Abstract
Galactose and N-acetyl-D-galactosamine-inhibitable lectin of Entamoeba histolytica has roles in pathogenicity and induction of protective immunity in rodent models of amoebiasis. Recently, the intermediate subunit of the lectin, Igl1, of E. histolytica has been shown to have hemolytic activity. However, the corresponding lectin is also expressed in a non-virulent species, Entamoeba dispar, and another subunit, Igl2, is expressed in the protozoa. Therefore, in this study, we compared the activities of Igl1 and Igl2 subunits from E. histolytica and E. dispar using various regions of recombinant Igl proteins expressed in Escherichia coli. The recombinant E. dispar Igl proteins had comparable hemolytic activities with those of E. histolytica Igl proteins. Furthermore, Igl1 gene-silenced E. histolytica trophozoites showed less hemolytic activity compared with vector-transfected trophozoites, indicating that the expression level of Igl1 protein influences the activity. These results suggest that the lower hemolytic activity in E. dispar compared with E. histolytica reflects the lower expression level of Igl1 in the E. dispar parasite.
Introduction
Amoebiasis due to infection with Entamoeba histolytica (E. histolytica) is a problematic parasitic disease in many countries. E. histolytica causes an estimated 50 million cases of dysentery, colitis and extraintestinal abscesses, resulting in 40,000 to 100,000 deaths annually [1]. Adherence of E. histolytica trophozoites to colonic mucins and host cells is mediated by a galactose (Gal)- and N-acetyl-D-galactosamine (GalNAc)-inhibitable lectin [2]. This lectin is a 260-kDa heterodimer of two glycoproteins: a transmembrane heavy subunit (Hgl, 170 kDa), one of the key molecules in amebic adherence, and a glycosylphosphatidylinositol (GPI)-anchored light subunit (Lgl, 35/31 kDa).
Another GPI-anchored 150-kDa intermediate subunit (Igl, 150 kDa) is non-covalently associated with the Hgl/Lgl dimer in different lipid raft-like domains and also contributes to adherence [3–5]. There are two isoforms of Igl, which are referred to as Igl1 and Igl2, and both contain multiple CXXC motifs with different localization in E. histolytica trophozoites [6, 7]. These two Igls are also found in Entamoeba dispar (E. dispar), which is morphologically indistinguishable from E. histolytica, but is non-pathogenic [8]. The expression level of Igl1 is about twice as high in E. histolytica HM-1:IMSS than in E. dispar SAW1734RclAR, whereas that of Igl2 is comparable in the two species, suggesting that Igl1 may be associated with the pathogenicity of E. histolytica [8]. In fact, Igl1 is recognized by sera from patients with amoebiasis and is also a vaccine candidate [9, 10].
Igl is a parasitic lectin that binds to p-aminophenyl-β-D-galactopyranoside-Sepharose gel in a Gal-affinity column [5]. E. histolytica Igl has also been detected, in addition to Hgl and Lgl, in the protein fraction that binds to GalNAc bovine serum albumin-coated magnetic beads [11]. Recently, while exploring the lectin domain of Igl, we found that Igl1 of E. histolytica possesses both hemolytic and cytotoxic activities [12]. However, it is unclear whether Igl1 of E. dispar and Igl2 of both species have the same activity. Therefore, in this study, we compared the hemolytic activities of E. dispar Igls with those of E. histolytica Igls in vitro. We also attenuated expression of Igl1 in E. histolytica utilizing a gene-silencing technique and evaluated the effect on hemolytic activity, since E. dispar has lower expression of Igl1 compared with E. histolytica [8].
Materials and methods
Expression and refolding of recombinant Igl proteins and Ni column purification of the proteins
Recombinant EhF-Igls, EdF-Igls, EhNM-Igl1, EhM-Igl1, EhC-Igl1 or EdC-Igl1 proteins with a His-tag at the N-terminus were expressed in Escherichia coli BL21 Star(DE3)pLysS cells (Invitrogen) or ECOS™ competent BL21(DE3) cells (Nippon Gene Co.), using the primers shown in Table 1. The proteins were further purified using a Ni column, as described in detail previously [9, 12].
Table 1. Oligonucleotide primers used in the study.
| Primer | Positiona | Sequence (5′ to 3′)b | Ref. |
|---|---|---|---|
| (for Eh Igls) | |||
| EhIgl-S14 | 40–59 | CCCTCGAGGATTATACTGCTGATAAGCT | [9, 12] |
| EhIgl2-S14 | 40–70 | CCCTCGAGGATTATACTGCTGATAAACTCATTAATAACC | [7] |
| EhIgl-S294 | 880–898 | CCCTCGAGACAGAAGAAAATAAATGTA | [9, 12] |
| EhIgl-S603 | 1807–1827 | CCCTCGAGGAAGGACCAAATGCAGAAGAT | [9, 12] |
| EhIgl-AS753 | 2244–2259 | CCCTCGAGTTATAGCCTTTGTTCAGTG | [9, 12] |
| EhIgl-AS1088c | 3247–3264 | CCCTCGAGTTAAATGCCTTTAGCTCCATT | [9, 12] |
| (for Ed Igls) | |||
| EdIgl-S14 | 40–59 | CCCTCGAGGAGTACAAAGCTGATAAACT | [8] |
| EdIgl2-S14 | 40–63 | CCCTCGAGGATTACAAAGCTGATAAACTCATC | [8] |
| EdIgl-S604 | 1810–1830 | CCCTCGAGGAAGGACCAAATGAAGAAGAT | * |
| EdIgl-AS1097c | 3274–3291 | CCCTCGAGTTAAATTCCTTTACTTCCATT | [8] |
a Nucleic acid numbering is based on the E. histolytica Igl1 and Igl2 gene sequences (AF337950 and XM_647302) and E. dispar Igl1 and Igl2 gene sequences (AB287423 and AB287424).
b Nucleotides added for cloning and translation termination are underlined.
c EhIgl-AS1088 and EdIgl-AS1097 are common for EhIgl2 and EdIgl2 respectively.
* This study.
SDS-PAGE and coomassie brilliant blue staining of purified recombinant proteins
Recombinant proteins (1 μg each) were mixed with SDS sample buffer (Invitrogen) and subjected to SDS-PAGE. The gel was treated with SimplyBlue Safe stain solution (Invitrogen) and incubated until blue bands appeared on the gel [12].
Hemolytic assays using recombinant lectins and measurement of released hemoglobin
Hemolytic assays and quantification of hemolytic activity were conducted as previously described [12]. Briefly, recombinant Igls (2 μM each, 50 μl) were mixed with 50 μl of horse red blood cell (HoRBC) solution at room temperature and images were taken at several time points. A Hemoglobin B Test Kit (Wako, Osaka, Japan) was used to measure the concentration of hemoglobin in supernatants of RBCs incubated with recombinant proteins or trophozoites for 8 h or 1 h. The results are expressed as the mean of 5 experiments with SD.
Culturing Entamoeba trophozoites
Trophozoites of E. histolytica HM-1:IMSS G3 [13] strain were cultivated axenically in Diamond BI-S-33 medium [14] and used for generating Igl gene-silenced trophozoites. Trophozoites of E. dispar SAW1734RclAR strain were grown monoxenically with Pseudomonas aeruginosa or with Crithidia fasciculata and trophozoites of E. dispar CYNO9:TPC strain were axenically cultured in YIGADHA-S medium [15].
Preparation of Igl1 gene-silenced Entamoeba histolytica trophozoites
Isolation of total RNA and mRNA from trophozoites and cDNA synthesis were performed as previously described [16]. For silencing of the Igl1 gene, the DNA fragment from 156- to 408-nt (gsIgl1A strain) or from 1- to 466-nt (gsIgl1B strain) in Igl1 was PCR-amplified from cDNA using Phusion DNA polymerase (New England Biolabs) and specific primer sets (gsIgl1A strain: sense, 5′-CGA GGC CTC ACT GGA AAT AAT AAG ACA TG-3′; antisense, 5′-GTC GGA GCT CAC CAT CAA CAG TAG TAG ACA TC-3′, gsIgl1B strain: sense, 5′-CGA GGC CTC ATG TTT ATT CTT CTT TTA TTC ATA TC-3′; antisense, 5′-GTC GGA GCT CGA CCA ACA CAA TTT TCT GCA TG-3′, containing Stu I and Sac I recognition sites, respectively). The fragments were digested with Stu I and Sac I and ligated into a Stu I/Sac I double-digested psAP-2-Gunma plasmid [17] using a Ligation-Convenience Kit (Nippon Gene Co., Tokyo, Japan). Lipofection of trophozoites and selection and maintenance of transformants were performed as previously described [18].
Real-time PCR analysis
Real-time PCR was essentially performed as previously described [8]. Briefly, total RNAs of E. histolytica and E. dispar trophozoites isolated using a RNeasy Plus Mini Kit (Qiagen) were used for cDNA synthesis with an ExScript™ RT Reagent Kit (Takara). Reaction mixtures for quantitative real-time PCR analysis were prepared using SYBR Premix Ex Taq II (Takara), specific primers, Rox dye, and the cDNAs. Forty cycles of amplification with recording of fluorescence intensity in each cycle were performed using StepOnePlus™ Real-Time PCR System (ABI). Expression levels of Igl genes were analyzed using the comparative CT method with actin as an internal standard. The experiments were repeated 3 times, including the steps of culture and isolation of RNA.
Antibodies
Purified human mAb XEhI-20 (anti-EhIgl1) and XEhI-B5 (anti-EhIgl2) were used for detection of Igls of E. histolytica [7]. Mouse mAb ED2-495 against recombinant Igl2 of E. dispar was obtained as described previously [8] and characterized before use. Mouse mAb ED2-1 specific for Igl2 of E. dispar was used as a control [8]. Pooled ascites rich in mouse mAbs were used in the study.
Dot blot analysis
Recombinant Igls (300 ng) were blotted on a nitrocellulose membrane and then air-dried. Filter strips were blocked with 3% bovine serum albumin in PBS and reacted with ED2-1 or ED2-495 for 30 min. After washing with PBS containing 0.05% Tween-20 (PBST), the strips were incubated with horseradish peroxidase (HRP)-labeled goat anti-mouse IgG antibody (MP Biomedicals) for 30 min. The strips were then washed with PBST and developed with a Konica Immunostaining HRP-1000 kit.
Immunoblot assay
Whole cell lysates (15 or 20 μg protein/well) were applied to a 5–20% gradient polyacrylamide gel (Atto Corp., Tokyo, Japan) and SDS-PAGE was conducted under reducing or non-reducing conditions, respectively. The proteins in the gel were transferred onto an Amersham™ Hybond™ P 0.45 PVDF membrane (GE Healthcare) that was then incubated with 5% skim milk in PBST for blocking. Mouse ascites (ED2-495) against E. histolytica Igls was diluted 500 times with PBST containing 5% skim milk. Rabbit antiserum against ATP sulfurylase (AS) of E. histolytica was prepared [16] and diluted 500-fold with 5% skim milk in PBST. Anti-mouse and anti-rabbit immunoglobulin F(ab’)2 fragments conjugated with HRP (Amersham) were diluted 3000 times with PBST and used as the secondary antibody. Immunoblot assays with XEhI-20 or XEhI-B5 [7] were performed using SDS-PAGE in a 5–20% gradient gel under non-reducing conditions, with 30 μg of XEhI-20 or XEhI-B5 used as the primary antibody and HRP-labeled goat anti-human IgG antibody (MP Biomedicals) diluted 1000 times with PBST as the secondary antibody. Immobilon™ Western (Millipore) was used as a substrate for visualization of the proteins. Detection of chemiluminescence and quantification of band intensities were performed by Ez-Capture MG and CS Analyzer ver. 3.0 (Atto Corp.), respectively.
Immunofluorescence assay
Sample preparation for IFA was performed as previously described [18, 19]. Briefly, after amoeba transformants were incubated on 5-mm round wells on glass slides, the cells were fixed with 4% paraformaldehyde in PBS for 10 min, washed four times with PBS, and permeabilized with 0.05% Triton X-100 in PBS for 5 min. After blocking with 3% bovine serum albumin in PBS, samples were reacted with ED2-495 (mouse IgG), XEhI-20 (human IgG), or XEhI-B5 (human IgG) diluted 1:50 in PBS and subsequently reacted with secondary antibody diluted 1:500 (Alexa Fluor® 488 goat anti-mouse or human IgG; Life Technologies) in PBS. Fluorescence images were obtained using a LSM510 Meta confocal Microscope (Zeiss) in lambda emission fingerprinting mode [20, 21].
Hemolytic assay using Entamoeba histolytica and Entamoeba dispar trophozoites
The assay was conducted as previously described [12, 22] with slight modifications. Briefly, vector-transfected (control) or Igl1 gene-silenced (gsIgl1A or gsIgl1B) E. histolytica or wild-type E. dispar (SAW1734RclAR strain or CYNO9:TPC strain) trophozoites prepared as described above were harvested and washed with PBS. Then 1×105 trophozoites were mixed with 1% HoRBCs in 100 μl PBS and incubated at 37°C for 1 h. The cell suspension was sedimented at 2000 rpm for 5 min and the concentration of hemoglobin in the supernatant was determined as described above.
Statistical analysis
Multiple comparisons were performed by ANOVA with a Dunn test, with P < 0.05 considered significant.
Results
Recombinant Igls
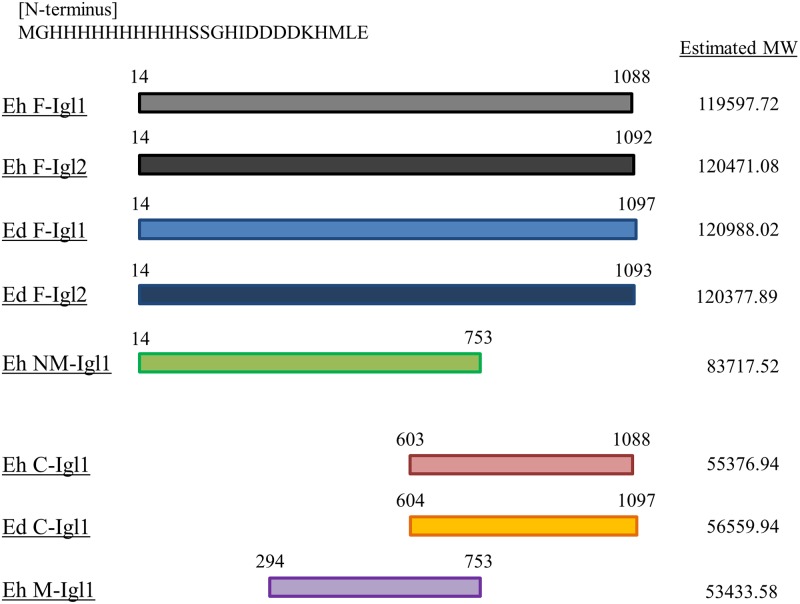
Full-length (EhF-Igl1: aa 14 to 1088 of E. histolytica Igl1, EhF-Igl2: aa 14 to 1092 of E. histolytica Igl2, EdF-Igl1: aa 14 to 1097 of E. dispar Igl1, EdF-Igl2: aa 14 to 1093 of E. dispar Igl2), N-terminal and middle (NM-Igl: aa 14 to 753 of E. histolytica Igl1), middle (M-Igl: aa 294 to 753 of E. histolytica Igl1), and C-terminal (C-Igl: aa 603 to 1088 of E. histolytica Igl1 and aa 604 to 1097 of E. dispar Igl1) regions of E. histolytica (Eh) and E. dispar (Ed) Igls with a His-tag at the N-terminus (Fig 1) were expressed in E. coli [12]. Recombinant proteins were purified using Ni columns, the buffer was changed to PBST, and purities were confirmed by SDS-PAGE (Figs 2A and 3A). The recombinant proteins were then used in further studies.
Fig 1. Recombinant Igl proteins used in the study.
Recombinant Igl proteins were constructed with a His-tag at the N-terminus. Full length (F-Igl), N-terminus and middle (NM-Igl), middle (M-Igl), and C-terminus (C-Igl) Igl1 and Igl2 of Entamoeba histolytica (Eh) and Entamoeba dispar (Ed) were expressed in E. coli and purified using Ni columns. Estimated molecular weights of each protein including the His-tag are shown [ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/)].
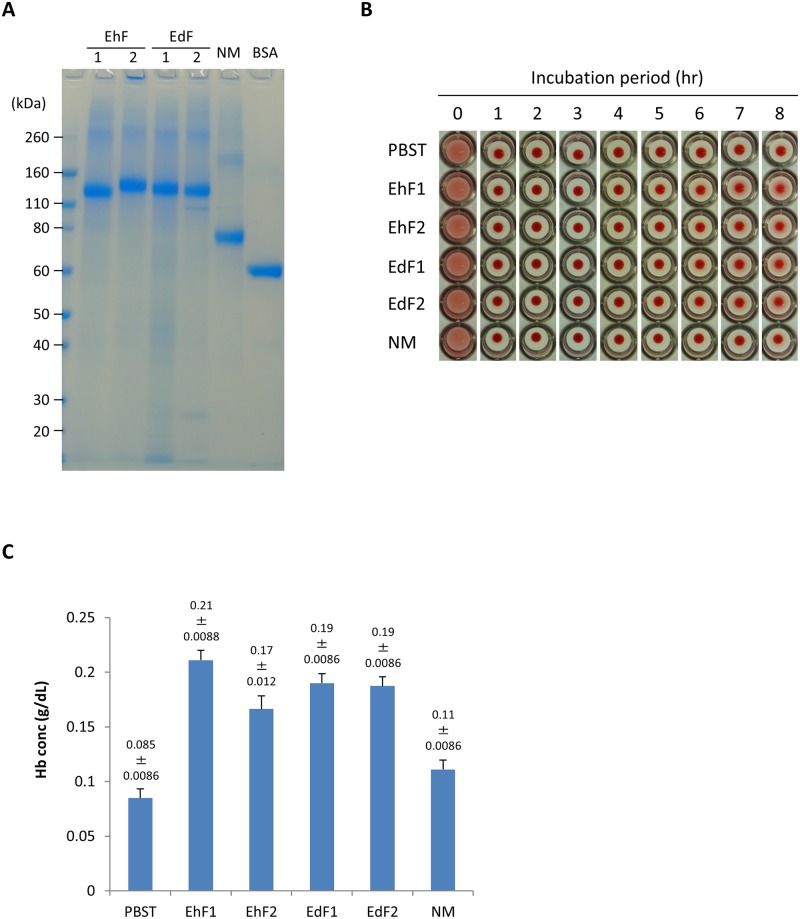
Fig 2. Time-course of hemolytic activities of EhF-Igl1, EhF-Igl2, EdF-Igl1, EdF-Igl2 and EhNM-Igl1 proteins.
Recombinant Igl proteins (2 μM, 50 μl) were incubated with 50 μl of 2% (v/v) HoRBCs in PBS for the indicated periods. A. Protein purity and amount were confirmed by SDS-PAGE using NuPAGE Novex Bis-Tris (4–12% gradient) gels with 1 μg of each protein. B. HoRBCs were incubated in a 96-well plate after the indicated periods with Igls. Representative images of 5 independent studies are shown. C. Concentrations of hemoglobin (Hb) released in the supernatant of samples incubated for 8 h. Data are the mean ± SD from 5 independent experiments. EhF-Igls and EdF-Igls showed significantly (**p< 0.01 by ANOVA with Dunn test) higher hemolytic activities than EhNM-Igl1 and PBST. EhF1: EhF-Igl1, EhF2: EhF-Igl2, EdF1: EdF-Igl1, EdF2: EdF-Igl2, NM: EhNM-Igl1.
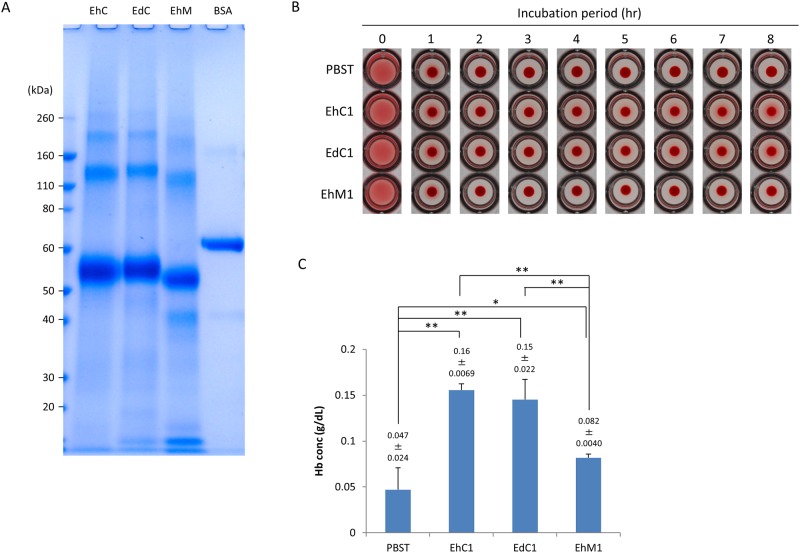
Fig 3. Time-course of hemolytic activities of EhC-Igl1, EdC-Igl1 and EhM-Igl1 proteins.
Recombinant Igl proteins (2 μM, 50 μl) were incubated with 50 μl of 2% (v/v) HoRBCs in PBS for the indicated periods. A. Protein purity and amount were confirmed by SDS-PAGE using NuPAGE Novex Bis-Tris (4–12% gradient) gels with 1 μg of each protein. B. HoRBCs were incubated in a 96-well plate after the indicated periods with Igls. Representative images of 5 independent studies are shown. C. Concentrations of hemoglobin (Hb) released in the supernatant of samples incubated for 8 h. Data are the mean ± SD from 5 independent experiments. *p< 0.05, **p< 0.01 by ANOVA with Dunn test. EhC1: EhC-Igl1, EdC1: EdC-Igl1, EhM1: EhM-Igl1.
Hemolytic activities of recombinant proteins against horse red blood cells (HoRBCs)
We recently showed that EhF-Igl1 has hemolytic activity [12]. EhF-Igl1 and EhF-Igl2 have 83–84% amino acid sequence identity. EdF-Igl1 has 75–76% amino acid sequence identity with EhF-Igl1, and EdF-Igl2 has 73–74% amino acid sequence identity with EhF-Igl2 [8]. To evaluate whether EhF-Igl2, EdF-Igl1 and EdF-Igl2 also have hemolytic activity, HoRBCs (2% v/v) in PBS were mixed with EhF-Igl1, EhF-Igl2, EdF-Igl1, EdF-Igl2 or EhNM-Igl1 (Fig 2B). EhNM-Igl1 was used as a low activity control because it has less hemolytic activity than EhC-Igl1, but a similar molecular weight to EhF-Igls and EdF-Igls [12]. Samples were mixed in U-bottom 96-well plates and incubated at room temperature for up to 8 h to evaluate the hemolytic activities (Fig 2B) based on the concentration of released hemoglobin [Hb] in the supernatant after 8 h (Fig 2C). EhF-Igls and EdF-Igls had significantly higher hemolytic activities than PBST ([Hb] 0.085±0.0086 g dL-1) and EhNM-Igl1 (0.11±0.0086 g dL-1).
The hemolytic activity of EhF-Igl1 resides in the C-terminus of the protein [12]. To assess whether the C-terminus of EdF-Igl1 also has this activity, we conducted an assay of EhC-Igl1 and EdC-Igl1 (Fig 3), using EhM-Igl1 as a weakly active control. EdC-Igl1 (0.15±0.022 g dL-1) and EhC-Igl1 (0.16±0.0069 g dL-1) had equivalent activity (Fig 3C), and EhM-Igl1 showed slightly higher activity (0.082±0.0040 g dL-1) than PBST (0.047±0.024 g dL-1) in an ANOVA test. These results show that EdF-Igl1 has similar hemolytic activity to that of EhF-Igl1 and that the C-terminus of EdF-Igl1 has a role in this activity.
Hemolytic activities of Entamoeba dispar strains
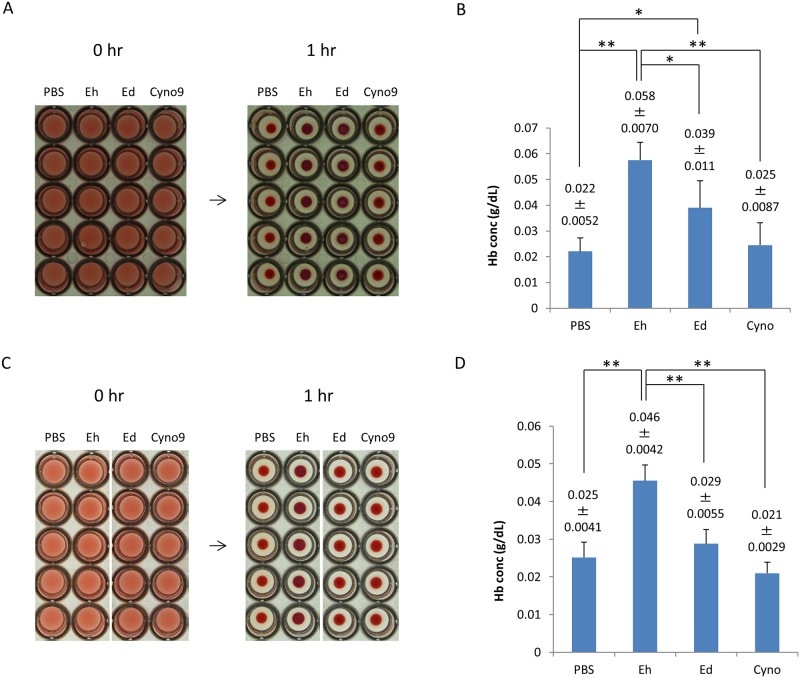
We have reported that E. histolytica trophozoites have hemolytic activity and that the activity can be blocked by an antibody recognizing M/C-Igl of EhIgl1 [12]. To assess whether E. dispar trophozoites also have this activity, the trophozoites were incubated with HoRBCs for 1 hr. The trophozoites of E. dispar SAW1734RclAR strain were cultured with Pseudomonas aeruginosa or with Crithidia fasciculata monoxenically because they were difficult to culture axenically. Trophozoites of another E. dispar strain, CYNO9:TPC, were able to be cultured axenically and the hemolytic activity of this strain was also assessed. As shown in Fig 4, SAW1734RclAR trophozoites cultured with Pseudomonas aeruginosa had hemolytic activity (Fig 4A and 4B) while trophozoites of the same strain cultured with Crithidia fasciculata and CYNO9:TPC strain did not have activity under the test conditions. The activity in SAW1734RclAR trophozoites cultured with Pseudomonas aeruginosa is due to contamination of hemolysin expressed in Pseudomonas aeruginosa [23].
Fig 4. Hemolytic activities of trophozoites of Entamoeba dispar strains.
A and B. E. histolytica (Eh), E. dispar SAW1734RclAR cultured with Pseudomonas aeruginosa (Ed) or E. dispar CYNO9:TPC (Cyno9) trophozoites (1×105) were incubated with HoRBCs at 37°C for 1 h. A. Images of HoRBCs in a 96-well plate just after mixing with trophozoites (0 hr) and after incubation for 1 h with trophozoites (1 hr). B. Released hemoglobin (Hb) concentration in the supernatant of the mixture of trophozoites and HoRBCs after incubation for 1 h at 37°C. C and D. E. histolytica (Eh), E. dispar SAW1734RclAR cultured with Crithidia fasciculata (Ed) or E. dispar CYNO9:TPC (Cyno9) trophozoites (1×105) were incubated with HoRBCs at 37°C for 1 h. C. Images of HoRBCs in a 96-well plate just after mixing with trophozoites (0 hr) and after incubation for 1 h with trophozoites (1 hr). D. Released hemoglobin (Hb) concentration in the supernatant of the mixture of trophozoites and HoRBCs after incubation for 1 h at 37°C. Data are the mean ± SD from 5 independent experiments. **p< 0.01, *p< 0.05 by ANOVA with Dunn test.
Hemolytic activity of Igl1 gene-silenced Entamoeba histolytica
Recombinant Igl proteins from E. histolytica and E. dispar showed similar hemolytic activities. However, expression of Igl1 in trophozoites of E. histolytica is about twice as high as that in E. dispar trophozoites [8]. At the same time, E. dispar trophozoites did not have hemolytic activity (Fig 4). To evaluate whether the expression level of Igl1 affects hemolytic activity, we generated Igl1 gene-silenced (gsIgl1) E. histolytica strains and conducted the hemolytic assay.
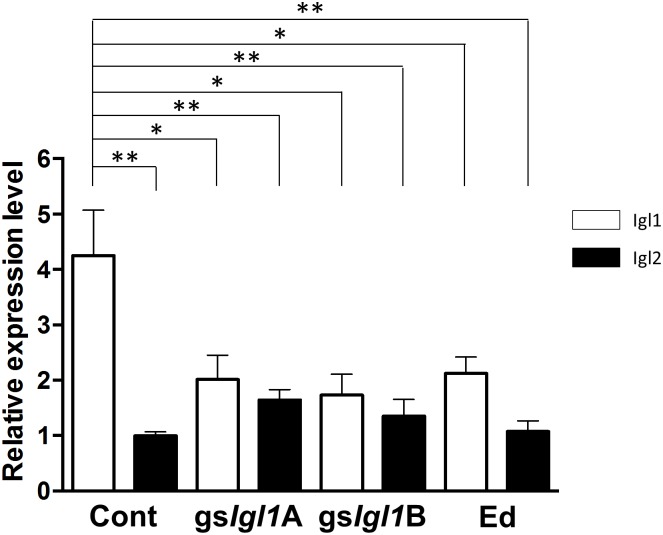
Expression levels of Igl1 and Igl2 in the gene-silenced E. histolytica trophozoites were evaluated quantitatively by real-time PCR (Fig 5). E. dispar trophozoites expressed about a half the level of Igl1 compared with E. histolytica trophozoites (Cont), as described previously [8]. Both Igl1 gene-silenced E. histolytica strains (gsIgl1A and gsIgl1B) showed significantly lower expression of Igl1, but not Igl2, compared with vector control trophozoites (Fig 5). We could not generate an Igl2 gene-silenced E. histolytica strain for unknown reason. Interestingly, Igl2 expressions in Igl1-silenced E. histolytica strains were rather high compared with that in vector control trophozoites (Fig 5).
Fig 5. Real-time PCR analysis of Igl genes from E. histolytica and E. dispar.
Expression levels of Igl1 (open bars) and Igl2 (filled bars) in trophozoites from E. histolytica strain with an empty vector (Cont), E. histolytica gsIgl1 strains (gsIgl1A and gsIgl1B) and E. dispar SAW1734RclAR strain (Ed) were compared using actin as an internal standard. Expression levels are shown as values relative to the mean expression level of Igl2 from Cont. Vertical bars indicate the S.E. of the mean from 3 experiments. *p< 0.05, **p< 0.01 by ANOVA with Dunn test.
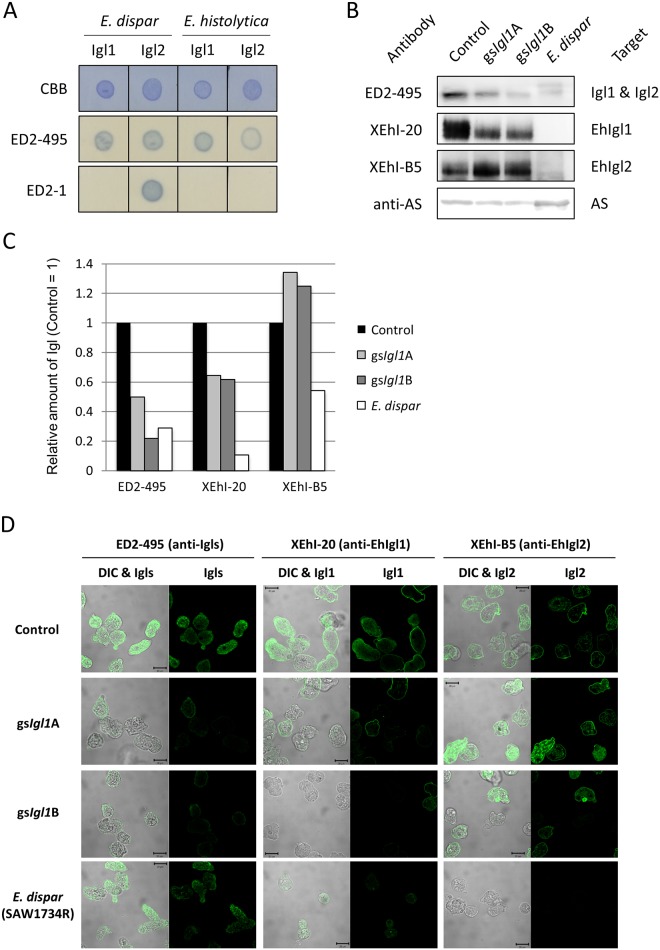
Downregulation of Igl1 was further confirmed by Western blotting and cell staining (Fig 6). Western blotting by mAb ED2-495, which recognizes both Igl1 and Igl2 of E. histolytica and E. dispar (Fig 6A), showed 50–80% reduction of Igl expression in gsIgl1A and gsIgl1B strains, with this reduction due mainly to downregulation of Igl1 (Fig 6B and 6C). Expression levels of Igl2 in gsIgl1A and gsIgl1B strains were higher than in vector control trophozoites, in agreement with the real-time PCR results shown in Fig 5. Downregulation of Igl1 in gsIgl1 strains was also confirmed using an immunofluorescence assay (Fig 6D). These results indicate that establishment of the gsIgl1 strains was successful.
Fig 6. Establishment of Igl-attenuated Entamoeba histolytica trophozoites.
A. Dot blot analysis of reactivity of ED2-495 against E. histolytica and E. dispar Igls. B. Western blot of Igls in control, gene-silenced E. histolytica and E. dispar trophozoites. C. Relative quantification of Igls expressed in control, gene-silenced E. histolytica and E. dispar trophozoites. D. Suppression of Igl1 protein expression by gene-silencing confirmed by IFA. Control: vector transfected E. histolytica trophozoite, gsIgl1: Igl1 gene-silenced E. histolytica trophozoite, AS: ATP sulfurylase, DIC: differential interference contrast. Amino acid sequence alignment of ATP sulfurylase between E. histolytica and E. dispar is shown in S1 Fig.
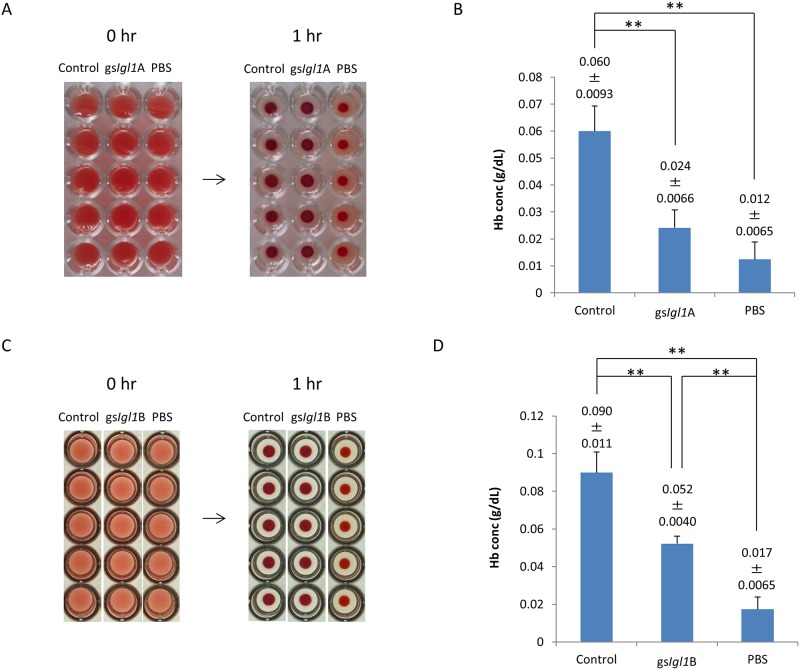
After incubation of vector control or gsIgl1A trophozoites with HoRBCs for 1 h, the rim of the accumulated samples became vague in treatment with control trophozoites compared with gsIgl1A-treated samples (Fig 7A). For quantitative evaluation, supernatants of the incubated samples were collected and assayed for released Hb (Fig 7B). The hemolytic activity of gsIgl1A trophozoites (0.024±0.0066 g dL-1) was significantly lower than that of control trophozoites (0.060±0.0093 g dL-1). PBST treatment gave a [Hb] of 0.012±0.0065 g dL-1. E. histolytica gsIgl1B trophozoites also had a lower hemolytic activity than control trophozoites (Fig 7C and 7D), indicating that lower hemolytic activity reflects lower Igl1 expression in the parasites.
Fig 7. Hemolytic activities of Igl1-attenuated Entamoeba histolytica trophozoites.
Vector transfected or Igl1 gene-silenced E. histolytica trophozoites (1×105) were incubated with HoRBCs at 37°C for 1 h. A and C. Images of HoRBCs in a 96-well plate just after mixing with trophozoites (0 hr) and after incubation for 1 h with trophozoites (1 hr). B and D. Released hemoglobin (Hb) concentration in the supernatant of a mixture of trophozoites and HoRBCs after incubation for 1 h at 37°C. Data are the mean ± SD from 5 independent experiments. **p< 0.01 by ANOVA with Dunn test. Control: vector transfected E. histolytica trophozoite, gsIgl1A and gsIgl1B: Igl1 gene-silenced E. histolytica trophozoite.
Discussion
The E. histolytica lectin consists of three subunits, Hgl, Lgl and Igl, of which Hgl and Igl have lectin activities [24]. Recently, we found that EhIgl1 had hemolytic and cytotoxic activities [12]. Since E. dispar, a non-virulent species, also has an Igl1 subunit homologue, it is of interest to determine whether EdIgl1 has these activities. In this study, we showed that EdIgl1 has similar hemolytic activity to that of EhIgl1, with the site of this activity residing at the C-terminus in both proteins. There are two isoforms of Igl, and therefore, we also evaluated the hemolytic activities of EhIgl2 and EdIgl2. All EhIgls and EdIgls had hemolytic activity in our assay. This is the first study to show that E. dispar Igls and E. histolytica Igl2 lectins have hemolytic activities.
Factors related to the virulence of Entamoeba spp. remain unclear, despite several detailed studies [25–28]. Among the potential factors, both Hgl and Lgl lectin subunit expression are lower in E. dispar compared with E. histolytica [29]. Low expression of Lgl1 was also found in an avirulent E. histolytica Rahman strain compared with the highly virulent E. histolytica HM-1:IMSS strain [30]. Expression of dominant negative Hgl or Lgl in the E. histolytica HM-1:IMSS strain and antisense inhibition of expression of Lgl in the same strain gave a less virulent strain [30–32]. Antisense inhibition of expression of EhCP5, an amoebic cysteine protease, in the HM-1: IMSS strain resulted in reduced virulence [33–35]. EhCP5 is missing in E. dispar [36] and is expressed at a lower level in the Rahman strain compared to the HM-1:IMSS strain [25]. Antisense inhibition of amoebapore expression in the HM-1:IMSS strain also decreases amoebic virulence [37]. Thus, many virulence-related molecules have been identified, but there may be additional factors related to amoebic virulence [38].
The Igl subunit also has vital roles in the pathogenicity of the parasite, including attachment to host cells and killing activities [12, 39, 40]. EhF-Igl1 and EhF-Igl2 have 83–84% amino acid sequence identity, while the amino acid sequence identity of EhF-Igl1 and EdF-Igl1 is 75–76% and that of EhF-Igl2 and EdF-Igl2 is 73–74% [8]. EhC-Igl1 and EdC-Igl1 have 76–77% amino acid sequence identity, with conserved cysteine residues. The Igls also have different expression levels in each species, with Igl1 having higher expression in E. histolytica than in E. dispar, but Igl2 having similar expression in the two species [8]. To assess whether the difference in level of Igl1 proteins affects the hemolytic activity of E. histolytica, we generated Igl1 gene-silenced E. histolytica trophozoites and compared the activity with vector-transfected E. histolytica trophozoites. Interestingly, a 40% reduction of Igl1 protein expression led to a significant decrease in hemolytic activity.
This observation correlates with the weaker cytotoxicity of non-virulent E. dispar in a Gal/GalNAc lectin-mediated manner in vitro [41]. Erythrophagocytosis of E. dispar was observed in another study [42]. More importantly, E. dispar is pathogenic in experimental animals [43–46] and in humans [47, 48]. These effects have recently been reviewed [49]. The mechanism of the pathogenicity to E. dispar is still unclear, but it is possible that the incidence of infection by Entamoeba spp. is related to the expression levels of Igls and other related proteins. Further studies are needed to evaluate this possibility.
One of the interesting observations in this study was increased expression of Igl2 in both of Igl1-silenced strains. By contrast, decreased level of Igl2 expression has been observed by short hairpin RNA-mediated knockdown of Igl1 [50]. The discrepancy may be due to the difference of methods used to prepare transfectants.
Supporting information
Asterisks indicate the same amino acids.
(TIF)
Acknowledgments
The authors would like to thank Dr. Yoshinori Mitsui (Department of Parasitology, NEKKEN) for critical advice and Ms. Kyoko Masuda for technical support. Dr. Satoshi Kaneko and Dr. Yoshito Fujii (Department of Eco-epidemiology, NEKKEN) supported this study. We would also like to thank Dr. Seiki Kobayashi (Department of Infectious Diseases, Keio University School of Medicine) for supplying E. dispar.
Data Availability
All relevant data are within the paper.
Funding Statement
The Joint Usage/Research Center on Tropical Disease, Institute of Tropical Medicine, Nagasaki University (to KK and HT, 27-Ippan-7 and 28-Ippan-5), a Grant-in-Aid for Young Scientists (B) (to KK, KAKENHI: JP 23790460), and Grants-in-Aid for Scientific Research (C) (to KK, KAKENHI: JP 16K08761) from the Japan Society for the Promotion of Science (JSPS) and JSPS KAKENHI (to HT, grant nos. 23117009, 26460516 and 16H05819).
References
- 1.Stanley SL Jr. Amoebiasis. Lancet. 2003; 361: 1025–1034. doi: 10.1016/S0140-6736(03)12830-9 [DOI] [PubMed] [Google Scholar]
- 2.Petri WA Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol. 2002; 56: 39–64. doi: 10.1146/annurev.micro.56.012302.160959 [DOI] [PubMed] [Google Scholar]
- 3.Laughlin RC, McGugan GC, Powell RR, Welter BH, Temesvari LA. Involvement of Raft-Like Plasma Membrane Domains of Entamoeba histolytica in Pinocytosis and Adhesion. Infect Immun. 2004; 72: 5349–5357. doi: 10.1128/IAI.72.9.5349-5357.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welter BH, Goldston AM, Temesvari LA. Localisation to lipid rafts correlates with increased function of the Gal/GalNAc lectin in the human protozoan parasite. Entamoeba histolytica, Int J Parasitol. 2011; 41: 1409–1419. doi: 10.1016/j.ijpara.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng XJ, Tsukamoto H, Kaneda Y, Tachibana H. Identification of the 150-kDa surface antigen of Entamoeba histolytica as a galactose- and N-acetyl-D-galactosamine-inhibitable lectin. Parasitol Res. 1998; 84: 632–639. [DOI] [PubMed] [Google Scholar]
- 6.Cheng XJ, Hughes MA, Huston CD, Loftus B, Gilchrist CA, Lockhart LA, et al. Intermediate subunit of the Gal/GalNAc lectin of Entamoeba histolytica is a member of a gene family containing multiple CXXC sequence motifs. Infect Immun. 2001; 69: 5892–5898 doi: 10.1128/IAI.69.9.5892-5898.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachibana H, Cheng XJ, Tsukamoto H, Itoh J. Characterization of Entamoeba histolytica intermediate subunit lectin-specific human monoclonal antibodies generated in transgenic mice expressing human immunoglobulin loci. Infect Immun. 2009; 77: 549–556. doi: 10.1128/IAI.01002-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tachibana H, Cheng XJ, Kobayashi S, Okada Y, Itoh J, Takeuchi T. Primary structure, expression and localization of two intermediate subunit lectins of Entamoeba dispar that contain multiple CXXC motifs. Parasitology. 2007; 134: 1989–1999. doi: 10.1017/S0031182007003459 [DOI] [PubMed] [Google Scholar]
- 9.Tachibana H, Cheng XJ, Masuda G, Horiki N, Takeuchi T. Evaluation of recombinant fragments of Entamoeba histolytica Gal/GalNAc lectin intermediate subunit for serodiagnosis of amebiasis. J Clin Microbiol. 2004; 42: 1069–1074. doi: 10.1128/JCM.42.3.1069-1074.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min X, Feng M, Guan Y, Man S, Fu Y, Cheng X, et al. Evaluation of the C-Terminal Fragment of Entamoeba histolytica Gal/GalNAc Lectin Intermediate Subunit as a Vaccine Candidate against Amebic Liver Abscess. PLoS Negl Trop Dis. 2016; 10: e0004419 doi: 10.1371/journal.pntd.0004419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCoy JJ, Mann BJ. Proteomic analysis of Gal/GalNAc lectin-associated proteins in Entamoeba histolytica. Exp Parasitol. 2005; 110: 220–225. doi: 10.1016/j.exppara.2005.02.013 [DOI] [PubMed] [Google Scholar]
- 12.Kato K, Yahata K, Dhoubhadel BG, Fujii Y, Tachibana H. Novel hemagglutinating, hemolytic and cytotoxic activities of the intermediate subunit of Entamoeba histolytica lectin. Sci Rep. 2015; 5: e13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006; 2: e48 doi: 10.1371/journal.ppat.0020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978; 72: 431–432. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi S, Imai E, Haghighi A, Khalifa SA, Tachibana H, Takeuchi T. Axenic cultivation of Entamoeba dispar in newly designed yeast extract-iron-gluconic acid-dihydroxyacetone-serum medium. J Parasitol. 2005; 91: 1–4. doi: 10.1645/GE-3386 [DOI] [PubMed] [Google Scholar]
- 16.Makiuchi T, Mi-ichi F, Nakada-Tsukui K, Nozaki T. Novel TPR-containing subunit of TOM complex functions as cytosolic receptor for Entamoeba mitosomal transport. Sci Rep. 2013; 3: e1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi-ichi F, Makiuchi T, Furukawa A, Sato D, Nozaki T. Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica. PLoS Negl Trop Dis. 2011; 5: e1263 doi: 10.1371/journal.pntd.0001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci USA. 2009; 106: 21731–21736. doi: 10.1073/pnas.0907106106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakada-Tsukui K, Saito-Nakano Y, Ali V, Nozaki T. A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica. Mol Biol Cell. 2005; 16: 5294–5303. doi: 10.1091/mbc.E05-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson KA, Snyder DS, Goodell MA. Skeletal muscle fiber-specific green autofluorescence: potential for stem cell engraftment artifacts. Stem Cells. 2004; 22: 180–187. doi: 10.1634/stemcells.22-2-180 [DOI] [PubMed] [Google Scholar]
- 21.Inomoto C, Umemura S, Egashira N, Minematsu T, Takekoshi S, Itoh Y, et al. Granulogenesis in non-neuroendocrine COS-7 cells induced by EGFP-tagged chromogranin A gene transfection: identical and distinct distribution of CgA and EGFP. J Histochem Cytochem. 2007; 55: 487–493. doi: 10.1369/jhc.6A7110.2007 [DOI] [PubMed] [Google Scholar]
- 22.Tovy A, Hertz R, Siman-Tov R, Syan S, Faust D, Guillen N, et al. Glucose starvation boosts Entamoeba histolytica virulence. PLoS Negl Trop Dis. 2011; 5: e1247 doi: 10.1371/journal.pntd.0001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostroff RM, Vasil AI, Vasil ML. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol. 1990; 172: 5915–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frederick JR, Petri WA Jr. Roles for the galactose-/N-acetylgalactosamine-binding lectin of Entamoeba in parasite virulence and differentiation. Glycobiology. 2005; 15: 53R–59R. doi: 10.1093/glycob/cwj007 [DOI] [PubMed] [Google Scholar]
- 25.Thibeaux R, Weber C, Hon CC, Dillies MA, Avé P, Coppée JY, et al. Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLoS Pathog. 2013; 9: e1003824 doi: 10.1371/journal.ppat.1003824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer L, Chadee K. The immunopathogenesis of Entamoeba histolytica. Exp Parasitol. 2010; 126: 366–380. doi: 10.1016/j.exppara.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 27.Gilchrist CA, Petri WA Jr. Using differential gene expression to study Entamoeba histolytica pathogenesis. Trends Parasitol. 2009; 25: 124–131. doi: 10.1016/j.pt.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: Potential implications for amebic pathogenesis. Infect Immun. 2006; 74: 340–351. doi: 10.1128/IAI.74.1.340-351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai DR, Kobayashi S, Kain KC. Entamoeba dispar: Molecular characterization of the galactose/N-acetyl-D-galactosamine lectin. Exp Parasitol. 2001; 99: 226–234. doi: 10.1006/expr.2001.4672 [DOI] [PubMed] [Google Scholar]
- 30.Ankri S, Padilla-Vaca F, Stolarsky T, Koole L, Katz U, Mirelman D. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol Microbiol. 1999; 33: 327–337. [DOI] [PubMed] [Google Scholar]
- 31.Vines RR, Ramakrishnan G, Rogers JB, Lockhart LA, Mann BJ, Petri WA Jr. Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a β2 integrin motif. Mol Biol Cell. 1998; 9: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz U, Ankri S, Stolarsky T, Nuchamowitz Y, Mirelman D. Entamoeba histolytica expressing a dominant negative N-truncated light subunit of its gal-lectin are less virulent. Mol Biol Cell. 2002; 13: 4256–4265. doi: 10.1091/mbc.E02-06-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ankri S, Stolarsky T, Mirelman D. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol Microbiol. 1998; 28: 777–785. [DOI] [PubMed] [Google Scholar]
- 34.Ankri S, Stolarsky T, Bracha R, Padilla-Vaca F, Mirelman D. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect Immun. 1999; 67: 421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moncada D, Keller K, Ankri S, Mirelman D, Chadee K. Antisense inhibition of Entamoeba histolytica cysteine proteases inhibits colonic mucus degradation. Gastroenterology. 2006; 130: 721–730. doi: 10.1053/j.gastro.2005.11.012 [DOI] [PubMed] [Google Scholar]
- 36.Jacobs T, Bruchhaus I, Dandekar T, Tannich E, Leippe M. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol Microbiol. 1998; 27: 269–276. [DOI] [PubMed] [Google Scholar]
- 37.Bracha R, Nuchamowitz Y, Leippe M, Mirelman D. Antisense inhibition of amoebapore expression in Entamoeba histolytica causes a decrease in amoebic virulence. Mol Microbiol. 1999; 34: 463–472. [DOI] [PubMed] [Google Scholar]
- 38.MacFarlane RC, Singh U. Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot Cell. 2007; 6: 2139–2146. doi: 10.1128/EC.00174-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng XJ, Tachibana H, Kaneda Y. Protection of hamsters from amebic liver abscess formation by a monoclonal antibody to a 150-kDa surface lectin of Entamoeba histolytica. Parasitol Res. 1999; 85: 78–80. [DOI] [PubMed] [Google Scholar]
- 40.Cheng XJ, Kaneda Y, Tachibana H. A monoclonal antibody against the 150 kDa surface antigen of Entamoeba histolytica inhibits adherence and cytotoxicity to mammalian cells. Med Sci Res. 1997; 25: 159–161. [Google Scholar]
- 41.Dodson JM, Clark CG, Lockhart LA, Leo BM, Schroeder JW, Mann BJ. Comparison of adherence, cytotoxicity, and Gal/GalNAc lectin gene structure in Entamoeba histolytica and Entamoeba dispar. Parasitol Int. 1997; 46: 225–235. [Google Scholar]
- 42.Talamás-Lara D, Chávez-Munguía B, González-Robles A, Talamás-Rohana P, Salazar-Villatoro L, Durán-Díaz Á, et al. Erythrophagocytosis in Entamoeba histolytica and Entamoeba dispar: a comparative study. Biomed Res Int. 2014; 2014: 626259 doi: 10.1155/2014/626259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomes MA, Melo MN, Macedo AM, Pena GP, Caliari MV, Silva EF. Characterization of Entamoeba histolytica and Entamoeba dispar by biological, biochemical, and molecular parameters. Arch Med Res. 2000; 31: S249–250. [DOI] [PubMed] [Google Scholar]
- 44.Costa AO, Gomes MA, Rocha OA, Silva EF. Pathogenicity of Entamoeba dispar under xenic and monoxenic cultivation compared to a virulent E. histolytica. Rev Inst Med Trop S Paulo. 2006; 48: 245–250. [DOI] [PubMed] [Google Scholar]
- 45.Dolabella SS, Serrano-Luna J, Navarro-García F, Cerritos R, Ximénez C, Galván-Moroyoqui J.M., et al. Amoebic liver abscess production by Entamoeba dispar. Ann Hepatol. 2012; 11: 107–117. [PubMed] [Google Scholar]
- 46.Guzmán-Silva MA, Santos HL, Peralta RS, Peralta JM, de Macedo HW. Experimental amoebic liver abscess in hamsters caused by trophozoites of a Brazilian strain of Entamoeba dispar. Exp Parasitol. 2013; 134: 39–47. doi: 10.1016/j.exppara.2013.01.015 [DOI] [PubMed] [Google Scholar]
- 47.de Martinez AM, Gomes MA, Viana JdaC, Romanha AJ, Silva EF. Isoenzyme profile as parameter to differentiate pathogenic strains of Entamoeba histolytica in Brazil. Rev Inst Med Trop S Paulo. 1996; 38: 407–412. [DOI] [PubMed] [Google Scholar]
- 48.Graffeo R, Archibusacci CM, Soldini S, Romano L, Masucci L. Entamoeba dispar: A rare case of enteritis in a patient living in a nonendemic area. Case Rep Gastrointest Med. 2014; 2014: 498058 doi: 10.1155/2014/498058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliveira FM, Neumann E, Gomes MA, Caliari MV. Entamoeba dispar: Could it be pathogenic. Trop Parasitol. 2015; 5: 9–14. doi: 10.4103/2229-5070.149887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linford AS, Moreno H, Good KR, Zhang H, Singh U, Petri WA Jr. Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 2009; 9: 38 doi: 10.1186/1471-2180-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Asterisks indicate the same amino acids.
(TIF)
Data Availability Statement
All relevant data are within the paper.