SUMMARY
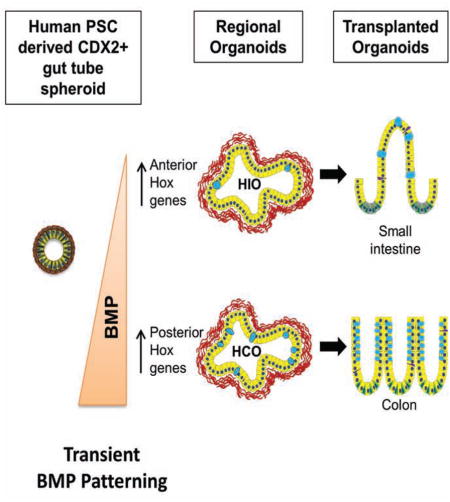
Gastric and small intestinal organoids differentiated from human pluripotent stem cells (hPSCs) have revolutionized the study of gastrointestinal development and disease. Distal gut tissues such as cecum and colon, however, have proven considerably more challenging to derive in vitro. Here we report the differentiation of human colonic organoids (HCOs) from hPSCs. We found BMP signaling is required to establish a posterior Satb2+ domain in developing and postnatal intestinal epithelium. Brief activation of BMP signaling is sufficient to activate a posterior HOX code and direct human PSC-derived gut tube cultures into HCOs. In vitro, HCOs express colonic markers and contained colon-specific cell populations. Following transplantation into mice, HCOs undergo morphogenesis and maturation to form tissue that exhibits molecular, cellular, and morphologic properties of human colon. Together these data show BMP-dependent patterning of human hindgut into HCOs, which will be valuable for studying diseases including colitis and colon cancer.
Graphical Abstract
INTRODUCTION
The epithelium of the gastrointestinal tract is derived from the definitive endoderm, a germ layer which forms during gastrulation. The process of gut tube morphogenesis transforms the definitive endoderm into a primitive gut tube with a foregut, midgut and hindgut. The midgut gives rise to the small and proximal large intestine and the hindgut gives rise to the distal large intestine and rectum (Zorn and Wells, 2009). The small intestine is further subdivided into the duodenum, jejunum and ileum (Jeejeebhoy, 2002), whereas the large intestine is subdivided in to the cecum, colon and rectum (Jeejeebhoy, 2002). While there are numerous studies of development of the small intestine (Korinek et al., 1998; Ratineau et al., 2003; Roberts et al., 1995; Sherwood et al., 2011; Walker et al., 2014; Walton et al., 2012), less is known about development of human large intestine/colon. Furthermore, diseases affecting this region of the gastrointestinal (GI) tract such as colitis, colon cancer, polyposis syndromes and Irritable Bowel Syndrome are prevalent (Molodecky et al., 2012; Siegel et al., 2014; Zbuk and Eng, 2007). Animal models of polyposis syndromes and intestinal cancer are limited since polyps and tumors preferentially form in the small intestine and rarely in the colon or rectum (Haramis et al., 2004; He et al., 2004; Moser et al., 1990).
We previously described a method in which hPSCs can be differentiated into intestinal tissue using a step-wise approach that mimics embryonic intestinal development. PSCs were first differentiated into definitive endoderm using Activin A, then into posterior gut tube structures call spheroids using FGF4 and activation of canonical Wnt, then 3-dimentional growth of spheroids where they form human intestinal organoids (HIOs) (Spence et al., 2011). HIOs have a small intestinal identity and have proven extremely useful for modeling small intestinal biology (Bouchi et al., 2014; Finkbeiner et al., 2015; Watson et al., 2014; Xue et al., 2013). However, PSC-derived large intestinal organoids have not been developed, and given the prevalence of disease in the large intestine, such a system would allow for interrogation of development and disease mechanisms in this region of the GI tract.
To develop a method for generating large intestinal organoids, we first identified Satb2 as a definitive marker of the presumptive large intestinal epithelium in frogs, mice, and humans. Using Satb2 as a marker, we show that BMP signaling is required for specification of posterior gut endoderm of frogs and mice, consistent with the known role of BMP in posterior-ventral development (Kumar et al., 2003; Roberts et al., 1995; Sherwood et al., 2011; Tiso et al., 2002; Wills et al., 2008). Moreover, stimulation of BMP signaling in PSC-derived gut tube cultures for 3 days is sufficient to induce a posterior HOX code and the formation of SATB2-expressing colonic organoids. Human colonic organoids (HCOs) had a marker profile and cell types consistent with large intestine. Furthermore, HCOs, but not HIOs, formed colonic enteroendocrine cells (EEC) in response to expression of NEUROG3, demonstrating that HCOs were functionally committed to the colonic region. In addition, HCOs engrafted under the kidney capsule of immunocompromised mice and grown in vivo for 8–10 weeks, maintain their regional identify, formed tissues with colonic morphology, contained colon-specific cell types, had zones of proliferation and differentiation, and well-formed smooth muscle layers. Lastly, RNA-seq analysis demonstrated that HIOs and HCOs underwent substantial maturation and express regional markers consistent with a small and large intestinal identity respectively. In summary, we identified an evolutionarily conserved BMP-HOX pathway in frogs and mice and used this to direct hindgut patterning and formation of human colonic organoids.
RESULTS
SATB2 expression marks the gut endoderm of posterior embryonic and adult intestine
To generate large intestinal organoids from human PSCs, we first identified markers that distinguish different domains developing embryonic gut tube. Consistent with previous reports, Gata4 marked the gut endoderm from the posterior foregut to the yolk stalk of e9.5 mouse embryos (Figure S1A) (Aronson et al., 2014; Battle et al., 2008; Beuling et al., 2008a; Beuling et al., 2007a; Beuling et al., 2007b; Beuling et al., 2010; Beuling et al., 2008b; Bosse et al., 2007; Kohlnhofer et al., 2016; Patankar et al., 2012a; Patankar et al., 2012b; Sherwood et al., 2009; Walker et al., 2014). At later stages of development (e11.5-e16.5), Gata4 continued to distinctly mark the anterior but not the posterior intestine (Figure S1B–D,I–J). This expression domain remains intact into adulthood in both mice (not shown) and humans (Figure S1K–L).
In order to identify markers of the posterior fetal intestine, we mined public expression databases such as GNCPro™, TiGER and Human Protein Atlas for colon enriched genes (described in the Materials and Methods section) and found Satb2, a CUT-class of homeobox genes (Holland et al., 2007) (Gyorgy et al., 2008), as a potential marker of large intestine. Satb2 protein was first detected in the posterior endoderm of mouse embryos at e9-9.5 and formed a discreet expression boundary with Gata4 at the yolk stalk (Figure S1A), suggesting that Satb2 also marks the caudal midgut. This is a broader expression domain than previously identified (Dobreva et al., 2006). Satb2 expression continued to mark the posterior intestinal endoderm throughout development (e11.5-16.5) (Figure S1B,C,E,F,H,J,) and in the postnatal colon in mice (not shown) and humans (Figure S1L). In humans, GATA4 and SATB2 mark proximal and distal regions of the human fetal and adult intestinal tract respectively (Bernstein et al., 2010; Fagerberg et al., 2014) (Wang et al., 2015) (Figure S2A–C). We conclude that Gata4 and Satb2 expression boundaries are established early during development of mouse and marks future boundaries of the developing small and large intestine in mice and humans.
BMP signaling is required for Satb2 expression in the hindgut endoderm of mouse and frog embryos
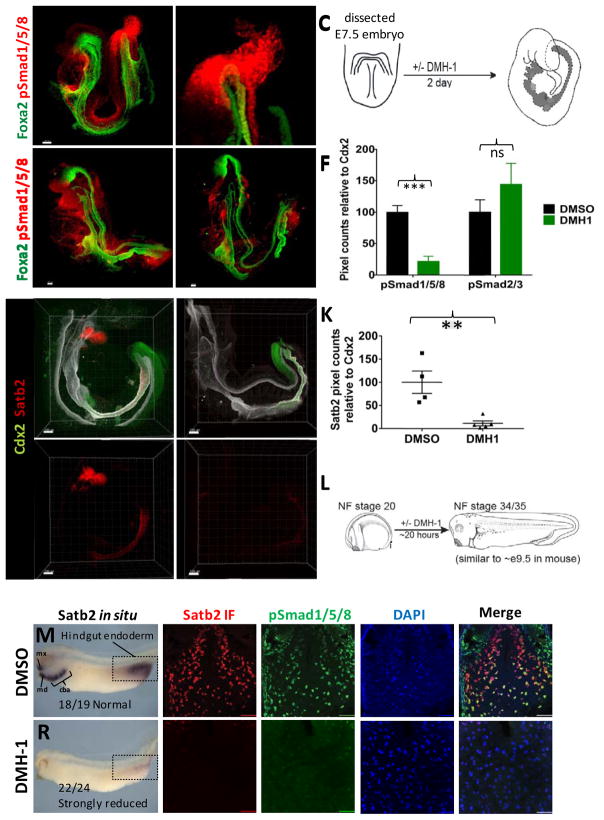
We next used Satb2 as a marker to identify pathways that promote posterior intestinal fate in mouse and frog embryos. We first focused on BMP signaling and observed that BMP signaling was highly active in the endoderm and mesoderm of the posterior gut tube of e8.5 mouse embryos as measured by phosphorylated Smad1/5/8 (pSMAD1/5/8) (Figure 1A–B). Moreover, inhibition of BMP signaling with the inhibitor DMH-1 (Figure 1C) resulted in a significant reduction in pSmad1/5/8 levels and a loss of Satb2 expression in the posterior gut tube of cultured mouse embryos (Figure 1D–K). In addition, Satb2 expression was lost in the first brachial arch of DMH-1 treated embryos consistent with previous studies in Zebrafish (Sheehan-Rooney et al., 2013). DMH-1 had no impact on TGFβ signaling as measured by pSmad2/3 levels (Figure 1F). We further determined BMP is required for Satb2 expression in the hindgut and brachial arches of frog embryos (Figure 1L–V), and this was confirmed using the BMP-antagonist Noggin (not shown). Taken together these results revealed a conserved pathway in vertebrates whereby BMP signaling is required for defining the posterior most region of the developing gut tube that gives rise to the distal ileum and large intestine.
Figure 1.
Bmp signaling regulates Satb2 expression in mouse and frog embryos.
(A) Whole-mount pSmad158 (red) and Foxa2 (green) staining of e8.5 mouse embryo showing nuclear staining around the developing hindgut (n=6). (B) Inset of optical slices from boxed region in (A) showing pSmad1/5/8 staining in the hindgut mesoderm and endoderm (D, dorsal; V, ventral). (C) Schematic of mouse embryo isolated at the headfold stage and cultured for 2 days +/− Bmp inhibition with DMH-1. (D,E) Whole-mount pSmad1/5/8 (red) and Foxa2 (green) staining of DMSO (D) and DMH-1 (E) treated embryos after 48 hours of culture. (F) Quantification of pSmad1/5/8 and pSmad2/3 staining in relative to Cdx2 in embryos cultured in DMSO or DMH-1 (n=3 embryos per condition). (G–J) Whole-mount immunostaining of Cdx2 (green), Satb2 (red) and Foxa2 (white) of mouse embryos (n=6 for each condition) following 2 days of culture in DMSO (G,H) or DMH-1 (I,J). Arrows in H–J point to the approximate location of the yolk stalk (BA1, first brachial arch). (K) Quantification of Satb2 expression in mouse embryos treated with DMSO or DMH-1. (L) Schematic of Bmp inhibition in Xenopus tropicalis embryos. In situ hybridization of Satb2 in Xenopus tropicalis embryos treated with DMSO (M) or DMH-1 (R). The white dotted line in (M) and (R) depict the plane of section used subsequent analysis. Mx and md= maxillary and mandibular processes of first brachial arch. Cba= Caudal brachial arches. Immunofluorescence of Satb2 (red), pSmad1/5/8 (green), DAPI (blue), and color merged images from Xenopus tropicalis embryos treated with DMSO (N–Q) or DMH-1 (S–V). Scale bars for = 100 μm in G–H and 50 μm in all other panels. **p ≤ 0.01 and ***p ≤ 0.001 for 2 tailed t-test.
BMP signaling promotes posterior fate in human gut tube cultures
We next investigated if BMP signaling could be used to promote a posterior gut tube fate in humans using nascent CDX2+ gut tube spheroids derived from human PSCs as previously described (Spence et al., 2011). We either inhibited or activated BMP signaling using the BMP inhibitor NOGGIN or BMP2 respectively (Figure 2A) and monitored BMP signaling levels by accumulation of nuclear pSMAD1/5/8. Control cultures had low levels of pSMAD1/5/8 protein, which was lost upon addition of NOGGIN (Figure 2B–D). In contrast, addition of BMP2 caused a rapid accumulation of pSMAD158 in both epithelial and mesodermal cells suggesting both cell types respond to BMP signals similar to what we observed in mouse embryos (Figure 1A–B). The specificity of pSmad1/5/8 staining was confirmed using adult mouse colon, which showed pSmad1/5/8 staining restricted to the differentiated compartment of the upper crypt (Figure 2E), as previously reported (Hardwick et al., 2004; van Dop et al., 2009; Whissell et al., 2014). Further analysis of organoids revealed that 3 days of BMP2 treatment was sufficient to induce high levels of SATB2 protein in the epithelium compared to NOGGIN and control cultures (Figure 2F–I). This suggests that a short pulse of BMP activity is sufficient to pattern spheroid endoderm into a posterior gut tube fate.
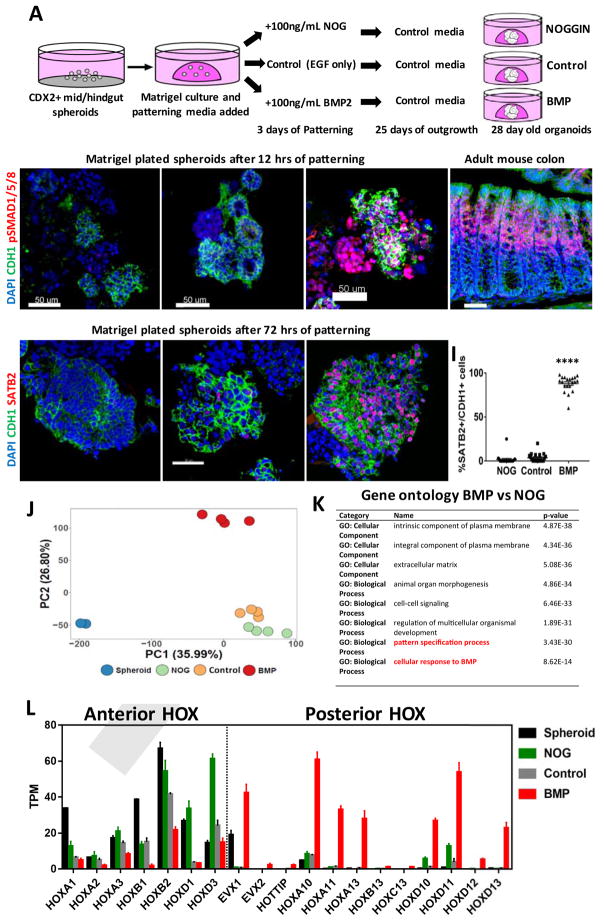
Figure 2.
BMP2 induces SATB2 and a posterior HOX code in human gut tube spheroids.
(A) Schematic of gut tube spheroid patterning protocol. (B–D) BMP signaling levels as measured by pSMAD1/5/8 (red) staining of spheroids treated with NOGGIN (B), no treatment (C) and BMP2 (D) for 12 hours. (E) pSmad1/5/8 staining of adult mouse colon showing increased BMP signaling at to the top of crypts. (F–H) SATB2 expression in spheroids treated with NOGGIN (F), no treatment (G) and BMP2 (H) for 72 hours. (I) Quantification of the percentage of SATB2+ CDH1+ epithelium following patterning. (J) Principal component analysis of nascent spheroids and spheroids after 3 days of patterning. (K) Gene ontology analysis of differentially expressed genes between BMP vs NOG treated spheroids. (L) Graph of TPM (Transcripts per million) values of spheroids before and after patterning. Samples analyzed were spheroids before patterning (n=2), and NOGGIN, Control and BMP2 treated spheroids 3 days after patterning (n=4 for each group). For quantification in I, 20 organoids from at least 3 experiments were examined. Error bars represent SD. Scale bars = 50 μm. ****p ≤ 0.0001 determined by 2 tailed t-test comparing NOGGIN+Control treated spheroids and BMP2 treated spheroids.
To identify BMP-mediated transcriptional networks that ultimately confer posterior identity in the gut, we performed RNA-seq analysis on human gut tube cultures treated with BMP2 or noggin for 3 days. Principal component analysis showed that BMP2-treated cultures clustered separately from NOGGIN and control treated organoids (Figure 2J) and that BMP signaling affects multiple biological processes including organ morphogenesis, cell-cell signaling, pattern-specification (Figure 2K). The most definitive regulators of A-P patterning are HOX genes, and we found that BMP activation resulted in broad down regulation of anterior HOX genes and up regulation of posterior HOX genes (Figure 2L). In particular we saw BMP-mediated increases in multiple paralogs of HOX10, 11, 12 and 13 groups. These results demonstrate that BMP signaling broadly regulates A-P hox code during patterning of the human gut tube and suggest a mechanism by which the distal GI tract is initially specified.
BMP signaling acts downstream of SHH to induce a posterior HOX code
Previous studies suggest that Sonic Hedgehog (Shh) acts upstream of Bmp4 and Hox13 expression during posterior gut patterning in chick embryos (Figure S3A) (Roberts et al., 1995). However, the relative epistatic relationship between BMP and Hox13 (Figure S3B) was not investigated due to embryonic lethality caused by Bmp4 overexpression in the midgut and hindgut (De Santa Barbara et al., 2005; Roberts et al., 1995). We used human gut tube cultures to model the epistatic relationship of SHH-BMP-HOX13 during posterior gut tube patterning. Activation of hedgehog signaling with the smoothened agonist SAG led to a concentration dependent activation of the BMP signaling target gene MSX2 and the mesenchymal HOX factors, HOXA13 and HOXD13 (Figure S3C). The ability of HH signaling to activate HOXA13 was dependent on BMP and was blocked by NOGGIN (Figure S3D–E). Moreover, BMP2 was still able to induce HOXA13 expression in the presence of the HH inhibitor Cyclopamine (Figure S3F–G), confirming that BMP signaling functions downstream of SHH as previously reported (Shyer et al., 2015; Walton et al., 2012; Walton et al., 2009; Walton et al., 2016). In addition, SAG-mediated activation of HOX13 was only a fraction of that mediated by BMP2 (Figure S3C), suggesting that BMP may have HH-independent activity. Consistent with this, activation of SHH signaling during BMP patterning did not increase SATB2 expression (Figure S4A). Experiments in Xenopus confirmed this epistatic relationship between SHH and BMP (data not shown) suggesting that this mechanism is evolutionarily conserved. Taken together our data suggest that BMP signaling is sufficient to activate the posterior HOX code and does so downstream of HH signaling.
BMP-derived organoids cultured in vitro maintain a distal identity
We next investigated if 3 days of BMP treatment is sufficient to confer stable regional identity following extended culture of organoids for 25 days (Figure 3). Levels of ONECUT1 (a marker of proximal small intestine) were highest in NOGGIN and control treated organoids and absent in BMP2 treated organoids (Figure 3A–D). Conversely, SATB2 was absent in the epithelium of NOGGIN and control treated organoids but broadly expressed in nearly all of the CDX2+ epithelial cells of BMP2 treated organoids (Figure 3E–H, Figure S4A). Importantly, modulation of BMP signaling had similar proximal-distal patterning effects on multiple human PSC lines, including embryonic stem cell lines H1 and H9 and induced pluripotent stem cell lines (IPSC 54.1 and IPSC 72.3) (shown below). We frequently observed non-epithelial SATB2 expression in NOGGIN and control organoids (data not shown) possibly due to the presence of other cell types that are known to be present in HIOs in vitro (Spence et al., 2011). Examination of HOXB13 and HOXD13, which is expressed in posterior epithelium and mesenchyme respectively, further revealed that BMP treated organoids maintained posterior patterning following prolonged culture in vitro (Figure S4B–C).
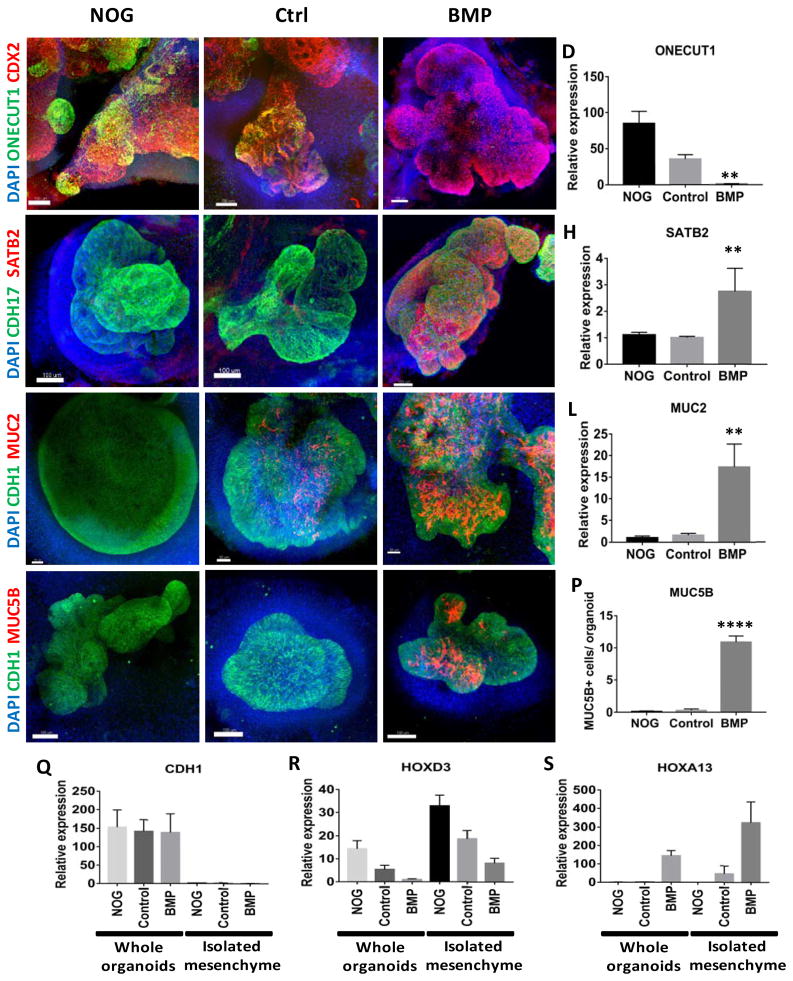
Figure 3.
Regional patterning is maintained in human intestinal organoids following prolonged in vitro culture.
(A–D) Whole-mount immunofluorescence and QPCR analysis with the proximal marker ONECUT1 (green) of 28 day old organoids that resulted from the initial 3 day treatment of spheroids with NOGGIN, control, or BMP2. Staining with CDX2 (red) and DAPI (blue) were also used to detect the epithelium and mesenchyme. (E–H) Expression of the posterior marker SATB2 (red) detected by IF and by QPCR. (I–L) Analysis of the pan-goblet cell marker MUC2 (red) by IF and by QPCR. (M–P) Analysis of the colon-specific goblet cell marker MUC5B (red) by IF. The number of MUC5B+ cells was quantified in (P). (Q–S) Analysis of patterning markers in isolated mesenchyme cultures relative to whole organoids. QPCR analysis of CDH1 (Q), the proximal HOX gene HOXD3 (R), and the distal HOX gene HOXA13 (S) in whole organoids and in mesenchyme cultures derived from NOGGIN, control, or BMP2 treated organoids. CDH1 was only observed in whole organoids that contained epithelial cells. Error bars represent SEM. For IF minimum of 10 organoids from at least 3 different experiments were examined for each condition. For QPCR a minimum of 5 biological replicates from 2 separate experiments were examined. Scale bars = 100 μm. **p ≤ 0.01 and ****p ≤ 0.0001 determined by 2 tailed t-test comparing NOGGIN+Control treated organoids and BMP2 treated organoids.
Goblet cells are distributed in a low-to-high gradient from proximal small intestine to distal large intestine (Rodriguez-Pineiro et al., 2013), and we investigated if our patterned organoids reflected this gradient. At 28 days revealed that BMP2 treated organoids had higher numbers of goblet cells compared to NOGGIN and Control organoids, as visualized by intracellular MUC2 staining (Figure 3I–L). We further confirmed the regional identity of goblet cells using the marker MUC5B, which is expressed by a subset of goblet cells in the colon but not in the small intestine (van Klinken et al., 1998). MUC5B staining was absent in NOGGIN and control treated 28-day organoids but was present in BMP2 treated organoids (Figure 4M–P). Goblet cell morphology became more mature in older organoids (44 day) (Figure S4D–I), where we observed goblet cells in the process of secreting mucus into the lumen of the organoids (Figure S4J–L). The ability to observe mucus secretion in BMP treated organoids suggests that this organoid system would be useful to study mucus secretion and the roles of mucus in intestinal pathophysiology.
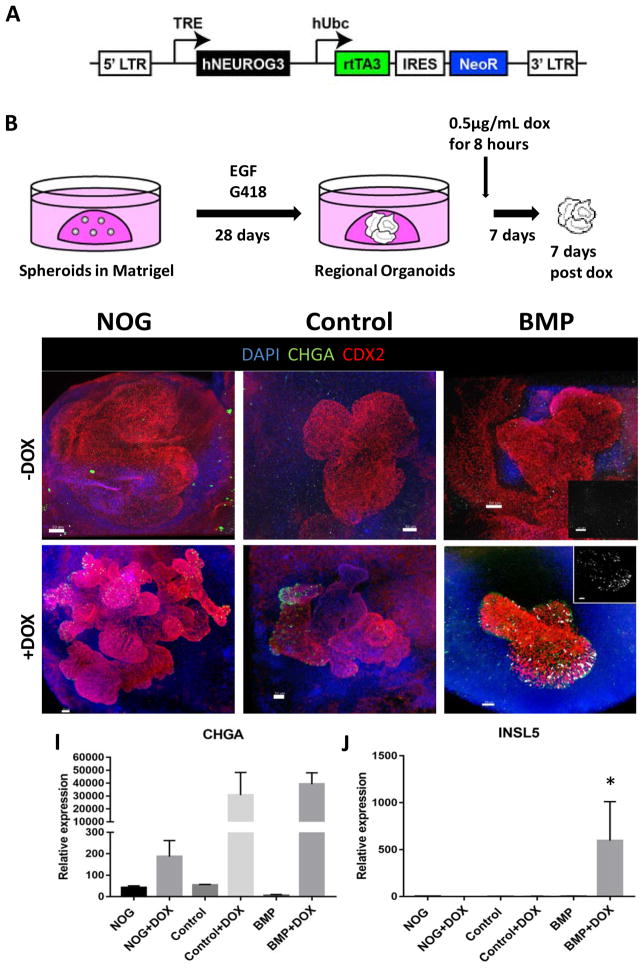
Figure 4.
HCOs but not HIOs gave rise to colon-specific enteroendocrine cells in response to expression of the proendocrine transcription factor NEUROGENIN 3.
(A–B) Schematic of the doxycycline inducible NEUROG3 lentiviral construct used to generate the IPSC72.3 inducible NEUROG3 line, and the doxycycline induction protocol. Whole-mount staining with Chromagranin A (green), CDX2 (red) and INSL5 (white) of 35 day old organoids patterned with NOGGIN (C,F), Control (D,G) or BMP2 (E,H). (C–E) Untreated organoids (-Dox) and (F–H) organoids with expressed NEUROG3 (+Dox) are shown. Insets in E and H show a magnified view of INSL5 staining. (I, J) QPCR analysis of NEUROG3 induction of enteroendocrine cells in HIOs and HCOs as measured by CHGA (I) and for INSL5 (J) expression. For IF, a minimum of 10 organoids were examined per condition. For QPCR, data is representative of 2 different experiments with NOGGIN (n=3), Control (n=3) or BMP (n=6) treated organoids. Error bars represent SEM. Scale bars = 50 μm. *p ≤ 0.05 determined by 2 tailed t-test comparing NOGGIN+Control treated organoids and BMP2 treated organoids.
While the regional pattern of organoids is stable after 28 days in culture, we wanted to investigate if early patterning was fully established after the initial 3 day treatment. To do so, we shifted 3 day NOGGIN-treated spheroids to BMP2-containing media for 3 days and conversely shifted 3 day BMP treated spheroids to NOGGIN-containing media for 3 days. Proximal organoids generated with NOGGIN did not express SATB2 in response to BMP2 demonstrating that proximal fate was stable following 3 days of patterning (Figure S4A). In the converse experiment, while 3 days of BMP2 treatment was sufficient to induce a stable distal fate, a subset of organoids lost SATB2 expression in response to NOGGIN treatment (Figure S4A). These data suggest that the posterior gut tube retains plasticity, consistent with the observation that the colonic endoderm of midgestation rat embryos is more regionally plastic than the small intestinal endoderm (Ratineau et al., 2003).
Patterning of organoid mesenchyme by BMP signaling
While stimulation of BMP signaling conferred regional identity to organoid epithelium, we also observed pSMAD1/5/8 in the non-epithelial compartment of BMP2 treated organoids during patterning, and upregulation of posterior HOX factors known to be expressed in the mesenchyme. To determine if mesenchymal patterning was stable, or required continued patterning input from epithelium, we isolated and expanded mesenchymal cell cultures for 2–3 weeks and analyzed them for expression of regional HOX genes. Mesenchymal cultures were lacking E-cadherin expressing cells, suggesting that they were primarily comprised of mesenchyme (Figure 3Q). Analysis of HOXD3, which is enriched in proximal intestinal mesenchyme (Yahagi et al., 2004), confirmed that the mesenchyme from NOGGIN and control treated organoids have a stable proximal identity while BMP treated organoids had decreased expression of HOXD3 (Figure 3R) and high levels HOXA13 (Figure 3S), which continues to be expressed in human colon fibroblasts (Higuchi et al., 2015). Taken together, these data suggest that early modulation of BMP signaling patterns both the epithelium and the mesenchyme and that mesenchymal patterning is stable even in the absence of epithelium.
Induction of colonic enteroendocrine cells is restricted to BMP2 treated organoids
The development of several ECC subtypes is regionally restricted to specific segments of small and large intestine. For example, expression of the protein INSL5 is restricted to colonic EECs (Burnicka-Turek et al., 2012; Thanasupawat et al., 2013). As a functional test of colonic identity, we determined if experimental induction of the colonic EEC marker INSL5 was restricted to BMP2-treated distal organoids. To do this we inducibly expressed the proendocrine transcription factor NEUROG3 using an iPSC line harboring a doxycycline (DOX) inducible NEUROG3 expression cassette (Figure 4A) as previously described (McCracken et al., 2017; McCracken et al., 2014). We performed a 6-hour pulse of DOX and after an additional 7 days in culture observed a robust induction of EECs as measured by CHGA positive cells (Figure 4B–I). However, we only observed INSL5 positive cells in BMP2 treated organoids and confirmed this by QPCR analysis (Figure 4C–H,J). Given that INSL5-expressing cells are only in the colon, our data strongly suggest that BMP2-treated organoids are functionally committed to the colonic fate. The expression of distal markers like SATB2, MUC5B and HOXA13 and the competence to generate colon specific ECCs support the conclusion that BMP2 treated organoids are colonic, and thus will be referred to as human colonic organoids (HCOs).
Regional identity of patterned organoids is maintained in vivo
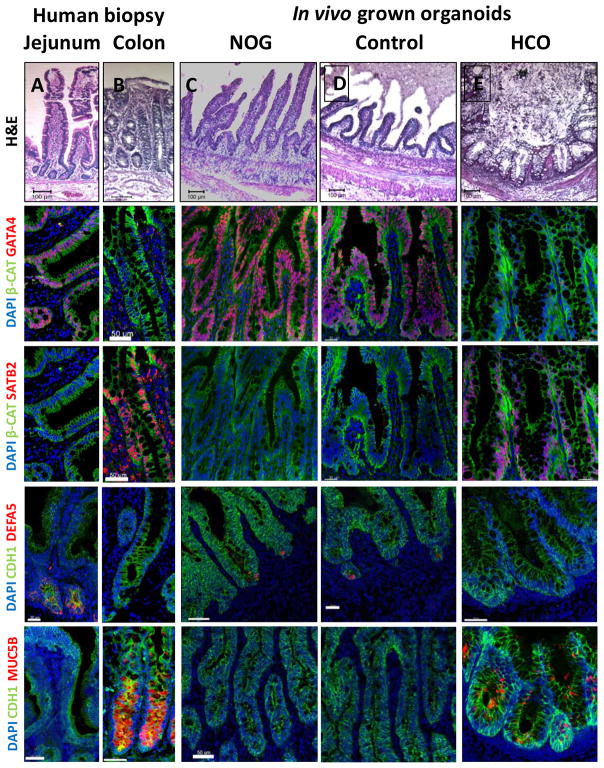
Previous studies of mouse and human fetal intestine have demonstrated that regional identity and tissue morphology of different regions of the intestine were maintained following orthotopic transplantation and growth in immunocompromised mice (Duluc et al., 1994; Savidge et al., 1995). To determine if HIOs and HCOs that were patterned in vitro would maintain regional identity and grow into small and large intestinal tissue, we transplanted them under the mouse kidney capsule for 6–10 weeks, which we previously demonstrated results in HIO maturation into small intestinal tissue (Watson et al., 2014). We observed that the engraftment of NOGGIN and control HIOs was more efficient than HCOs (Figure S5A). Consistent with their regional identity, transplanted HIOs and HCOs developed into mature tissues that morphologically resembled either small or large intestine, respectively (Figure 5A–E). The epithelium of NOGGIN and control organoids formed well-defined crypts and tall villi, comparable to human small intestine. In contrast BMP2-treated organoids contained crypts but lacked villi, similar to colon.
Figure 5.
HIOs and HCOs maintained regional identity following transplantation in vivo.
(A–E) H&E staining of biopsies from human jejunum and colon and of NOGGIN-derived HIOs, control HIOs, and BMP2-derived HCOs that were transplanted underneath the mouse kidney capsule and grown for 8–10 weeks in vivo. The samples of the same conditions were stained with the proximal intestinal marker GATA4 (F–J), the distal intestinal marker SATB2 (K–O), the Paneth cell marker DEFA5 (P–T), and the colon-specific goblet cell marker MUC5B (U–Y). Note that although GATA4 and SATB2 double staining was done in different channels but on the same slides for panels (F–O), they are shown as individual pseudocolored (red) images. For human biopsies n=2. For transplanted NOGGIN treated organoids n=12, for control organoids n=7, and for BMP2 treated organoids n=16. Scale bars= 50 μm.
In addition to their morphological resemblance to either small or large intestine, transplanted HIOs and HCOs expressed distinct regional markers and contained regionally enriched cell types. For example, the majority of the epithelium of NOGGIN and control HIOs expressed the proximal marker GATA4 and did not express the large intestinal marker SATB2 (Figure 5F–I, K–N, Figure S5B–E). Conversely HCO epithelia were uniformly SATB2+ but did not express GATA4 (Figure 5J,O, Figure S5B–E). In addition, Paneth cells expressing DEFA5 were present in the crypts of NOGGIN and control HIOs, but were absent HCOs (Figure 5P–T, Figure S5F) similar to the human colon (Wehkamp et al., 2006). The colonic goblet cell marker MUC5B (van Klinken et al., 1998), was expressed by a subset of goblet cells of HCOs but is not detectible in NOGGIN or control HIOs (Figure 5U–Y, Figure S5G). Additionally, MUC2+ goblet cell numbers were higher in HCOs compared HIOs consistent with the abundance of goblet cells seen in the human colon (Figure S5H–L). The patterning markers, the presence of MUC5B expressing goblet cells, and the absence of Paneth cells all support the conclusion that transplanted HCOs have colonic epithelium.
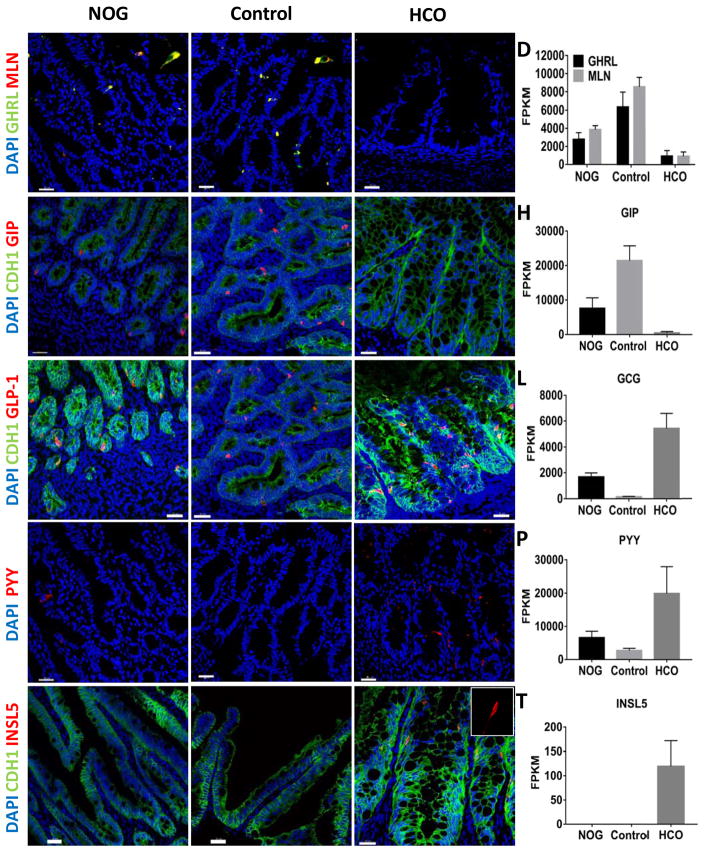
In vivo matured HIOs and HCOs express regional enteroendocrine hormones
There are at least 12 major EEC subtypes that are found in different regions of the gastrointestinal tract and we analyzed HIOs and HCOs for the presence of regional EECs. Ghrelin and Motilin are found predominantly in the proximal intestine, and correspondingly these hormones were largely expressed in NOGGIN and control HIOs but not HCOs (Figure 6A–D). Similarly, GIP, which is found in K-cells of the small intestine but is absent in the colon, were found in NOGGIN and control HIOs but not in HCOs (Figure 6E–H). We then examined presence of distally enriched EECs in HCOs by analyzing for expression of GLP-1 and PYY, which are more abundant in the colon. We observed higher numbers of GLP-1 and PYY cells and higher expression of preproglucagon and PYY in HCOs than in HIOs (Figure 6I–P). In addition, we found expression of the colon specific hormone INSL5 (Burnicka-Turek et al., 2012; Thanasupawat et al., 2013), only in HCOs (Figure 6Q–T).
Figure 6.
In vivo grown organoids express region specific hormones.
Analysis of expression of the regionally expressed hormones (A–D) Ghrelin (GHRL), Motilin (MLN), (E–H) GIP, (I–L) GLP-1, (M–P) PYY and (Q–T) INSL5 in HIOs and HCOs grown for 8–10 weeks underneath the mouse kidney capsule. The proximally enriched hormones GHRL, GIP and MLN were enriched in NOGGIN and control HIOs (A–H). The distally enriched hormones GLP-1 and PYY were enriched in BMP2-derived HCOs (I–O). The colon specific hormone INSL5 was only present in HCOs (Q–T). Data is representative of at least 5 transplanted organoids per condition. Insets in (A) and (B) show GHRL and MLN double positive cells. (D, H, L, P, T) FPKM values for GHRL, MLN, GIP, GLP1, PYY, and INSL5 are from RNA-seq data. FPKM values represent 3 biological replicates per condition. Scale bars= 30 μm.
Analysis of stem and progenitor cells in HIOs and HCOs in vitro and in vivo
To determine if in vitro-derived HIOs and HCOs express markers of stem and progenitor cells, we used the H9-BAC-LGR5-eGFP transgenic line that has been described previously (McCracken et al., 2014; Watson et al., 2014). Examination of LGR5-eGFP and SOX9 expression in in vitro grown organoids revealed expression in broad epithelial domains similar to the expression patterns in Lgr5-eGFP mice as early as e13.5 (Shyer et al., 2015) (Figure S6A–O). Consistent with in vivo maturation, the expression domains of LGR5-eGFP, SOX9 and KI67 became restricted to the inter-villus region of HIOs and the base of rudimentary crypts in HCOs following transplantation (Figure S6P–X). Given that Sox9 and Lgr5 mark intestinal and colonic stem cells capable of forming enteroids and colonoids in mice (Gracz et al., 2010; Ramalingam et al., 2012) we investigated if the epithelium of transplanted organoids could be isolated and used to generate enteroids and colonoids. Both HIOs and HCOs gave rise to cultures of epithelial organoids that grew and could be passaged (Figure S6Y–A′). Moreover, HCO-derived epithelial cultures expressed the colonic markers CKB, FXYD3, SATB2, and HOXB13 but did not express the proximal small intestine markers PDX1 or GATA4 suggesting that regional identity was maintained (Figure S6B′–D′). These data suggest that HIOs and HCOs grown in vitro and in vivo contain regional progenitor and stem cells.
Global transcriptional analysis of HIOs and HCOs
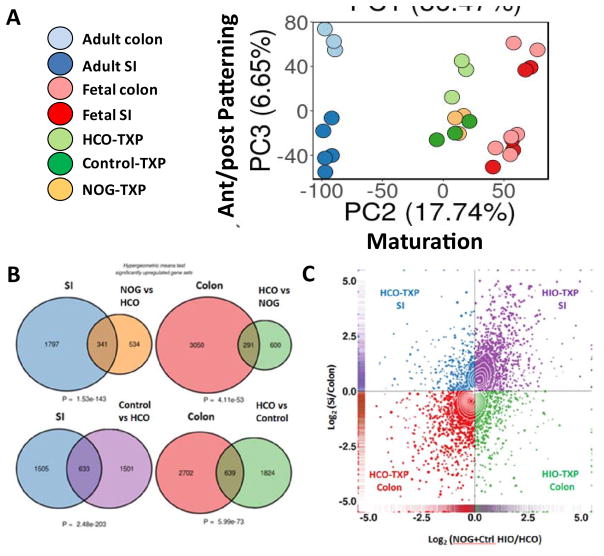
In order to broadly interrogate the regional identity and maturation of HIOs and HCOs, we performed RNA-seq analysis of HIOs and HCOs grown in vivo and compared them with published data sets of human fetal and adult small and large intestines. Principal component analysis revealed that primary tissues isolated from adult and fetal intestine clustered together along principal component 1 (PC1) axis, which accounted for 36.5% of the cumulative variation among samples (Figure S7A). A GO analysis revealed that this variation was due to cell types that were present only in primary tissues and not our PSC-derived transplants. For example 6 of the top 10 biological processes present in human primary tissues and absent in transplants were related to immune cells (Figure S7B–C). The second principal component (PC2) accounts for 17.7% of cumulative variation and separates the samples according to maturity (Figure 7A). This component revealed that transplanted organoids are more mature than human fetal intestine and fetal colon but not as mature as adult colon and intestine. The third principal component (PC3) accounts for 6.7% of cumulative variation and separates the samples according to regional identity, and shows that HCOs are more similar to colon whereas HIOs cluster with small intestine (Figure 7A). Interestingly, human fetal samples did not cluster based on regional identity (small intestine vs colon) suggesting that these samples may not have been cleanly isolated from the indicated region of the GI tract.
Figure 7.
Global transcriptional analysis of HIOs and HCOs and comparison with human small intestine and colon.
(A) Principal component analysis human adult and fetal small intestine and colon compared with transplanted HIOs and HCOs. (B) Hypergeometric means test comparing human adult small intestine with HIOs and human adult colon with HCOs. (C) 4-way scatter plot comparing transcripts that were differentially expressed in human small intestine and colon compared to HIOs and HCOs.
We next used hypergeometric means test to determine the probability that HIOs and HCOs share similar patterns of region-specific gene expression small intestine and colon (Figure 7B). A total of 341 transcripts are expressed in the small intestine and in NOGGIN treated HIOs as compared to colon or BMP2 treated HCOs, a proportion that is exceedingly unlikely by chance alone (P = 1.5 × 10−143). Similarly, the gene set that is up-regulated in the control HIOs shares a highly significant degree of similarity with the gene set up-regulated in adult small intestine relative to the adult colon (P = 2.5 × 10−203). Conversely, the gene set up-regulated in HCOs are highly enriched for genes that are up-regulated in the colon relative to the small intestine (P = 4.1 × 10−53 and P = 6.0 × 10−73, respectively). This analysis concluded that HIO patterning is most similar to human small intestine and HCO patterning is colonic. To further explore the nature of HIOs (NOG and control treated) and HCOs, we conducted differential expression analysis (adult small intestine vs. adult colon; HIOs vs. HCOs). We generated 4-way scatter plot, which also demonstrated that a high proportion of genes up-regulated in the colon were also up-regulated in HCOs and the majority of genes up-regulated in the small intestine were also up-regulated in HIOs (Figure 7C, Table S1). Taken together, these data suggest we have developed a robust method to differentiate PSCs into human colonic tissue.
DISCUSSION
Historically, the classification of foregut, midgut, and hindgut are based on the development of the anterior and posterior intestinal portals and the source of mesenteric blood supply (Uppal et al., 2011). An alternative definition of midgut and hindgut have been proposed, in which the midgut is the portion of the intestine derived from the portion anterior to the umbilicus and the hindgut derives posterior to the umbilicus (Johnston, 1913; Savin et al., 2011). In either case, the historic reliance on anatomical landmarks, and lack of more precise molecular markers to distinguish fore, mid and hindgut, have made it difficult to develop methods to generate these cell/tissues in vitro from PSCs. Therefore, identification of markers that clearly demarcate regions of developing mid and hindgut is essential.
We used a combination of CDX2, GATA4, ONECUT1 and SATB2 to identify that distinct molecular boundaries are established at early stages of mid and hindgut development in Xenopus, mouse and humans. Interestingly, GATA4 and SATB2 expression domains form a boundary at the yolk stalk/presumptive umbilical cord in mice, and this boundary is maintained throughout development and in the adult intestine. The fact that GATA4 expression marks the intestine anterior to the umbilicus, and SATB2 expression marks the domain posterior to the umbilicus, suggests that the umbilicus is the boundary between the midgut and hindgut (Johnston, 1913; Savin et al., 2011).
While ONECUT1 expression in HIOs and SATB2 expression in HCOs is consistent with their proximal and distal identify respectively, GATA4 was not as robustly expressed in proximal HIOs in vitro as would be expected given its embryonic expression (data not shown). In contrast, GATA4 was robustly expressed following in vivo maturation of HIOs and in enteroids generated from patient biopsies (data not shown). This could suggest that factors involved in expression of GATA4 are absent in our culture conditions or that maturation in vivo is required for epithelial expression of GATA4. This data also suggests that high levels of GATA4 expression may be dispensable for early regionalization of the intestine, consistent with intestinal Gata4 knockout mice that retain normal Onecut factor expression (Battle et al., 2008). In addition, a small subset of BMP treated organoids lost CDX2 expression and activated expression of the bladder markers Keratin 13 and Uroplakin 1a (data not shown). This is consistent with BMP organoids having a hindgut fate since urothelial tissue is derived from the hindgut/cloaca (Georgas et al., 2015).
SATB2 is expressed throughout development of the distal ileum and large intestine, however it is not known if SATB2 is required for development of the distal intestine. Mouse knockout studies have focused on craniofacial and cortical neuronal development since mutations in SATB2 has been implicated in Cleft Palate associated with 2q32-q33 deletions and Glass Syndrome (FitzPatrick et al., 2003). However, there is indirect evidence that SATB2 may play a role human colonic physiology. SATB2 has been identified in Genome Wide Association Studies as an ulcerative colitis susceptibility gene (McGovern et al., 2010). In addition, loss of SATB2 expression has been shown to be associated with poor prognosis in colorectal cancer patients (Eberhard et al., 2012). Future studies with HCOs may allow identification of SATB2 targets in the developing colon, which could provide insight into the pathology of ulcerative colitis and colorectal cancer.
Several studies in model organisms have implicated the BMP signaling pathway in patterning endoderm during hindgut development (Kumar et al., 2003; Roberts et al., 1995; Sherwood et al., 2011; Tiso et al., 2002; Wills et al., 2008). Consistent with this, we have demonstrated that posterior patterning of human definitive endoderm is dependent on BMP signaling, as inhibition of BMP abrogates the ability of WNT and FGF to promote a posterior endoderm fate (McCracken et al., 2014). However, it is not surprising that BMP signaling plays other temporally distinct roles during intestinal development. For example, after the establishment of proximal-distal regional domains, BMP signaling functions to establish the crypt-villus axis in the intestine and colon (Li, 2005). Thus, a temporal requirement for patterning allows the embryo to use the same signaling pathway for multiple purposes gut development, as has been reported in Drosophila midgut (Driver and Ohlstein, 2014; Guo et al., 2013). In a humans disease context, mutations in BMPR1A are associated with a subset of patients with Juvenile Polyposis Syndrome. The HCO system was highly amenable for identifying the HOX code that is downstream of BMP during early development and it could be interesting to determine if hamartomatous polyps with BMPR1A mutations have altered HOX gene expression.
We previously reported the in vitro directed differentiation and in vivo transplantation of HIOs (Spence et al., 2011; Watson et al., 2014), which were small intestinal. Given the physiology and pathological conditions that affect the large intestine, it was imperative to develop a colonic model system to interrogate pathophysiological questions specific to the colon. Developmentally, this system has allowed us to investigate fundamental questions about how regional identity is established. HIOs and HCOs develop tissue specific cell types, such as Paneth cells in the HIOs and colon-specific goblet cells in HCOs. Moreover HIOs and HCOs have a distinct set of EECs that are normally enriched in the small and large intestine, respectively. Regionalized organoids should provide a platform for future studies of how different regions of the intestine give rise to regionalized stem cells. In addition, generation of HCOs will allow for modeling of diseases that affect the colon such as ulcerative colitis and colorectal cancer.
CONTANCT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to the Lead Contact James M. Wells (james.wells@cchmc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human biopsy tissue
Human jejunum and colon biopsy tissue was collected with informed consent from donors and under the approval of the Institutional Review Board of Cincinnati Children’s Hospital Medical Center (protocol 2012-2858). Samples were used as controls for immunofluorescence staining of regional markers.
Animals
Wild type CD1 mice were purchased from Charles River labs and were used for timed matings and analysis of fetal intestine. Immune deficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice, 8–16 weeks old were used in transplantation experiments (obtained from the Comprehensive Mouse and Cancer Core Facility, Cincinnati, Ohio). All mice were housed in the animal facility at the Cincinnati Children’s Hospital Medical Center (CCHMC) in accordance with NIH Guidelines for the Care and Use of Laboratory Animals. Animals were maintained on a 12hr light-dark cycle with access to water and standard chow ad libitum. Healthy animals were used for all experiments. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of CCHMC (protocols 2013-0006, 2016-0004, 2016-0014, and 2016-0059). Wildtype outbred Xenopus tropicalis (purchased from Nasco) were used for frog studies. Adult Xenopus tropicalis animals were housed according to Cincinnati Children’s Hospital Institutional Animal Care And Use Committee (IACUC) guidelines (protocol IACUC2016-0059).
Cell lines and maintenace
Human ESC lines H1 (WA01) and H9 (WA09) were purchased from WiCell (NIH approval number NIHhESC-10-0043 and NIHhESC-10-0062). The H1 line is male and the H9 line is female. The IPSC72.3 line was generated by the CCHMC Pluripotent Stem Cell Facility, was approved by the CCHMC institutional review board and was previously characterized (McCracken et al., 2014). IPSC72.3 had a normal male karyotype and differentiated into endoderm, mesoderm, and ectoderm lineages in an in vivo teratoma assay. Human embryonic stem cells and induced pluripotent stem cells were grown in feeder-free conditions in six-well Nunclon surface plates (Nunc) coated with Matrigel (BD Biosciences) and maintained in mTESR1 media (Stem Cell Technologies) at 37°C with 5% CO2. Cells were checked daily for differentation and were passaged every 4 days using Dispase solution (Thermo-Fisher Scientific). All lines were checked for karyotype and routinely checked for mycoplasma.
METHOD DETAILS
Experimental Design
To ensure reproducibility of the method for gut tube spheroid generation (Munera and Wells, 2017; Watson et al., 2014), the same method was used by at least 9 different investigators in the Wells lab and by 2 different investigators in the Helmrath lab. Induction of posterior HOX factors in HCOs was observed by 6 different investigators. No specific methods were used for randomization and investigators were not blinded to the identity of samples. No statistical methods were utilized to determine sample size.
BMP inhibition in frog and mouse embryos
Xenopus tropicalis embryo culture and small molecule treatments were performed as previously described (Rankin et al., 2012; Rankin et al., 2015). DMH-1 (Sigma D8946) was dissolved in DMSO, and used at final concentration of 20 μM; equal concentrations of DMSO vehicle were used on sibling embryos. Inhibitor treatment experiments were repeated twice with similar effects on the markers analyzed. For Xenopus in-situ hybridization analyses, DIG-labeled antisense RNA probes were generated using linearized full-length cDNA plasmid templates (X.tropicalis satb2 was purchased from ATCC, clone 7720194; HinDIII, T7 for probe). In-situ hybridization was performed as described previously (Sive et al., 2000). Briefly, embryos were fixed in MEMFA (100 mM MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 3.7% (v/v) formaldehyde) at 4°C overnight and then stored in 1 00% ethanol at −20°C. Fixed embryos were then re-hydrated, permeabilized with proteinase K, washed with 0.1M tri-ethanolamine (pH 7.0–8.0) and then neutralized with acetic anhydride. Embryos were then re-fixed in PBS+3.7% formaldehyde for 20 minutes, and pre-hybridized in hybridization buffer (50% Formamide, 5X SSC pH7,1 mg/mL Torula Yeast RNA (Sigma R3629-5G), 100 ug/mL heparin (Sigma H3393-50KU) , 1X Denhardt’s , 0.1% Tween-20, 10mM EDTA pH 8) for 3 hours at 65°C. Embryos were then incubated in DIG-probe solution (0.5ug/mL) overnight at 65°C, washed extensively in 2X and 0.2X SSC buffers, blocked in blocking buffer (maleic acid buffer, MAB, 100mM maleic acid,150mM NaCl, 20% Heat-inactivated lamb serum (Gibco 16070-096), 2% Roche Blocking reagent (Sigma 11-096-176-001)) for 2 hours at room temperature, and incubated in anti-DIG-alkaline phosphatase (Sigma11093274910) diluted 1:5,000 in blocking buffer overnight at 4°C. After extensive washing in MAB, embryos were incubated in alkaline phosphatase buffer (100mM TRIS-Cl (pH 9.5), 50mM MgCl2, 100mM NaCl, 0.1% Tween-20, 2mM levamisole) 2x15 minutes, and then in BM Purple chromogenic substrate (Sigma 11442074011) at 18°C for 24 to 48 hours. The staining reaction was stopped by washing in MAB+10mM EDTA and then embryos were fixed in Bouin’s fixative (LabChem,Inc LC11790-1) overnight to fix the stain. Finally, embryo pigment was bleached in 1%H2O2, 5% Formamide, 0.5X SSC.
For mouse whole embryo cultures, e7.5 embryos were cultured in a 1:1 mixture of Ham’s F12 medium and whole embryo culture rat serum (Harlan Labs) containing N-2 Supplement (Invitrogen). Vessels were placed on a roller culture apparatus (BTC Engineering, Cambridge, UK) and maintained for 2 days at 37°C and gassed with 20% O2 and 5% CO2. BMP signaling was inhibited by treatment with 5 μM DMH-1, with DMSO serving as a vehicle control. For wholemount immunofluorescence staining, embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Embryos were then dehydrated in m ethanol and stored at −20°C. To permeabilize embryos for staining, embryos were incubated in Dent’s bleach (4:1:1 Methanol, DMSO, 30% H2O2) for 2 hours at room temp on a Labnet Gyrotwister. Embryos were then rehydrated in a methanol plus PBS series (100%, 75%, 50%, 25%, 0%). Following rehydration, samples were processed as detailed below for organoid wholemounts.
Generation of human gut tube cultures
Human intestinal organoids were generated and maintained as previously described (Munera and Wells, 2017; Watson et al., 2014). Human embryonic stem cells and induced pluripotent stem cells were grown in feeder-free conditions in six-well Nunclon surface plates (Nunc) coated with Matrigel (BD Biosciences) and maintained in mTESR1 media (Stem Cell Technologies). For induction of definitive endoderm (DE), human ES or iPS cells were passaged with Accutase (Invitrogen) and plated at a density of 100,000 cells per well in a Matrigel-coated, Nunclon surface 24-well plate. For Accutase split cells, 10 μM Y27632 compound (Sigma) was added to the media for the first day. After the first day, media was changed to mTESR1 and cells were grown for an additional 24 hours. Cells were then treated with 100 ng/mL of Activin A for 3 days as previously described (Spence et al., 2011). DE was then treated with hindgut induction medium (RPMI 1640, 2 mM L-glutamine, 2% decomplemented FBS, penicillin-streptomycin and 100 ng/mL Activin A) for 4 d with 500 ng/mL FGF4 (R&D) and 3 μM Chiron 99021 (Tocris) to induce formation of mid-hindgut spheroids.
Patterning of human gut tube cultures
Spheroids were collected from 24-well plates pooled and plated in Matrigel (BD) with a minimum of 30 spheroids plated per well. To generate proximal HIOs, spheroids were overlayed with intestinal growth medium (Advanced DMEM/F-12, N2, B27, 15 mM HEPES, 2 mM L-glutamine, penicillin-streptomycin) supplemented with 100 ng/mL EGF (R&D) alone, or 100 ng/mL EGF with 100 ng/ml NOGGIN (R&D). To generate HCOs, spheroids were overlayed with 100 ng/mL EGF plus 100 ng/mL BMP (R&D). For SHH experiments, 1 μM SAG (Tocris), 5 μM SAG or 2.5 μM Cyclopamine (Tocris) were added to control media for initial 3 days after which RNA samples were collected. Media was changed at 3 days with only EGF being maintained in the media for all patterning conditions. Media was then changed twice weekly thereafter. HIOs and HCOs were replated in fresh Matrigel every 14 days at a density of 5–10 organoids per well.
Generation of NEUROG3 inducible line
To generate a doxycycline inducible NEUROG3 line, we transduced IPSC 72.3 cells with pINDUCER21-NEUROG3 lentivirus and selected using 250 g/mL of G418. Both the IPSC 72.3 cell line and the inducible NEUROG3 have been described previously (McCracken et al., 2014). Stably transduced cells were differentiated into mid/hindgut spheroids and then patterned into HIOs or HCOs. Spheroids were grown for 28 days and were pulsed with 0.5ug/mL of doxycycline for 8 hrs. At day 35, organoids were collected and were analyzed by QPCR and IF. Multiple wells of organoids (10–20 organoids) were collected for each biological replicate.
Growth of organoid mesenchyme
Mesenchymal cells from organoids which attach to the bottom of the 24-well plate and grow in 2 dimensions. To expand mesenchymal cells from organoids, DMEM 10%FBS + L-glutamine + penicillin-streptomycin was added to wells from which organoids had been harvested at 14 days. Media was changed twice weekly for a total of 2–3 weeks until near 100% confluence was achieved. Once confluent, mesenchymal cells were collected for QPCR analysis.
Transplantation of HIOs and HCOs
Transplantation studies were done with approval from the CCHMC IACUC (protocols 2013-0006 and 2016-0014). NSG mice were kept on antibiotic chow (275 p.p.m. Sulfamethoxazole and 1,365 p.p.m. Trimethoprim; Test Diet). Food and water was provided ad libitum before and after surgeries. A single HIO, matured in vitro for 28 days, was removed from Matrigel, washed with cold phosphate-buffered saline (DPBS; Gibco), and embedded into purified type I collagen (rat tail collagen; BD Biosciences) 12 hours before surgery to allow for formation of a solidified gel plug. These plugs were then placed into standard growth media overnight in intestinal growth medium (Advanced DMEM/F-12, B27, 15 mM HEPES, 2 mM L-glutamine, penicillin-streptomycin) supplemented with 100 ng/mL EGF (R&D). HIOs were then transplanted under the kidney capsule as previously reported (Watson et al., 2014). Briefly, the mice were anesthetized with 2% inhaled isoflurane (Butler Schein), and the left side of the mouse was then prepped in sterile fashion with isopropyl alcohol and povidine-iodine. A small left-posterior subcostal incision was made to expose the kidney. A subcapsular pocket was created and the collagen-embedded HIO was then placed into the pocket. The kidney was then returned to the peritoneal cavity and the mice were given an IP flush of Zosyn (100 mg/kg; Pfizer Inc.). The skin was closed in a double layer and the mice were given a subcutaneous injection with Buprenex (0.05 mg/kg; Midwest Veterinary Supply). At 8–10 weeks following engraftment, the mice were then humanely euthanized and kidney transplants were removed.
Culture of crypts from transplanted organoids
Transplants were gently opened to expose lumen and pinned flat on a Sylgard-coated Petri dish with the mucosal surface exposed. Villi and mucus were removed from the mucosal surface by gently scratching the surface with curved forceps. The tissue was then washed several times with the ice-cold chelation buffer (PBS + 2% sorbitol, 1% sucrose, 1% BSA). The mucosa was then covered in 2mM EDTA chelation buffer and incubated for 30 minutes on ice. Mucosa was then washed with chelation buffer (without EDTA) several times to remove the EDTA. Crypts were harvested by gently scraping the mucosal surface using curved forceps. Crypts were then transferred to a 15 ml conical tube, filtered through a 150 micron mesh and spun down at 150 × g for 10 minutes. The pellet containing the crypts was then resuspended in Matrigel and plated in a 24 well plate. Cultures were maintained in WENR + gastrin + nicotinamide + A83-01 + SB202190 media defined by Sato and colleagues (Sato et al., 2011).
Immunofluorescence Staining
Tissues were fixed for 1–3 hours in 4% PFA on ice depending on the size of the tissue. Organoids and transplant engraftments were frozen in OCT. OCT sections were blocked using donkey serum (5% serum in 1X PBS plus 0.5% Triton-X) for 30 min and incubated with primary antibody overnight at 4 °C. Slides were then washed 3X with 1X PBS plus 0.5% Triton-X (PBST) and incubated in secondary antibody with DAPI in blocking buffer for 2 h at room temperature. Please see Table S2 for a list of antibodies and respective dilutions. Slides were then washed 2X with 1X PBS plus 0.5% Triton-X followed by a final wash in 1X PBS. Coverslips were then mounted using Fluoromount-G® (SouthernBiotech). Images were captured on a Nikon A1 confocal microscope and analyzed using Imaris Imaging Software (Bitplane).
For whole-mount staining of organoids (and mouse embryos), fixed organoids were washed with PBS and then permeabilized in PBST overnight at 4 °C on a rocking platform. Organoids were then blocked for 6–8 hours on a rocking platform, incubated in primary antibody overnight at 4 °C on a rocking platform, washed 5X with PBST, incubated in secondary antibody overnight at 4 °C on a rocking platform, washed 2X with PBST followed by a final wash in PBS. Organoids were then dehydrated by washing 3 times in 100% Methanol and cleared using Murray’s clear (2 parts benzyl benzoate and 1 part benzyl alcohol). Images were captured on a Nikon A1 confocal microscope using Z-correction and analyzed using Imaris Imaging Software (Bitplane).
Quantification of Immunofluorescence images
Image quantitation of whole embryos was done by splitting images into separated channels and then measuring pixel area using ImageJ (NIH). Pixel area was determined for each channel, the ratio between channels was determined and the ratio for control treated embryos was represented as 100. Quantitation of in vitro and in vivo grown organoids was done on sections from which images were captured as explained above. The number of CDX2, GATA4 and SATB2 positive nuclei were quantified using the spot function in Imaris following calibration with human biopsy samples.
RNA isolation and QPCR
RNA was extracted using Nucleospin® RNA extraction kit (Macharey-Nagel) and reverse transcribed into cDNA using Superscript VILO (Invitrogen) according to manufacturer’s protocols. QPCR primers were designed using the qPrimerDepot webased tool (primerdepot.nci.nih.gov). Primer sequences are listed in Table S3. QPCR was performed using Quantitect SYBR® Green PCR kit (Qiagen) and a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems). For spheroids and organoids 2–3 wells were collected for each biological replicate.
Identification of the posterior marker SATB2
To identify markers of large intestine, we first used GNCPro http://gncpro.sabiosciences.com/gncpro/expression_grapher.php to identify transcription factors upregulated in colon (compared to other tissues) based on the University of Tokyo database. Based on this search, SATB2 was the 6th ranked gene in colon. To verify that SATB2 is indeed upregulated in the colon, we searched SATB2 expression using the TiGER database (http://bioinfo.wilmer.jhu.edu/tiger/db_gene/SATB2-index.html). To further confirm the expression of SATB2 in the colon, and to examine protein expression across numerous tissues, we used the Human Protein Atlas (http://www.proteinatlas.org/search/satb2). A similar approach was used to identify other markers of large intestine/colon.
RNA-seq processing
RNA library construction and RNA sequencing was performed by the Cincinnati Children’s Hospital DNA Sequencing Core, using an Illumina HiSeq2500 platform. The quality of the Illumina sequencing run was evaluated by analyzing FASTQ data for each sample using FastQC version 0.10.1 http://www.bioinformatics.babraham.ac.uk/projects/fastqc to identify features of the data that may indicate quality problems (e.g. low quality scores, over-represented sequences, inappropriate GC content, etc.). No major issues were identified by the QC analysis. We used the software package Tuxedo Suite for alignment, differential expression analysis, and post-analysis diagnostics. Briefly, we aligned reads to the reference transcriptome (UCSC hg19) using TopHat version 2.0.13 and Bowtie version 2.2.5 (Langmead et al., 2009). We used default parameter settings for alignment, with the exception of: “–b2-very-sensitive” to maximize the accuracy of the read alignment, as well as “–no-coverage-search” and “–no-novel-juncs” limiting the read mapping to known transcripts. Cufflinks version 2.2.1 (Trapnell et al., 2012) was used for RNA abundance estimation. UCSC hg19.fa was used as the reference genome sequence and UCSC hg19.gtf was used for transcriptome annotation. We applied the following parameters in Cufflinks: “–multi-read-correct” to adjust expression calculations for reads that map in more than one locus, and “–compatible-hits-norm” and “–upper-quartile–norm” for normalization of expression values. Normalized TPM and FPKM tables were generated using the CuffNorm function. RNA sequence assembly and transcriptional analysis was conducted using the 64-bit Debian Linux stable version 7.10 (“Wheezy”) platform.
Differential expression analysis
All plots and statistical analyses were conducted in R version 3.3.1 (2016-06-21). Plots were generated using the R package ‘ggplot2’ (Ginestet, 2011). Differential expression analysis and statistical tests of Cufflinks output were completed with the R package ‘SeqRetriever’ ‘SeqRetriever’ version 0.6 https://github.com/hilldr/SeqRetriever. Hypergeometric means testing was used to evaluate relative enrichment of shared gene expression signatures between groups using the R package ‘GeneOverlap’ http://shenlab-sinai.github.io/shenlab-sinai/. Gene ontology analysis was done using lists of differentially expressed genes which were analyzed in TOPPGENE Suite (https://toppgene.cchmc.org/).
QUANTIFICATION AND STATISTICAL ANALYSIS
For animal experiments such as those in Figure 1 and Figure S1, “n” represents the number of animals which were analyzed. For experiments involving patterned spheroids and in vitro grown organoids, “n” represents the number of biological replicates (2–3 wells were collected for each replicate). For experiments involving transplanted organoids, “n” represents the number of individual animals that were transplanted with a single organoid. Quantification of data are represented as mean ± SEM with the exception of Figure 2L in which data is represented as mean ± SD. Data for NOGGIN HIOs and Control HIOs was combined for comparisons with HCOs based on their similarities in RNA and protein expression. To directly compare HIOs and HCOs, t tests with 2-tailed distribution and two-sample equal variance were run in Microsoft Excel. This analysis assumes equal variance which we confirmed by F test.
DATA AND SOFTWARE AVAILABILITY
The accession number for data generated for this paper is ArrayExpress accession E-MTAB-5658 and includes the 3 day patterning data shown in figure 2 as well as the data from patterned organoids which were transplanted in vivo shown in Figure 7 and Figure S7.
Adult small intestine and large intestine RNA-seq data were downloaded from the public database E-MTAB-1733. These data sets represent whole organ tissue which includes the epithelium and muscle layers. Accession numbers for the small intestine samples: ERR315344, ERR315381, ERR315409, ERR315442, ERR315461. Accession numbers for the large intestine samples: ERR315348, ERR315357, ERR315484. For Figure S2B, processed FPKM data was downloaded from https://github.com/hilldr/Finkbeiner_StemCellReports2015. These data include adult duodenum (ERS326992, ERS326976) and small intestine samples listed above from E-MTAB-1733 as well as human fetal intestinal (also whole organ) samples from GSE18927. Accession numbers for human fetal small intestine are GSM1059508, GSM1059521, GSM1059486, GSM1059507, GSM1059517, GSM1220519. For Figure S2C, data was obtained from GEO accession GSE66749 platform GLP5175. The following samples were used: GSM1385160, GSM1385161, GSM1385162, GSM1385163, GSM1385164, GSM1385165, GSM1385166, GSM1385167, GSM1385168, GSM1385169, GSM1385170, GSM1385171, GSM1614646, GSM1614646. Sample values were determined using the GEO2R “profile graph” function and searching for GATA4 and SATB2 by their ID numbers (3086100 and 2594089 respectively).
The complete RNA-seq FASTQ processing pipeline and analysis scripts are available at https://github.com/hilldr/Munera2017.
Supplementary Material
Acknowledgments
We thank the members of the Wells and Zorn laboratories for reagents and feedback. We acknowledge core support from the Cincinnati Digestive Disease Center Award (P30 DK0789392), CCHMC Confocal Imaging Core and Pluripotent Stem Cell Facility. We would like to thank Matt Kofron and the Confocal Imaging Core for training and guidance and Chris Mayhew and Amy Pitstick from the Pluripotent Stem Cell Facility for cultures and reagents. We would also like to thank Jaime Schweitzer, Mariana Louza-Stevens and Laura Runck for technical help. This work was supported by National Institutes of Health grants R01DK092456, U19 AI116491, U18EB021780 (J.M.W.), R01DK102551 (M.H.M.), U01DK103117 (M.H.M, N.F.S. and J.M.W), NIDDK070858 (A.M.Z.) and a postdoctoral fellowship from the Crohn’s and Colitis Foundation of America (J.O.M.).
Footnotes
Author Contributions
J.O.M. and J.M.W. conceived the study and experimental design, performed and analysed experiments and co-wrote the manuscript. N.S., C.W., M.M, J.E.V.,N.F.S, and M.H. performed kidney transplantation experiments. S.A.R. and A.M.Z. performed Xenopus experiments. J.C.H., A.L.Z., J.R.H. and K.S. performed experiments. P.C., D.H. and J.R.S. performed bioinformatic analysis. J.M.S performed mouse whole embryo culture experiments. All authors contributed to the writing or editing of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aronson BE, Aronson SR, Berkhout RP, Chavoushi SF, He A, Pu WT, Verzi MP, Krasinski SD. GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27. Bba-Gene Regul Mech. 2014;1839:1273–1282. doi: 10.1016/j.bbagrm.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Bondow BJ, Iverson MA, Adams SJ, Jandacek RJ, Tso P, Duncan SA. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135:1676–1686. e1671. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Bosse T, aan de Kerk DJ, Piaseckyj CM, Fujiwara Y, Katz SG, Orkin SH, Grand RJ, Krasinski SD. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008a;322:179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Bosse T, Buckner MA, Krasinski SD. Co-localization of Gata4 and Hnf1 alpha in the gastrointestinal tract is restricted to the distal stomach and proximal small intestine. Gastroenterology. 2007a;132:A586–A586. [Google Scholar]
- Beuling E, Bosse T, de Kerk DA, Piaseckyj CM, Fujiwara Y, Orkin SH, Krasinski SD. Fog cofactors partially mediate Gata4 function in the adult mouse small intestine. Gastroenterology. 2007b;132:A692–A693. [Google Scholar]
- Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA, et al. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut. 2010;59:888–895. doi: 10.1136/gut.2009.204990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Kerkhof IM, Piaseckyj CM, Dawson PA, Pu WT, Grand RJ, Krasinski SD. The absence of GATA4 in the distal small intestine defines the ileal phenotype. Gastroenterology. 2008b;134:A83–A84. [Google Scholar]
- Bosse T, Fialkovich JJ, Piaseckyj CM, Beuling E, Broekman H, Grand RJ, Montgomery RK, Krasinski SD. Gata4 and Hnf1alpha are partially required for the expression of specific intestinal genes during development. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1302–1314. doi: 10.1152/ajpgi.00418.2006. [DOI] [PubMed] [Google Scholar]
- Bouchi R, Foo KS, Hua H, Tsuchiya K, Ohmura Y, Sandoval PR, Ratner LE, Egli D, Leibel RL, Accili D. FOXO1 inhibition yields functional insulin-producing cells in human gut organoid cultures. Nat Commun. 2014;5:4242. doi: 10.1038/ncomms5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnicka-Turek O, Mohamed BA, Shirneshan K, Thanasupawat T, Hombach-Klonisch S, Klonisch T, Adham IM. INSL5-deficient mice display an alteration in glucose homeostasis and an impaired fertility. Endocrinology. 2012;153:4655–4665. doi: 10.1210/en.2012-1161. [DOI] [PubMed] [Google Scholar]
- De Santa Barbara P, Williams J, Goldstein AM, Doyle AM, Nielsen C, Winfield S, Faure S, Roberts DJ. Bone morphogenetic protein signaling pathway plays multiple roles during gastrointestinal tract development. Dev Dyn. 2005;234:312–322. doi: 10.1002/dvdy.20554. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Driver I, Ohlstein B. Specification of regional intestinal stem cell identity during Drosophila metamorphosis. Development. 2014;141:1848–1856. doi: 10.1242/dev.104018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duluc I, Freund JN, Leberquier C, Kedinger M. Fetal endoderm primarily holds the temporal and positional information required for mammalian intestinal development. J Cell Biol. 1994;126:211–221. doi: 10.1083/jcb.126.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlen M, Ericson Lindquist K, Jirstrom K. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931–938. doi: 10.1038/bjc.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, et al. Identification of SATB2 as the cleft palate gene on 2q32–q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Georgas KM, Armstrong J, Keast JR, Larkins CE, McHugh KM, Southard-Smith EM, Cohn MJ, Batourina E, Dan H, Schneider K, et al. An illustrated anatomical ontology of the developing mouse lower urogenital tract. Development. 2015;142:1893–1908. doi: 10.1242/dev.117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J R Stat Soc a Stat. 2011;174:245–245. [Google Scholar]
- Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24- expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol-Gastr L. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201:945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci. 2008;27:865–873. doi: 10.1111/j.1460-9568.2008.06061.x. [DOI] [PubMed] [Google Scholar]
- Haramis APG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, Offerhaus GJ, Van Deventer SJ, Peppelenbosch MP. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126:111–121. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang JW, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Higuchi Y, Kojima M, Ishii G, Aoyagi K, Sasaki H, Ochiai A. Gastrointestinal Fibroblasts Have Specialized, Diverse Transcriptional Phenotypes: A Comprehensive Gene Expression Analysis of Human Fibroblasts. Plos One. 2015;10 doi: 10.1371/journal.pone.0129241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PWH, Booth HAF, Bruford EA. Classification and nomenclature of all human homeobox genes. Bmc Biol. 2007;5 doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeejeebhoy KN. Short bowel syndrome: a nutritional and medical approach. CMAJ. 2002;166:1297–1302. [PMC free article] [PubMed] [Google Scholar]
- Johnston TB. Extroversion of the Bladder, complicated by the Presence of Intestinal Openings on the Surface of the Extroverted Area. J Anat Physiol. 1913;48:89–106. [PMC free article] [PubMed] [Google Scholar]
- Kohlnhofer BM, Thompson CA, Walker EM, Battle MA. GATA4 regulates epithelial cell proliferation to control intestinal growth and development in mice. Cell Mol Gastroenterol Hepatol. 2016;2:189–209. doi: 10.1016/j.jcmgh.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LH. BMP signaling inhibits intestinal stem cell self-renewal through antagonizing Wnt signaling. Gastroenterology. 2005;128:A702–A702. [Google Scholar]
- McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH, Wells JM. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A Dominant Mutation That Predisposes to Multiple Intestinal Neoplasia in the Mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Munera JO, Wells JM. Generation of Gastrointestinal Organoids from Human Pluripotent Stem Cells. Methods Mol Biol. 2017;1597:167–177. doi: 10.1007/978-1-4939-6949-4_12. [DOI] [PubMed] [Google Scholar]
- Patankar J, Obrowsky S, Hoefler G, Battle M, Kratky D, Levak-Frank S. Intestinal Deficiency of Gata4 Protects from Diet-Induced Hepatic Steatosis by Suppressing De Novo Lipogenesis and Gluconeogenesis in Mice. J Hepatol. 2012a;56:S496–S496. doi: 10.1016/j.jhep.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar JV, Obrowsky S, Doddapattar P, Hoefler G, Battle M, Levak-Frank S, Kratky D. Intestinal GATA4 deficiency protects from diet-induced hepatic steatosis. J Hepatol. 2012b;57:1061–1068. doi: 10.1016/j.jhep.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD, Magness ST. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol. 2012;302:G10–20. doi: 10.1152/ajpgi.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Gallas AL, Neto A, Gomez-Skarmeta JL, Zorn AM. Suppression of Bmp4 signaling by the zinc-finger repressors Osr1 and Osr2 is required for Wnt/beta-catenin-mediated lung specification in Xenopus. Development. 2012;139:3010–3020. doi: 10.1242/dev.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Thi Tran H, Wlizla M, Mancini P, Shifley ET, Bloor SD, Han L, Vleminckx K, Wert SE, Zorn AM. A Molecular atlas of Xenopus respiratory system development. Dev Dyn. 2015;244:69–85. doi: 10.1002/dvdy.24180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratineau C, Duluc I, Pourreyron C, Kedinger M, Freund JN, Roche C. Endoderm- and mesenchyme-dependent commitment of the differentiated epithelial cell types in the developing intestine of rat. Differentiation. 2003;71:163–169. doi: 10.1046/j.1432-0436.2003.t01-1-710203.x. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Johnson RL, Burke AC, Nelson CE, Morgan BA, Tabin C. Sonic Hedgehog Is an Endodermal Signal Inducing Bmp-4 and Hox Genes during Induction and Regionalization of the Chick Hindgut. Development. 1995;121:3163–3174. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pineiro AM, Bergstrom JH, Ermund A, Gustafsson JK, Schutte A, Johansson ME, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol. 2013;305:G348–356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Morey AL, Ferguson DJ, Fleming KA, Shmakov AN, Phillips AD. Human intestinal development in a severe-combined immunodeficient xenograft model. Differentiation. 1995;58:361–371. doi: 10.1046/j.1432-0436.1995.5850361.x. [DOI] [PubMed] [Google Scholar]
- Savin T, Kurpios NA, Shyer AE, Florescu P, Liang H, Mahadevan L, Tabin CJ. On the growth and form of the gut. Nature. 2011;476:57–62. doi: 10.1038/nature10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan-Rooney K, Swartz ME, Lovely CB, Dixon MJ, Eberhart JK. Bmp and Shh signaling mediate the expression of satb2 in the pharyngeal arches. PLoS One. 2013;8:e59533. doi: 10.1371/journal.pone.0059533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Maehr R, Mazzoni EO, Melton DA. Wnt signaling specifies and patterns intestinal endoderm. Mech Dev. 2011;128:387–400. doi: 10.1016/j.mod.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis : a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanasupawat T, Hammje K, Adham I, Ghia JE, Del Bigio MR, Krcek J, Hoang-Vu C, Klonisch T, Hombach-Klonisch S. INSL5 is a novel marker for human enteroendocrine cells of the large intestine and neuroendocrine tumours. Oncol Rep. 2013;29:149–154. doi: 10.3892/or.2012.2119. [DOI] [PubMed] [Google Scholar]
- Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Tubbs RS, Matusz P, Shaffer K, Loukas M. Meckel’s diverticulum: a review. Clin Anat. 2011;24:416–422. doi: 10.1002/ca.21094. [DOI] [PubMed] [Google Scholar]
- van Dop WA, Uhmann A, Wijgerde M, Sleddens-Linkels E, Heijmans J, Offerhaus GJ, Weerman MAV, Boeckxstaens GE, Hommes DW, Hardwick JC, et al. Depletion of the Colonic Epithelial Precursor Cell Compartment Upon Conditional Activation of the Hedgehog Pathway. Gastroenterology. 2009;136:2195–2203. doi: 10.1053/j.gastro.2009.02.068. [DOI] [PubMed] [Google Scholar]
- van Klinken BJ, Dekker J, van Gool SA, van Marle J, Buller HA, Einerhand AW. MUC5B is the prominent mucin in human gallbladder and is also expressed in a subset of colonic goblet cells. The American journal of physiology. 1998;274:G871–878. doi: 10.1152/ajpgi.1998.274.5.G871. [DOI] [PubMed] [Google Scholar]
- Walker EM, Thompson CA, Battle MA. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol. 2014;392:283–294. doi: 10.1016/j.ydbio.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, Gumucio DL. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A. 2012;109:15817–15822. doi: 10.1073/pnas.1205669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Grosse AS, Hu CB, Czerwinski M, Richards N, Gumucio DL. Epithelial Hedgehog signals direct mesenchymal villus patterning through BMP. Dev Biol. 2009;331:489–489. [Google Scholar]
- Walton KD, Whidden M, Kolterud A, Shoffner SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S, et al. Villification in the mouse: Bmp signals control intestinal villus patterning. Development. 2016;143:427–436. doi: 10.1242/dev.130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, Kern F, Ning G, Hong Y, Khor CC, et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522:173–178. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CL, Mahe MM, Munera J, Howell JC, Sundaram N, Poling HM, Schweitzer JI, Vallance JE, Mayhew CN, Sun Y, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, Bevins CL. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- Whissell G, Montagni E, Martinelli P, Hernando-Momblona X, Sevillano M, Jung P, Cortina C, Calon A, Abuli A, Castells A, et al. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- Wills A, Dickinson K, Khokha M, Baker JC. Bmp signaling is necessary and sufficient for ventrolateral endoderm specification in Xenopus. Dev Dyn. 2008;237:2177–2186. doi: 10.1002/dvdy.21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, Shah YM. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi N, Kosaki R, Ito T, Mitsuhashi T, Shimada H, Tomita M, Takahashi T, Kosaki K. Position-specific expression of Hox genes along the gastrointestinal tract. Congenit Anom (Kyoto) 2004;44:18–26. doi: 10.1111/j.1741-4520.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Zbuk KM, Eng C. Hamartomatous polyposis syndromes. Nat Clin Pract Gastr. 2007;4:492–502. doi: 10.1038/ncpgasthep0902. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.