Abstract
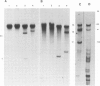
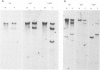
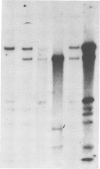
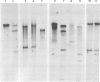
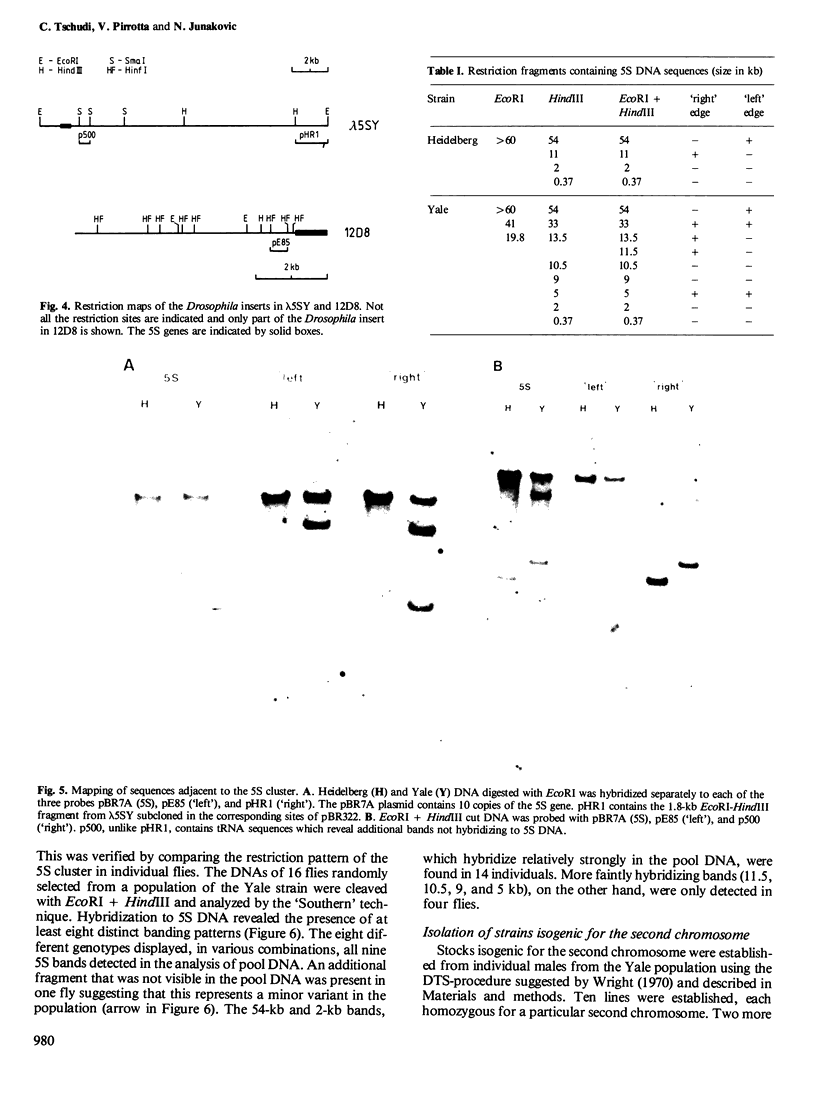
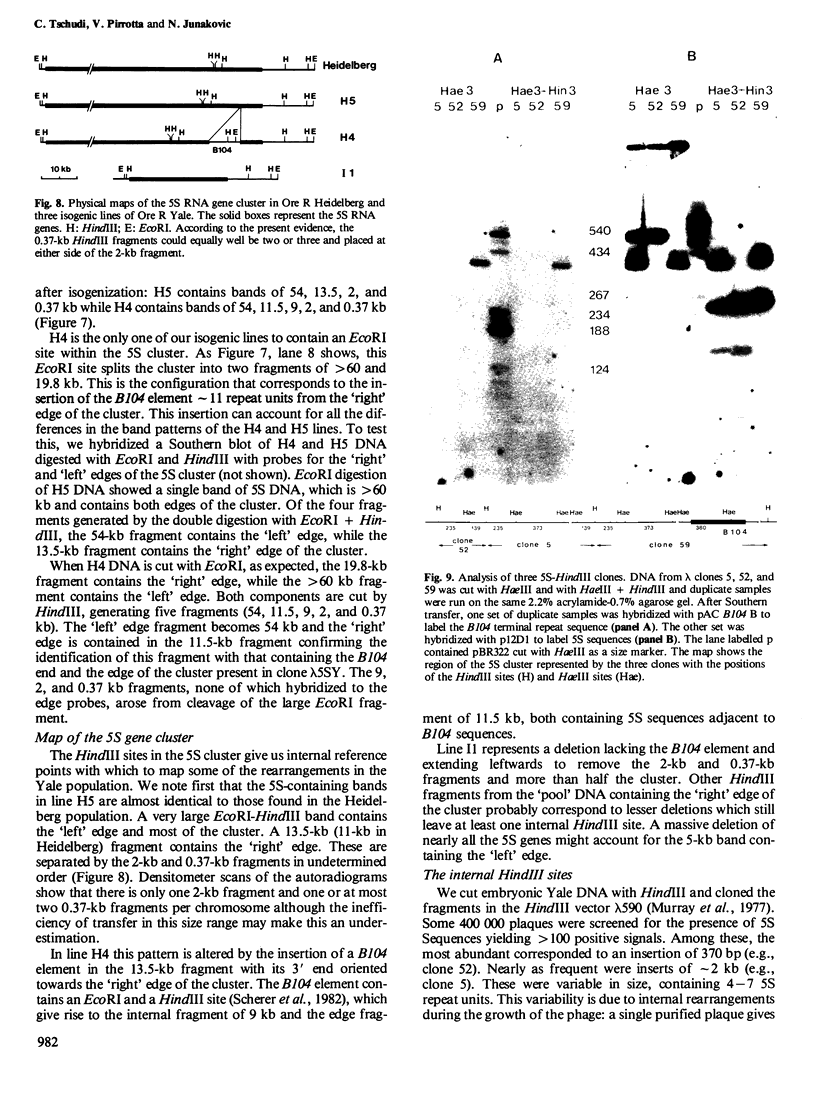
The organization of the 5S RNA gene cluster of Drosophila melanogaster is different in two Oregon R stocks that have been separated for a number of years. The Oregon R Yale population contains various different arrangements of the cluster. One of these is due to the insertion of a B104 element near one end of the cluster. Other arrangements lack the B104 insertion and have instead a variety of deletions originating in the vicinity of the B104 insertion site and removing from 0 to 60% of the 5S RNA genes without affecting nearby tRNA genes. In contrast, the Oregon R Heidelberg population has no B104 element in the 5S gene cluster and no heterogeneity in the arrangement of the cluster. We propose that transposable elements inserted at a genomic locus generate heterogeneity in a population at that locus due to excision of the element with and without accompanying deletions of flanking sequences. As a consequence, a fly population would accumulate a large number of deletions scattered throughout the genome in as many loci as contain transposable elements. We show further that D. melanogaster contains a large redundancy of 5S RNA genes since the 60% deletion of the cluster shows no visible phenotype when homozygous or when heterozygous against a total deletion of the entire 5S gene cluster.
Keywords: Drosophila, 5S RNA genes, transposable elements, deletion formation
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artavanis-Tsakonas S., Schedl P., Tschudi C., Pirrotta V., Steward R., Gehring W. J. The 5S genes of Drosophila melanogaster. Cell. 1977 Dec;12(4):1057–1067. doi: 10.1016/0092-8674(77)90169-6. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Green M. M. Controlling element mediated transpositions of the white gene in Drosophila melanogaster. Genetics. 1969 Feb;61(2):429–441. doi: 10.1093/genetics/61.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. M. The genetics of a mutable gene at the white locus of Drosophila melanogaster. Genetics. 1967 Jul;56(3):467–482. doi: 10.1093/genetics/56.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Addison W. R., Gillam I. C., Grigliatti T. A., Tener G. M. Hybridization of tRNAs of Drosophila melanogaster to the region of the 5S RNA genes of the polytene chromosomes. Chromosoma. 1981;82(3):385–397. doi: 10.1007/BF00285764. [DOI] [PubMed] [Google Scholar]
- Hershey N. D., Davidson N. Two drosophila melanogaster tRNAGly genes are contained in a direct duplication at chromosomal locus 56F. Nucleic Acids Res. 1980 Nov 11;8(21):4899–4910. doi: 10.1093/nar/8.21.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovemann B., Sharp S., Yamada H., Söll D. Analysis of a drosophila tRNA gene cluster. Cell. 1980 Apr;19(4):889–895. doi: 10.1016/0092-8674(80)90080-x. [DOI] [PubMed] [Google Scholar]
- Junakovic N. Variability in the molecular organization of the 5S RNA genes among strains of Drosophila melanogaster. Nucleic Acids Res. 1980 Aug 25;8(16):3611–3622. doi: 10.1093/nar/8.16.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli E., Schmidt T. The localization of tRNA4Glu genes from Drosophila melanogaster by "in situ" hybridization. Nucleic Acids Res. 1978 May;5(5):1465–1478. doi: 10.1093/nar/5.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Procunier J. D., Dunn R. J. Genetic and molecular organization of the 5S locus and mutants in D. melanogaster. Cell. 1978 Nov;15(3):1087–1093. doi: 10.1016/0092-8674(78)90292-1. [DOI] [PubMed] [Google Scholar]
- Saigo K., Millstein L., Thomas C. A., Jr The organization of Drosophila melanogaster histone genes. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):815–827. doi: 10.1101/sqb.1981.045.01.100. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Tschudi C., Pirrotta V. Sequence and heterogeneity in the 5S RNA gene cluster of Drosophila melanogaster. Nucleic Acids Res. 1980 Feb 11;8(3):441–451. doi: 10.1093/nar/8.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimber D. E., Steffensen D. M. Localization of 5S RNA genes on Drosophila chromosomes by RNA-DNA hybridization. Science. 1970 Nov 6;170(3958):639–641. doi: 10.1126/science.170.3958.639. [DOI] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]