Abstract
A main requisite in the phagocytosis of ingested material is a coordinated series of maturation steps which lead to the degradation of ingested cargo. Photoreceptor outer segment (POS) renewal involves phagocytosis of the distal disk membranes by the retinal pigment epithelium (RPE). Previously, we identified melanoregulin (MREG) as an intracellular cargo-sorting protein required for the degradation of POS disks. Here, we provide evidence that MREG-dependent processing links both autophagic and phagocytic processes in LC3-associated phagocytosis (LAP). Ingested POS phagosomes are associated with endogenous LC3 and MREG. The LC3 association with POSs exhibited properties of LAP; it was independent of rapamycin pretreatment, but dependent on Atg5. Loss of MREG resulted in a decrease in the extent of LC3-POS association. Studies using DQ™-BSA suggest that loss of MREG does not compromise the association and fusion of LC3-positive phagosomes with lysosomes. Furthermore, the mechanism of MREG action is likely through a protein complex that includes LC3, as determined by colocalization and immunoprecipitation in both RPE cells and macrophages. We posit that MREG participates in coordinating the association of phagosomes with LC3 for content degradation with the loss of MREG leading to phagosome accumulation.
Keywords: Melanoregulin, Autophagy, Retinal pigment epithelium, Phagocytosis, Microtubule-associated protein 1 light chain 3
Introduction
Phagocytosis is an evolutionarily conserved process by which phagocytes engulf external particles, enveloping them to form internal phagosomes [1]. Macroautophagy (hereafter referred to as autophagy) is also an evolutionarily conserved lysosomal catabolic process necessary for the degradation of cytoplasmic material in order to eliminate protein aggregates and limit bacterial proliferation, thus contributing to cell homeostasis during periods of stress including nutrient deprivation. Components of the autophagic pathway cooperate with phagocytosis in non-canonical autophagic processes [2, 3]. Amongst these, the hybrid autophagy-phagocytic pathway termed microtubule associated protein 1 light chain 3 (LC3) associated phagocytosis (LAP) is activated in macrophages upon bacterial challenge [4] as well as in epithelial cells during entosis [5, 6]. In the case of bacterial challenge by Escherichia coli for example, components of the autophagy pathway directly conjugate LC3 to phagosomal membranes encompassing bacteria in the absence of classic double membrane phagophore structures. The absence of LAP in these cells results in increased production of proinflammatory cytokines and decreased anti-inflammatory cytokines [7]. LC3 is also recruited to single membrane entotic vacuoles, macropinosomes, and phagosomes harboring dead cells [5, 6]. LAP utilizes the Vps34/beclin1 and Atg5/12/16 l conjugation systems resulting in lipidation of LC3 directly onto the single membrane (nascent) phagosomes with the LC3-decorated phagosome fusing with lysosomes for degradation. This autophagosome independent, LC3-associated degradative event occurs under nutrient replete conditions and is thus independent of the upstream mammalian target of rapamycin (mTOR)-mediated activation of the ULK1 complex. Several lines of evidence suggest that the convergence of the phagocytic and autophagic pathways results in enhanced clearance of engulfed material as degradative processes are synergistically utilized to accelerate phagosome maturation and increase degradation of internalized pathogens or debris [4, 8].
LAP appears to be required for the daily clearance of ingested material in the retinal pigment epithelium (RPE). Vertebrate photoreceptor cells maintain their health and normal physiological function through the life-long renewal of their outer segments. Diurnal phagocytosis by the RPE serves as a homeostatic regulator; in addition to the daily degradation of engulfed photoreceptor outer segment (POS) proteins, it is also responsible for the breakdown of POS-derived lipid components, as well as recycling of visual pigments [9, 10]. RPE cells are one of the most phagocytic cells known in nature; in a synchronized burst of activity, each of these post-mitotic cells phagocytosis distal tips of photoreceptors, each of which shed over 5 % of their outer segment mass daily [11–14]. Autophagy-dependent processes are particularly vital for maintaining homeostasis for long-lived post-mitotic cells like the RPE whose catabolic cascade is challenged with the daily burden of POS phagocytosis, LDL and oxLDL endocytosis and the clearance of intracellular debris. Progressive dysfunction of the degradative capacity of the RPE has been implicated in numerous pathways of retinal disease [15–18] with decreased LC3II resulting in accelerated aging and degeneration of the RPE [19, 20]. Studies by Reme et al. [21, 22] over 30 years ago identified autophagic structures and a diurnal pattern of autophagy-dependent processes during phagocytosis, and subsequently, additional studies have described the role of autophagy in the maintenance of RPE and photoreceptor integrity [22–27]. Chen et al. (2012) provided evidence that autophagy increases in the presence of all-trans retinal and plays a protective role in the RPE in vivo [28]. Autophagy-associated proteins were found to follow a bimodal expression profile, with shifts in photoreceptor autophagy proteins that changed during light and dark, while changes in RPE autophagy protein levels appeared to be sensitive to phagocytosis of POSs [29]. Kim et al. (2013) described a decrease in photoreceptor response to light and decreased chromophore levels in Atg5-deficient RPE cells. They further show that RPE-mediated phagocytosis of photoreceptor outer segments is associated with LC3 and inhibited upon Atg5 knockout; however, the molecular details of this process remain elusive [10].
A critical aspect of phagosome maturation is association with and subsequent degradation by lysosomes. Our previous studies suggest that an intracellular sorting protein, melanoregulin (MREG), plays a role in this process in the RPE. MREG, a 28 kDa peripheral membrane protein is the product of the Mreg dsu gene [30]. The loss of this gene product was originally shown to rescue the pigmentation phenotype of dilute, ashen, and leaden mice, and it is also involved in keratinocyte development [31] and regulation of melanosome size [32]. In RPE cells, loss of MREG results in arrest of POS-phagosome maturation leading to the accumulation of opsin-positive phagosomes and the lipofuscin components A2E/A2PE in aged mice [33] as well as increased basolateral laminin [34].
Thus, using the RPE cell as a model of MREG-mediated phagosome degradation in the current study, we explored the hypothesis that POS phagosomes annex components of the autophagic machinery that are recognized by MREG for lysosomal degradation. These studies provide the first evidence that RPE cells utilize an MREG-mediated LC3-associated phagocytic pathway for digestion of POS. We show that single membrane POS containing phagosomes recruit endogenous LC3 in an ATG5-dependent manner that is dependent on MREG but independent of the mTOR pre-initiation complex. This LC3–POS association was observed by the detection of endogenous LC3, in polarized human fetal cells, ARPE-19 cells as well as in Mreg+/+ mouse RPE. The LC3-positive phagosomes follow a classic phagosome maturation profile [35]; they distribute in a perinuclear fashion with LC3-positive phagosomes observed basally, 30 min after POS pulse. In the absence of MREG, in the Mregdsu/dsu mouse RPE or upon MREG knockdown in human RPE cells, virtually no LC3 was associated with ingested OSs. MREG appears to be an LC3 binding partner based on immunoprecipitation (IP) and GST pull-down studies. We thus propose that a protein complex containing MREG is involved in the formation of LC3-decorated POS.
Results
Autophagic Proteins are Diurnally Expressed in RPE
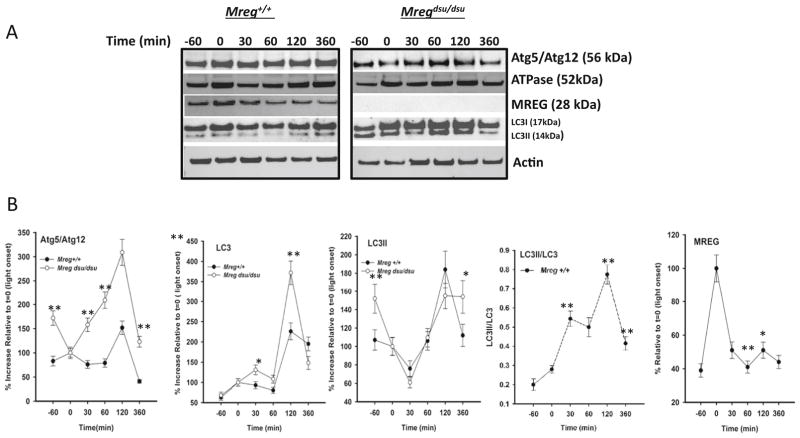
The continuous renewal of photoreceptor outer segment disks daily puts an enormous burden on the degradative pathways within RPE cells. Dysfunction in any aspect of phagocytosis can lead to the accumulation of phagosomes containing undigested POSs. Our previous studies show that loss of the intracellular sorting protein, MREG, results in A2E/A2PE accumulation [33] suggesting a requirement for MREG in complete POS degradation. The phagocytosis of the distal disks from mammalian rod photoreceptor cells by the RPE is controlled by a circadian rhythm and peaks at dawn, or light onset [36]. In the previous studies, we counted the number of phagosomes in the RPE of Mregdsu/dsu and Mreg+/+ retinas fixed at different times of day [33]. In the Mregdsu/dsu, we observe a threefold increase in phagosomes beginning 2 h after light onset compared with age-matched Mreg+/+ RPE [33]. This increase is in contrast to the early time points (30 and 60 min after light onset) when there was no increase in phagosomes in Mregdsu/dsu versus Mreg+/+ RPE.
In light of recent studies suggesting that phagocytes often utilize a hybrid phagocytic-autophagy pathway [4–6, 10], we analyzed the levels of proteins associated with degradative processes in RPE lysates from Mregdsu/dsu and Mreg+/+ with emphasis on autophagic proteins (Fig. 1a). In control mice, MREG levels appear to follow a bimodal pattern with highest levels at light onset and a second peak beginning 2 h later (Fig. 1a, b). In control mice, total LC3 was also elevated 2 h after light onset with a corresponding increase in its lipidated form, LC3II, as compared to t=0. Atg5/Atg12 protein components of the upstream conjugation system responsible for LC3 lipidation (Fig. 1a, b) also increase at 2 h, as does the vacuolar H-ATPase proton pump (Fig. 1a). In the Mregdsu/dsu RPE, Atg5/12 and LC3 were increased relative to the levels seen in the Mreg+/+ as was the lipidated form of LC3, LC3II at virtually all time points relative to light onset. Autophagic flux defined here at a ratio of LC3II/LC3 was elevated at 30 min after light onset with an increase observed after 2 h in the Mreg+/+ RPE.
Fig. 1.
Loss of melanoregulin is associated with LC3 accumulation. a Diurnal expression profile of proteins associated with hybrid autophagy–phagocytosis degradation processes. RPE cell lysates were prepared from 6-month-old Mreg dsu/dsu and Mreg+/+ mice at the times indicated after light onset (t=0) and immunoblotted for proteins indicated. A representative immunoblot is shown for each time point. b Quantitation of Mreg dsu/dsu and Mreg+/+ mouse RPE immunoblots. All blots were normalized to β-actin as a loading control. Results are average of three independent RPE lysate samples, each sample preparation consisting of six mouse eyes. Error bars indicate ±SEM, (*p<0.01 or **p<0.005, n=18)
LC3 Recruitment to POS in Human RPE Requires Atg5 and MREG but is not Induced by Rapamycin
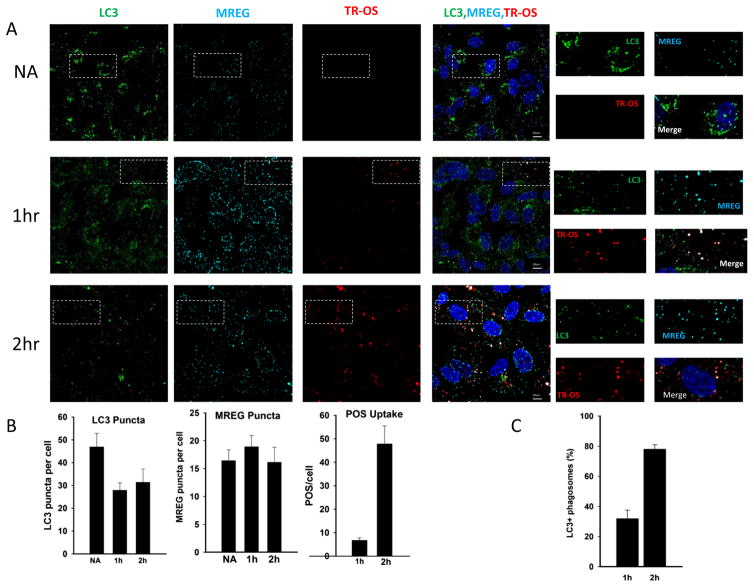
Human RPE cells in culture retain phagocytic activity allowing us to define the relationship between POS maturation, LC3, and MREG. To dissect the process of RPE phagocytosis further, ARPE19 (C2) cells were fed Texas Red (TR)-POS for up to 2 h. Within the first hour, a fraction (approximately 25 %) of the ingested POS colocalized with LC3 and a sub-fraction of these with MREG (Fig. 2a, 1 h time point), with an increase in LC3 decorated TR-OS by 2 h (Fig. 2a, 2 h time point). High magnification images (SFig. 1) depict the TR-OS as enveloped by LC3 and in some cases MREG. Over the time course of TR-OS uptake, there was an initial decrease in LC3 puncta (Fig. 2b) and an increase in TR-OS (Fig. 2b) with no statistically significant change in MREG puncta (Fig. 2b).
Fig. 2.
Ingested OSs colocalize with LC3 in human RPE cells. a LC3 associates with ingested TR-OS. ARPE19 (C2) cells unchallenged (labeled NA) or challenged with TR-OS for 1 or 2 h were washed, external fluorescence quenched with trypan blue, fixed, and stained for LC3 (Cell Signaling) and for MREG. Cells were imaged and codistribution analyzed using a binary submask Pearson’s coefficient 0.68. Individual channels are indicated. The white dotted boxes delineate the regions magnified to the right of the merged images. Individual LC3 decorated TR-OS are shown in S. Fig. 1. b Average TR-OS, LC3 puncta, and MREG puncta per cell at 1 and 2 h of TR-OS challenge. c LC3 association with TR-OS at 1 and 2 h, Pearson coefficient=0.74. The percentage of TR-OS that are also LC3 positive is indicated
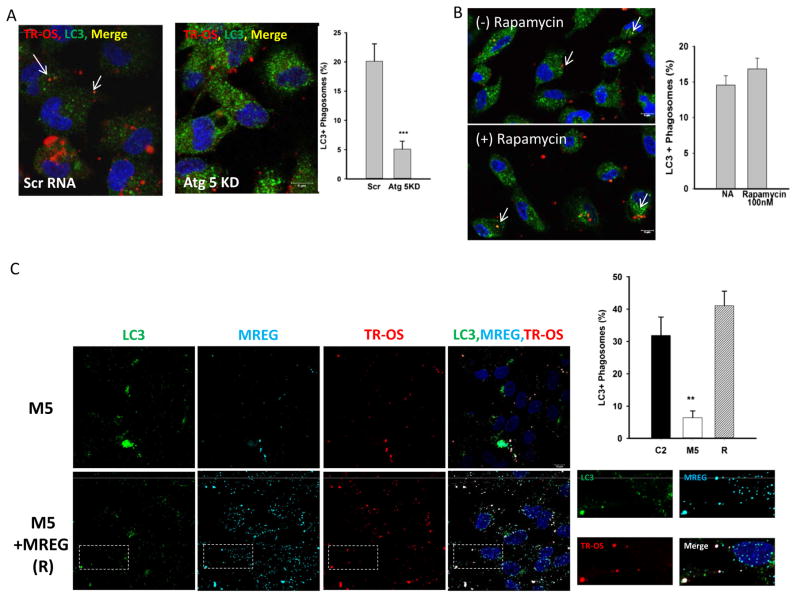
To determine the relationship between LC3-TR-OS association and canonical autophagy, the effect of rapamycin and Atg5 levels on LC3-OS association was evaluated. When RPE Atg5 expression was silenced by over 70 % with small interfering RNA (siRNA) (SFig. 2B), there was a 75 % decrease in the extent of LC3 associated with TR-OS (Fig. 3a); 20 % of the phagosomes in the control cells associated with LC3 and only 5 % in the Atg5 KD cells (Fig. 3a). Individual LC3 and TR-OS images show elevated LC3 expression (SFig. 2A and D) as expected, while the loss of Atg5 expression had no effect on TR-POS uptake (SFig. 2C). When RPE cells were treated with rapamycin (100 nM) prior to TR-POS challenge, there was no change in the extent of LC3-OS association (Fig. 3b). Rapamycin activity was confirmed as loss of phospho-S6 by immunoblot (SFig. 2F). Thus, LC3 association with POS is rapamycin independent and Atg5 dependent. These two independent lines of evidence suggest that our in vitro phagocytosis system recapitulates the properties of LC3 associated phagocytosis in a manner similar to that observed ex vivo using GFP-LC3 [10].
Fig. 3.
LC3 association with phagosomes is dependent on Atg5 but independent of rapamycin. a LC3 association with TR-OS is decreased upon Atg5 knockdown (see SFig. 2). Atg5 knock down RPE cells challenged with TR-OS for 2 h, were washed, external fluorescence quenched with trypan blue, fixed, and stained for LC3. Cells were imaged and codistribution analyzed using a binary submask Pearson’s coefficient 0.68. Error bars represent ±SEM, (***p<0.001). b TR-OS codistribution with LC3 is unaffected by rapamycin. ARPE19 cells incubated with 100 nM rapamycin for 4 h prior to 2 h TR-OS challenge and remained in the media for the duration of the study. Cells were imaged and codistribution analyzed using a binary submask. c M5 (MREG knockdown) and M5 cells transfected with MREG, (these cells are designated (R)) challenged with TR-OS for 1 h were fixed, stained for LC3 and MREG. Cells were imaged and codistribution analyzed using a binary submask Pearson’s coefficient 0.64. The image quantitation data in this figure is an average of 40 cells per field, with 10 fields analyzed in two independent experiments. Error bars represent ±SEM (**p<0.005). C2 data plotted is from images shown in Fig. 2. Western blot analysis showing MREG levels in C2, M5, and MREG rescue experiments (S. Fig. 3)
Since MREG was found to co-distribute with the LC3-positive phagosomes, we sought to determine the role of MREG in LC3 association with POS. MREG levels were knocked down by over 90 % in ARPE19 cells transduced with MREG small hairpin RNA (shRNA) (M5 cells) compared to control shRNA (C2) cells [34] (Fig. 2a and SFig. 3A). M5 cells were challenged with TR-OS for up to 2 h as described by [37]. The extent of LC3 association with TR-OS decreased in the absence of MREG, from 40 % in C2 (Figs. 2a and 3c) at 1 h to less than 5 % in M5 (Fig. 3a, c). TR-OS uptake was unaffected by MREG deletion with no statistically significant difference (p>0.01) in the number of POS per cell in M5 and C2 cells (SFig. 3B) and as described previously in Mregdsu/dsu RPE [33]. We further confirmed that there was no change in lysosomal pH in the C2 and M5 cells for up to 5 h after challenge (SFig. 3C). Because shRNA can give off-target affects, to further confirm that LC3-OS association requires MREG, MREG levels in M5 cells were restored to approximately wild-type levels (SFig. 3A) in a series of rescue experiments (labeled R in Fig. 3c). When TR-OS were added to MREG-rescue cells, a substantial increase in the percent of LC3 associated with the TR-OS was observed; from 10 % in M5 to over 60 % in R cells (Fig. 3c). These studies show for the first time that MREG participates in the association of LC3 with phagosomes in a process that has the properties of LC3 associated phagocytosis; LC3 association in an Atg5 dependent, Ulk1-independent manner.
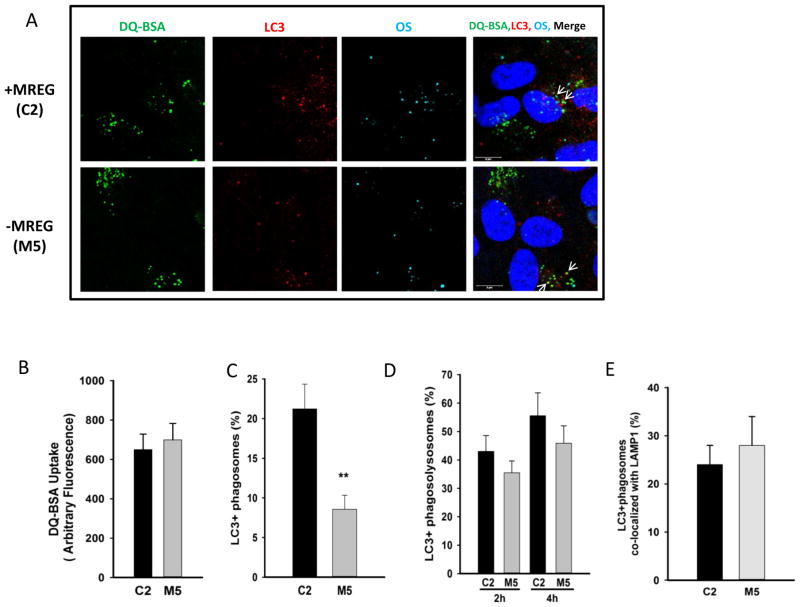
Critical in the degradation of ingested OSs is the association and fusion of phagosomes with lysosomes. To determine if MREG is required for the association of LC3-positive phagosomes with lysosome, we used DQ™-BSA as a reporter of lysosomal activity. As lysosomal enzymes separate the protein strands, Green-DQ™-BSA dequenches, leading to a rise in green fluorescence. When C2 and M5 cells were incubated with DQ™-BSA for 1 h, equivalent amounts were incorporated (Fig. 4a (DQ-BSA panel) and Fig. 4b). The DQ™-BSA containing cells were subsequently fed AF647-POS from the apical chamber. In the absence of MREG (M5, MREG knockdown cells), fewer LC3-positive phagosomes were observed with only 7±2 % of the ingested OS labeled with LC3 compared to 22±3 % in the C2 cells (Fig. 4a, c). The association of the LC3-positive phagosomes with lysosomes was determined through analysis of the triple co-localization between AF647-POS, LC3, and dequenched DQ™-BSA in the polarized C2 and M5 cells. LC3-positive phagosomes matured to phagolysosomes, detected as the percent of LC3-positive phagosomes colocalized with DQ™-BSA to the same extent in both C2 and M5 cells (Fig. 4a, d (merged panel)). Further confirmation that successfully fused LC3-POS phagosomes were delivered to lysosomes was provided with LAMP1-POS triple colocalization studies (Fig. 4e). Even though LC3 association with POS was reduced by over 60 % in the absence of MREG, the LC3-positive POS in the M5 cells still associated with lysosomes.
Fig. 4.
LC3-OS-lysosome association is independent of MREG. a DQ™-BSA containing C2 and M5 cells were fed Alexa Fluor 647 labeled-POS (AF647-POS) at a density of ~10 particles per cell directly to the media on the apical side for 1 h, at 37 °C. The assay was terminated and extracellular fluorescence quenched. Cells were fixed and stained for LC3 as described in the methods. A representative image of each channel is shown, with colocalization indicated with arrows. b Uptake of DQ™-BSA Green in C2 (black square) and M5 (white square) ARPE19 cells. C2 or M5 cells were incubated with 10 μg/ml DQ™-BSA in the apical chamber for 1 h at 37 °C, washed, fixed, and imaged in three different fields with ~40 cells per field. Error bars represent ±SEM. There is no statistically significant difference between DQ™-BSA uptake in C2 and M5 cells. c Loss of MREG led to decreased LC3-POs association. DQ™-BSA containing C2 (black square) and M5 (white square) cells were fed Alexa Fluor 647 labeled-POS (AF647-POS) at a density of ~10 particles per cell directly as described above. The assay was terminated and extracellular fluorescence quenched. Cells were fixed and stained for LC3 as described in the methods. Cells from each coverslip were imaged in three different fields (n=3 fields, ~40 cells per field). d LC3-positive phagosomes associate with Green DQ-BSA (lysosomal) in the absence of MREG. Cells from each coverslip were imaged in three different fields (n=3 fields, ~40 cells per field). Colocalized areas (DQ™-BSA, LC3, and AF647-POS) had a Pearson’s coefficient above 0.50. e LC3-positive phagosomes associate with LAMP1 (lysosomes) in the absence of MREG. Cells from each coverslip were imaged in three different fields (n=3 fields, ~40 cells per field). Colocalized areas (LAMP1, LC3, and AF647-POS) had a Pearson’s coefficient above 0.50
LC3 Associates with Opsin-Positive Phagosomes in Human Fetal Cells
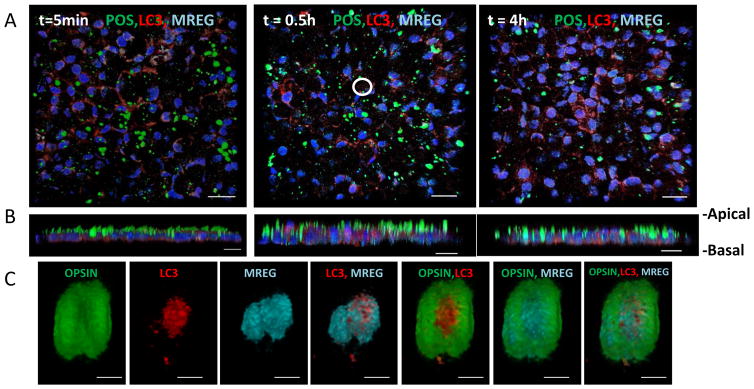
In the next series of studies, we expanded our analyses to include human fetal RPE cells. Using polarized human fetal RPE cell cultures, we followed the temporal and spatial characteristics of LC3 association with OS phagosomes. Primed hfRPE cells were fed unlabeled-OS (from apical chamber) for 20 min (pulse phase), the cells were washed and subsequently chased for 5 min, 30 min, or 4 h. OS was detected as opsin-positive structures using anti-opsin mAb-4D2. As indicated in Fig. 5a, LC3 associated with opsin-positive phagosome. Opsin degradation over the 4-h chase period was confirmed by immunoblot analysis (SFig. 4A). Numerous LC3–opsin-positive phagosomes also codistributed with MREG (Fig. 5a–c). A representative image of LC3 and opsin positive as well as LC3–opsin–MREG-positive phagosomes is shown in Fig. 5a (30 min chase). A corresponding higher magnification and volume reconstruction of the LC3–opsin–MREG-positive phagosomes is shown in Fig. 5c. Such colocalized populations of phagosomes were analyzed relative to the distance from the nucleus to define the spatial characteristic of LC3-OS association in polarized RPE.
Fig. 5.
LC3 associates with POS in human fetal RPE cells a POS pulse/chase time course. hfRPE cells were pulsed with POS for 20 min and phagocytosis was allowed to continue for the time points indicated; t=0 h, no POS addition, t=0.5 h chase, and t=4 h chase. Cells were fixed and stained; POS are visualized green (anti-opsin 4D2), LC3 is visualized red (anti-LC3), and MREG visualized, and pseudo colored cyan (anti-MREG) staining is as described in Methods. Nuclei are stained blue. Images shown are 3D volume reconstruction of multiple Z planes (coronal view). The white circle in the t=0.5 h panel represents region form which the 3D volume reconstructions are shown in b. b 3D volume reconstruction of multiple Z planes (sagital view) of no POS addition, t=0.5 h chase and t=4 h chase. Scale bar equal 10 micron. c Spatial association of LC3 and MREG with POS. 3D volume view of single POS taken up within hfRPE at t=30 min. Cells were fixed and phagosomes stained for proteins indicated in order from left to right: labeled for opsin, labeled for LC3, labeled for MREG, merge of LC3 and MREG images, merge of opsin and LC3 images, merge of opsin and MREG images, and merge of opsin, LC3, and MREG images. Scale bar=1 micron
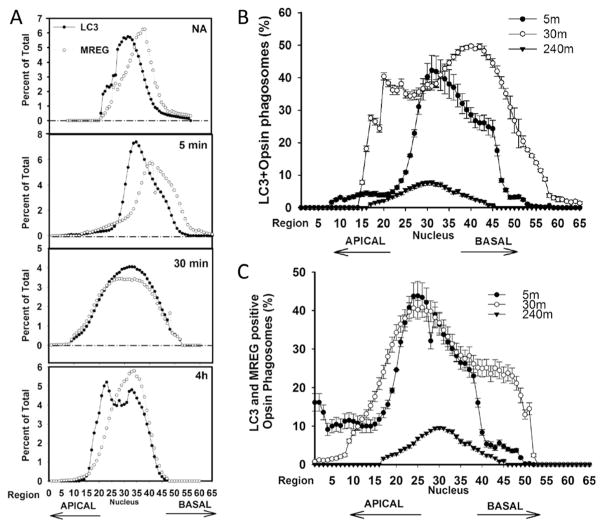
After a 5 min chase, the bulk of the LC3 and MREG was detected in the basal region (representative experiment; Fig. 6a) with distribution towards the nucleus and apical region after 4 h. We also analyzed the percent of the total POS that was LC3 positive. After a 30 min chase, over 40 % of the POS was LC3 positive; they distributed not only within a region at the level of the nucleus but also as a more distinct basal pool (Fig. 6b). It appears that most of the LC3-positive OSs also contained MREG, given that LC3–opsin–MREG-positive structures were observed apically representing 20 % of the total POS in this region (Fig. 6c, indicated with arrow) with 40 % of the POS in an area peripheral to the nucleus. After the 30 min chase, the LC3–opsin–MREG-positive phagosomes appeared to also localize basally.
Fig. 6.
MREG and LC3 redistribute within RPE during phagosomes maturation. a Time course of LC3 and MREG distribution in RPE after 20 min OS pulse. The percent of the total intracellular LC3 or MREG is indicated. b LC3-associated phagosomes are more abundant in basal region. LC3–opsin-positive structures were quantified in each Z-slice at various time points indicated. Percent of total POS (opsin-positive structures) that are LC3 positive as a function of position in RPE is indicted. Data are average of three independent experiments each experiment consisted of imaging at least ten regions. Error bars represent ±SEM. c LC3–MREG-associated phagosomes are more abundant in basal region. Percent of total POS (opsin-positive structures) that are LC3 and MREG positive as a function of position in RPE is indicted. Data are average of three independent experiments each experiment consisted of imaging at least ten regions. Error bars represent ±SEM
LC3 Associates with MREG In Vitro and In Vivo
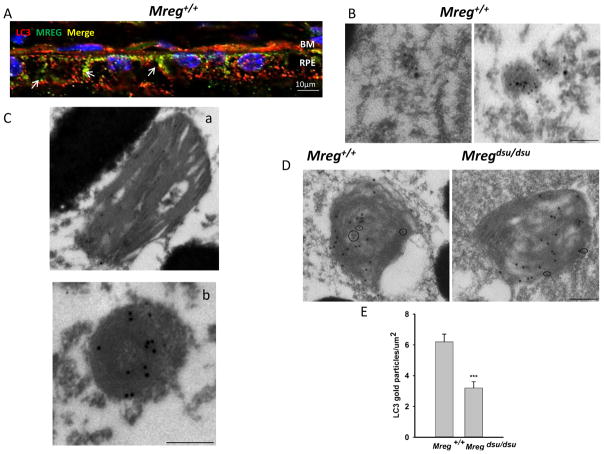
To determine if LC3 association with POS was due to an LC3–MREG association, we analyzed the distribution of MREG and LC3 in Mreg+/+ RPE. MREG-positive structures (shown in green) as well as LC3-positive puncta (shown in red) were observed both apically and basolaterally in the mouse RPE, with a fraction of the LC3 colocalized with MREG (Fig. 7a). We also analyzed the intracellular disposition of LC3 and MREG by double immunogold labeling of Mreg+/+ RPE. At 3 h after lights on, MREG (small gold particles) and LC3 (large gold particles) containing vesicles were observed in the cytosol of the RPE, although not all LC3-positive vesicles contained MREG (Fig. 7b). At this same time point, numerous structures containing OS debris was labeled with both MREG and LC3, based on morphology we have identified these structures as phagosomes (containing identifiable disk membranes; Fig. 7c, panel a) and phagolysosomes (containing identifiable stacked membranes that have curled up and thus no longer appearing disk-like; Fig. 7c, panel b). Thus, the association of LC3 with phagosomes, as observed in cultured human RPE cells (Figs. 2, 3, and 5), was also detected in vivo, in mouse RPE. Quantification of double immunogold labeling by opsin (large gold particles) and LC3 (small gold particles) antibodies in the Mregdsu/dsu and Mreg+/+ RPE showed fewer LC3-positive phagosomes in the Mregdsu/dsu RPE (Fig. 7d, e).
Fig. 7.
LC3 associates with phagosomes and MREG in epithelial cells a MREG and LC3 codistribute in Mreg+/+ (C57Bl6/J) RPE. Eyecups prepared from Mreg+/+ mice (6 months old, 6 h after light onset) were fixed and stained with anti-MREG (mAb, Abnova), shown in green and anti-LC3 rabbit polyclonal (Cell Signaling), shown in red. Pearson’s coefficient 0.74. RPE Retinal Pigment Epithelium; BM Bruch’s Membrane. Scale bar=10 μm. b LC3 and MREG are associated with intracellular vesicles. Scale bar is 100 nm. c LC3 localizes to disk membrane containing structures in mouse RPE. Retinal sections were labeled with anti-MREG mAb165 and anti-LC3 mAb (Abcam) conjugated to gold particles (large-MREG) and (small-LC3). Scale bar is 250 nm. d LC3 localizes to opsin-positive phagosomes. Retinas from C57Bl6/J mice (6 month old, 6 h after light onset) were prepared by embedment in L.R. white resin. Sections were labeled with anti-opsin mAb 4D2 and anti-LC3 mAb (Abcam) conjugated to gold particles. Large particles (opsin) and small particles (LC3). Scale bar is 250 nm. e Phagosomes in Mreg +/+ and Mregdsu/dsu RPE were identified based on RHO ab labeling. The LC3b immunogold labeling was quantified in RHO-positive phagosomes and expressed as gold particles per micrometer squared of phagosome section. (Panel d is a representative image). p<0.01
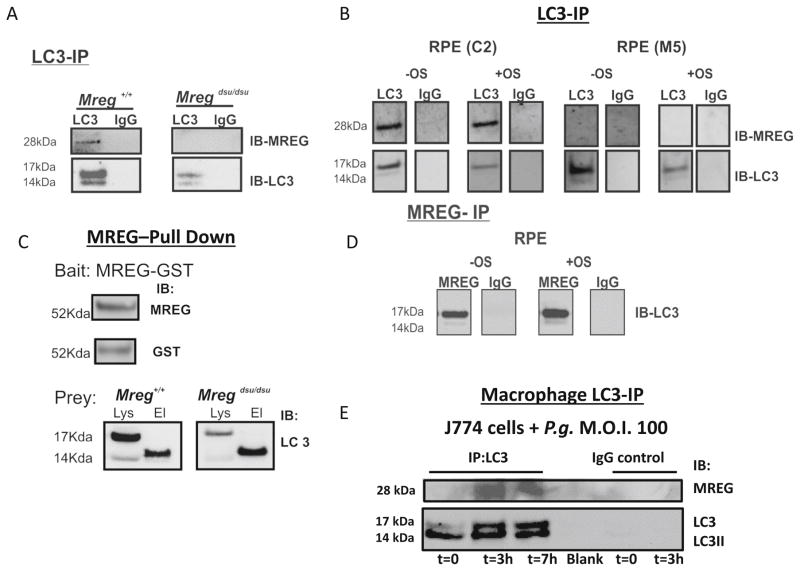
MREG association with LC3 containing complexes was further confirmed biochemically; when primary RPE cell lysates isolated from Mregdsu/dsu and Mreg+/+ mice were immunoprecipitated with an anti-LC3 antibody, a 28 kDa band immunoreactive with anti-MREG Ab was detected in addition to LC3 and LC3II (Fig. 8a). In human ARPE19 cells, IP with anti-LC3 antibody isolated a complex containing MREG, both in the presence and absence of OS challenge (Fig. 8b). LC3-IP of stable MREG knockdown cells-M5 [34] resulted in no detectable MREG but LC3 and LC3II are seen as expected. IgG controls showed no detectable protein binding (Fig. 8a, b). When RPE lysates from Mregdsu/dsu and Mreg+/+ mice were used as the prey in MREG-GST pull downs, LC3II was primarily detected (Fig. 8c). Immunoprecipitation of MREG containing complexes from ARPE19 cells (+/− OS challenge) with an anti-MREG mAb 165 (Fig. 8d) isolated a complex containing LC3. Collectively, this set of results suggests that MREG is likely an LC3 binding partner and contributes to the formation of LC3-associated phagosomes containing OS in the RPE, both in vivo and in vitro.
Fig. 8.
LC3 immunoprecipitates with MREG in mouse RPE and ARPE19 cells. a. Immunoprecipitation (IP) studies were carried out with lysates prepared from Mreg +/+ and Mreg dsu/dsu RPE cells (6-month-old animals, sacrificed 3 h after light onset). Proteins were immunoprecipitated with a polyclonal anti-LC3 antibody and then immunoblotted (IB) using an anti-MREG mAb or as a control anti-LC3. MOPC1 was used as a non-specific IgG control. The blots shown are representative of three independent experiments. b Immunoprecipitation (IP) studies were carried out with lysates prepared from C2 and M5 RPE cells isolated either at t=0 (no OS addition, designated, −OS) or after 2 h outer segment feeding (designated, +OS). Proteins were immunoprecipitated with a polyclonal anti-LC3 antibody and then immunoblotted (IB) using an anti-MREG mAb or as a control anti-LC3. MOPC1 was used as a non-specific IgG control. The blots shown are representative of three independent experiments. c MREG-GST pulls down LC3 from mouse RPE lysates. Top right; the GST fusion protein (MREG-GST) bait was detected as a 52 Kda fusion protein by both anti-MREG and anti-GST antibodies. For pull down assays, MREG-GST attached to the glutathione resin was incubated with lysates prepared from Mreg +/+ and Mreg dsu/dsu RPE cells (6-month-old animals, sacrificed 3 h after light onset) at 4 °C overnight as detailed in Methods. Interaction of MREG-GST and LC3 was verified by immunoblot (IB) analysis with anti-LC3 in the elution (labeled El). The blots shown are representative of three independent experiments. d Immunoprecipitation studies were carried out with lysates prepared from ARPE19 cells fed OS for 2 h as described in the Methods. Protein complexes were immunoprecipitated with anti-MREG mAb 165 antibody and then immunoblotted (IB) using an anti-LC3 antibody. e MREG immunoprecipitates with anti-LC3 in macrophages upon bacterial challenge. J774A.1 cells challenged with P. gingivalis strain ATCC 33277 (M.O.I=100) for 3 and 7 h at 37 °C. Cells were washed, lysates prepared, and LC3 containing complexes were immunoprecipitated with anti-LC3 and IB probed with anti-MREG or anti-LC3
We have previously shown that MREG is also expressed in cells of the immune system [38], thus the association of MREG with LC3 was further evaluated using murine macrophages (J774 cells) challenged with Porphyromonas gingivalis. In these cells, IP with anti-LC3 antibody isolated a complex containing MREG, only upon challenge with P. gingivalis (Fig. 8d). Furthermore, the immunoprecipitated protein complex also contained LC3 and LC3II as expected. Collectively, these biochemical studies suggest that the association of MREG with LC3 is not limited to RPE cells but likely includes other phagocytic cells as well.
Discussion
Our studies provide the first evidence that POS degradation by the RPE utilizes an MREG-mediated association of ingested POS with LC3. The association of endogenous LC3 with opsin-rich phagosomes in the RPE was documented in hfRPE cells and ARPE19 cells as well as mouse RPE cells. Previously, the association between endogenous LC3 and ingested POS had not been documented. In a series of studies, Kim et al. (2013) demonstrated colocalization between opsin and GFP-LC3, in in vivo and in vitro systems in which the fluorescently tagged LC3 is overexpressed or is in addition to expression of endogenous LC3 [10]. In those studies, over 90 % of the ingested POS was GFP–LC3 positive. The studies presented herein suggest that this level of LC3–POS association is an overestimation most likely due to the overexpression of LC3 in the fluorescently tagged form. We routinely observe less that 50 % of the ingested POS decorated with LC3 after a 1 h challenge. If LC3-associated phagocytosis in the RPE follows the paradigms defined in macrophages upon bacterial challenge, then it is not unexpected to find that not all but only a fraction of the phagosomes is decorated with LC3 [4, 8]. LAP may serve as a parallel clearance mechanism, one that may be enhanced under stress. Furthermore, it is not unreasonable to propose that the extent of LAP is linked to the available pool of LC3 and is thus indirectly affected by the status of basal autophagy in these phagocytic cells.
Our in vitro and in vivo studies suggest that LC3 association with ingested POS requires the intracellular sorting protein MREG. Loss of this protein both in vivo and in vitro results in delayed phagosome digestion manifest as the accumulation of opsin-positive structures. MREG is a membrane associated, 28 kDa protein, localized to intracellular RPE vesicles [32, 33] and to phagosomes upon POS challenge in both mouse and human RPE cells (Figs. 3, 4, 5, and 6). In the absence of MREG in ARPE19 M5 cells as well as in the RPE of Mregdsu/dsu mice, although POS uptake was normal [33] (SFig. 2), there was virtually no LC3 associated with POS. When MREG levels were restored to control levels in rescue experiments, the extent of LC3-positive OS was similar to the in control (+MREG) cells (Fig. 3c). Phagosomes accumulated in the RPE of Mregdsu/dsu mice [33] and in aged mice, the loss of MREG coincides with the accumulation of lipofuscin components, A2E/A2PE [33]. Collectively, our studies suggest that the Mregdsu/dsu mouse can serve as a useful model in which to understand the relative contribution of LC3-associated phagocytosis to the degradation of ingested POS by the RPE.
As with other phagocytic processes, POS containing nascent phagosomes undergo a coordinated series of fusion events leading to the modification of their limiting membrane and of contents during phagosome maturation. The polarized structure of RPE cells necessitates the movement of phagosomes from the apical membrane toward the basal membrane where they are degraded in phagolysosomes [35]. The molecular details of this phagosome maturation process include acidification of the OS phagosomal lumen necessary for Cathepsin D (Cat-D)-mediated degradation of opsin [39, 40]. Direct interactions between OS-containing degradative compartments have also been documented. Bosch et al. [41] identified two types of degradative opsin- and Cat-D-positive compartments in the RPE of C57Bl6/J (Mreg+/+) mice. These two “lysosomal” pools were temporally separated, with a second step of degradation observed over 2 h after light onset. The Atg5/Atg12 components of the upstream conjugation system responsible for LC3 lipidation (Fig. 1a, b) also increase at 2 h, as do the vacuolar ATPase (Fig. 1b) and LAMP1 (data not shown). The bimodal pattern of autophagyprotein expression we observed was similar albeit not identical to that recently described by Yao et al. (2014) [29]. In C57Bl6/J mice, these authors observe an increase in LC3II/LC3 (autophagic flux) at 5.5 h after lights on [29], with no statistically significant difference in this ratio between their 2.5 and 5.5 h time points. In our studies, we observe the highest levels of LC3II/LC3 at 2 h after light onset with the levels elevated over those observed at light onset (t=0) for up to 6 h. The details of the differences in magnitude of the LC3II/LC3 ratios can only be evaluated with a comparison of identical time points and lighting conditions. Furthermore, both MREG and LC3 levels are increased at 2 h. In the Mregdsu/dsu mouse, the levels of LC3 are increased relative to controls (Fig. 1a, b). Studies in [26, 42] over 30 years ago identified autophagic structures and a diurnal pattern of autophagy-dependent processes during phagocytosis. Collectively, these independent series of studies coupled with the fact that it is advantageous for the post-mitotic RPE to utilize processes that optimize phagosome degradation suggest that these early studies may have been looking at aspects of LAP.
Immuno-EM studies of mouse RPE as well as codistribution analyses by confocal microscopy reveal that LC3 and MREG localize to the same intracellular structures and are often associated with POSs. The labeling of the phagosome lumen with LC3 we observe is also consistent with LC3 labeling in autophagosomes, most likely due to membrane exchange [43, 44]. Furthermore, the association of LC3 with the lysosome is not entirely unexpected since both LAP and canonical autophagy converge at the lysosome association step, such as the stage at which LC3 recycles from the phagosome is unknown. It is, however, predicted that LC3 delipidation likely involves an Atg4-mediated event [45]. The molecular mechanism by which MREG mediates LC3 association is likely through a protein complex containing LC3. Both immunoprecipitation studies and GST-Pull down assays (Fig. 8a–d) suggest that these proteins interact not only in cultured RPE cells but in mouse RPE. MREG is thus the first LC3 binding partner shown to be required for LAP in a phagocyte.
It is clear that defects in autophagy as well as the age-dependent decreases in autophagy-related processes result in cellular dysfunction contributing to disease progression [46–49]. Autophagy-dependent processes are particularly vital to maintain homeostasis for long-lived post-mitotic cells like the RPE whose catabolic cascade is challenged with the daily burden of OS phagocytosis, LDL and oxLDL endocytosis and the clearance of intracellular debris. Progressive dysfunction of the degradative capacity of the RPE has been implicated in numerous pathways of age-related macular degeneration [15–18] with reduced autophagic function resulting in accelerated aging and degeneration of the RPE [19, 20]. Numerous studies have described the role of autophagy in the maintenance of RPE and photoreceptor integrity [22, 24–27, 29, 49]. Herein, we describe the contribution of hybrid autophagy- and phagocytosis-dependent processes on OS degradation and provide mechanistic insight into the role of MREG in these processes.
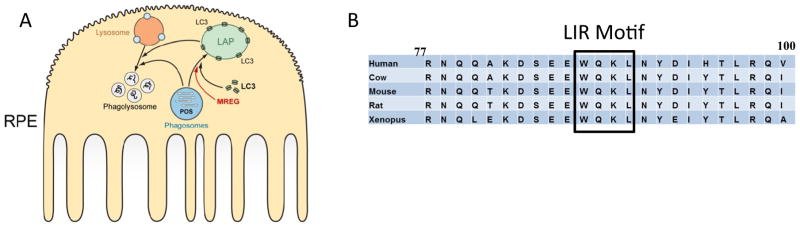
Our schematic (Fig. 9) summarizes our current knowledge regarding MREG’s involvement in the formation of LC3-positive phagosomes in the RPE. We posit that MREG participates in the association of LC3 with ingested OS, consistent with this role is the prediction that MREG binds an LC3 containing protein complex, as suggested by our IP and GST-pull down studies (Fig. 8 and SFig. 5) as well as by the identification of an LC3-interacting region (LIR) [50] predicted in human MREG (Fig. 9b). Whether MREG’s role is through direct interaction with LC3 through this domain is unknown and currently under investigation. Once decorated with LC3, the LC3-positive phagosomes can be transported to lysosomes likely in an MREG-independent manner based on DQ-BSA studies (Fig. 4). The LC3 and MREG are predicted to be recycled and not degraded by lysosomal proteases. We predict that the requirement for MREG is likely early in the phagosome maturation process.
Fig. 9.
Schematic representation of LC3 associated phagocytosis in RPE and the role of MREG in this process. a Our studies suggest that the maturation of phagosomes (depicted in blue) requires LC3 in a process that is mediated by MREG (shown in c) to form an LC3-associated phagosome (LAP). The contents of the LAP are subsequently degraded in phagolysosomes. b LC3 interacting region of MREG. Computational analysis-based recognition motif software was used to predict the region of LC3 association with MREG as residues 87–90 in MREG. This region is conserved between the species indicated
LAP was first identified in macrophages, in which it is stimulated in response to pathogenic challenge. In those cells, the up-regulation of this hybrid degradative process with elements of both autophagy and phagocyte maturation is proposed to be a mechanism by which the macrophage clears toxic debris. Our immunoprecipitation studies confirm the association of MREG with LC3 upon bacterial challenge (Fig. 8e) with P. gingivalis, with specificity for the bacteria as opposed to TR-OS (SFig. 5D) This observation is particularly significant given that P. gingivalis is known to traffic to LC3-positive structures [51]. We propose that the RPE cell may utilize LAP in a manner similar to the macrophage, with up-regulation of this process in response to environmental stress or toxic degradative load. Further studies delineating the precise contribution of LAP to POS degradation are critical in understanding the relationship between LAP, photoreceptor renewal, cell stress, and pathophysiological conditions. Furthermore, a detailed understanding of the upstream signaling events that trigger and regulate LAP will provide an important avenue for future therapies. Lastly, our macrophage studies suggest a more global role for MREG in LC3-associated degradative processes while confirming specificity for phagocyte-specific uptake.
Materials and Methods
Antibodies and Reagents
Antibodies used were rabbit anti-Z01 and Alexa Fluor 594 donkey anti-rabbit IgG (Life Technologies Inc), rabbit anti-phospho S6 and rabbit anti-LC3 (2775S, Cell Signaling), rabbit anti-ATG5, mouse anti-MREG and anti-opsin antibody 4D2–Dy-light 488 (Novus Biologicals), mouse anti-LAMP1 (Ab25630), rabbit anti-LC3 and rabbit anti-V-ATPase (AbCam), rabbit anti-ATG5 (Novus Biologicals), rabbit anti-MREG (Abnova), Alexa Flu-or 594 anti-rabbit, Alexa Fluor 488 anti-rabbit, AlexaFluor-488 anti-mouse (Invitrogen), goat anti-Actin, rabbit anti-goat IgG (H + L), and goat anti-rabbit IgG (H+L) (Santa Cruz Biotechnology). DQ™-BSA-Green, Texas red (mixed isomer), and Alexa Fluor 647 succinimidyl ester were purchased from Invitrogen, Cytoseal from Electron Microscopy Sciences and Hoescht 33258 from Anaspec, Inc.
Animals
Melanoregulin (MREG) is the product of the Mreg-dsu gene locus [previously known as dilute suppressor (dsu) [30]]. Mregdsu/dsu mice carry the Mregdsu allele (mouse accession number Q6NV65), in which the deletion of the first two exons results in an effective null allele [30]. Mregdsu/dsu mice (on C57BL6/J genetic background) used in these studies were originally maintained and propagated at the NCI, National Institutes of Health, [generous gifts from Drs. N. Jenkins and N. Copeland (Texas Medical Center)]. Both Mregdsu/dsu and Mreg+/+ [C57Bl6/J mice, (obtained from the Jackson Laboratory)] mice were housed under standard cyclic light conditions: 12-h light/12-h dark and fed ad libitum. Both female and male mice were use in these studies. All procedures involving animals were approved by the University of Pennsylvania Institutional Animal Care and Use Committee and were in accordance with the Association for Research in Vision and Ophthalmology guidelines for use of animals in research.
Cell Culture and Transfection
Human Fetal RPE Cells (hfRPE)
hfRPE monolayers were cultured on T25 flasks (Passage 0; P0) as previously described [52]. Briefly, hfRPE cells were trypsinized from a T25 flask and seeded into 12-well transwells at ≈1.25×105 cells/well (passage 1; P1) as described [52]. P1 hfRPE cells were cultured for approximately 4 weeks to reach maturity (transepithelial resistance >500Ω cm2) prior to experimentation. TER was measured with Epithelial Volt-Ohm Meter (EVOM2) (WPI, Sarasota, FL) at RT. Cells were maintained at 37 °C at 5 % CO2 with fluid renewal twice a week in media consisting of Advanced MEM (Life Technologies), 5 % heat inactivated fetal bovine serum (Sigma), 5 % Penicillin–Streptomycin (Life Technologies, Inc), and 5 % Glutamax (Life Technologies, Inc).
ARPE-19 Cells
(CRL-2302, ATCC) were grown in DMEM/F12+10 % FBS at 37 °C. M5 (stable MREG knockdown) and C2 (control) cells were generated and maintained as described [34]. For rescue experiments, the M5 cells were transiently transfected (48 h) using lipofectamine and MREG as described [38]. Transwell (12 mm, 0.4-μm pore size) filters were coated with 10 μg/cm2 purified mouse laminin (BD, Biosciences) prior to plating. The basolateral chamber was filled with 1.5 ml of DMEM:F12 containing 1 % FBS. Individual filters were seeded with 1.6 × 105 cells in a total volume of 0.5 ml of media containing 1 % FBS (3.3×105 per ml), in the apical chamber. Media was changed bi-weekly and cells were cultured for 14 days at 37 °C, 5 % CO2 [53]. The transepithelial resistance (TER) reached a peak of 40Ω cm2 after 14 days in culture under these conditions. The monolayer was identified by immunofluorescence using rabbit anti-ZO1 followed by Alexa Fluor 594 donkey anti-rabbit IgG secondary antibody and Hoescht 33258 labeling.
For Atg5 siRNA experiments, cells were seeded at a density of 1×105 cells per well on 6-well plates. Cells were transfected using Lipofectamine 2000 [per manufacturer’s instructions, (Invitrogen)] with 100 pmol of ATG5 siRNA I (Cell Signaling Technologies # 6345S) or 100 pmol of Silencer Negative Control siRNA (Invitrogen # AM4611) and cells analyzed 72-h post transfection. In rapamycin studies, cells were kept under regular growth conditions (10 % FBS) or challenged with 100 nM rapamycin for 4 or 24 h.
Preparation and Labeling of POS
Photoreceptor outer segments (POSs) purified from frozen dark-adapted bovine retinas [54] were labeled with either Texas–Red (TR) or Alexa Fluor 647 succinimidyl ester [37]; briefly, POS was incubated (1 h at RT in the dark) with 0.01 mg/ml Texas Red (mixed isomer, Life Technologies Inc.) or 0.05 mg/mg (1:20) Alexa Fluor 647 succinimidyl ester (Invitrogen). Free dye was routinely removed by dialysis for 4 h or occasionally overnight at 4 °C against 4 l of 3 mM KCL, 1.8 mM KH2PO4, 138 mM NaCl, and 8 mM Na2HPO4·7H20, pH of 7.4 (PBS). The labeled POS was checked for morphology and clumping prior to the start of each phagocytosis assay.
POS Phagocytosis Assay
Cells (ARPE19, C2, M5, or R) were incubated with Texas red outer segments (TR-OS) at a density of 10 particles/cell. Phagocytosis was allowed to continue for up to 4 h at 37 °C with the assay terminated at indicated time points with removal of the media. Cells were washed three times in 1 mM MgCl2 and 0.2 mM CaCl2 (PBS-CM) and extracellular TR-OS fluorescence quenched with 0.2 % Trypan Blue (10 min. at 37 °C, 5 % CO2). After four successive washes in PBS-CM, coverslips were fixed and processed for immunofluorescence.
DQ™-BSA assay
LC3-Phagolysosome formation was monitored with DQ™-BSA essentially as described in [55]. Polarized C2 or M5 cells was incubated with 10 μg/ml DQ™-BSA-Green in the apical chamber for 1 h at 37 °C, and then washed three times in media. Alexa Fluor 647-labeled-POS (AF647-POS) were added at a density of ~10 particles per cell directly to the media on the apical side for 2 h at 37 °C. The assay was terminated with the extracellular fluorescence quenched as described above. After three successive washes in PBS-CM, the inserts were fixed in 4 % PFA and processed for immunofluorescence.
POS Pulse Chase Studies
Prior to POS pulse chase experiments, the photoreceptor naive hfRPE cells were primed with ten particles of POS per cell (ratio of 10:1, POS to cell) for 48 h. Pulse chase studies were begun with the removal of the existing media and primed cells rinsed three times with growth media. POS at ratio of 10:1 were added to the apical chamber in fresh growth media and incubated at 37 °C for 20 min (pulse). After 20 min pulse, cells were rinsed three times with growth media, fresh growth media was added, and phagosome degradation was allowed to continue for up to 4 h (chase). The assay was terminated at designated time points with the removal of media, cells washed three times (37 °C PBS-CM), and filters immediately processed for immunofluorescence microscopy.
Immunofluorescent Labeling
POS Phagocytosis Assays and DQ™-BSA Studies
Inserts were permeabilized with ice cold methanol (−20 °C, 10 min) followed by a PBS wash and incubation for 30 min in PBS at 4 °C to rehydrate. Inserts were blocked in 4 % BSA in PBS containing 0.05 % Triton TX-100 (PBST) for 60 min at 37 °C. After blocking, inserts were incubated at 37 °C for 60 min with anti-LC3 rabbit polyclonal antibody 1:150. After the incubation with primary antibody, inserts were washed 3× in PBST and incubated with secondary antibody, Alexa Fluor 594 donkey anti-rabbit IgG at 1:1000 and Hoechst 33258 at 1:10,000 (37 °C for 60 min). In preparation for microscopy, the filters were cut from the inserts and mounted in Cytoseal Z-stack images of 1 μm apical to basolateral sections were captured on a Nikon A1R laser scanning confocal microscope with a 60× water objective (NA 1.2) using Nikon’s Elements software 4.1. Autolysosome formation induced by 100 nM rapamycin was used as a positive control (data not shown).
Pulse-Chase Studies
At the indicated time points for the pulse-chase studies, the transwell filters were fixed in 4 % formaldehyde/PBS for 10 min at RT followed by three PBS washes. Cells were permeabilized with ice-cold methanol (10 min at −20 °C) followed by three washes in PBST. After blocking in 4 % BSA/PBST (37 °C for 60 min), filters were incubated with anti-LC3 rabbit polyclonal antibody (1:300) and anti-MREG mAb (1:300). After the incubation with primary antibody, the filters were washed three times in PBST and then incubated with mAb anti-opsin antibody 4D2–Dy-light 488 (Novus) 1:500 along with secondary antibody Alexa Fluor 594 donkey anti-rabbit IgG (Life Technologies, Inc.), Alexa Fluor 647 donkey anti-mouse IgG (Life Technologies, Inc.) at 1:1000, and Hoechst 33258 (Anaspec, Inc.) at 1:10,000 at 37 °C for 10 min. Three successive PBST washes followed the incubation with secondary antibodies. In preparation for microscopy, the filters were removed from their holders and were mounted in Cytoseal (Electron Microscopy Sciences) into glass slides and cover slipped.
Immunohistochemistry was performed on frozen sections of 4- to 6-month-old mouse retinas [38]. Eyecups were fixed in 4 % paraformaldehyde in PBS (pH 7.4) overnight at 4 °C, cryoprotected in 30 % sucrose, and embedded in OCT. The 7-μm sections were successively stained with anti-LC3 rabbit antibody (Cell Signaling) at 1:250 dilution and anti-MREG mouse monoclonal antibody MO1 at 1:250 dilution, followed by secondary antibodies Alexa Fluor 594 anti-rabbit and Alexa Fluor 488 anti-mouse, respectively, at 1:1000 dilution. Controls were incubated with secondary antibodies only. Nuclei were visualized with Hoechst 33258. Images were captured on a Nikon A1R live cell confocal imaging system. Data were analyzed using Nikon Elements AR Software ver. 3.2.
Imagining and Quantitation
POS Phagocytosis Assays and DQ™-BSA Studies
Cells from each coverslip were imaged in three different fields (n=3 fields, −/+s.e.m, ~40 cells per field) and colocalization analysis performed using NIS-Elements 4.0 software. The %POS colocalized with LC3 and/or DQ™-BSA-Green or LAMP1 was quantified to determine the LC3-positive phagosomes and phagolysosomes. Colocalized areas were defined by a Pearson’s coefficient above 0.50.
Pulse-Chase Studies
Images were captured on a Nikon A1R laser scanning confocal microscope with a 60× water objective (NA 1.2) using Nikon’s Elements software 4.1. Image analysis was performed by defining the intensity threshold for the individual image channels within each Z plane. Then restrictions were applied for object area, object size, and circularity range. Individual cells and ROI’s generation were based on segmentation of both nuclear and cytoplasmic staining. Object counting (using applied restrictions) in each cell was used to determine number of puncta per cell for each channel per Z plane. Colocalized populations of puncta were determined by sorting the appropriate intersection of pixel area and intensity across each channel per Z plane with colocalized areas having a Pearson’s coefficient above 0.50. Standard statistics were applied to the cellular puncta populations for each time point.
RPE Extraction
Mregdsu/dsu and Mreg+/+ mice were sacrificed at specific times relative to the onset of light (−60, 0, 30, 60, 120, and 360 min) with four mice of each genotype for each time point. Extracted eyes were placed in ice-cold PBS. To isolate the RPE layer, the lens, optic nerve, excess muscle and fat, iris epithelium, and neural retina were removed under a dissection microscope. The eyecup was carefully everted with forceps in order to expose the RPE layer. RPE cells were then gently separated from the posterior eyecup. The entire procedure was performed on ice. For mice sacrificed at 60 min before lights on, the RPE layer was extracted in the dark under red light.
Immuno-electron Microscopy
Ultrathin sections (70 nm) were obtained from LR white-embedded mouse (Mregdsu/dsu and Mreg+/+) retinas fixed in 4 % PFA, 0.5 % Glutaraldehyde in 0.1 M cacodylate buffer (EM grade reagents, from Electron Microscopy Sciences). They were double-labeled with LC3 and MREG antibodies, generated in rabbit and mouse, respectively, (see above) as described previously [56]. Rabbit IgG antibodies and mouse IgG antibodies, conjugated to either 10 or 15-nm gold particles, were used as secondary antibodies (Jackson Laboratories). Images were acquired on a Zeiss EM 910, and quantifications were made using ImageJ software (Rasband, W.S., Image J, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2014.)
Immunoprecipitation
RPE Cells and Mouse Lysates
LC3 containing complexes were isolated by immunoprecipitation [Pierce Direct IP Kit (Thermo Scientific 26148)]. Ten micrograms of Rabbit polyclonal Ab to LC3 (Abcam 48394) and MOPC-21 purified immunoglobulin (Sigma M5284) were used for coupling to AminoLink Plus Coupling Resin. Columns were incubated on a rotator at room temperature for 120 min to resuspend the slurry. After 2 h of incubation, resin was washed six times with wash buffer. ARPE19 cells were seeded on 100× 100 mm plates a few days before treatment. Cells were 90 % confluent for experiments. To starve cells, we used medium containing 0.5 % FBS overnight at 37 °C and 5 % CO2. To treat cells with POS, 30,000.00 particles were used for 100×100 mm plates for 3 h in incubator at 37 °C and 5 % CO2. Five hundred microliters of lysis buffer was used for both treatments with 1000 ug of protein from each treatment used for immunoprecipitation. After samples were added to antibody-coupled resin, columns were incubated with shaking overnight at 4 °C. Bound proteins were eluted with elution buffer according to manufacturer’s protocol [Pierce Direct IP Kit (Thermo Scientific 26148)].
Murine Macrophages (J774 Cells)
J774A.1 cells, a mouse macrophage cell line (ATCC TIB-67), were maintained at 37 °C at 5 % CO2 with fluid renewal twice a week in media consisting of RPMI 1640 (Life Technologies), 5 % heat inactivated fetal bovine serum (Sigma), and 1 % L-Glutamine (Sigma). The cells were challenged with P. gingivalis strain ATCC 33277 (purchased from the ATCC (Manassas, VA, USA) and grown as described [57] for up to 7 h at 37 °C. Cells were washed, c lysates prepared, and LC3 containing complexes isolated by immunoprecipitation [Pierce Direct IP Kit (Thermo Scientific 26148)] with anti-LC3 exactly as described above for RPE cells.
Generation of MREG-GST and GST Pull Down Binding Assays
MREG-GST constructs were generated as follows: Site directed mutagenesis (Genetailor site directed mutagenesis system, Life Technologies, Inc.) using oligonucleotide primers 5′-cacaggaaacagtatccatggcccctatactaggtt-3′ and 5′-aacctagtataggggccatggatactgtttcctgtg-3′ was performed to insert an NcoI site adjacent to the initiation codon of the GST gene in pGEX2T. PCR using primers 5′-ccctcgctctccatgggctgag-3′ and 5′-ctgagcctctcccatgggacttggaaacggaa-3′ was performed to amplify the MREG ORF in pCMV6-XL5 MREG (cat# SC312525, Origene, Inc.) with an NcoI site replacing the stop codon. The mutated pGEX2T and the MREG PCR amplicon were digested with NcoI and ligated together. Proper insert orientation was selected by restriction mapping; insert and junction sequences were verified by double-stranded DNA sequencing. The expression and purification of GST fusion proteins have been described previously [58]. Briefly, Escherichia coli BL21 (DE3) cells transformed with GST–polypeptide constructs in pGEX-2 T vector [38, 58] were grown in LB medium containing 50 μg/ml ampicillin. Cells were harvested 2 h after induction with 0.1 mM isopropyl β-D-thiogalactopyranoside (IPTG), spun down, washed in PBS, and sonicated in the presence of 35 mM octyl glucopyranoside (OG) in PBS (pH 7.2). Cellular debris was removed by centrifugation, and the GST pull-downs performed as described previously [33] using Pierce GST Protein Interaction Pull-Down Kit (Product number 21516).
Immunoblotting
Cleared RPE lysates were prepared as described [33]. Briefly, ARPE-19, hfRPE cells, or isolated mouse RPE were lysed in RIPA buffer (150 mM NaCl, 1 % Triton-X-100, 0.5 % Na-deoxycholate, 0.1 % SDS, 50 mM Tris, pH 8) containing protease inhibitor cocktail (Roche Applied Science) and phosphatase inhibitor cocktail (Sigma), homogenized, and sonicated. Debris was pelleted at 13,500 rpm for 15 min at 4 °C and supernatant collected. Equal amounts of protein were separated on 4–12 % SDS-PAGE gels (Invitrogen) under reducing conditions and transferred onto nitrocellulose membranes using an iBlot Gel Transfer System (Invitrogen). Blots were probed with anti-Atg5 (1:2000), anti-MREG (1:500), or anti-LC3 (1:1000) antibody and anti-actin (1:5000) as a loading control. Membranes were subsequently incubated with the appropriate peroxidase-conjugated secondary antibody (1:2500 for LC3, ATG5, and MREG and 1:5000 for actin; Thermo Scientific) for 1 h at room temperature. Blots were developed by enhanced chemiluminescence (ECL-West-Pico) on a Kodak Image Station 4000 MM. Relative band intensity was determined using Kodak Molecular Imaging Software v 4.0.0 (Eastman Kodak Company).
Measurement of Lysosomal pH
Lysosomal pH was determined using a ratiometric lysosomal pH indicator dye (Lysosensor Yellow/Blue DND-160, Invitrogen) as described [59, 60].
Statistical Analyses
Data were analyzed using SigmaStat version 3.1. Data are reported as mean±SD or SEM as indicated. Statistical analysis used a 1-way ANOVA Dunn’s test or student’s t test as indicated, results with p<0.01 (*) or p<0.005 (**) were considered significant and p<0.001 (***) highly significant.
Supplementary Material
Acknowledgments
This work was supported by grants from PHS; R01EY010420, R01DE022465, and R21EY018705 to KBB, R01 EY013434 to CHM, Vision Research Core Grant EY001583 (KBB and CHM) and P30EY00331 and R01EY07042 to DSW. DSW is an RPB Jules and Doris Stein Professor. The authors would like to thank Dr. Anuradha Dhingra and Mr. Frank P. Stefano for their expert technical assistance as Managers of the PDM-Live Cell Imaging Core.
Abbreviations
- POS
Photoreceptor outer segment
- RPE
Retinal pigment epithelium
- TR
Texas red
- LC3
Microtubule-associated protein 1 light chain 3
- IP
Immunoprecipitation
- MREG
Melanoregulin
- IEM
Immuno-electron microscopy
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12035-014-8920-5) contains supplementary material, which is available to authorized users.
Contributor Information
Laura S. Frost, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA
Vanda S. Lopes, UCLA School of Medicine, Jules Stein Eye Institute, Los Angeles, CA 90095, USA
Alvina Bragin, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Juan Reyes-Reveles, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Jennifer Brancato, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Art Cohen, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Claire H. Mitchell, Department of Anatomy and Cell Biology, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA
David S. Williams, UCLA School of Medicine, Jules Stein Eye Institute, Los Angeles, CA 90095, USA
Kathleen Boesze-Battaglia, Department of Anatomy and Cell Biology, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
References
- 1.Oczypok EA, Oury TD, Chu CT. It’s a cell-eat-cell world: Autophagy and phagocytosis. Am J Pathol. 2013;182:612–622. doi: 10.1016/j.ajpath.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and noncanonical autophagy: Variations on a common theme of self-eating? Nat Rev Mol Cell. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 3.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florey O, Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 2012;22:374–380. doi: 10.1016/j.tcb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev. 2009;227:203–220. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- 8.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 9.Young RW, Droz B. The renewal of protein in retinal rods and cones. J Cell Biol. 1968;39:169–184. doi: 10.1083/jcb.39.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, Ferguson TA. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young RW, Bok D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 15.Beatty S, Kohl M, Phil M, Henson D, Boulton M. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 16.Hollyfield JG, Bonilha VL, Rayborn ME, Yang X, Shadrach KG, Lu L, Ufret RL, Salomon RG, Perez VL. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14:194–198. doi: 10.1038/nm1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopitz J, Holz FG, Kaemmerer E, Schutt F. Lipids and lipid peroxidation products in the pathogenesis of age-related macular degeneration. Biochimie. 2004;86:825–831. doi: 10.1016/j.biochi.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 18.Chen PM, Gombart ZJ, Chen JW. Chloroquine treatment of ARPE-19 cells leads to lysosome dilation and intracellular lipid accumulation: Possible implications of lysosomal dysfunction in macular degeneration. Cell Biosci. 2011;1:10. doi: 10.1186/2045-3701-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaaraniranta K, Salminen A, Eskelinem EL, Kopitz J. Heat shock proteins as gatekeepers of proteolytic pathwyas-implications for age-related macular degeneration. Ageing Res Rev. 2009:1280139. doi: 10.1016/j.arr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Terman A, Gustafsson B, Brunk UT. Autophagy, Organelles and ageing. J Pathol. 2007;211:134–143. doi: 10.1002/path.2094. [DOI] [PubMed] [Google Scholar]
- 21.Reme C, Wirz-Justice A, Rhyner A, Hofmann S. Circadian rhythm in the light response of rat retinal disk-shedding and autophagy. Brain Res. 1986;369:356–360. doi: 10.1016/0006-8993(86)90550-0. [DOI] [PubMed] [Google Scholar]
- 22.Reme CE, Wolfrum U, Imsand C, Hafezi F, Williams TP. Photoreceptor autophagy: Effects of light history on number and opsin content of degradative vacuoles. Invest Ophthalmol Vis Sci. 1999;40:2398–2404. [PubMed] [Google Scholar]
- 23.Frost LS, Mitchell CH, Boesze-Battaglia K. Autophagy in the eye: Implications for ocular cell health. Exp Eye Res. 2014;124c:56–66. doi: 10.1016/j.exer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunchithapautham K, Rohrer B. Autophagy is one of the multiple mechanisms active in photoreceptor degeneration. Autophagy. 2007;3:65–66. doi: 10.4161/auto.3431. [DOI] [PubMed] [Google Scholar]
- 25.Kunchithapautham K, Rohrer B. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy. 2007;3:433–441. doi: 10.4161/auto.4294. [DOI] [PubMed] [Google Scholar]
- 26.Reme C. Autophagy in rods and cones of the vertebrate retina. Dev Ophthalmol. 1981;4:101–148. [PubMed] [Google Scholar]
- 27.Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: Possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Sawada O, Kohno H, Le YZ, Subauste C, Maeda T, Maeda A. Autophagy protects the retina from light-induced degeneration. J Biol Chem. 2013;288:7506–7518. doi: 10.1074/jbc.M112.439935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Jia L, Shelby SJ, Ganios AM, Feathers K, Thompson DA, Zacks DN. Circadian and non-circadian modulation of autophagy in photoreceptors and retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2014;55:3237–3246. doi: 10.1167/iovs.13-13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Sullivan TN, Wu XS, Rachel RA, Huang JD, Swing DA, Matesic LE, Hammer JA, 3rd, Copeland NG, Jenkins NA. dsu functions in a MYO5A-independent pathway to suppress the coat color of dilute mice. Proc Natl Acad Sci U S A. 2004;101:16831–16836. doi: 10.1073/pnas.0407339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu XS, Masedunskas A, Weigert R, Copeland NG, Jenkins NA, Hammer JA. Melanoregulin regulates a shedding mechanism that drives melanosome transfer from melanocytes to keratinocytes. Proceedings of the National Academy of Sciences of the United States of America; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachel RA, Nagashima K, O’Sullivan TN, Frost LS, Stefano FP, Marigo V, Boesze-Battaglia K. Melanoregulin, product of the dsu locus, links the BLOC-pathway and OA1 in organelle biogenesis. PLoS One. 2012;7:e42446. doi: 10.1371/journal.pone.0042446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damek-Poprawa M, Diemer T, Lopes VS, Lillo C, Harper DC, Marks MS, Wu Y, Sparrow JR, Rachel RA, Williams DS, Boesze-Battaglia K. Melanoregulin (MREG) modulates lysosome function in pigment epithelial cells. J Biol Chem. 2009;284:10877–10889. doi: 10.1074/jbc.M808857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frost LS, Lopes VS, Stefano FP, Bragin A, Williams DS, Mitchell CH, Boesze-Battaglia K. Loss of melanoregulin (MREG) enhances cathepsin-D secretion by the retinal pigment epithelium. Vis Neurosci. 2013;30:55–64. doi: 10.1017/S0952523813000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc Natl Acad Sci U S A. 2003;100:6481–6486. doi: 10.1073/pnas.1130432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaVail MM. Circadian nature of rod outer segment disc shedding in the rat. Invest Ophthalmol Vis Sci. 1980;19:407. [PubMed] [Google Scholar]
- 37.Mao Y, Finnemann SC. Essential diurnal Rac1 activation during retinal phagocytosis requires αvβ5 integrin but not tyrosine kinases focal adhesion kinase or Mer tyrosine kinase. Mol Biol Cell. 2012;23:1104–1114. doi: 10.1091/mbc.E11-10-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boesze-Battaglia K, Song H, Sokolov M, Lillo C, Pankoski-Walker L, Gretzula C, Gallagher B, Rachel RA, Jenkins NA, Copeland NG, Morris F, Jacob J, Yeagle P, Williams DS, Damek-Poprawa M. The tetraspanin protein peripherin-2 forms a complex with melanoregulin, a putative membrane fusion regulator. Biochemistry. 2007;46:1256–1272. doi: 10.1021/bi061466i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deguchi J, Yamamoto A, Yoshimori T, Sugasawa K, Moriyama Y, Futai M, Suzuki T, Kato K, Uyama M, Tashiro Y. Acidification of phagosomes and degradation of rod outer segments in rat retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1994;35:568–579. [PubMed] [Google Scholar]
- 40.Woessner JF., Jr Specificity and biological role of cathepsin D. Adv Exp Med Biol. 1977;95:313–327. doi: 10.1007/978-1-4757-0719-9_19. [DOI] [PubMed] [Google Scholar]
- 41.Bosch E, Horwitz J, Bok D. Phagocytosis of outer segments by retinal pigment epithelium: phagosome-lysosome interaction. J Histochem Cytochem. 1993;41:253. doi: 10.1177/41.2.8419462. [DOI] [PubMed] [Google Scholar]
- 42.Reme CA, Sulser M. Diurnal variation of autophagy in rod visual cells in the rat. Graefes Archiv Opthalmologie. 1977;203:261–270. doi: 10.1007/BF00409832. [DOI] [PubMed] [Google Scholar]
- 43.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA, Wu XS, Rovira II, Fergusson MM, Weigert R, Connelly PS, Daniels MP, Komatsu M, Cao L, Finkel T. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med. 2013;19:1281–1287. doi: 10.1038/nm.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 46.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 49.Krohne TU, Stratmann NK, Kopitz J, Holz FG. Effects of lipid peroxidation products on lipofuscinogenesis and autophagy in human retinal pigment epithelial cells. Exp Eye Res. 2010;90:465–471. doi: 10.1016/j.exer.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Birgisdottir AB, Lamark T, Johansen T. The LIR motif—Crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 51.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 52.Maminishkis A, Chen S, Jalickee S, Banzon T, Shi G, Wang FE, Ehalt T, Hammer JA, Miller SS. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 54.Boesze-Battaglia K, Albert AD. Phospholipid distribution among bovine rod outer segment plasma membrane and disk membranes. Exp Eye Res. 1992;54:821–823. doi: 10.1016/0014-4835(92)90040-y. [DOI] [PubMed] [Google Scholar]
- 55.Vazquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 56.Lopes VS, Gibbs D, Libby RT, Aleman TS, Welch DL, Lillo C, Jacobson SG, Radu RA, Steel KP, Williams DS. The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum Mol Genet. 2011;20:2560–2570. doi: 10.1093/hmg/ddr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kinane JA, Benakanakere MR, Zhao J, Hosur KB, Kinane DF. Porphyromonas gingivalis influences actin degradation within epithelial cells during invasion and apoptosis. Cell Microbiol. 2012;14:1085–1096. doi: 10.1111/j.1462-5822.2012.01780.x. [DOI] [PubMed] [Google Scholar]
- 58.Boesze-Battaglia K, Goldberg AF, Dispoto J, Katragadda M, Cesarone G, Albert AD. A soluble peripherin/Rds C-terminal polypeptide promotes membrane fusion and changes conformation upon membrane association. Exp Eye Res. 2003;77:505–514. doi: 10.1016/s0014-4835(03)00151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Lu W, Reigada DNJ, Laties AM, Mitchell CH. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(−/−) mice: Pharmacologic approaches and functional recovery. Invest Ophthalmol Vis Sci. 2008;49:772–780. doi: 10.1167/iovs.07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guha S, Coffey EE, Lu W, Lim JC, Beckel JM, Laties AM, Boesze-Battaglia K, Mitchell CH. Approaches for detecting lysosomal alkalinization and impaired degradation in fresh and cultured RPE cells: Evidence for a role in retinal degenerations. Exp Eye Res. 2014;126:68–76. doi: 10.1016/j.exer.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.