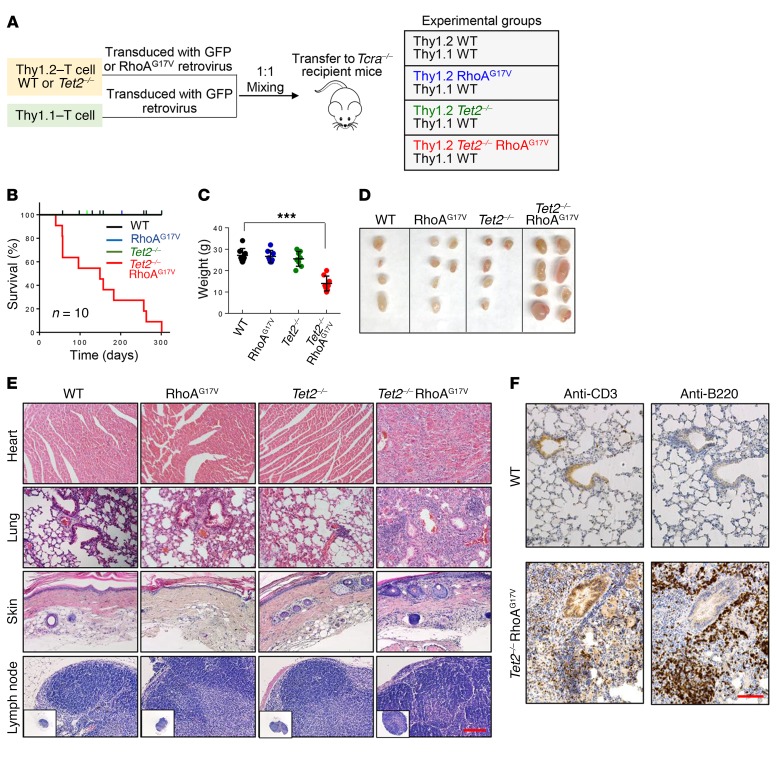
To determine the functional consequences of TET2 loss and RhoAG17V expression in mature T cells (particularly CD4+ and CD8+ T cells), we performed competitive adoptive T cell transfer experiments using both WT and Tet2-KO (Tet2–/–) mice (Figure 1A). We isolated T cells from spleens and lymph nodes from WT and Tet2–/– mice (Thy1.2+) (25) and then transduced them with retroviruses encoding either GFP or FLAG-tagged RhoAG17V-IRES-GFP in vitro to yield 4 experimental groups of mice (WT, RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V). In parallel, congenic Thy1.1+ T cells were prepared from B6.PL-Thy1α/CyJ mice and further transduced with a retrovirus encoding GFP to serve as an internal control. We subsequently mixed Thy1.1+ and Thy1.2+ T cells at a 1:1 ratio and injected them into TCRα-deficient recipient mice that lack the majority of endogenous TCRα+ T cells (Figure 1A). Flow cytometric analysis showed a retroviral transduction efficiency of 45% to 70% in all the experimental groups, as indicated by GFP signals (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI92026DS1). The deletion of Tet2 and heterologous expression of FLAG-RhoAG17V were further confirmed by Western blotting (Supplemental Figure 1B).
Figure 1. Recipient mice transferred with Tet2–/– RhoAG17V T cells have an inflammatory phenotype.
(A) Schematic representation of the experimental design for competitive adoptive T cell transfer experiments. Thy1.2+ T cells transduced with the corresponding retroviruses were mixed at a 1:1 ratio with control Thy1.1+ T cells and transferred into TCRα-deficient (Tcra–/–) recipient mice by retro-orbital injection to characterize the functional consequences following Tet2 deletion (Tet2–/–) and/or RhoAG17V expression. (B) Kaplan-Meier survival curves for recipient mice transferred with Thy1.2+ WT (black), RhoAG17V (blue), Tet2–/– (green), or Tet2–/– RhoAG17V (red) T cells (n = 10 mice per group; 3 independent experiments). Only the mice transferred with Tet2–/– RhoAG17V T cells showed lethal activity, with a median survival of 21.3 weeks. (C) Weight measurement of the surviving recipient mice 20 weeks after T cell transfer (n = 7 mice; 2 independent experiments). ***P < 0.001, by ANOVA with Dunnett’s post-hoc correction. (D) Representative images of lymph nodes from recipient Tcra–/– mice 20 weeks after adoptive transfer of WT, RhoAG17V, Tet2–/–, or Tet2–/RhoAG17V T cells. (E) H&E staining of heart, lung, and skin tissues and lymph nodes isolated from recipient mice transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells (20 weeks after adoptive transfer). Scale bar: 100 μm. Original magnification: ×4. (F) Immunohistochemical staining for anti-CD3 (T cell marker) and anti-B220 (B cell maker) in lung tissues isolated from 20-week-old recipient mice transferred with WT or Tet2–/– RhoAG17V T cells. Scale bar: 100 μm.
We first compared the survival of recipient mice in 4 experimental groups (Figure 1B). Mice transferred with WT, RhoAG17V, or Tet2–/– T cells remained viable, with no gross abnormalities during the 300-day experimental window. After a follow-up period of approximately 1 year, no obvious phenotypes were observed in those 3 groups of mice. In striking contrast, the survival of mice transferred with T cells harboring both genetic lesions (Tet2–/– RhoAG17V) was significantly reduced, with a median survival of 21.3 weeks (red curve, Figure 1B). Moribund mice transferred with Tet2–/– RhoAG17V T cells displayed substantial weight loss (Figure 1C), severe skin ulcers on the tail, paws, and ears, accompanied by pruritus (Supplemental Figure 1C) and lymphomegaly (Figure 1D), as is typically seen in patients with AITL. Histopathological analysis of major organs derived from the Tet2–/– RhoAG17V group indicated severe infiltration of both T and B lymphocytes (as indicated by anti-CD3 and anti-B220 staining, respectively) in heart, lung, and skin, but these pathological changes remained very mild or absent in the other 3 groups (Figure 1, E and F). A thorough scrutiny of lymph nodes and spleens isolated from the Tet2–/– RhoAG17V recipient mice further revealed a marked increase in the numbers of follicles and activated germinal centers (marked by anti-CD35 staining; Supplemental Figure 1D). Furthermore, we observed the most notable increase in the population of germinal center B cells (B220+GL7+Fas+) (Supplemental Figure 1E) and elevated expression of proinflammatory IL-6 (Supplemental Figure 1F) in the Tet2–/– RhoAG17V group, thereby recapitulating immune-related phenotypes that are commonly observed in PTCL patients (11, 32). Together, these data indicate that TET2 loss and RhoAG17V expression cooperate to mount an immunoinflammatory response that ultimately causes death of recipient mice.
TET2 loss and RhoAG17V expression confer a proliferative advantage in CD4+, but not CD8+, T cells.
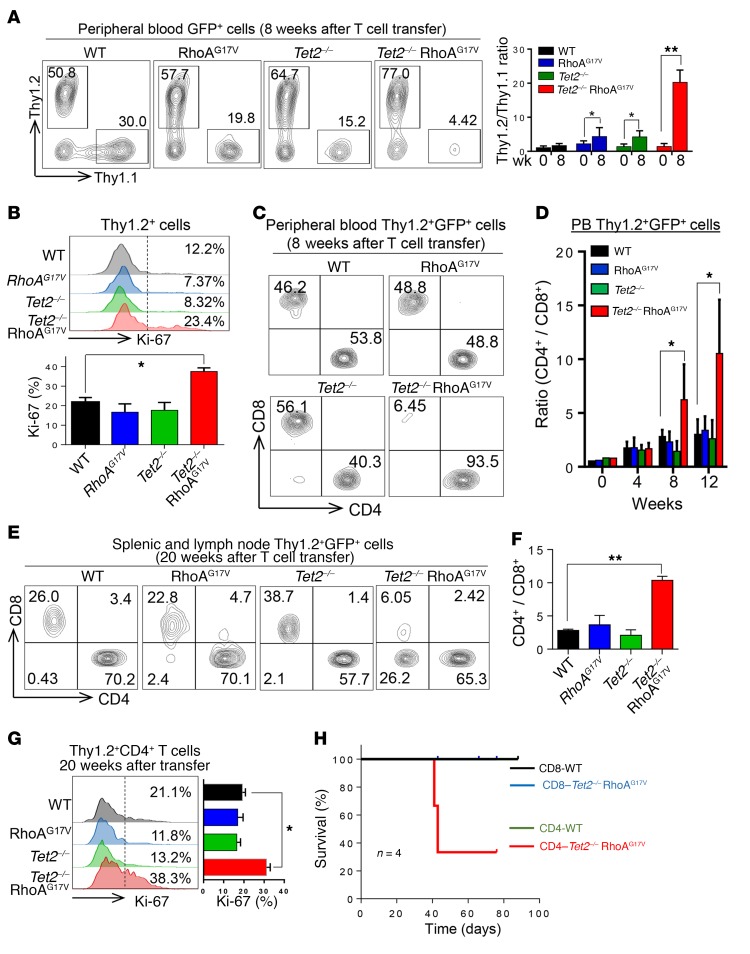
Although the majority of recipient mice transferred with Tet2–/– RhoAG17V T cells showed skin ulcers and weight loss at least 12 to 15 weeks after the transfer, the growth advantage of double-mutated T cells could be readily detected as early as 8 weeks after adoptive transfer. Therefore, we examined the abundance of transferred Thy1.2+ cells compared with Thy1.1+ control cells in the peripheral blood of recipient mice at this relatively early time point. Compared with WT mice, the groups with a single genetic lesion (RhoAG17V or Tet2–/–) showed a modest increase in the population of Thy1.2+ cells (~3- to 4-fold higher than Thy1.1+ cells), whereas Thy1.2+ T cells with both genetic lesions (Tet2–/– and RhoAG17V) showed the highest growth advantage (77.0% vs. 4.42%; >20-fold higher than normal Thy1.1+ cells) (Figure 2A). Ki-67 staining further confirmed a significantly higher proliferative rate of adoptively transferred Thy1.2+ Tet2–/– RhoAG17V T cells in the peripheral blood of recipient mice (Figure 2B). We also visualized this scenario, although to a lesser extent (~3.3-fold increase in the Thy1.2/Thy1.1 ratio), in spleens and lymph nodes isolated from recipient mice transferred with Tet2–/– RhoAG17V T cells (Supplemental Figure 2A).
Figure 2. TET2 loss and RhoAG17V expression cause abnormal expansion of CD4+ T cells.
(A) Peripheral blood analysis of GFP+ Thy1.1+ and Thy1.2+ T cell populations in recipient mice 8 weeks after transfer. Bar graph shows the ratio of GFP gated Thy1.2+ and Thy1.1+ T cells before (week 0) and 8 weeks after adoptive T cell transfer (n = 4 mice; 2 independent experiments). (B) Ki-67 staining of Thy1.2+ T cells in the peripheral blood of recipient mice 8 weeks after adoptive cell transfer. Bar graph shows the statistical analysis of Ki-67+ Thy1.2+ cells in each group (n = 3 mice; 3 independent experiments). (C) Peripheral blood analysis of Thy1.2 and GFP double-positive CD4+ and CD8+ T cells in the peripheral blood of recipient mice 8 weeks after adoptive transfer. (D) Statistical analysis of peripheral blood (PB) Thy1.2 and GFP double-positive CD4+/CD8+ T cell ratio at the indicated time points after T cell transfer (n = 3 mice; 3 independent experiments). (E) Representative FACS plots and (F) quantification of the ratio of CD4+/CD8+ T cells (gated by Thy1.2+GFP+) in peripheral lymphoid tissues (spleens and lymph nodes) derived from the indicated recipient mice (n = 3 mice; 3 independent experiments). (G) Representative flow cytometric profiles of Ki-67 staining of peripheral Thy1.2+CD4+ T cells isolated from recipient mice transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells 20 weeks after adoptive transfer (n = 3 mice, 3 independent experiments). Bar graph shows statistical analysis of the percentage of Ki-67+ cells. (H) Kaplan-Meier survival curves of recipient mice transferred with CD4+ or CD8+ T cells (WT vs. Tet2–/– RhoAG17V; n = 4 mice; 1 experiment). Data represent the mean ± SD. *P < 0.05 and **P < 0.01, by 2-tailed Student’s t test (A, D, and G) and ANOVA with Dunnett’s post-hoc correction (B and F).
It has been reported that TET2 mutations are detected in CD4+ and CD8+ T cells and HSPCs, but the RhoAG17V mutation is primarily identified in CD4+, but not in CD8+, T cells in AITL patients (6). This clinical feature motivated us to further determine which T cell subsets acquired a proliferative advantage. After analyzing the relative populations of CD4+ and CD8+ T cells in the peripheral blood of recipient mice, we observed a pronounced increase in CD4+ T cells (gated for Thy1.2+GFP+), beginning 8 weeks after T cell transfer, in the Tet2–/– RhoAG17V group, but not in the other 3 groups (WT, RhoAG17V, or Tet2–/–) (Figure 2, C and D). Flow cytometric analysis indicated a dominance of CD4+ T cells (>90%) in the Thy1.2+GFP+ gated population in the peripheral blood from the Tet2–/– RhoAG17V group (Figure 2C), suggesting a CD4+ T cell–specific growth advantage after depletion of TET2 and expression of the RhoAG17V mutant. A similar trend was observed in peripheral lymphoid organs derived from mice transferred with Tet2–/– RhoAG17V T cells (8 weeks after transfer, Supplemental Figure 2B; or 20 weeks after transfer, Figure 2, E and F). Thy1.2+CD4+ T cells isolated from those lymphoid organs showed a nearly 2-fold increase in Ki-67 staining (Figure 2G).
The proliferative advantage of Tet2–/– RhoAG17V CD4+, but not CD8+, T cells was further independently validated in vitro (Supplemental Figure 2, C–F). We transduced CD4+ or CD8+ T cells isolated from WT or Tet2–/– mice with retroviruses expressing either GFP (as a control) or RhoAG17V–internal ribosome entry site–GFP (RhoAG17V-IRES-GFP) and then compared their proliferation using a CellTrace proliferation assay. Upon anti-CD3 and anti-CD28 stimulation for 2 to 4 days, Tet2–/– RhoAG17V CD4+ T cells showed the most remarkable increase in proliferation. Consistent with a previous study of TET-depleted lymphocytes (33), Tet2–/– CD4+ T cells also showed a moderate enhancement of proliferation (Supplemental Figure 2, C and D). By comparison, genetic lesions in TET2 and/or RhoA did not seem to perturb the proliferation of transduced CD8+ T cells (Supplemental Figure 2, E and F), further confirming a CD4+ T cell–specific effect. Furthermore, we performed adoptive T cell transfer experiments using purified CD4+ or CD8+ T cell subsets (WT vs. Tet2–/– RhoAG17V). Only mice transferred with Tet2–/– RhoAG17V CD4+ T cells, but not CD8+ T cells, showed a reduction in survival (Figure 2H), which echoed our observation in recipient mice transferred with Tet2–/– RhoAG17V T cells (Figure 1B). Collectively, these findings suggest a strong and unique synergistic effect of TET2 loss and RhoAG17V expression on CD4+, but not CD8+, T cells. This agrees well with the mutation signatures observed in patients with AITL, i.e., TET2 and RhoA mutations coexist in CD4+, but not CD8+, T cells (6).
The survival and cell-cycle progression of CD4+ T cells are altered as a result of TET2 loss and RhoAG17V expression.
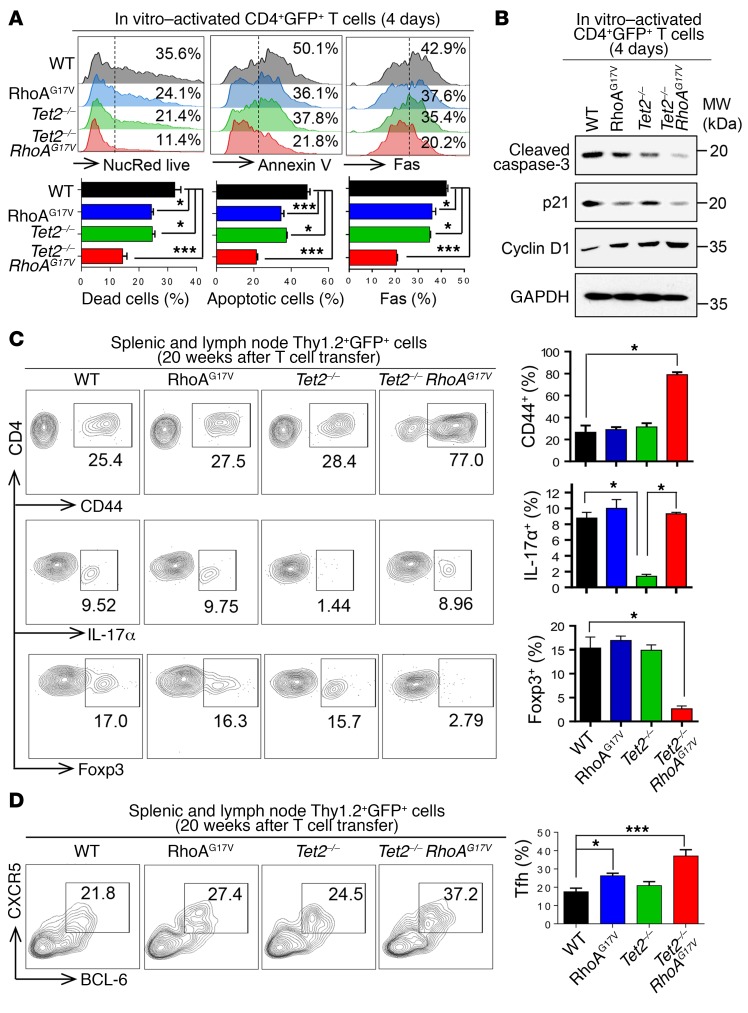
To further assess how Tet2 deletion and RhoAG17V expression affect the fate of CD4+ T cells, we monitored cell death and apoptosis in the 4 groups (WT, RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V) of in vitro–cultured CD4+ T cells. Compared with WT T cells, we detected up to a 2- to 3-fold reduction (NucRed staining) in cell death and apoptosis (annexin V staining) in Tet2–/– RhoAG17V CD4+ T cells (Figure 3A). In parallel, Fas, a key ligand involved in regulating CD4+ apoptosis upon T cell receptor (TCR) activation (34), was downregulated by over 2-fold in the Tet2–/– RhoAG17V group (Figure 3A). Indeed, Western blotting further confirmed the significant reduction in the amounts of cleaved caspase 3 in the cell lysates of Tet2–/– RhoAG17V T cells (Figure 3B). Cell-cycle analysis further revealed an increase in Tet2–/– RhoAG17V CD4+ T cells in the S phase (Supplemental Figure 3A), which agrees with the accelerated proliferation phenotype described above (Figure 2). These cell-cycle alterations were associated with the downregulation of p21 and upregulation of cyclin D1 (Figure 3B). We also observed the changes in protein levels of cleaved caspase 3, p21, and cyclin D1 in Tet2–/– and RhoAG17V CD4+ T cells, but to a much lesser extent, again implying a synergy between TET2 and RhoA in regulating related signaling pathways.
Figure 3. TET2 depletion and RhoAG17V expression suppress cell death and disrupt peripheral T cell homeostasis.
(A) Flow cytometric analysis of NucRed live), annexin V, and Fas staining in in vitro–activated GFP+CD4+ T cells (day 4). Representative flow cytometric plots for each indicated group. Bar graphs show quantification of the percentages of cells with positive staining (n = 3 independent experiments). (B) Immunoblots showing the detection of cleaved caspase-3, p21, and cyclin D1 on day 4 following in vitro activation of CD4+ T cells (WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V). GAPDH served as a loading control. MW, molecular weight. (C) Representative flow cytometric plots of Thy1.2+GFP+CD4+ T cells isolated from spleens and lymph nodes from recipient mice adoptively transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells (20 weeks after transfer). Bar graphs show quantification of the percentages of CD4+ T cells (n = 3 mice, 1 experiment) with positive staining for CD44, IL-17α (Th17 marker), or Foxp3 (Treg marker). (D) Representative flow cytometric plots of Thy1.2+, GFP+, and CD4+ gated Tfh cells isolated from spleens and lymph nodes from recipient mice adoptively transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells (20 weeks after transfer). Bar graph shows quantification of the percentages of CXCR5+BCL-6+ Tfh cells (n = 3 mice; 1 experiment). Data represent the mean ± SD. *P < 0.05 and ***P < 0.001, by ANOVA with Dunnett’s method for multiple comparisons.
TET2 loss and RhoAG17V expression result in a peripheral T cell imbalance.
To identify the major CD4+ T cell subsets responsible for the induction of immunoinflammatory responses in mice transferred with Tet2–/– RhoAG17V T cells, we sought to examine whether TET2 loss and RhoAG17V mutation would lead to an imbalance of effector and regulatory T cells. We analyzed the effector CD4+ T cell population in each group and observed a 3-fold increase in the population of CD4+CD44+ T cells (Thy1.2+GFP+) only in the Tet2–/– RhoAG17V group, indicating the activation of transferred CD4+ T cells (Figure 3C). Given the strong systematic inflammatory response, we first tested whether the increase of CD4+CD44+ T cells in the Tet2–/– RhoAG17V mice led to an aberrant production of IFN-γ. Compared with the WT group with a low population of CD4+CD44+ T cells, we detected an IFN-γ increase of only approximately 1.25-fold in the double-mutation group (Supplemental Figure 3B). Similarly, we did not observe a significant difference in the serum levels of IFN-γ between the WT and double-mutant recipient mice (Supplemental Figure 3C), suggesting that other mechanisms (e.g., a Th17/Treg imbalance) might account for the observed inflammatory phenotype. Compared with WT, we detected a decrease of 85% in the Th17 subset (marked by IL-17α) among all Thy1.2+GFP+CD4+ T cells isolated from recipient mice transferred with Tet2–/– T cells, a result that is consistent with previous findings (35). Most interestingly, the additional introduction of the RhoAG17V mutation into Tet2–/– T cells (Tet2–/– RhoAG17V) rescued the decline of Th17 cells in the recipient mice (middle panel, red bar, Figure 3C), a trend that was further validated in vitro by directing naive CD4+ T cell differentiation toward the Th17 lineage (Supplemental Figure 3D). This phenotypic change agreed with the alterations in the expression of RAR-related orphan receptor γ (Rorc), which encodes RORγt, as a key transcriptional factor to mediate Th17 lineage specification (36). Compared with WT, Rorc expression dropped by 80% in Tet2–/– T cells, but was restored to a level comparable to WT levels in Tet2–/– RhoAG17V CD4+ T cells (Supplemental Figure 3E). Thus, RhoAG17V seemed to antagonize the Th17-suppressive effect associated with TET2 loss.
In parallel, we detected an approximately 80% reduction of Foxp3+ Tregs in recipient mice transferred with Tet2–/– RhoAG17V T cells (Figure 3C). Foxp3 is the lineage-determining transcription factor required for the maintenance of Tregs (37). Tregs are well known for their immunosuppressive role: they downregulate effector T cell function, maintain tolerance to self-antigens, and prevent autoimmune disease (37). TET2 depletion in mice has been previously shown to reduce Foxp3 gene expression, with a subsequent decrease in Tregs (38–40). Our in vitro–directed T cell differentiation study further confirmed this (Supplemental Figure 3F). Notably, the additional introduction of RhoAG17V in Tet2–/– T cells led to a further reduction in the expression of Foxp3+ as well as TGF-β, a key growth factor that initiates and regulates Foxp3 expression (41) (Supplemental Figure 3G). Conversely, we observed a significant increase (~1.7-fold) in the population of CXCR5+BCL-6+ follicular T helper (Tfh) cells in peripheral lymphoid tissues from mice transferred with Tet2–/– RhoAG17V T cells (Figure 3D), which recapitulated the Tfh-like phenotypes observed in patients with AITL (42).
Clearly, Tet2 deletion, together with RhoAG17V expression, resulted in aberrant CD4+ T cell activation and an imbalance of peripheral T cell homeostasis. The resultant reduction in Tregs and increase in Tfh cells may account for the immunoinflammatory phenotypes observed in the recipient mice following adoptive T cell transfer.
TET2 loss and RhoAG17V expression remodel the transcriptional network and disrupt FoxO1-mediated signaling.
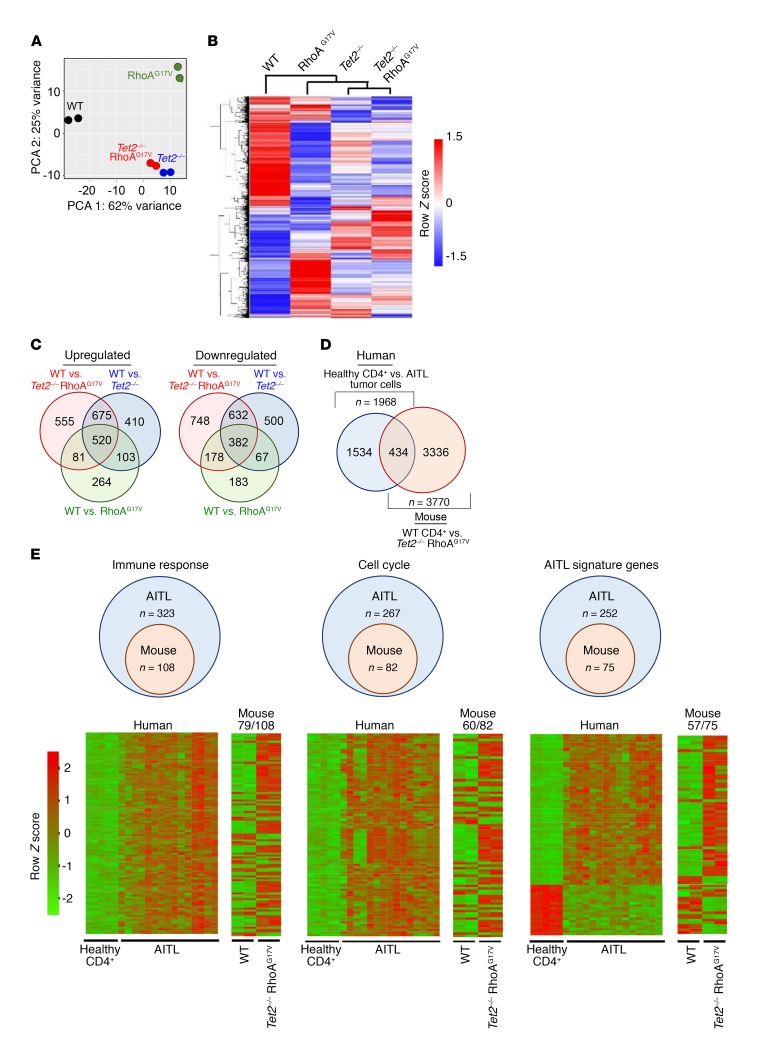
To further dissect the molecular mechanisms underlying the observed immune phenotypes, we performed transcriptome analysis of Thy1.2+GFP+CD4+ cells that were isolated from recipient mice using RNA-sequencing (RNA-seq) (Supplemental Figure 4, A–C). We obtained 42–96 million RNA-seq reads uniquely mapped to the transcriptome for a sample from each of the 4 groups with 2 replicates (Supplemental Figure 4A). Replicates had Pearson correlations close to 1.00 (Supplemental Figure 4B), indicating good reproducibility. The high quality of RNA-seq data is also indicated by the fact that all samples had good read coverage in the gene body (Supplemental Figure 4C). Principle component analysis (PCA) and cluster analysis of gene expression data sets showed that the Tet2–/– RhoAG17V group was most closely related to the Tet2–/– group, but was substantially distinct from either the WT or RhoAG17V group (Figure 4, A–C, and Supplemental Table 1), indicating that TET2 loss is the major determinant responsible for changes in gene expression in Tet2–/– RhoAG17V CD4+ T cells. Since the DNA methylation status is intimately involved in transcriptional regulation, we next measured the global 5mC and 5hmC levels in the 4 groups using a well-established dot blot assay. As expected, we found a reduction of 5hmC levels in both Tet2–/– and Tet2–/– RhoAG17V cells, but not in WT or RhoAG17V cells (Supplemental Figure 4D). The global 5mC levels in all the groups remained unaltered (Supplemental Figure 4E). Given these findings, we conclude that the decrease in global 5hmC resulting from TET2 depletion might be one of the causes for transcriptional alterations in the Tet2–/– RhoAG17V group.
Figure 4. Dysregulation of gene expression and DNA hydroxymethylation following TET2 depletion and/or RhoAG17V expression in CD4+ T cells.
(A) PCA of gene expression of Thy1.2+GFP+CD4+ T cells isolated from the spleens and lymph nodes of recipient mice transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells. (B and C) Heatmap (B) and Venn diagram (C) displaying gene expression for all genes differentially expressed in each indicated mutant group relative to WT. (D) Venn diagram depicting the overlap of DEGs between mouse Tet2–/– RhoAG17V CD4+ T cells (relative to WT CD4+ T cells) and human AITL tumor cells (relative to normal CD4+ T cells from healthy donors). (E) Comparison of overlapping DEGs between AITL patients (vs. healthy CD4+ T cells) and murine Tet2–/– RhoAG17V CD4+ T cells (vs. WT CD4+ T cells). Diagram shows the numbers of overlapping genes in the murine transcriptomes that fell into the same enriched gene categories centered by DEGs identified from patients with AITL. Heatmap shows the expression profiles of DEGs (human and mouse) under the indicated functional categories. Each row represents 1 DEG identified from human (5 healthy donors vs. 15 AITL patients) or mouse (2 cases for both the WT and double-mutation groups) comparative transcriptome analysis, while each column represents 1 sample. The number of genes (vs. the total number of genes) showing the same differential expression trend (up- or downregulation) as that in the human RNA-seq results is indicated under the mouse heatmaps.
By comparing the differentially expressed genes (DEGs) among the 4 groups (relative to WT), we found that the Tet2–/– RhoAG17V group displayed the most pronounced changes in gene expression, with 555 and 748 genes specifically up- or downregulated, respectively (Figure 4C and Supplemental Table 1). To further examine whether Tet2–/– RhoAG17V CD4+ T cells from the recipient mice recapitulated the gene expression signatures representative of human AITL tumor cells, we compared murine DEGs identified from our own study (mouse Tet2–/– RhoAG17V vs. WT CD4+ T cells) with human DEGs (8) from existing transcriptome data sets (human AITL tumors vs. CD4+ T cells from healthy donors) (Figure 4, D and E). Our unbiased comparative analysis revealed a total of 434 overlapping DEGs between the mouse and human RNA-seq data sets (Figure 4D and Supplemental Table 1), which covered 22% and 11.5% of total DEGs identified in the human and mouse data sets, respectively. We further classified the overlapping DEGs according to gene clusters identified from previous human AITL RNA-seq analyses (8): 33.4% (108 of 323; mouse DEGs/human DEGs), 30.7% (82 of 267), and 29.8% (75 of 252) of overlapping DEGs fell into the functional categories of immune response, cell cycle, and AITL tumor signature gene sets (8, 43), respectively (Figure 4E and Supplemental Table 1). The majority of these overlapping genes (>73%) displayed the same trend in expression (up- or downregulation) in both the human and mouse data sets (heatmap shown in Figure 4E; genes listed in Supplemental Table 1). Collectively, the mouse Tet2–/– RhoAG17V CD4+ T cells used in our study at least partially recapitulated the gene expression signatures of human AITL tumor cells.
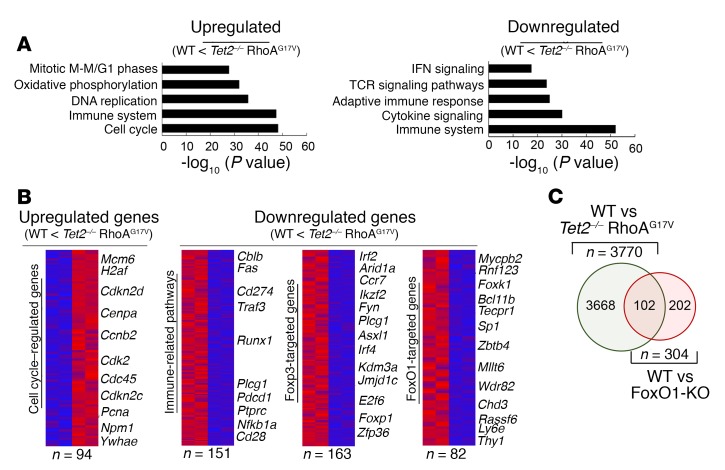
Gene set enrichment analysis (GSEA) of the WT and Tet2–/– RhoAG17V groups revealed that upregulated genes were enriched in genes implicated in the cell cycle and DNA replication (e.g., cyclin family genes and cyclin-dependent kinase inhibitors; Figure 5, A and B), a finding that resonates with the phenotypic changes in cell proliferation (Figure 2 and Supplemental Figure 2) and the cell cycle (Figure 3 and Supplemental Figure 3A). On the other end of the DEG spectrum, downregulated genes were most abundantly found under the categories of immune system and TCR signaling pathways (Figure 5, A and B), including negative regulators of TCR or NF-κB signaling (e.g., casitas B-lineage lymphoma B [Cblb], TNF receptor–associated factor 3 [Traf3], nuclear factor κ B subunit 1 [Nfkb1a], and protein tyrosine phosphatase, receptor type, C [Ptprc]) and some lymphoma-associated genes (e.g., phospholipase C, γ 1 [Plcg1], programmed cell death 1 [Pdcd1], and CD274 antigen [Cd274]) that have been recently reported in adult T cell lymphoma (44). In line with our aforementioned observations of a decrease in Foxp3+ expression and a reduction in Treg numbers, we observed a marked reduction in the expression of Foxp3-targeted genes in Tet2–/– RhoAG17V cells (Figure 5B).
Figure 5. CD4+ T cells with genetic lesions in both TET2 and RhoA (Tet2–/– RhoAG17V genotype) display distinct gene expression signatures.
(A) GSEA of upregulated and downregulated genes between WT and Tet2–/– RhoAG17V CD4+ T cells. (B) Heatmaps of subgroups of upregulated and downregulated genes highlighted in the GSEA analysis. (C) Venn diagram depicting the overlap of downregulated genes during the comparison of Tet2–/– RhoAG17V or FoxO1-KO relative to WT CD4+ T cells.
When comparing the gene expression profiles between WT and Tet2–/– RhoAG17V CD4+ T cells, we further noticed that a substantial fraction of downregulated genes (Figure 5B) was associated with FoxO1, a forkhead family of transcriptional factors involved in regulating T cell functions (45, 46). Nonetheless, the change in the expression of FoxO1 itself, as revealed below by quantitative PCR (qPCR) (Figure 6A), was less than 20%. We also compared the list of downregulated DEGs in the Tet2–/– RhoAG17V group with the published DEG list representative of FoxO1-KO CD4+ T cells (45) and surprisingly discovered 102 overlapping DEGs (approximately one-third of 304 DEGs in FoxO1-KO vs. WT; red circle; Figure 5C) between the 2 conditions. These findings strongly imply that FoxO1 might constitute one of the key targets subjected to modulation by TET2- and RhoA-associated pathways in CD4+ T cells.
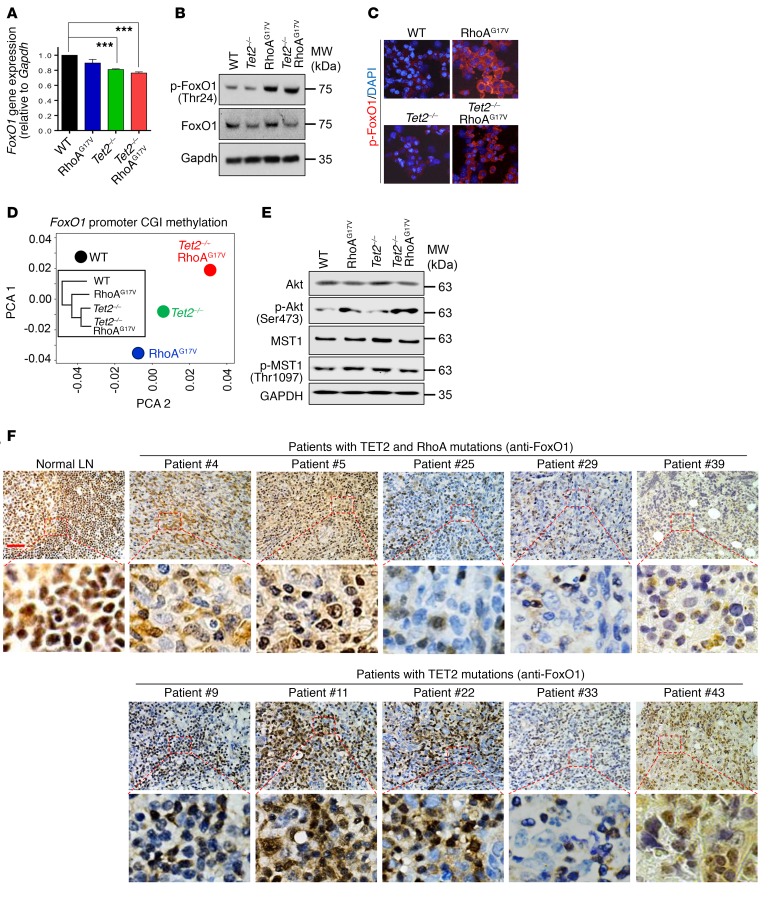
Figure 6. Deletion of Tet2 and expression of RhoAG17V synergistically regulate FoxO1 activity.
(A) Real-time reverse transcription PCR (RT-PCR) analysis of FoxO1 gene expression (normalized to Gapdh) in Thy1.2+GFP+CD4+ T cells isolated from peripheral lymphoid tissues of recipient mice. The mice were adoptively transferred with WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V T cells. Results are presented as the fold change relative to WT (arbitrarily set to 1). Data from 5 independent experiments are shown. ***P < 0.001, by 2-tailed Student’s t test. (B) Immunoblot analysis showing the expression of total and phosphorylated FoxO1 in GFP+CD4+ T cells (WT, RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V) that were in vitro–activated for 4 days. GAPDH served as a loading control. (C) Representative confocal images showing immunostaining of p-FoxO1 in WT, RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V in vitro–activated GFP+CD4+ T cells. (D) PCA and cluster analysis (inset) of the DNA methylation status of the CpG island (CGI) at the FoxO1 promoter in GFP+CD4+ T cells from mice of the indicated groups. The Tet2–/– and Tet2–/– RhoAG17V groups were most closely related. (E) Immunoblot analysis showing the expression of total and phosphorylated Akt and MST1 in GFP+CD4+ T cells (in vitro–activated for 4 days) from mice of the indicated groups. GAPDH served as a loading control. (F) Immunohistochemical analysis of FoxO1 distribution in normal human lymph nodes (top left) and lymphoma biopsies isolated from patients with AITL bearing both TET2 and RhoA mutations (top) or TET2 mutations alone (bottom). Scale bar: 50 μm. Original magnification: ×40.
Dual suppression of FoxO1 activity by TET2 loss and RhoAG17V expression.
To further understand how TET2 loss and RhoAG17V expression perturbed FoxO1 function, we measured the mRNA and protein levels as well as phosphorylation of FoxO1 in WT, RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V CD4+ T cells (Figure 6). We found that the loss of TET2 alone resulted in a decrease of approximately 20% in the expression of FoxO1 at a transcriptional level (Figure 6A), with a subsequent reduction in total protein levels of FoxO1, but not a reduction in phosphorylated FoxO1 (p-FoxO1) levels (Figure 6B). The RhoAG17V T cells did not show significant changes in FoxO1 gene expression, but had increased p-FoxO1, as revealed by both Western blotting (Figure 6B) and immunostaining (Figure 6C). The combined effects of TET2 loss and RhoAG17V expression, as anticipated, were a significant reduction in both the mRNA and protein levels of FoxO1, with an accompanying upregulation of its phosphorylation (Figure 6, A–C). Previous studies have shown that p-FoxO1 is transported from the nucleus to the cytoplasm and then becomes a target for proteasomal degradation to be fully inactivated (47). These results led us to hypothesize that FoxO1 is subjected to dual modulation by TET2-mediated DNA demethylation and RhoA-associated signaling pathways. Specifically, TET2 might regulate the transcription of FoxO1 through demethylation of the regulatory genomic regions (promoter and/or enhancer) of FoxO1, while RhoA is involved in regulating the phosphorylation and subsequent subcellular localization of FoxO1.
To test this hypothesis, we first determined the DNA methylation status at the promoter of FoxO1 using locus-specific bisulfite sequencing in the 4 experimental groups (WT, RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V). The PCA and cluster analysis showed close DNA methylation patterns between Tet2–/– and Tet2–/– RhoAG17V CD4+ T cells, suggesting that TET2 is the major epigenetic regulator affecting DNA methylation at FoxO1 promoters (Figure 6D). Among a total of 122 CpG sites within a CpG island of the FoxO1 promoter analyzed in our assay, we detected increased DNA methylation (>1.5-fold) in 26, 37, and 68 CpG sites in RhoAG17V, Tet2–/–, and Tet2–/– RhoAG17V CD4+ T cells, respectively, when compared with WT (Supplemental Figure 5A). The majority of CpG sites showed a moderate increase (between 1.5- and 5-fold) in DNA methylation upon Tet2 deletion and/or RhoAG17V expression, which might be associated with the 20% decrease in FoxO1 gene expression in Tet2–/– and Tet2–/– RhoAG17V CD4+ T cells (Figure 6A).
Next, to identify potential upstream factors that might control FoxO1 phosphorylation, we focused on Akt and MST1, two protein kinases that are most prominently known to regulate this process (48). We performed Western blotting to determine the total protein and phosphorylation levels of Akt and MST1. MST1 and p-Akt have been reported to mediate FoxO1 phosphorylation and its subsequent nucleus-to-cytosol translocation (48). As shown in Figure 6E, compared with the WT and Tet2–/– groups, RhoAG17V and Tet2–/– RhoAG17V cells showed higher levels of p-Akt, whereas the total Akt protein levels remained largely unaltered. Of note is that the elevated phosphorylation of FoxO1 correlated with its more abundant accumulation in the cytosol in RhoAG17V and Tet2–/– RhoAG17V CD4+ T cells (Figure 6C). With regard to MST1, we observed no difference in the total or phosphorylated protein levels among the 4 groups, thus ruling out its participation in upregulating p-FoxO1. These results suggest that RhoAG17V is at least one of the causes that alters the activity of Akt and subsequently upregulates FoxO1 phosphorylation, a finding that is consistent with an earlier study performed in Jurkat T cells (8).
To explore whether altered FoxO1 subcellular localization and/or expression can be used as clinically relevant immunophenotypic and molecular features in human primary AITL tumor samples, we performed immunohistochemical analysis of FoxO1 in AITL tumor specimens (a total of 43 cases), as well as in lymph nodes from healthy donors. In normal lymph nodes, the IHC immunostaining of FoxO1 was detected almost exclusively in the nuclei, with strong signals (left, Figure 6F). The FoxO1 staining patterns in the majority of AITL tumor specimens fell into 1 of the following 4 categories (Figure 6F, Supplemental Figure 5B, and Supplemental Table 2): (a) negative or low staining in both the cytosol and nuclei (e.g., cases 8, 30, and 42); (b) variegated staining in both the cytosol and nuclei (e.g., cases 5, 16, and 19); (c) dominant staining in the cytosol (e.g., cases 4 and 6); or (d) dominant staining in the nuclei (e.g., cases 9, 10, and 23). To further analyze the TET2 and RhoA mutation status in these patients, we performed targeted amplicon sequencing. Among the 43 patients with AITL, 22 patients (~51%) had the RhoAG17V mutation (co-occurrence with TET2 mutations) (Supplemental Table 2), and all patients had TET2 mutations, but with varying mutation frequencies across the TET2 coding regions (Supplemental Figure 5C and Supplemental Table 2). Notably, 17 of 43 patients displayed high mutation frequencies within the TET2 catalytic domain (TET2CD) (Supplemental Figure 5C and Supplemental Table 2), with 15 of 17 also having the RhoAG17V mutation (Supplemental Figure 5D). We next asked whether there is a correlation between FoxO1 immunostaining and the mutation status in 43 AITL patients. Overall, AITL tumor cells from most patients (20 of 22) bearing both TET2 and RhoAG17V mutations showed low nuclear and/or cytosolic FoxO1 staining (Figure 6F, Supplemental Figure 5B, and Supplemental Table 2). Nonetheless, AITL patients with TET2 mutations alone showed heterogeneous FoxO1 staining patterns (with 16 patients showing strong FoxO1 staining and 5 patients showing relatively weak FoxO1 staining) (Figure 6F, Supplemental Figure 5B, and Supplemental Table 2), possibly due to the different TET2 mutation frequencies and the existence of additional unidentified mutations and pathways in regulating the function of FoxO1 in those AITL patients. Among all the AITL samples, not a single AITL patient was found to have RhoAG17V alone, which was consistent with previous exome sequencing results that suggest the exclusive coexistence of TET2 and RhoA mutations in patients with AITL (6).
In aggregate, our data indicate that TET2 loss and RhoAG17V synergized together to modulate FoxO1 activity by suppressing the transcription of FoxO1 and promoting its phosphorylation through epigenetic regulatory and RhoA-mediated signaling pathways.
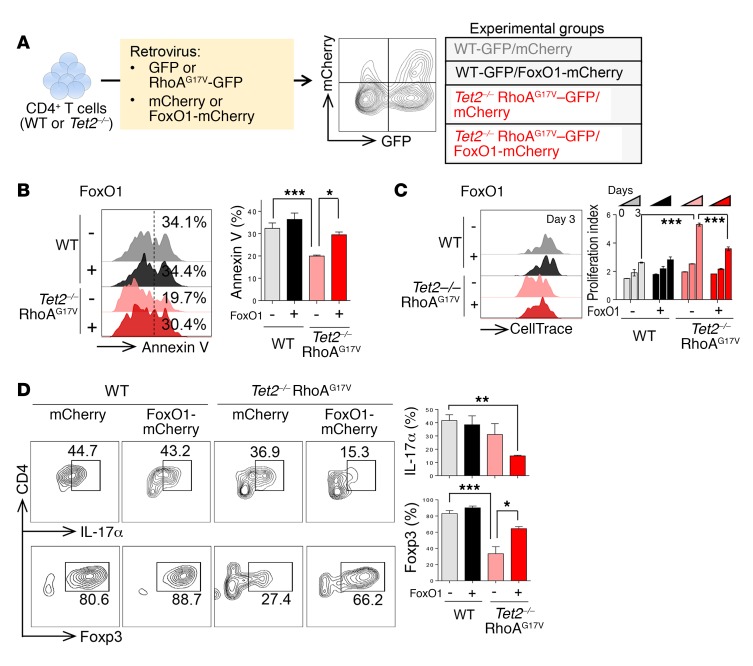
Restoring FoxO1 levels rescues Tet2–/–RhoAG17V–associated abnormalities in CD4+ T cells. Our findings described above converged on FoxO1 inactivation as a potential key molecular event that ultimately causes aberrant CD4+ T cell proliferation, survival, and polarization. To further test this in vivo, we transduced WT T cells and Tet2–/– RhoAG17V CD4+ T cells (GFP+ population) with retroviruses encoding mCherry (control) or HA-FoxO1-IRES-mCherry (Figure 7A and Supplemental Figure 6, A and B) and assessed cell proliferation and apoptosis of GFP/mCherry double-positive cells in 4 groups (WT-mCherry, WT-FoxO1, Tet2–/– RhoAG17V–mCherry, and Tet2–/– RhoAG17V–FoxO1). FoxO1 expression, as independently confirmed by flow cytometry (Supplemental Figure 6A) and Western blotting (Supplemental Figure 6B), antagonized the effects of genetic lesions in TET2 and RhoA by promoting apoptosis (red vs. pink bars; Figure 7B) and suppressing the proliferation (Figure 7C) of Tet2–/– RhoAG17V CD4+ T cells during 3 days of in vitro stimulation. As a control, we observed no significant alterations of cell apoptosis or proliferation in WT CD4+ T cells that similarly expressed FoxO1 (black vs. gray bars; Figure 7, B and C). To examine the effect of FoxO1 expression on CD4+ T cell polarization in vitro, we differentiated transduced WT and Tet2–/– cells toward the Th17 or Treg lineages in vitro with their corresponding set of cytokines. While FoxO1 expression exerted negligible effects on WT CD4+ T cells, it clearly corrected the imbalance of Th17 cells and Tregs induced by Tet2 deletion and RhoAG17V, as reflected in the upregulation of Foxp3 expression and the concomitant downregulation of IL-17α expression (Figure 7D). These data demonstrate that FoxO1 reexpression in Tet2–/– RhoAG17V cells indeed restored proper CD4+ effector T cell function.
Figure 7. Reexpression of FoxO1 rescues the abnormalities of Tet2–/– RhoAG17V CD4+ T cells.
(A) Experimental scheme of FoxO1 reexpression in WT or Tet2–/– RhoAG17V CD4+ T cells. GFP was used as a marker to monitor the expression of RhoAG17V or related controls, whereas mCherry signals indicated the expression of FoxO1-mCherry or mCherry as a control. GFP and mCherry double-positive cells were gated for further immunophenotypic analysis. (B and C) Representative flow cytometric plots of annexin V (B) and CellTrace (C) staining for in vitro–activated CD4+ T cells (WT, WT-FoxO1, Tet2–/– RhoAG17V, or Tet2–/– RhoAG17V–FoxO1). Bar graphs in B and C show statistical analyses of the flow cytometric data (n = 3 independent experiments). (D) In vitro–differentiated CD4+ T cells stained for the Treg marker Foxp3 and the Th17 marker IL-17α. WT or Tet2–/– naive T cells were transduced with retroviruses encoding mCherry or FoxO1-mCherry and then subjected to directed differentiation toward Tregs or Th17 cells. Bar graphs show immunostaining quantification (n = 3 independent experiments). Data represent the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, by ANOVA with Tukey’s post-hoc correction.
Discussion
The recent exosome sequencing and integrative genomic studies of lymphoma samples have led to the discovery of recurrent mutations in epigenetic regulators (e.g., TET2, DNA methyltransferase 3 α [DNMT3A], and isocitrate dehydrogenase [NADP(+)] 2, mitochondrial [IDH2]) and RhoA in PTCLs (6–8). In particular, the coexistence of somatic mutations in TET2 and RhoA is present in over 60% of patients with AITL and constitutes an aggressive subtype of PTCL that is often associated with autoimmune-like manifestations (1, 2, 10, 11). This breakthrough heralds the advent of a molecular era in the mechanistic dissection of AITL and other types of PTCL that share similar genetic defects. Here, we show that genetic depletion of TET2, together with RhoAG17V expression, results in mature CD4+ T cell abnormalities characterized by increased cell proliferation, decreased apoptosis or cell death, aberrant activation of TCR signaling, and an imbalance among Th17 cells, Tregs, and Tfh cells. The disruption of T cell homeostasis might account for the immunoinflammatory-like phenotypes observed in adoptive T cell transfer mouse models, which resemble the inflammatory background in patients with AITL (2, 11).
In the present study, we have collected compelling evidence to support the notion that mutational cooperativity between TET2 and RhoA is required to disrupt normal CD4+ T cell function both in vitro and in vivo through inactivation of FoxO1. Our study showcases a previously unappreciated cooperation between an epigenetic regulator and the RhoA GTPase signaling pathway to maintain normal CD4+ T cell activity (Figure 8). TET2 is an epigenetic regulator and dioxygenase that catalyzes DNA methylation oxidation to mediate DNA demethylation (12). TET2 mutations are reported in a wide spectrum of hematological disorders and neoplasms, including myelodysplastic syndromes, acute myeloid leukemia and chronic myelomonocytic leukemia, and both B and T cell lymphomas (17–22). Genetic disruption of TET2 in mice has been shown to promote HSPC self-renewal and biased differentiation toward the myeloid lineage (20, 24–26). However, TET2 loss of function in general is insufficient to cause a malignant transformation of blood cells (23), indicating that secondary mutation(s) might be needed to promote the progression of hematological malignancies (49–51). Indeed, with respect to mature T cell function, genetic lesions in both TET2 and RhoA, but not TET2 loss or the G17V RhoAG17V mutation alone, lead to the most pronounced perturbations of the proliferation of CD4+ T cells, including the CD4+CD44+ T cell subsets that are regarded as cells of origin of some types of PTCLs, and may contribute to tumor cell growth and inflammatory responses (52). In parallel, we also observed an overt Th17/Treg/Tfh functional imbalance in the double-mutation group, which might account for the inflammatory phenotype seen in recipient mice transferred with Tet2–/– RhoAG17V T cells. Mechanistically, TET2 is involved in remodeling the transcriptional network in CD4+ T cells through its epigenetic regulatory function, whereas RhoA is more devoted to controlling TCR-associated signaling pathways in CD4+ T cells.
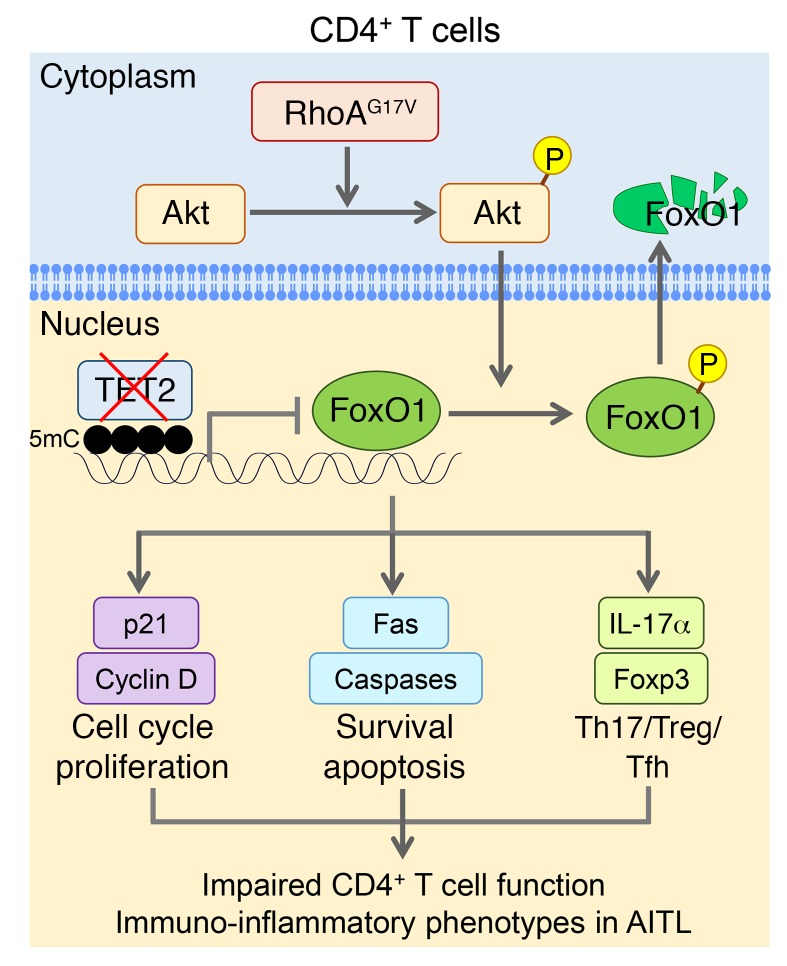
Figure 8. Tentative model depicting the dual modulation of FoxO1 through cooperative actions of TET2 and RhoA in murine CD4+ T cells.
TET2-medaited 5mC oxidation is responsible for maintaining the proper methylation status of the FoxO1 promoter to regulate its transcription. RhoA GTPase signaling is required to balance the phosphorylation (P) state of Akt so as to control the phosphorylation of its downstream effector, FoxO1, which is indispensable for T cell proliferation and survival. Both pathways cooperatively work together to regulate the FoxO1-medaited signaling. Genetic defects in TET2 or RhoA alone may not exert an overtly detrimental effect on FoxO1. However, in patients with AITL or other types of PTCLs, TET2 loss of function and expression of the RhoA mutant (G17V) synergistically disrupt the transcriptional and posttranslational modification networks of CD4+ T cells, as most notably exemplified by combined FoxO1 inactivation through hypermethylation at its promoter region and elevated phosphorylation to drive the cytosolic translocation of nuclear FoxO1 to expedite its degradation. The inactivation of FoxO1 results in abnormal CD4+ T cell proliferation and survival and imbalanced T cell homeostasis to elicit an aberrant immune response and to further drive AITL disease progression.
This functional synergy is best illustrated in the dual regulation of FoxO1, as illustrated in Figure 8. FoxO1 is one of the key transcriptional factors that maintain T cell proliferation and survival (53). TET2 loss leads to alterations in the methylation status of the FoxO1 promoter (hypermethylation) to negatively regulate the transcription of FoxO1 (by ~20%). Although we cannot rule out the possibility that TET2 might also control DNA demethylation at distal regulatory regions (e.g., enhancer) of FoxO1, the alteration of DNA methylation at the promoter due to TET2 loss is certainly associated with the transcriptional regulation of FoxO1 in CD4+ T cells, but the causal effect remains to be established. On the other hand, the inactivating mutation in RhoA GTPase (G17V) ultimately causes elevated phosphorylation of FoxO1 to promote its nucleus-to-cytosol translocation for proteasomal degradation. A genetic defect in TET2 or RhoA alone is insufficient to cause abnormal phenotypes, but the combined effects can have an overtly detrimental effect on CD4+ T cells. Loss of FoxO1 has been known to reduce the expression of apoptotic genes and Fas, which agrees with the apoptosis-suppressive phenotype we observed in Tet2–/– RhoAG17V CD4+ T cells (Figure 3A). Furthermore, FoxO1 positively controls Foxp3 gene expression and maintains the proper functioning of Tregs to suppress autoimmunity (48, 53). It has been shown that FoxO1 is a negative regulator of Th17 by inhibiting RORγt activity (54) and that inactivation of FoxO1 is essential for Tfh cell differentiation (55). Therefore, FoxO1 inactivation arising from genetic defects in both TET2 and RhoA could lead to an imbalance in effector T cells to induce inflammatory-like phenotypes such as those we observed in recipient mice after adoptive T cell transfer. Although FoxO1 itself remains an intractable transcriptional factor to be targeted, other key regulatory factors within the FoxO1-mediated transcriptional network might provide alternative candidate pools for developing improved antilymphoma therapy. Overall, the inactivation of FoxO1 that resulted from TET2 loss and RhoAG17V provides an additional basis for devising new diagnoses, prognoses, and therapeutics for lymphoma.
In both our in vitro and in vivo studies, we did not observe malignant transformation of mature CD4+ T cells. Nonetheless, the gene expression profiles of murine Tet2–/– RhoAG17V CD4+ T cells characterized in the current study are reminiscent of those of human AITL tumor cells. By comparing the transcriptomes of mouse CD4+ T cells harboring an AITL-like genetic background with those of human AITL tumor cells, we observed similar transcriptional output patterns (e.g., AITL signature genes [ref. 43] and critical genes involved in the immune response and cell cycle) in both cell types, thus attesting to the disease relevance of our experimental system. The overexpression of RhoAG17V on the Tet2–/– background led to acute inflammation-like phenotypes in the mice, causing relatively early death of the animals (median survival of 21.3 weeks). This might have prevented us from observing any overt malignant transformation or tumor formation during the course of the in vivo studies. Nonetheless, our in vivo model based on adoptive T cell transfer at least partially recapitulates the immune-related clinical manifestations commonly seen in AITL patients, and thus our study is one of the few to establish a clinically relevant in vivo model for the mechanistic study of AITL-associated symptoms. In the near future, it is imperative to generate inducible, genetically modified RhoAG17V-knockin plus Tet2–/– mice that might attenuate acute inflammation and provide a prolonged time window for potential malignant transformation, thereby enabling the generation of an improved mouse model that faithfully mimics human PTCLs.
Methods
Mouse T cell isolation.
T cells were isolated using a Dynabeads Untouched Mouse T Cell Kit (Thermo Fisher Scientific). Mouse CD4+ or CD8+ T cells were purified using a mouse CD4+ or CD8+ T Cell Isolation Kit (Miltenyi Biotec). In brief, spleens and lymph nodes were isolated from WT and Tet2–/– mice and ground with the back of a syringe plunger to release splenocytes and lymphocytes into the culture dish. The homogenized cell suspensions were filtered with a cell strainer (nylon mesh with 70-μM pores) to remove debris. The suspended cells were centrifuged, and the red blood cells were lysed with ammonium chloride potassium (ACK) lysis buffer. The residual cells were then purified using the T cell isolation kits according to the manufacturers’ instructions. The purity of the isolated T cells was assessed by flow cytometry.
Adoptive T cell transfer.
Isolated total T cells (Thy1.1+ or Thy1.2+) and CD4+ or CD8+ T cells were mixed together at the desired ratio in 200 μl PBS and retro-orbitally injected into 6- to 8-week-old male and female TCRα-deficient mice (purchased from The Jackson Laboratory, 1 million cells/mouse). The reconstitution efficiency of T cells was assessed by examining the GFP or mCherry fluorescence signals in peripheral blood from the recipient mice 4 weeks after adoptive cell transfer.
RNA-seq library preparation and bioinformatic analysis.
Poly A–tailed mRNA was enriched using a Poly(A)Purist Kit (Thermo Fisher Scientific) and followed by RNA-seq library preparation using an Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs) according to the manufacturer’s instructions. RNA-seq was performed using the Illumina HiSeq 2500 or NextSeq 500 with the 75-bp pair-ended running mode. The pair-ended reads were mapped against mm10 using STAR with default parameters, and the RefSeq gene annotation was obtained from the UCSC’s genome database (September 7, 2015). The number of reads mapped to each gene was counted using HTSeq (-m intersection-nonempty, -s no, -t exon, -i gene_id; http://www.huber.embl.de/users/anders/HTSeq/), with uniquely mapped reads as input. The raw read counts across all genes were used for calculating reads per kb per million (RPKM) values. To identify the differentially expressed genes among the experimental groups, we performed negative binomial tests for pairwise comparisons between WT and RhoAG17V, Tet2–/–, or Tet2–/– RhoAG17V by using the Bioconductor package DESeq2, with a corrected P value of 0.05 or less and fold-change thresholds of greater than 2 or less than 0.5. The functional enrichment analyses of significantly differentially expressed genes between 2 conditions were performed using GSEA with a preranked function. The Pearson correlations between biological replicates were 0.99 and 1.0, indicating high reproducibility of sequencing experiments, whereas the Pearson correlations between conditions ranged from 0.96 to 0.98. For the heatmaps, row-wise scaled RPKM values across all samples were plotted using the function heatmap.2 in the R package gplots (www.r-project.org).
Locus-specific DNA methylation analysis.
Purified genomic DNA was subjected to bisulfite conversion using the MethylCode Bisulfite Conversion Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The primers targeted to the bisulfite-converted FoxO1 locus were designed using MethPrimer (http://www.urogene.org/methprimer/) with default settings. The targeted amplicons were amplified using a PyroMark PCR Kit (QIAGEN), followed by library construction using a Nextera XT DNA Library Prep Kit (Illumina). The libraries were sequenced with a NextSeq 500 (Illumina) using a 75-bp pair-ended sequencing mode. The amplicon sequences were mapped to the FoxO1 promoter locus using BSMAP on the default setting (https://sites.google.com/a/brown.edu/bioinformatics-in-biomed/bsmap-for-methylation). Methylation ratios were calculated using model-based analysis of bisulfite sequencing data (MOABS) (56).
Patients’ samples and custom amplicon sequencing.
AITL patients’ samples were collected from patients with AITL at the MD Anderson Cancer Center (USA), Fujian Medical University (China), and Zhejiang Cancer Hospital (China). Genomic DNA was isolated from AITL patients’ tumor samples using a RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific). Targeted amplicon sequencing libraries were prepared using a TruSeq Custom Amplicon Kit (Illumina) and sequenced with the NextSeq 500 (Illumina). The mutation status of TET2 and RhoA was analyzed with an issac variation caller (https://github.com/sequencing/isaac_variant_caller) and the VarScan v2.2.3 pileup2snp function. For each sample, the Burrows-Wheeler Aligner (BWA) version 0.7.12 was used to map all sequencing reads to hg19. Picard was used to sort the align file. The issac variation caller configured with exome/target sequencing parameters call single nucleotide variants (SNVs) for each site (depth ≥20). SNV annotations were done using snpEFF software.
Accession numbers.
The RNA-seq data sets are deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE90976).
Statistics.
A minimum of 3 biological replicates were analyzed for all quantitative analysis. A 2-tailed Student’s t test was used to determine statistical significance between 2 groups. For multiple group comparisons, ANOVA with Dunnett’s or Tukey’s post-hoc correction was used. A P value of less than 0.05 was considered statistically significant. Error bars represent the SD.
Study approval.
Experiments and handling of mice were conducted following federal, state, and local guidelines under an IACUC protocol and with approval of the IACUC of the Institute of Biosciences and Technology at Texas A&M University. All patients provided informed consent under a research protocol approved by the IRBs of the MD Anderson Cancer Center, Zhejiang Cancer Hospital, and Fujian Medical University. These IRB-approved protocols were carried out in accordance with the Declaration of Helsinki principles.
Author contributions
YH directed and oversaw the project. YH and AR conceived the project. SZ performed the adoptive T cell transfer experiment and mouse phenotype analysis. SZ and HZ performed in vitro T cell activation experiments. SZ, WH, and MJY performed histological analysis of AITL patients’ samples. YH, YZ, and ML generated all the libraries for next-generation sequencing analysis. JL and DS performed RNA-seq bioinformatics analysis, locus-specific DNA methylation analysis, and amplicon-targeted sequencing analysis. ML performed the dot-blot analysis and immunofluorescence imaging experiment. JZ performed Western blotting. MM and SI contributed to the flow cytometric profiling and cell-sorting procedures. HYY, SZ, YY, and MJY provided the AITL patients’ samples and clinical input. AR provided intellectual input for the project. YH and YZ wrote the manuscript, and all other authors participated in discussions, data interpretation, and manuscript editing.
Supplementary Material
Acknowledgments
We thank Margaret Goodell at the Baylor College of Medicine (Houston, Texas, USA) for her advice and sharing of reagents. We are grateful to Jianjun Shen and the MD Anderson Next-Generation Sequencing Core at Smithville for their high-throughput sequencing work (CPRIT RP120348 and RP170002). This work was supported by grants from the Cancer Prevention and Research Institute of Texas (RR140053, to YH); an Innovation Award from the American Heart Association (16IRG27250155, to YH); the John S. Dunn Foundation Collaborative Research Award (to YH); the Translational Research Program (TRP) of the Leukemia and Lymphoma Society (6464-15, to AR); the NIH (R01GM112003, to YZ); the Welch Foundation (BE-1913, to YZ); the American Cancer Society (RSG-16-215-01-TBE, to YZ); and by an allocation from the Texas A&M University Start-up Funds (to YH and DS). MJY was partially supported by grants from the National Cancer Institute, NIH (R01 CA164346 and R01 CA200703) and Developmental Research Awards in Leukemia (SPORE CA100632) and CPRIT RP140402.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(8):2998–3012.https://doi.org/10.1172/JCI92026.
Contributor Information
Shengbing Zang, Email: shengbingzang@163.com.
Jia Li, Email: jli@ibt.tamhsc.edu.
Haiyan Yang, Email: haiyanyang1125@163.com.
Hongxiang Zeng, Email: zeng@ibt.tamhsc.edu.
Wei Han, Email: whan@ibt.tamhsc.edu.
Jixiang Zhang, Email: jixiang.zhang@ibt.tamhsc.edu.
Minjung Lee, Email: mlee@ibt.tamhsc.edu.
Sevinj Isgandarova, Email: sisgandarova@ibt.tamhsc.edu.
Yaling Yang, Email: yyang8@mdanderson.org.
Yubin Zhou, Email: yzhou@ibt.tamhsc.edu.
Anjana Rao, Email: arao@liai.org.
M. James You, Email: mjamesyou@mdanderson.org.
Deqiang Sun, Email: dsun@ibt.tamhsc.edu.
Yun Huang, Email: yun.huang@ibt.tamhsc.edu.
References
- 1.Foss FM, Zinzani PL, Vose JM, Gascoyne RD, Rosen ST, Tobinai K. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756–6767. doi: 10.1182/blood-2010-05-231548. [DOI] [PubMed] [Google Scholar]
- 2.Bajor-Dattilo EB, Pittaluga S, Jaffe ES. Pathobiology of T-cell and NK-cell lymphomas. Best Pract Res Clin Haematol. 2013;26(1):75–87. doi: 10.1016/j.beha.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlin L, Coiffier B. The changing landscape of peripheral T-cell lymphoma in the era of novel therapies. Semin Hematol. 2014;51(1):25–34. doi: 10.1053/j.seminhematol.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Cortés JR, Palomero T. The curious origins of angioimmunoblastic T-cell lymphoma. Curr Opin Hematol. 2016;23(4):434–443. doi: 10.1097/MOH.0000000000000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstock DM, et al. A roadmap for discovery and translation in lymphoma. Blood. 2015;125(13):2175–2177. doi: 10.1182/blood-2015-01-623777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 7.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo HY, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371–375. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 9.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cortes JR, Ferrando A, Palomero T. Molecular pathogenesis of malignant lymphomas: PTCL, NOS AITL. In: Lenz G, Pasqualucci L, eds. Malignant Lymphomas: Biology and Molecular Pathogenesis. Berlin, Germany: De Gruyter ; 2016;16:311-22. [Google Scholar]
- 11.Jaffe ES, Nicolae A, Pittaluga S. Peripheral T-cell and NK-cell lymphomas in the WHO classification: pearls and pitfalls. Mod Pathol. 2013;26 Suppl 1:S71–S87. doi: 10.1038/modpathol.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 19.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23(7):1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 22.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojtek AB, Cooper JA. Rho family members: activators of MAP kinase cascades. Cell. 1995;82(4):527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 29.Cleverley SC, Costello PS, Henning SW, Cantrell DA. Loss of Rho function in the thymus is accompanied by the development of thymic lymphoma. Oncogene. 2000;19(1):13–20. doi: 10.1038/sj.onc.1203259. [DOI] [PubMed] [Google Scholar]
- 30.Tybulewicz VL, Henderson RB. Rho family GTPases and their regulators in lymphocytes. Nat Rev Immunol. 2009;9(9):630–644. doi: 10.1038/nri2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagata Y, et al. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood. 2016;127(5):596–604. doi: 10.1182/blood-2015-06-644948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheikha A, Aljanadi M, Alshehri M, Raziuddin S. Peripheral T-cell lymphomas - cytokine profile and characteristic interleukin-4 and tumor-necrosis-factor alpha-deficiency. Int J Oncol. 1994;4(1):91–95. doi: 10.3892/ijo.4.1.91. [DOI] [PubMed] [Google Scholar]
- 33.Muto H, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J. 2014;4:e264. doi: 10.1038/bcj.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waring P, Müllbacher A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol Cell Biol. 1999;77(4):312–317. doi: 10.1046/j.1440-1711.1999.00837.x. [DOI] [PubMed] [Google Scholar]
- 35.Ichiyama K, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42(4):613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448(7152):480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 37.Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241(1):260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue X, et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med. 2016;213(3):377–397. doi: 10.1084/jem.20151438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang R, et al. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity. 2015;43(2):251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasidharan Nair V, Song MH, Oh KI. Vitamin C Facilitates Demethylation of the Foxp3 Enhancer in a Tet-Dependent Manner. J Immunol. 2016;196(5):2119–2131. doi: 10.4049/jimmunol.1502352. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Konkel JE. TGF-beta and ‘adaptive’ Foxp3(+) regulatory T cells. J Mol Cell Biol. 2010;2(1):30–36. doi: 10.1093/jmcb/mjp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Leval L, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952–4963. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 43.Piccaluga PP, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res. 2007;67(22):10703–10710. doi: 10.1158/0008-5472.CAN-07-1708. [DOI] [PubMed] [Google Scholar]
- 44.Kataoka K, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 45.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30(3):358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33(6):890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouyang W, Li MO. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. 2011;32(1):26–33. doi: 10.1016/j.it.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du X, et al. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J Immunol. 2014;192(4):1525–1535. doi: 10.4049/jimmunol.1301060. [DOI] [PubMed] [Google Scholar]
- 49.Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48(9):1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scourzic L, et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia. 2016;30(6):1388–1398. doi: 10.1038/leu.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cutucache CE, Herek TA. Burrowing through the Heterogeneity: Review of Mouse Models of PTCL-NOS. Front Oncol. 2016;6:206. doi: 10.3389/fonc.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12(9):649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lainé A, et al. Foxo1 Is a T Cell-Intrinsic Inhibitor of the RORγt-Th17 Program. J Immunol. 2015;195(4):1791–1803. doi: 10.4049/jimmunol.1500849. [DOI] [PubMed] [Google Scholar]
- 55.Stone EL, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 2015;42(2):239–251. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun D, et al. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 2014;15(2):R38. doi: 10.1186/gb-2014-15-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.