Abstract
Several recent studies have provided insights into the genetic regulation of blood pressure. A new study extends these findings by coupling genome-wide association study data with functional validation approaches to identify and explore loci associated with blood pressure, generating a genetic risk score model to predict future cardiovascular risk
REFERS TO
Warren, H.R. et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. http://dx.doi.org/10.1038/ng.3768
Hypertension is the biggest single contributing factor to disease and mortality worldwide1,2 and is a primary modifiable risk factor for renal, cardiovascular and cerebrovascular disease3. Findings from a new study by Warren et al. 4 expand our current understanding of the genetic basis of blood pressure variation by coupling data from large-scale genome-wide association studies (GWAS) with biological insights into gene function and underlying cardiovascular risk. The generation of a prediction model could be useful to assess the risk of cardiovascular disease in patient cohorts To investigate the genetic basis of blood pressure variation, Warren et al. performed a The GWAS analysis of systolic blood pressure (SBP), diastolic blood pressure (DBP), and pulse pressure (PP) on >140,000 unrelated individuals of European ancestry. Following the analysis of genetic association with multiple-levels of validation, 107 loci were identified. Of these loci, 24 were associated with SBP, 41 associated with DBP, and 42 with PP; many of the loci were associated with more than one blood pressure-related trait. Among those identified, multiple loci were associated with genes known to function in blood pressure homeostasis including ACE (which encodes angiotensin-converting enzyme), CACNA2D2 (which encodes a voltage-dependent calcium channel auxiliary subunit), MME (which encodes metallo-endopeptidase/neutral endopeptidase), ADRA2B (which encodes the adrenergic β 2B receptor), and PDE5 (which encodes phosphodiesterase 5a). Moreover, 27 of the validated loci showed genome-wide association with other cardiovascular disease-associated traits including coronary artery disease and myocardial infarction as well as cardiovascular risk factors. Finally, an Ingenuity pathway analysis showed an enrichment of pathways implicated in cardiovascular disease, including the α adrenergic pathway, the CXCR4 chemokine signalling pathway, the endothelin system, and angiotensin-receptor pathways. The genetic association analysis thus identified genetic loci that are known to be associated with hypertension.
To complement the association analysis, functional and transcript expression data were used to demonstrate that genes associated with the validated loci are enriched in 31 tissue and cell types with the greatest enrichment in arteries. Further expression analysis of 78 of the curated genes demonstrated clustering in vascular smooth muscle cells, aortic fibroblasts, and endothelial cells, indicating that the majority of the genes identified are associated with tissues and cells that are important in cardiovascular regulation. When possible, expression data were corroborated with previously reported studies describing the importance of different genes in blood pressure regulation and other functional data.
Of interest, the researchers generated a genetic risk score to evaluate the impact of the combination of all 107 loci on blood-pressure level and the risk of hypertension. Application of the genetic risk score to an independent cohort of patients demonstrated that individuals >50 years of age in the lowest quintile of the risk scores had a SBP approximately 9–10 mmHg lower than those in the highest quintile. The ability to lowering blood pressure by 9–10 mmHg is possible through dietary modifications alone5, and reductions in blood pressure of a similar magnitude have been shown to significantly reduce cardiovascular morbidity and mortality in hypertensive patients6. The assessment of a genetic risk score, utilizing these and other hypertension-associated loci, could therefore be useful in early life to predict the development of hypertension-associated disease and direct dietary or lifestyle modifications, which could minimize the development of disease in later life. Similarly, the genetic risk score was associated with increased risk of stroke, coronary heart disease, and all cardiovascular outcomes
Despite these encouraging findings, the value of DNA sequence alone for precision medicine in hypertension and other common and complex cardiovascular and renal diseases is likely limited7. These diseases are heavily influenced by environmental factors including lifestyle. Unfortunately, environmental factors are difficult to quantify or even identify, and the clinical or biological significance of the numerous DNA sequence variations associated with these diseases remains uncertain in most cases and likely involves interaction with environmental factors (FIG. 1). Analysis of the products of interactions between the genome and the environment in tissues relevant to the disease could be highly valuable in overcoming these limitations (FIG. 1). Functional readouts from interactions between the genome and environment could take the form of epigenetically modified DNA, changes in the profiles of RNA, proteins and small metabolites, and changes in physiological function 7. The functional and expression analysis for select loci, including the analysis of histone marks and long-range chromatin interactions, as reported by Warren and colleagues4 is a useful step in this direction. A concerted effort to systematically analyze the functional genomic landscape in tissues directly relevant to the disease and in patients that are deeply and rigorously phenotyped will be essential for advancing the precision medicine in hypertension and other common and complex cardiovascular and renal diseases.
Figure 1.
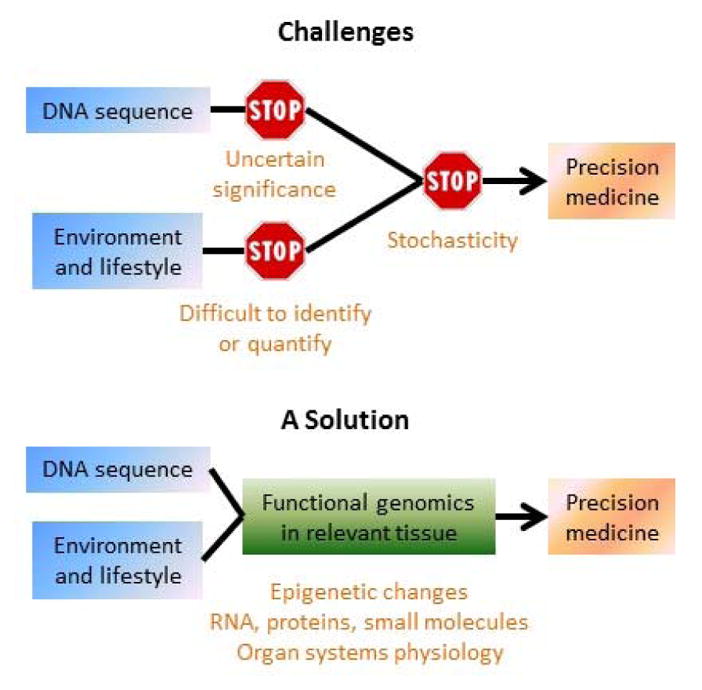
Functional genomics in disease-relevant tissues is essential for advancing the precision medicine for common and complex diseases.
This new report by Warren and colleagues has therefore identified a number of previously unrecognized genetic loci associated with hypertension and validated a number of others. Several recent association analyses, performed on similar-sized samples or even larger numbers of subjects, have also provided useful insight into blood pressure regulation and demonstrated new genetic variants important in hypertension8,9,10. The work by Warren et al.4 coupled observations from the genetic association arm of the study with functional and expression data to provide greater relevance to the observed genetic associations. Moreover, the generation of a genetic risk score and its application to an independent group of individuals demonstrates the potential application of identified genetic loci for use in a precision medicine approach that could be used to beneficially impact public health.
Acknowledgments
D.L.M.’s research is supported by NIH Grants DK96859 and HL116264 and AHA 15SFRN2391002. M.L.’s research is supported by NIH Grants HL116264, HL121233, HL125409, GM066730, HL082798 and AHA 15SFRN2391002.
Footnotes
Competing interests
The authors declare no competing interests
Bibliography
- 1.Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–201: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulter NR, Prabhakaran D, Caulfield M. Hypertension. Lancet. 2015;386:801–812. doi: 10.1016/S0140-6736(14)61468-9. [DOI] [PubMed] [Google Scholar]
- 3.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 4.Warren HR, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017 doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacks FM, et al. Effects on blood pressure of reduced dieeary sodium and the dietary approaches to stop hypertension (DASH) diet. N Eng J Med. 2001;344:3–10. doi: 10.1056/NEJM200101043440101. [DOI] [PubMed] [Google Scholar]
- 6.The SPRINT Research Group. A randomized trial of intensive versus standard blood-pressure control. N Eng J Med. 2015;373:2103–2116. doi: 10.1056/NEJMoa1511939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotchen TA, Cowley AW, Jr, Liang M. Ushering Hypertension into a New Era of Precision Medicine. JAMA. 2016;315:343–344. doi: 10.1001/jama.2015.18359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surendran P, et al. Trans-ancestry meta-analysis identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehret GB, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]


