Abstract
Mammalian and bacterial cells sense and exert mechanical forces through the process of mechanotransduction, which interconverts biochemical and physical signals. This is especially important in contact-dependent signaling, where ligand-receptor binding occurs at cell-cell or cell-ECM junctions. By virtue of occurring within these specialized junctions, receptors engaged in contact-dependent signaling undergo oligomerization and coupling with the cytoskeleton as part of their signaling mechanisms. While our ability to measure and map biochemical signaling within cell junctions has advanced over the past decades, physical cues remain difficult to map in space and time. Recently, supported lipid bilayer (SLB) technologies have emerged as a flexible platform to measure and manipulate membrane receptor mechanotransduction, allowing one to mimic cell-cell junctions. Changing the lipid composition and underlying substrate tunes bilayer fluidity, and lipid and ligand micro- and nano-patterning spatially control positioning and clustering of receptors. Patterning metal gridlines within SLBs introduces corrals that confine lipid mobility and introduce mechanical resistance. Here we review fundamental SLB mechanics and how SLBs can be engineered as tunable cell substrates for mechanotransduction studies. Finally, we highlight the impact of this work in understanding the biophysical mechanisms of cell adhesion.
Keywords: supported lipid bilayer, mechanotransduction, receptor clustering, molecular tension probe, adhesion
Graphical abstract
1. Introduction
Sensitivity to mechanical forces is a common feature that is shared by the vast majority of organisms ranging from bacteria to mammals. It is fundamental to developmental processes, disease, and normal physiology. Cells transduce mechanical forces into biochemical signaling events in a bidirectional manner through the process of mechanotransduction. Cell surface receptors and cytoskeletal proteins sense and exert piconewton forces, which influence downstream biochemical responses through a wide range of processes with different molecular mechanisms. For example, mechanical forces may change the rates of reactions by accelerating or decelerating bond lifetimes [1]. Forces can also confine proteins, thus enhancing local concentration and binding interactions. Alternatively, forces can unfold specific protein domains, which exposes cryptic binding sites or activates functions which is common in stretch sensitive ion channels [2–4]. The implications of mechanical forces in cell signaling are vast. Mechanical forces regulate hearing, cell migration and adhesion, embryo development, lineage commitment, heart disease, cancer metastasis, and the immune response [5–9]. Even small differences in molecular mechanics can lead to distinct outcomes. In the immune system, for example, piconewton (pN) differences in receptor mechanotransduction have been shown to attenuate downstream cell signaling [10, 11]. Therefore, to engineer effective cell and material-based therapies, it is critical to understand how cells interact physically with their environment and how mechanical forces contribute to signaling.
The most common model system to study these events is in adhesion, the process of cell-cell and cell-extracellular matrix attachment. In adhesion, cells transmit and sense the mechanical properties of neighboring cells and the extracellular matrix (ECM). Focal adhesions (FAs) structurally and mechanically link the cell and the matrix. These protein-rich assemblies connect the actin cytoskeleton to integrin receptors which physically connect to the underlying matrix [12]. Integrin receptors are dimeric proteins which can assume a folded low affinity state or an open, high affinity state. Integrins have been shown to pull on their ligands and exert traction forces, forces parallel to the plane of adhesion, on the matrix [13–16]. Cell-cell adhesions are more structurally and functionally diverse, ranging from primarily mechanical linkages such as adherens junctions and desmosomes to tight junctions, which control transport between cells, and immune cell synapses, which bring cells in physical contact for the initiation of an immune response. Integrin receptors including the LFA receptor have also been shown to be key players in cell-cell adhesion, but the primary mediators cell-cell adhesion are cadherin receptors, which form adherens junctions (AJs). Cadherins are tissue-specific calcium-dependent adhesion proteins that form dimers with adjacent (cis) and opposing (trans) cadherins. Cadherins indirectly link to the actin cytoskeleton, allowing force generation across cell-cell adhesions [17, 18]. In both cell-cell and cell-matrix adhesions, forces originate through the cytoskeleton. Actomyosin contractility is the primary mechanism of receptor-mediated forces, but actin also generates dynamic forces through treadmilling, the process of polymerizing and depolymerizing which exerts mechanical forces directly on the cell membrane [19, 20]. Actin cytoskeleton remodeling can also drive receptor translocation in clustering, which reinforces adhesion [21].
Adhesion sites are often modeled using ECM or cell-adhesion molecule modified substrates. Geometry and mechanics are adjusted by patterning immobilized ligands on substrates of varying rigidity, from sparsely crosslinked polymers to glass. However, the specific events in mechanotransduction remain fundamentally challenging to study. Whereas biochemical signaling can be manipulated by knock-down assays or by inhibitory drugs, mechanotransduction is linked to substrate rigidity and cannot easily be altered without fundamentally changing the system, including the density of ligands. Thus, despite the advances in scaffolding, the precise role of mechanical forces in adhesion assembly remains poorly understood.
Recently, several studies have attempted to bridge this gap by using supported lipid bilayer (SLB) technologies to spatially control the generation of mechanical forces [22–27]. SLBs are biomimetic phospholipid membranes that self-assemble on planar glass substrates (Figure 1A). They initially gained attention for their ability to form hybrid cell-cell interfaces and have been particularly useful in modeling antigen-presenting cells to study immune cell synapse formation during T cell activation [28, 29]. SLBs can be formed by either vesicle fusion, in which unilamellar vesicles adhere to the substrate, rupture, and fuse into a plane, or by Langmuir deposition, in which individual leaflets of the bilayer are sequentially added [30–32]. A thin layer of water separates the glass from the lower leaflet, allowing both leaflets to maintain their fluidity [33] (Figure 1A). Lipids freely diffuse in the XY-plane, and the diffusion coefficient is controlled by the bilayer’s phase [34] (Figure 1B). A high bending modulus confines diffusion to the plane of the substrate. Thus, the physical properties of SLBs closely mimic those of the plasma membrane, and cell-SLB interfaces recapitulate the fluid interface between adjacent cells that physically engage, serving as hybrid cell-cell junctions.
Figure 1.
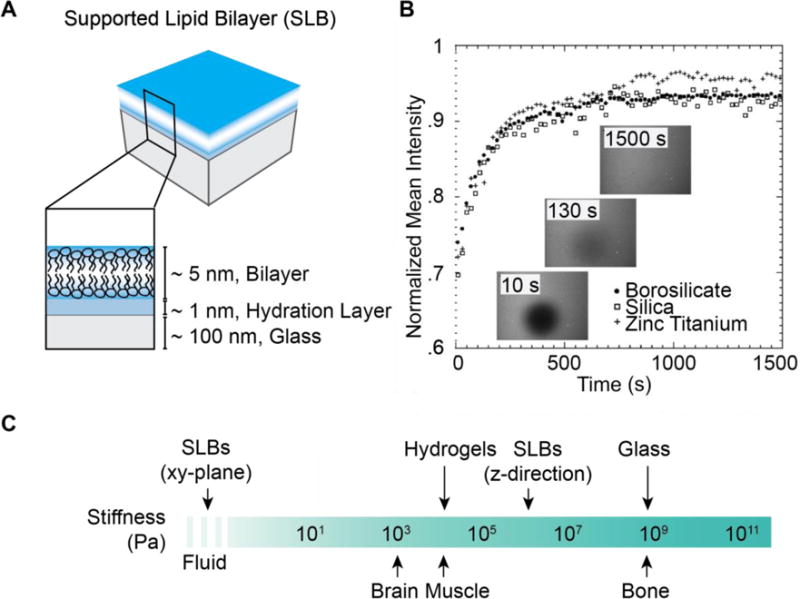
Supported Lipid Bilayer (SLB) design and mechanics. (A) SLBs contain a bilayer separated from a rigid substrate by a thin layer of water. (B) Representative FRAP of labeled lipids illustrating SLB lateral fluidity. Lipids in SLBs freely diffuse within the plane on three representative substrates. Following photobleaching, diffusion causes photobleached lipids to be diluted and the average fluorescence to increase. The disappearance of a visible bleached region indicates total recovery and a fluid bilayer. (Reprinted with permission from [34]. Copyright 2009, American Chemical Society). (C) SLB stiffness in comparison to tissue, hydrogels, and glass substrates. SLBs are anisotropic, behaving like fluids in the XY-plane, but stiffer than hydrogels in the Z-direction. Inspired by [146].
An important advantage of the SLB platform is the ability to manipulate ligand mechanics to study mechanotransduction. Therefore, SLBs have recently emerged as a platform to probe receptor signaling events in both cell-cell and cell-matrix adhesion. Because fluid bilayers cannot support lateral traction forces, signaling pathways proceed in the absence of mechanotransduction in the direction tangential to the membrane. By adjusting the fluidity of the bilayers or by patterning barriers as sites of force generation, resisting forces can be selectively introduced [35, 36]. In this review, we describe SLB biophysics and various methods to manipulate SLB mechanics and to measure signaling outcomes. We present this material alongside a discussion of literature that applies this platform to characterize integrin and cadherin mechanotransduction. Note that a number of reviews have fully described the SLB technologies and their use in studying cell biology [37, 38]. Nevertheless, our focus is to emphasize recent work that pertains to the study of cell mechanobiology.
1.1 Receptor Mechanics
In adhesion complexes, receptors serve as a mechanical linkage between the cell and the underlying matrix or an adjacent cell. Thus, these sites regulate signaling not only through binding, but also through force transduction. Mechanical forces adjust downstream cell signaling by modulating bond lifetime. For an idealized bond with a single energy barrier, the Bell model states that mechanical forces alter off rate, which reduces bond lifetime [1]. In this scenario, bond lifetime, τ, can be described as:
in which EA is the bond enegy, τ0 is bond lifetime at zero external forces, k is the Boltzmann constant, and T is temperature. In the case of applied force, this equation is modified:
In which γ is a structural parameter and f is the force applied to the bond. Receptor-ligand interactions vary in their response to forces. While most bonds will display a reduced lifetime with the application of pN forces, certain receptors form catch bonds. Catch bonds are an exception in which mechanical forces strengthen adhesion by lengthening bond lifetime. Many adhesion proteins, most notably the integrin family, have been shown to form catch bonds with their ligands [39, 40]. The general form of the Bell model can be applied to understand how forces drive the presentation of cryptic sites or the stabilization of weak interactions.
1.2 Advantages of Supported Lipid Bilayers
Many signaling pathways are contact-dependent and initiated at the cell membrane when a receptor interacts with a ligand presented on an opposing cell surface or ECM. Signaling responses are regulated in part by the biophysical properties of interaction, including bond lifetimes, receptor spatial organization, clustering, and mechanics at these interfaces [35, 36, 41–45]. SLBs provide a convenient model to study and perturb these membrane-mediated interactions and signaling pathways.
SLBs are a reductionist platform. Although the cell membrane includes a rich variety of proteins and lipids that segregate into complex domains, SLBs allow the isolation of a few receptors of interest to study receptor-receptor (cis) and receptor-ligand (trans) interactions. Furthermore, SLBs recapitulate the geometry of juxtacrine interactions, in which ligands and receptors are expressed on adjacent cells and physical contact between the cells is necessary to trigger signaling. Contact-dependent signaling pathways require surface anchoring of ligands and soluble ligand molecules often fail at initiating downstream receptor signaling cascades. For example, surface-bound ligands are required for integrin-mediated cell adhesion [46]. T cell triggering requires surface presentation of antigen and the formation of a physical junction between the T cell and the antigen presenting cell [43, 47]. By the incorporation of ligands or transmembrane proteins into an SLB, the native 2D binding geometry can be sufficiently mimicked to initiate a downstream response.
Although rigid surfaces can also be functionalized to present ligands in a planar geometry, SLBs offer a distinct advantage in their lateral fluidity, which permits clustering and transport [22]. Super-resolution imaging reveals that many receptors exist in nanoscale clusters on the cell membrane prior to signaling [48]. Upon receptor-ligand binding, hundreds to thousands of receptors associate together in microclusters, leading to signal amplification, increased specificity, and response-time coordination [49, 50]. Whereas individual receptors typically are not connected with the cytoskeleton, clustered receptors can associate with the cytoskeleton, providing a direct linkage between the extracellular proteins and the cell’s force generating machinery. Thus, receptor clustering reinforces cytoskeletal coupling and strengthens the force of adhesion [51]. In the case of unligated receptor clustering, cluster lifetime is reduced compared to the lifetime of ligand-bound receptor clusters [24]. In many cases, clusters are actively transported across the membrane, their translocation corresponding to the amplitude of biochemical signaling [35, 36, 52]. These mechanisms demonstrate the importance of ligated receptor lateral transport, which can only be captured on fluid substrates.
In addition, SLBs offer several experimental advantages. The bilayer’s 2D geometry permits quantitative analysis of receptor diffusion and oligomerization. These can be easily measured with fluorescence recovery after photobleaching (FRAP), fluorescence imaging, and fluorescence correlation spectroscopy (FCS) of tagged lipids or proteins [32]. The planar geometry of cell-SLB interactions can also be easily imaged with total internal reflection microscopy (TIRF). In TIRF, an evanescent wave excites fluorophores in a thin ~150 nm slice at the surface, providing fluorescence images with improved signal-to-noise ratio compared to epifluorescence [53]. Time-lapse TIRM images can be collected on time scales compatible with receptor transport and downstream biochemical signaling.
2. Mechanics in Supported Lipid Bilayer Systems
2.1 Mechanics of Supported Lipid Bilayers
2.1.1 Supported Lipid Bilayer Mechanical Characterization
Bilayer mechanical properties are typically characterized by the compression modulus, Ka, the bending modulus, Kb, and the edge energy, ∧ Ka describes the bilayer’s resistance to changing area, whereas Kb measures the energy needed to curve a bilayer. Unilamellar SLBs and SLBs on rigid substrates are tightly confined to XY-plane. In these cases, Kb is not a relevant parameter. However, fluctuations in the z-direction in stacked and cushioned SLBs depend on Kb. For small membrane deformations, Ka and Kb are linearly related, with Kb scaling with bilayer thickness. Ka exhibits phase-dependent behavior. Liquid disordered (lD) (fluid) SLBs have a low compression modulus of 0.12 N/m. Liquid ordered (lo) regions of the bilayer behave stiffly for small deformations, with a compression modulus of approximately 1.1 N/m. When further deformed, lipid interactions are disrupted, which causes the SLB to behave as a soft material with a compression modulus of 0.05 N/m [54]. ∧ quantifies the bilayer’s resistance to pore formation; it is the energy cost due to exposed fatty acid chains at a pore. A contributes to the bilayers ability to self-heal; positive edge energy indicates that pores will only form under the application of tension. Thus, Λ contributes to the stability of an SLB under receptor mediated forces. For 100 mol% DOPC SLBs, the edge tension, Λ per length, is 27.7 pN [55].
Deforming the SLB over a nanoscale pore using AFM allowed the measurement of an apparent SLB “spring constant” [56]. In fluid and gel-phase membranes, the apparent “spring constant” was found to be 0.0039 N/m and 0.015 N/m, respectively. For pore sizes below 100 nm, the restoring force decreased with pore radius. For deformations between 4 and 10 nm, the apparent “spring constant” was linearly related to surface tension and Kb [56]. The value of the apparent “spring constant” of an SLB is useful for quantifying local membrane deformations, specifically in the case of cell mechanotransduction on cushioned and multilamellar SLBs.
Cell substrate mechanical properties are most commonly characterized by their Young’s Modulus, E, which measures the substrate stiffness and is defined as stress (force per area) over strain (deformation). As this parameter is not well defined for membranes, direct comparison of SLB mechanical properties with those of conventional polymer supports is not simple. SLBs are anisotropic materials, rigid in the z-direction and minimally resistive in the lateral direction (2.2.1). The stiffness of SLBs in the z-direction is reflective of the mechanical properties of the underlying support. To obtain the elastic response of an SLB in the z-direction, Picas, et. al. developed a novel AFM-based method, PeakForce-Quantitative Nanomechanics [57]. SLBs on mica were oscillated vertically at 2 kHz and allowed to contact an AFM tip. At a loading force of 200 pN, the z-direction Young’s Modulus of SLBs was reported at 19.3 MPa for liquid phase and 28.1 MPa for gel phase SLBs [57]. Gel phase SLBs were effectively stiffer than fluid phase SLBs at all loading forces. These measurements indicate that in the vertical direction, bilayers supported on mica are stiffer than many biological tissues and hydrogels (kPa) but softer than glass (GPa) (Figure 1C). In contrast, polyethylenimine supported DMPC bilayers closely mimicked the stiffness of cells. The underlying polymer swelled to create a ~15 nm cushion between the lower leaflet of the SLB and the underlying mica substrate, leading to an effective Young’s Modulus of 32 – 47 kPa [58].
2.1.2. Lipid Extraction Under Force
In addition to the properties governing the reversible deformation of an SLB under tension, it is important to consider the irreversible destruction of bilayers under mechanical forces. Apart from specialized biological functions such as endocytosis, membranes can only undergo a few percent strain before rupture. In the case of adhesion receptor mechanobiology, the more important parameter is the force of lipid extraction rather than whole membrane rupture. The location of detachment can be determined by the relative energy gradient at the bond.
In this equation, Lm and Lb refer to lipid anchor and bond length, respectively, and Eb and Em are the energies of bond rupture and membrane failure, respectively. Given the case where the bond energies are similar, the likelihood of failure increases with hydrophobic tail length. Thus, given a constant bond length, the force of lipid extraction decreases with hydrocarbon chain length [59]. Wong, et. al. calculated that pulling a PEG-lipid from the bilayer into an aqueous environment would require 23 pN [60]. Leckband, et. al. measured an adhesion force of 80 pN required to an extract a lipid via biotin-streptavidin interaction [59]. For mica-supported POPC bilayers, 50 pN was required to extract a single POPE lipid using AFM. Cholesterol extraction in phase-separated SLBs using both AFM and molecular dynamics simulations revealed that extraction requires more force in lo regions than in ID regions. Benchmark receptor forces are provided in Section 4.2. SLBs are generally sufficiently stable to withstand short-term applied forces (~ 1 hr) by cells, but lipid extraction is noted at longer time scales. Yu, et. al. reported integrin endocytosis on SLBs and observed internalization 3 hrs following cell-substrate engagement [61]. B cells could extract antigen on viscoelastic plasma membrane sheets, but not on supported lipid bilayers which were more tightly coupled to the substrate [62].
2.2. Frictional and Mechanical Forces on Membranes and Receptor-Ligand Complexes
2.2.1 Diffusion and Viscous Drag in Supported Lipid Bilayers
Diffusion in an SLB is considered in two regimes: diffusion of lipids and similarly small molecules and diffusion of proteins and other large molecules. Lipid diffusion requires sufficiently large free volume and sufficiently high energy to disrupt neighboring tail interactions. In an SLB the diffusion coefficient, D, is determined by phase and substrate-SLB coupling. For larger species, the bilayer is treated as a continuous viscous media. Diffusion is attributed to the net sum of forces due to collisions with lipid molecules and the resisting frictional force, viscous drag that is imparted by the membrane [63]. The diffusion coefficient, D, and frictional coefficient, f, are inversely related by the Einstein Relation:
in which k is the Boltzmann constant and T is temperature. For an integral membrane protein in an SLB,
This assumes a cylindrical protein with radius α in a bilayer of height h. μ is the viscosity of the bilayer, μ′ is the viscosity of the surrounding media (μ ≫ μ′), U is the proteins velocity, and γ is Euler’s constant [63, 64].
Steric hindrance prevents direct transmembrane protein reconstitution in SLBs, so protein domains or ligands are more commonly tethered to lipid anchors. A useful model to quantify diffusion of tethered protein domains in SLBs is the the pleckstrin homology (PH) domain model, in which each PH domain attaches to single PIP3. Knight and Falke found that the PH domain protruded further into the surrounding media than into the SLB, but that the diffusion coefficient closely matched that of PIP3. This indicates that transport is regulated primarily by intra-membrane friction rather than by drag between lipid-tethered proteins and the surrounding media [65, 66]. These results logically follow from viscosity measurements, which suggest that SLBs are 200× more viscous than their surrounding aqueous environments [65].
In more complex scenarios, the diffusion coefficient depended on protein-lipid interactions. The diffusion coefficient of multimers inversely scaled with the number of bound lipids, and contributions of lipid binding and protein insertion into the hydrophobic core were additive for membrane penetrating proteins such that:
Here, N is the number of bound lipids, F is the frictional contribution of a single lipid, c is a constant, and P is the number of penetrating domains [67].
Typical diffusion coefficients for lipids in fluid and liquid-disorder SLBs are 1–4 μm2/s and .1 μm2/s, respectively [68]. The diffusion coefficient of 17 tethered protein domains with various degrees of lipid penetration ranged from .22 to 2.6 μm2/s. Corresponding frictional coefficients ranged from .39 for anti-biotin with only one lipid binding domain to 4.6 for a fusion construct with 6 bound lipids [67]. Biswas, et. al. estimated that extracellular domains of E-cadherin on a fluid SLB experienced an average of .5 fN viscous drag during lamellipodial retraction [26].
Although membrane-bound proteins only attach directly to one or few lipids, protein binding has been observed to alter lipid diffusion within a larger radius. Forstner, et. al. found that near the melting temperature, cholera toxin binding induced the formation of gel-phase islands in DMPC and DMOPC SLBs [69]. Molecular dynamics simulations of Kv1.2 ion channel in DOPC bilayers suggest that this layer includes approximately 50–100 lipids which diffuse with the protein [70]. At reasonably low concentrations, diffusion coefficients of membrane-bound proteins are concentration independent [71]. This rule breaks down when proteins are added in sufficiently high concentrations to form a monolayer [71, 72].
In cells, receptor-ligand transport is hindered by cytoskeletal barriers imposed on the bilayer. Spectrin forms a geometric mesh that supports the membrane, and actin can limit protein diffusion through the tether model, in which a protein is directly bound to the cytoskeleton, and the fence model, in which proteins diffusion is spatially limited by bulky cytoskeletal barriers [73, 74]. Edinin, et. al. demonstrated that these barriers dynamically confine protein diffusion, and that the mechanism of protein attachment to the membrane affects confinement [75]. Transferrin receptors tagged with beads and dragged across an NRK cell membrane using optical tweezers required 0.25 – 0.8 pN trapping force to cross cytoskeletal boundaries. At lower forces, receptors escaped the optical trap, suggesting the cellular barriers winning in a tug-of-war. Barriers behaved elastically with a spring constant of 3pN/μm, causing escaped receptors to quickly return to their original position [73, 74]. Similarly, E-cadherin dragged across the cell membrane with optical trap velocity of .6 um/sec followed the optical trap for .78 um, where it began to lag behind the displacement of the trap. The receptor escaped the trap at 1.32 um, corresponding to an optical trap force of .8 pN. Homogeneous SLBs cannot impart such forces on ligands and receptors, but several experimental techniques allow the experimenter to controllably pattern corrals (3.2.2), and these phenomena may affect receptor transport in the SLB-adhered cell.
2.2.2. Effects of Membrane Tension
Within both cells and cell-free systems, membrane tension has been shown to induce receptor-ligand transport. Smith, et. al. developed parallel fluid and non-fluid cell-free systems to assess adhesion of mobile versus immobile integrins. Giant unilamellar vesicles (GUVs) containing RGD-functionalized lipids equilibrated on SLBs containing mobile or immobile embedded integrin receptors, and a 2–4 pN vertical force was applied to the membrane using optical tweezers. Whereas immobile bonds stretched and ruptured, mobile linkages clustered beneath the GUV to resist detachment [76]. In the fluid system, which contained more GUV-surface linkages, each bond experienced negligible force (0.2fN), and the remodeling response was attributed to thermodynamic requirements to reduce free energy rather than to mechanics. Nevertheless, this work illustrates the concept that membrane tension can passively drive receptor reorganization at a juxtacrine interface. Accordingly, membrane tension applied by micropipette aspiration was sufficient to cause membrane flattening and passive E-cadherin recruitment in EAhy cells [77].
2.2.3. Force Generation by Cytoskeletal and Motor Proteins
Cytoskeletal proteins and motor proteins exert forces on membranes and their associated receptors. The cytoskeleton has two primary mechanisms of active force generation, polymerization and contractility. Actin and microtubules polymerize against the membrane, generating forces through a ratchet model. Thermodynamic fluctuations cause a transient space between the filament and the bilayer, allowing the insertion of a subunit. The extending polymer exerts pushing forces against the membrane. Footer, et. al. demonstrated that 8 parallel actin bundles can exert 1 pN force on a rigid wall, and actin comets have shown persistent polymerization at resisting forces of 4.3 nN [78, 79]. In podosomes, crosslinked actin polymerization against the membrane causes the cell to protrude into the substrate. These protrusions have been measured to exert an average of 94 nN or Formvar sheets [23, 80].
Actomyosin contractility directly pulls actin-bound receptors. Motor proteins including myosins walk along actin filaments, generating 3–4 pN per step per myosin head; this actomyosin contractility is responsible for receptor tension and traction forces in many systems [81–83]. A key question in the literature has been whether myosin can generate forces parallel to the membrane or only perpendicular. Long range traction forces are dissipated due to lipid diffusion, but recent work by Pyrpassopoulos, et. al. suggests that myosin motors can act in concert to generate low pN forces at a fluid interface [84]. Thus, while motor proteins generate large traction forces on rigid substrates, in-plane force generation and maintenance at fluid interfaces are more transient and require high cooperativity.
3. Supported Lipid Bilayers Technologies
3.1. Methods to Perturb Bilayer Mechanics
3.1.1. Tuning SLB Composition to Control Lateral Diffusion
The simplest way to manipulate SLB mechanics is to adjust lipid composition and packing. This can be accomplished by adjusting the degree of fatty acid unsaturation. Changes in phase are accompanied by changes in transport, both passive diffusion and active transport, due to the altered fluidity. Within an SLB, individual lipids interact via van der waals interactions, and their packing determines SLB phase. Below the melting temperature, SLBs are in gel phase with the lipid hydrocarbon tails rigidly arranged. Above the melting temperature, fatty acids rotate about their C-C bonds and exhibit long range coordinated motion. Lipids with longer hydrocarbon chains exhibit improved packing and reduced free volume, leading to slower diffusion [85].
In studies of mechanotransduction, adjusting SLB fluidity has two primary consequences. First, the kinetics of the system are altered. In a system with decreased fluidity, ligated receptor transport across the cell-SLB interface is slowed, potentially allowing for nucleation or for additional signaling molecules to bind. In addition, a gelphase or crowded membrane with low fluidity permits the generation of resisting forces. In a fluid system, lateral forces cannot be applied because there is no resistance [26]. An additional benefit of phase-controlled bilayers is the ability to better mimic the cell’s plasma membrane. SLBs provide a simplistic experimental platform, however they lack the complexity and richness of live cell membranes, which are separated into multiple domains and are comprised of hundreds of lipids and thousands of proteins. Adjusting the phase of a SLB can begin to capture the complexity of the plasma membrane and create a more physiologically relevant model.
Cholesterol biosynthesis is tightly regulated in part to modulate membrane fluidity, thickness, and integral protein activity. This is mediated by cholesterol’s rigid ring structure inserting into the membrane. Cholesterol is hypothesized to straighten saturated lipids, allowing them to pack more efficiently. This effect on lipid-lipid interactions likely more directly modulates bilayer fluidity than the introduction of cholesterol-lipid interactions (Figure 2A) [85, 86]. Accordingly, cholesterol is a common species used to modulate bilayers in vitro. Cholesterol containing membranes exhibit three states, gel phase, liquid ordered (lo) (at high concentrations of cholesterol and below Tm), and liquid disordered (ld) (at high temperatures and low cholesterol concentrations). In the lo phase, lipids exhibit strong tail interactions like in the gel phase, however they retain high fluidity. Cholesterol’s effects on bilayers are dependent on lipid composition, temperature, and cholesterol concentration. For example, DPPC membranes are homogenous at low concentrations of cholesterol and can exist in either the lo or gel state. At 10 mol% cholesterol, DPPC membranes phase separate into a cholesterol-depleted region and a cholesterol-rich liquid disordered region. However, the addition of 50 mol% cholesterol again gives rise to a homogeneous bilayer [87]. Thus, great care must be taken when doping bilayers with cholesterol to achieve the desired effect. In SLBs, 25 mol% cholesterol in DOPC membranes has been shown to reduce the diffusion coefficient of lipids and anchored proteins 4–5-fold [42].
Figure 2.
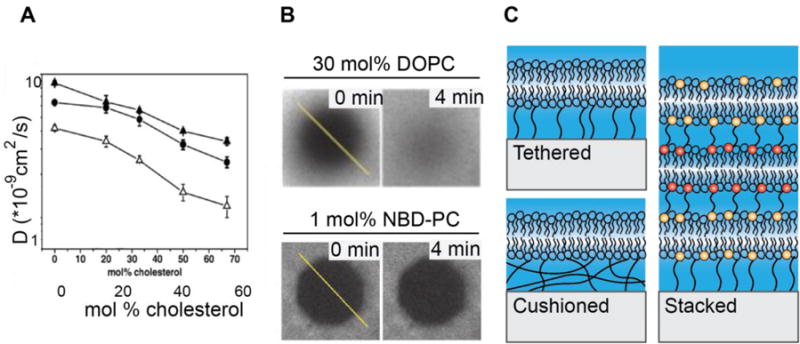
SLB phase and diffusion tuning. (A) Cholesterol reduces SLB fluidity in DmirPC (closed triangles), DOPC (cirlces), DEPC (open triangles) bilayers. Reprinted from [85] with permission of publisher. (B) Representative FRAP data from fluid and partially fluid DPPC bilayers containing, 30% DOPC and 1% NBD-PC, respectively. In DOPC SLBs, fluorescence almost fully recovers in 4 minutes, but in SLBs containing NBD-PC, the photobleached region persists, indicating low fluidity. (Reprinted from [26]. Copyright 2015, National Academy of Sciences). (C) Schematics of tethered, cushioned, and stacked SLBs.
As an alternate approach, SLB phase may be modulated by the addition of lipids with a bulky tail group (Figure 2B). 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) labeled with 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) emerged as a popular fluorescent probe in the study of membrane physiology; however differential scanning calorimetry revealed that as low as 1%, NBD-PC altered membrane physical properties [88]. Interestingly, the placement of NBD on the hydrocarbon tail determined the effect on phase. Harnessing the artifacts introduced by NBD-PC, Biswas, et. al. used NBD-PC to generate partially fluid bilayers to study adherens junction formation (Figure 2B) [26].
Sterically crowding the membrane with protein alters SLB fluidity without significantly changing the lipid composition. SLB functionalization with streptavidin is achieved by doping in a small amount of biotinylated lipid, typically biotin-DPPE. In kinetics assays, streptavidin binding saturated at 4 mol% biotin-DPPE with two biotinylated lipids binding each streptavidin. At concentrations below 4 mol% biotin-DPPE, streptavidin bound in a dose dependent manner. At 4 mol% biotin-DPPE, bound streptavidin forms a crystal monolayer [72, 89]. Whereas SLBs with low streptavidin coverage retained their fluidity, crowded streptavidin monolayers obstructed long-range diffusion [41]. Thus, the long-range diffusion coefficient is reduced by increasing the density of streptavidin molecules on the SLB. This method is particularly useful for probing the effects of lateral transport on receptors. By comparing Notch activation in cells on fluid, nonfluid, and rigid surfaces, Narui and Salaita identified that the Notch/Delta pathway is mechanosensitive and responds nonlinearly to ligand fluidity [41]. More recently, it was shown that platelets prefer to adhere to crowded membranes [90]. A nonfluid interface for cell adhesion was also recently fabricated by covalently linking ECM proteins to fluid lipids [91].
Selection of lipids with a transition temperature close to physiological conditions allows manipulation of bilayer fluidity without significantly changing SLB chemical composition. For example, 1-myristol-2-palmityol-sn-glycero-3-phosphocholine (MPPC) has a melting temperature of 35°C, allowing the lipids to be switched between gel phase and liquid phase in a temperature range compatible with live cell imaging. Demonstrating this, Adreasson-Ochsner fabricated micron sized 3-dimensional wells coated with supported MPPC bilayers containing E-cadherin ligands on the walls and base of the well. A single cell could spread in each well, and the differences in adhesion could be observed on chemically identical bilayers different only in their lateral fluidity [92]. A key nuance to this work is that the biology itself could be altered by the temperature change between fluid and non-fluid bilayers. However, given the small difference in temperature, these effects are likely minimal. The benefit to this method of adjusting phase is that it is ligand-concentration independent. Whereas membrane crowding with streptavidin also typically affects ligand binding and density, a simple assay with SLBs below and above their melting temperature avoids convolution with ligand presentation and circumvents the need to add a fluorescent lipid.
3.1.2. Stacked and Cushioned Bilayers
The physical properties of SLBs are closely linked with substrate mechanics and topology. SLBs are most often formed on silicon oxide glass, and a thin layer of water separates the lower leaflet from the glass. The exact effects of surface-lipid interactions are highly contested and preparation dependent, but evidence suggests that lipidsubstrate coupling can cause uneven leaflet lipid composition, drag between upper and lower leaflets, altered surface tension, and reduced fluidity [93–95]. According to the classic model developed by Evans and Sackmann, the frictional coefficient between the membrane and the substrate is inversely related to the thickness of the fluid layer of separation [96]. Therefore, increasing the thickness of the fluid supporting the bilayer will increase the mobility of the SLB. This insight has motivated the development of cushioned and stacked bilayers.
From a biomimetic standpoint, the effects of glass cannot be ignored. Whereas glass has a modulus on the order of GPa, atomic force microscopy measurements suggest that stiffness of the cell cortex which supports the cell membrane in vivo is one million times softer on the order of kPa [97]. Accordingly, creating SLBs on soft cortexlike supports is desirable. Here we discuss two converging approaches using SLB technologies: stacked bilayers in which multiple bilayers are fabricated on top of each other and cushioned bilayers in which the SLB is formed on a polymer support (Figure 2C). These systems physically decouple the SLB from the glass substrate, offering the potential for improved physiological relevance in cell studies. We anticipate these platforms to be extremely useful in elucidating the role of mechanics in cell signaling and cell differentiation.
Several fabrication methods for stacked SLBs have been attempted with varying levels of success. Covalent linking of lipids by NHS/EDC chemistry generated stacked bilayers, but the upper bilayer exhibited slowed diffusion and only ~75% of the lipids were mobile. This was hypothesized to be the result of nanoscale discontinuities in secondary SLB coverage, which were revealed by AFM [98]. Murray, et al. tethered biotinylated vesicles to streptavidin functionalized SLB and observed secondary bilayer formation at high vesicle concentrations. Diffusion was not significantly altered in the upper membrane compared to single layer SLBs [99]. Stacked SLBs stabilized with multiple favorable interactions improved quality, and SLBs connected with two positively charged bilayers with cholesterol functionalized DNA demonstrated high fluidity. However, these membranes remained challenging to characterize [100].
Recent advances have allowed the formation of homogeneous and heterogeneous SLBs with up to four layers. Zhu, et. al. demonstrated that the incorporation of 10% cationic or anionic lipids allowed the formation of homogeneous or patterned bilayers [101]. In patterned bilayers, the addition of saturated 1,2-distearoylsn-glycero-3-phosphocholine (DSPC) induced charged lipid phase separation. Phase separated domains aligned in each layer but were contingent upon bilayer fluidity. Stacked phase separated domains were also reported in phosphatidylcholine, cholesterol, and sphingomyelin containing membranes [102]. Together these data suggest an underlying physical explanation for aligned phases beyond electrostatic interaction. Kaizuka and Groves suggest the possible role of surface tension in preferentially aligning gel domains [102]. Patterned stacked bilayers require mechanical characterization but will undoubtedly emerge as a powerful tool to study adhesion.
Polymer “cushioned” and tethered bilayers provide an alternative method to decouple the SLB from the underlying glass [103]. These methods were developed with the goal of incorporating integral membrane proteins into SLBs; however they also offer potential for manipulating the mechanical microenvironment. Integral proteins incorporated into SLBs typically fail to maintain their fold and lateral mobility due to adsorption on the glass, steric hindrance to diffusion, and denaturation as proteins are dragged along the surface [104]. Polymer cushions or tethers lift the SLB off the glass, not only cushioning the bilayer, but also creating space for diffusing integral proteins. Whereas cytochrome b5 and annexin V were both immobile in SLBs formed directly on glass, a combination of BSA passivation and polymer tether incorporation raised the highly mobile fraction to 75% [105, 106]. Tethers must link the bilayer to the glass while not interacting with either the lipids or any incorporated proteins. In addition, the polymer of choice must be hydrophilic to support bilayer formation [105]. Therefore, PEG and chitosan have been popular choices for polymer tethers and cushions. Sterling, et. al. also fabricated actin-supported bilayers, in which the actin cushion attempted to better mimic the cortex; the results of this work emphasize that fluidity modulation is polymer specific [107]. Alternatively, cushioning the SLB with a thin cellulose cushion has been used to generate homogeneous bilayers with mobile integrins [104].
A major challenge in integral protein orientation is directing protein orientation. When proteins are reconstituted in vesicles for fusion with SLBs, their orientation scrambles. This skews diffusion measurements, because proteins with reversed orientation can be immobilized or denatured if their large extracellular domain interacts with the support. In mechanotransduction studies, these proteins would fail to interact with their binding partners. Recent work by Richards, et. al. suggested that protein orientation can be controlled by incorporating integral proteins using cell blebs, small, isolated vesicles from mammalian cells. Because these originate from the plasma membrane, proteins were oriented in their natural arrangement. Thermodynamically favorable downwards rupture preserved receptor orientation when vesicles fused during SLB formation [108].
Although the potential for polymer-cushioned bilayers as a platform to adjust the underlying substrate rigidity in mechanotransduction studies has not yet been explored, stacked and polymer-tethered bilayer technologies have been combined to generate robust surface-decoupled SLBs to study adhesion. Bilayers are linked by maleimide-thiol coupling of lipopolymer linkers [109]. To date, this is the only stacked bilayer system that has been applied as a novel cell substrate. 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine (POPC) bilayers were alternately doped with 5 mol% 1,2-dieasteroylsn-gllycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-200] (ammonium salt) (DSPE-PEG2000-Mal) or 5 mol% 1,2-dipalmitoyl-sn-glycero-3phosphothioethanol (Sodium Salt) (DPTE). A sucrose gradient promoted the sinking and fusion of giant unilamellar vesicles and malemeide-thiol coupling linked the upper and lower leaflets of adjacent bilayers. In agreement with the Evans-Sackmann model, the diffusion coefficient increased and the viscous drag coefficient decreased for each additional layer (Figure 2D) [96]. Lipopolymer-linked stacked bilayers coated with laminin behaved both viscoelastically and plastically. Individual lipids and attached receptors were laterally mobile but beads containing preclustered receptors became immobilized when attaching to the multi-bilayers [110]. Notably, AFM micrographs revealed that surface roughness increased with the addition of each layer; these effects were likely the result of decoupling between the upper bilayers and the glass [109].
3.2. Patterning to Control Receptor Mechanics
3.2.1. Lipid and Ligand Patterning
Ligand patterning is a powerful approach that has been widely applied in the study of adhesion and immune cell activation. On glass, block copolymer micelle nanolithography (BCML) allows the deposition of nanoparticles with precise control over particle density, ranging from 50 to 150 nm [111, 112]. By decorating these particles with ligand, BCML has been used to identify the critical pMHC density required to support T cell spreading, as well as to study the crosstalk between cell migratory behavior and ligand presentation [112]. To more closely mimic cell-cell interactions and to understand the role of ligand mobility, it is highly desirable to develop similar methods to control ligand geometry and density within supported lipid bilayers.
Until recently, patterning within bilayers has been limited to the ability to pattern blocks of membrane. Several methods have been developed in which lipids are either selectively added or removed in patterns. In one such method, a supported lipid bilayer formed on chromium grids was physically peeled off using a scanning probe tip and then refilled [113]. Alkaline conditions favored membrane removal. Neutralizing the pH promoted bilayer fusion and allowed these regions to be backfilled with a second lipid composition. SLB micro-voids could also be generated with deep UV illumination [114]. In a widely used method, SLBs are patterned with a polydimethylsiloxane (PDMS) stamp. PDMS stamps are inked with SLBs, which are contact transferred to a glass substrate. Combining this method with BSA barriers, it was possible to form bilayers with a lipid concentration gradient. Following stamping, vesicles with a second composition of lipids were added on top of the printed SLB and allowed to mix within compartments [115, 116]. The composition of the resulting membrane patches depended on the size of the stamp applied to each region defined by the BSA grid. Groves, et. al. achieved spatial control over HeLa and fibroblast cell adhesion on patterned SLBs by incorporating phosphatidylserine (PS), which promotes cell adhesion, into individual corrals on an SLB (Figure 3A) [116]. Using these methods, it would also be possible to selectively anchor adhesion ligands within distinct regions of the SLB by controlling which patches contain functionalized lipids or lipid ligands, such as glycolipids, which are commercially available and can support cell adhesion [117]. Examples include PIP, DNP, reactive lipids such as azide modified lipids and thiol reactive lipid headgroups.
Figure 3.
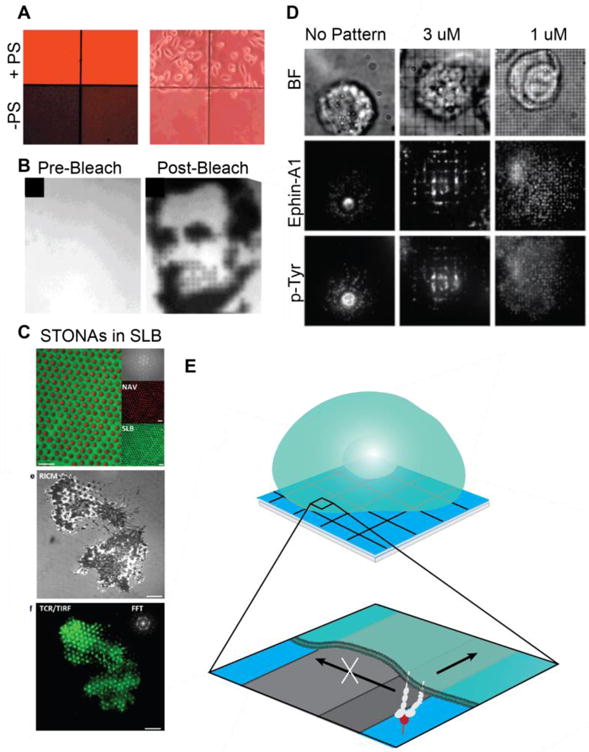
SLB composition and substrate patterning. (A) SLB composition controls HeLa cell adhesion. PS promotes HeLa cell adhesion, but few cells adhere on PS-free bilayers. Fluorescence image of SLBS (left). Phase contrast image of cell adhesion (right). (Reprinted from [116] with permission from publisher). (B) Photobleach printing on corralled bilayers demonstrates the ability to locally control SLBs. (Reprinted from [115] with permission from publisher. (C) STONAs containing nanodot-immobilized ligands embedded in an SLB; Representative image of T cells on a STONA patterned bilayer. Scale bars 4 um. (Reprinted from [123] with permission from publisher). (D) Spatiomechanical mutation of receptor transport and phosphorylation on corralled bilayers. Levels of tyrosine phosphorylation correlates with ephrin-A1 radial transport. (Reprinted from [35] with permission of publisher). (E) Diffusion barriers gate receptor transport and serve as sites of local force generation.
Another more common method to control the spatial arrangement of ligands within the supported lipid bilayer is by membrane protein photolithography. Optogenetic tools including caged and photoactivatable proteins are engineered with naturally occurring photoreceptors such as the LOV domain [118]. By shining the appropriate laser on the engineered proteins, the experimenter can spatiotemporally control protein accessibility and activation. In one approach, these domains can be linked to the protein of interest and then cleaved using photoactivation, therefore allowing the experimenter to control which ligands are accessible and which ligands remain caged. Combining this system with the supported lipid bilayer-T cell synapse model, DeMond, et. al. controlled T cell blast spreading and activation [119]. Membrane-bound MHC molecule IEK was loaded with MCC peptide fused to a light sensitive 6-nitroveratryloxycarbonyl (NVOC) blocking group. On bilayers with caged pMHC and ICAM-1 to support adhesion, T cell blasts crawled across the surface and failed to form Ts cell synapses. When the NVOC was cleaved with near-UV light, pMHC was exposed, causing T cells to adopt a round shape and to form immunological synapses.
Alternatively, light-sensitive linkers may be used to selectively remove ligands from the surface. Nakayama, et. al. developed a photoeliminative linker that can be used to both site-specifically add and remove proteins on a bilayer [120]. A photoeliminative 4-(4-(1-hydroxyethyl)-2-methoxy-5-nitrohenoxy)butanoic acid bridges a biotin group and a farnesyl group, which inserts into the bilayer. Prior to functionalization with streptavidin, biotin can be removed by UV-illumination through a patterned mask. Using a second round of UV irradiation at later time points, the protein may be released from the bilayer, providing a model to study adhesion and diffusion. To date, photoactivation of ligands has only been applied to continuous bilayers. On corralled bilayers, photobleaching of individual squares allows the generation of detailed patterns and images; the fraction of photobleached species per corral corresponding to the fractional area of the square exposed to light (Figure 3B) [121]. One can envision that combining these two methods could yield bilayers with precisely patterned ligands that maintain their lateral mobility but are confined to microscale patches.
Immobilized ligands can be incorporated into an SLB using gold nanoparticles (AuNPs) or nanodots. Lohmüller, et. al. used BCML to disperse AuNPs within the SLB plane. Ephrin A1 ligands were linked to AuNPs with thiolated DNA, and RGD was incorporated into the surrounding bilayer [122]. Thus, cells could engage with both laterally mobile and immobile ligands. In an alternate strategy, ligands were attached to size-tunable organic nanodot arrays (STONAs) surrounded by an SLB (Figure 3C) [123]. Beads deposited on a glass surface and coated in an aluminum mask of variable thickness determined STONA lattice spacing, ranging from 100 nm to 1800 nm. Following bead removal, a secondary aluminum-mask determined dot size, and nanodots were modified with biotinylated BSA for functionalization. An SLB was formed surrounding STONAs, giving rise to a ligand-island effect. T cells cultured on STONAs functionalized with anti-CD3 exhibited increased TCR clustering compared to T cells on homogeneous bilayers with equivalent ligand concentrations, and the tightness of adhesion was found to increase with ligand density. An unexplored consequence of ligand immobilization on STONAs is the development of a resisting force, which allows receptors and the cytoskeleton to locally apply tension. Simultaneous presentation of mobile ligands on the bilayer and immobile ligands on nanodot arrays will allow the relationship between ligand mobility and mechanics in cell signaling to be probed in parallel.
3.2.2. Diffusion Barriers
SLBs formed on substrates that are patterned with grids that prevent lipid diffusion separate into non-mixing microdomains. While such partitioned ‘corralled’ bilayers were initially developed for lithographic and electrostatic patterning, they have also become an indispensable tool in mechanobiology [35, 124–126]. Each individual region maintains its fluidity, but long-range diffusion is hindered by gridlines or by an energetic barrier to spreading. Thus, cell spreading and signaling on a corralled bilayer will be spatially mutated, and long-range receptor-ligand translocation is diminished (Figures 3D,E). In T cells, when a ligated receptor encounters a physical block, its speed is reduced and its translocation is deflected in an angle-dependent manner [127]. Grid lines also serve as sites of mechanical resistance. Cells cannot apply traction forces on homogeneous SLBs because the fluid bilayer fails to mount a resisting force; however lateral forces can be applied at barriers [23, 35]. In molecular mazes, which are similar to corralled SLBs, noncontiguous barriers are patterned onto glass, allowing the resulting bilayer to maintain its long-range fluidity while still presenting mechanical barriers [127]. These biochemically homogeneous platforms provide a direct method to probe biochemical versus mechanical signaling. A key challenge with diffusion barriers is the convolution of the effects due to blocked receptor transport versus mechanical force generation. Indeed, both clustering and mechanics are altered, resulting in spatiomechanical mutation. The exact forces that diffusion barriers impart are also unknown. Future studies will address this question using ratiometric MTFM on SLBs containing barriers.
Isolated membrane patches can be formed by scratching, blotting, stamping, or microfabrication. In the earliest example, tweezers were used to draw boundaries on a coverslip. Although SLBs in basic conditions remained partitioned at the scratch marks, bilayers in neutral and acidic conditions healed within hours, making them incompatible for cell imaging [128]. Following this work, Kung, et. al. formed BSA barriers by both the application of patterned BSA on a PDMS stamp and by removing lipids in a pattern using a PDMS stamp and then backfilling with BSA to generate walls [129]. Surprisingly, distinct membrane patches could also be formed using microcontact printing in the absence of barriers. SLBs applied in blocks with PDMS stamps maintained their shape because of the energetic barrier to spreading on glass and disrupting lipid tail interactions [115]. This method permits regions of ~5 μm to be fabricated. By using polymer-based lift off on slicon supported bilayers, Orth. et. al. were able achieve haptenated SLBs with 1 μm pattern precision [130]. SLBs separated by metal grids provide an optimal platform for studying receptor and cytoskeletal mechanotransduction due to their rigidity and ability to support cellular forces. A glass substrate is etched and coated with polymeric photoresist. Electron beam lithography exposes a grid pattern, and metal walls are deposited with electron beam evaporation. Grids are typically composed of chromium, but may also be aluminum or gold. The glass regions within the grid are exposed by sonication, and filled with an SLB formed by vesicle fusion [35, 124].
4. Methods to Measure Receptor Forces at Fluid Interfaces
4.1 Molecular Tension on Supported Lipid Bilayers
4.1.1 Ratiometric Tension Probes for Mobile Receptors
Molecular tension fluorescence microscopy (MTFM, previously reviewed by Jurchenko and Salaita [131]) is a method to optically image receptor mechanics at the living-nonliving interface. Previously, this method has been applied to map T cell receptor, epidermal growth factor receptor, and integrin forces with high spatiotemporal resolution [11, 15, 131–136]. In MTFM, an immobilized probe molecule comprised of a flexible linker and flanked by a fluorephore-quencher pair presents a ligand to a receptor of interest. In the resting (dark) state, the flexible linker is in a collapsed state, and the fluorophore and quencher remain in close proximity. When a receptor binds to the ligand and applies pN tension, the linker extends, causing separation of the fluorophore and quencher, which is accompanied by a significant increase in fluorescence. The flexible linker may be made of DNA, polymer, or protein and is selected based on its force-extension relationship. State-of-the-art probes yield greater than 100-fold increase in fluorescent intensity upon opening [11].
In typical MTFM, an increase in donor fluorescence serves as a quantitative reporter of quenching efficiency. This is valid because the immobilized probes have a fixed donor density and fluorescence intensity is directly proportional to quenching efficiency and quantum yield. When probes are attached to a fluid bilayer, the fluorescence intensity is proportional to probe density and quenching efficiency (force). Tension-mediate increases in donor fluorescence are convolved with increases in probe density due to ligated-receptor clustering (Figure 4A). Therefore, the application of MTFM to SLBs requires the separation of the signal contribution due to probe clustering versus probe opening.
Figure 4.
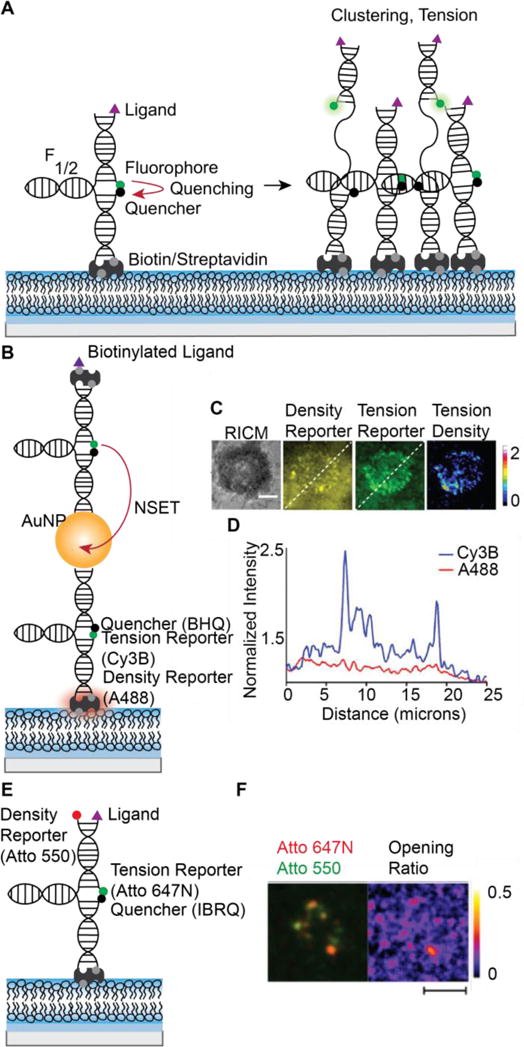
MTFM probes to map pN receptor tension at SLB-cell junctions. (A) On an SLB, fluorescence increases are attributed to both increased density (clustering) and probe opening (tension). Closed probe fluorescence is quenched. When receptors cluster or pull on probes, intensity increases. At F1/2, 50% probes will be open. (B) Nanoparticle-based ratiometric tension probes contain reporters for fluorescence and density. Closed probes are dual quenched by the BHQ molecular quencher and an AuNP and contain a second fluorophore on streptavidin. Labeled streptavidin serves as a probe density reporter. (C) Representative image of T cell tension and clustering. (D) Line scan of density reporter and tension reporter fluorescence. Scale bar 5 μm. (C,D reprinted from [136] with permission from publisher). (E) Ratiometric DNA tension probes containing a density reporter fluorophore on the hairpin strand. (F) Representative cell image of B cell receptors clustering and pulling on DNA tension probes. Scale bar 5 μm. (Reprinted from [137] with permission from publisher).
To address this problem, Ma, et. al. developed the first ratiometric MTFM probes, which allow the calculation of the contribution of signal due to tension, “tension density.” In this design, AuNPs are decorated with DNA tension probes and is attached to an SLB using biotin/streptavidin interaction. A second fluorophore is non-specifically attached to the streptavidin, and this “density reporter” fluorophore directly reports probe surface density. An additional benefit to this probe is the high signal-to-noise ratio; donor fluorescence is dual quenched by the molecular quencher and by nanoparticle surface-energy transfer (NSET) with the AuNP [136] (Figures 4B,C). Nowosad, et. al. published an alternate design in which the traditional MTFM probe is modified with a “density reporter” fluorophore on the hairpin strand. Tension is quantified by the “opening ratio” (Figures 4D,E) [137]. It would also be possible to achieve similar measurements by quantifying the “tension density” or FRET efficiency using DNA-FRET probes, which were previously applied to measure single receptor tension on glass [138].
4.1.2. Genetically Encoded Tension Probes
Although ratiometric MTFM is currently the only method to measure receptor mediated forces on a bilayer, other tension sensors could also be combined with SLBs to elucidate the role of fluidity in modulating the mechanics of accessory adhesome proteins. For example, Grashoff, et. al. introduced a genetically encoded tension sensor in 2014 [139]. This sensor contains two fluorescent proteins separated by a spider silk elastic domain, which measures pN tension across a protein. To date this probe has been inserted into a number of proteins including catenin, vinculin, α-actinin, and spectrin [17, 139, 140]. Combining cells transfected with genetically encoded tension sensors with SLBs and ratiometric MTFM, researchers will gain a more complete understanding of the role of traction forces and lateral fluidity in regulating adhesome mechanics.
4.2. Tension Measurements on Supported Lipid Bilayers
Integrin and cadherin molecule force maps have not yet been generated using ratiometric MTFM, but immune-cell receptor forces at the cell-SLB interface have been measured in both B and T cells. Both B and T cell receptors were capable of opening ratiometric tension probes on an SLB, but the measured tension was lower than that generated on glass. Ma et. al. reported T cell receptor tension of 4.7 pN on an SLB [136]. In contrast, Liu, et. al. demonstrated the ability of potent T cell receptors to unzip a 56 pN tension gauge tether (TGT) on glass [11]. Similarly, primary B cells could unfold a percentage of 7 pN probes on an SLB, but could not open 9 or 14 pN probes [137]. On glass, primary B cells were capable of unzipping 56 pN TGTs [10]. Importantly, MTFM measures the magnitude of tension rather than a force vector, and probe opening reflects net tension. On SLBs, where receptors cannot generate strong traction forces in the parallel to the bilayer, the majority of probe opening must be attributed to vertical forces. Thus, it is not surprising that tension measurements would be lower on an SLB compared to those on glass.
Ratiometric MTFM probes provide a method to characterize the relationship between pN mechanical forces and clustering in a variety of juxtacrine and cell-matrix interactions. Although these measurements have not yet been made, existing measurements on rigid substrates allow us to bound the range of forces. Note that force measurements are dependent on method, loading rate, and specific interaction parameters, so there a great deal of variability. Single molecule techniques report integrin bond strengths ranging 0.1 – 0.65 pN using magnetic tweezers to up to 100 pN using AMF and high loading rate. MTFM on glass suggests that integrins exert >100 pN forces; on 18 kPa elastomer substrates, the force per focal adhesion was measured to be 5.5 nN/um [135, 141–143]. Genetically encoded tension probes suggest that E-cadherin is under 1–2 pN constitutive tension, which increases at stressed cell-cell contacts. The AFM unbinding force for VE-cadherin was 35–55 pN [17, 144].
5. Mechanobiology of Adhesion Revealed Using Supported Lipid Bilayers
5.1. Lateral Fluidity Guides Cell Adhesion
The relationship between cell spreading and substrate stiffness has been well established; substrate stiffness can influence their lineage commitment and morphology [145–147]. In general, cells spread best on stiffer substrates, which support high traction forces that allow the cells to form larger contact areas [145, 148]. A related question is how cells respond to ligand mobility and tether flexibility. What are the effects of movable ligands versus rigidly anchored ligands? Many common substrates are limited in their ability to recapitulate the intrinsic flexibility of the ECM; therefore, these effects have been less studied than the stiffness response [149]. Recently, several novel substrates and mathematical models have been developed to probe these effects. SLBs are particularly well-suited for this line of research, because of their easily tunable architecture, fluidity, and functionalization.
Counterintuitively, increased fluidity does not always correspond with poor adhesion. Recall Smith, et. al.’s RGD-containing GUV’s that interacted with fluid and nonfluid receptors: receptor mobility increased the GUV’s ability to withstand external forces (2.2.1) [76]. Whereas nonmobile bonds stretched under tension, mobile receptor-ligand complexes reorganized under the GUV to distribute the forces [76]. Beyond the special case of catch bonds, nonmobile interaction lifetime would be shortened according to the Bell Model [1].
To test the effects of ligand fluidity on adhesion, Garcia, et. al. incorporated three peptides into DOPC and DPPC supported lipid monolayers (SLMs) and monitored the attachment and spreading of HAE amniontic endothelial cells, as well as THP-1 and M07 and hematopoietic protegenitor cells. HAE cells and both hemapoietic progenitor cells lines displayed decreased adhesion frequency and spreading on DOPC SLMs compared to DPPC SLMs [150]. DOPC and DPPC have the same headgroup, but DOPC is fluid at room temperature and DPPC is not, suggesting a preference for nonfluid bilayers. However, in a similar study using human mesenchymal stem cells (hMSCs), Kocer and Jonkheijm obtained opposite results (Figure 5A). On both SLBs, hMSC adhesion frequency increased with ligand density, however, cells always spread better on fluid DOPC SLBs than nonfluid DPPC SLBs. This effect was most pronounced at high ligand densities, where cell area double on DOPC SLBs compared to that on DPPC SLBs. These results suggest that hMSC adhesion is enhanced with increased ligand interaction and binding avidity obtained through ligand clustering on a fluid SLB [151]. Wong, et. al. probed hMSC differentiation substrates containing RGDs tethered to magnetic nanoparticles using PEG. Tethers were flexible at rest, but the application of a magnetic field restricted their flexibility. In contrast to SLB-culture, hMSCs exhibited increased spreading and focal adhesion assembly on less flexible tethers; these differences perpetuated over several days. These diverging results can likely be attributed to increased mechanotransduction by magnetic actuation and differences in signaling associated with long range versus short range translocation [152]. Attwood, et. al. found that human foreskin fibroblasts attached to RGD ligands on glass also preferred short tethers; increasing adhesion and cell area correlated decreasing tether length [153].
Figure 5.
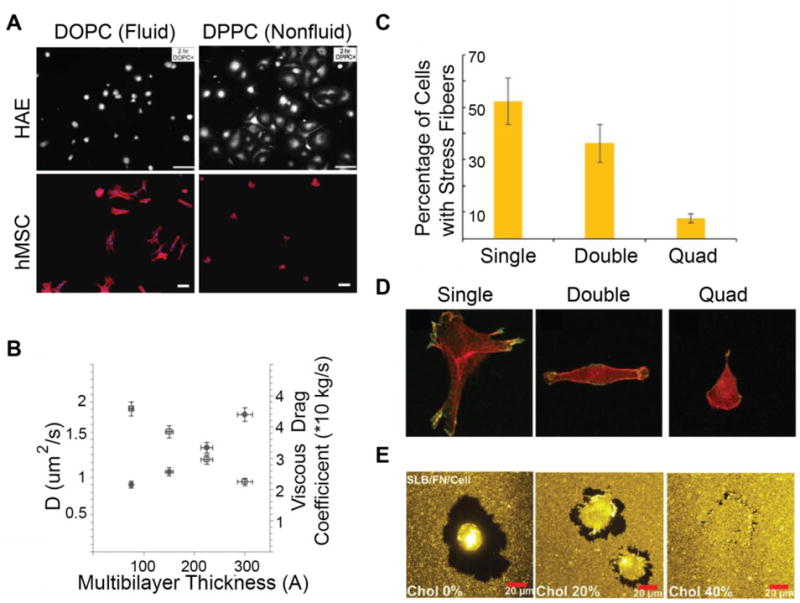
Cell adhesion on SLBs (A) HAE and hMSCs exhibit opposite adhesion trends on fluid and nonfluid SLBs, demonstrating the cell specificity of the response. Scale Bar 20 uM top, unspecified length, bottom. Reprinted from [150] and [151] with permission from publisher. (B) Mechanical characterization of stacked bilayers demonstrates that thicker bilayers are more viscous (Reprinted from [109] with permission from publisher). (C) Myoblasts on laminin coated lipopolymer-stacked bilayers exhibit fewer stress fibers with increasing SLB stack size. (D) Myoblast morphology is dependent on stack thickness. Cell spreading decreases with multibilayer thickness. 50 μm × 50 μm. (C,D reprinted from [110] with permission from publisher). E. Depletion zones decrease in size on viscous SLBs. (Reprinted from [91] with permission from publisher).
On lipopolymer-tethered stacked bilayers coated with a laminin network, cells exhibited reduced traction forces by both integrin and cadherin mediated adhesions with the addition of each plane [25, 110]. SLB fluidity increased with the number of layers (Figure 5B), and it was unusual that cells on a fluid SLB could generate any traction forces. These forces arose due to slowed cluster diffusion and leaflet coupling by lipopolymer tethers [110]. Cell stiffness, spread size, and adhesion size were all reduced with increasing stack layer and fluidity (Figure 5C,D). This trend was unsurprising given that the addition of each layer reduced substrate stiffness, and compliant substrates cannot develop strong traction forces. However, traction forces associated with rigid substrates are still not incompatible with ligand mobility. Pompe, et. al. propose a model of friction-controlled traction forces, in which focal adhesions are motile, and the friction of adhesion movement generates traction forces [154].
In Vafaei, et. al.’s SLB-ECM hybrid system, Huh-7.5 cells remodeled the local environment through a combination of packing flexible ECM proteins and lipid diffusion. Collagen and fibronectin were covalently coupled to fluid SLBs. Following coupling, lipids remained fluid with a diffusion coefficient of ~1 um^2/s, but ECM proteins did not diffuse, so adhered cells could generate traction forces. Huh-7.5 cells spread on the ECM functionalized-SLBs with lower area and rounder morphology than on ECM-functionalized glass due to reduced effective stiffness of the SLB platform. After 3 hours, cells showed ECM enrichment under the cell and depletion surrounding the cell (Figure 5E). As lipid fluidity was decreased with the addition of cholesterol, ECM depletion zones were also reduced, indicating that adhesion is modulated simultaneously by ligand flexibility and translation [91].
Kourouklis, et. al. took a synthetic approach to solving the same problem. Fluid amphiphilic block polymers containing RGD ligands were used to mimic the ECM, and fluidity was adjusted by changing the percent “lubricating” polymer, much like cholesterol composition could tune SLB fluidity [149]. Interestingly, 3T3 cells behaved nonlinearly. Cells on intermediate-fluidity substrates were consistently rounder and smaller with sparser focal adhesions compared to cells on substrates of higher and lower fluidity. Kourouklis, et. al. suggest that at low fluidity, cells generated traction forces on the substrate and adhered primarily through focal adhesions. At high fluidity, traction forces diffused, but receptors reinforced adhesion through clustering [149]. This result and corresponding model, along with Vafaei’s observation of enrichment allude to the cell-free case of adhesion strengthening through mobility.
Altogether these data indicate that the mobility response is specific. We propose that that it is regulated by a complex combination of fluidity and force sensing, molecular friction, adhesion mechanism, and cell and receptor-specific responses; the relationship between these factors is yet to be elucidated. Ligand lateral fluidity and flexibility are convoluted in many attempts to characterize this response. SLBs will be useful in separating the effects of these two parameters through phase tuning, corralling, and altered linker length.
5.2. Mechanobiology of Nascent Integrin Adhesions
While integrins and mature FAs have been extensively characterized using traction force microscopy (TFM), super resolution imaging, force probes, and single molecule force spectroscopy, nascent adhesions (NA) have been more challenging to study, because few methods have sufficient spatiotemporal and mechanical resolution to map their dynamics [12, 13, 15, 133, 155]. Recently SLBs have emerged as useful platform to probe NA formation and maturation into FA. Because fluid SLBs are traction force free, comparing the behavior of NAs on SLBs versus glass has allowed identification of the key biochemical and biomechanical signaling events in adhesion formation.
Initial integrin clustering and activation are independent of substrate mechanics and lateral forces; but mechanotransduction is required for the development of mature FAs from NAs. On glass, NAs containing integrins, paxillin, zyxin, and vinculin formed as actin polymerization extended lamellipodia at the leading edge of the cell. These clusters grew to 0.2 μm2 and the majority rapidly disassembled as actin passed over the NA and further extended the lamellipodia. On average, NA persisted for ~1 minute [24, 156]. A small fraction of NAs colocalized with actin and α-actinin tracks and matured rather than disassembling [156]. Nearly identical clusters formed on SLBs, but these NA persisted through the entire 15 minute observation window. NA size was independent of ligand density and activation state and was consistent on glass and SLBs [24]. NA formation on fluid SLBs and rigid glass indicated that NAs form independent of substrate rigidity. On fluid SLBs, mature adhesions failed to form without traction forces, but NAs stabilized due ligated integrin clustering. Further supporting this, myosin overexpression promoted NA maturation on glass, and myosin inhibition inhibited NA maturation but did not interfere with normal assembly or disassembly [24].
Cluster stabilization on SLBs allowed observation of previously undetected steps in NA recruitment and migration (Figure 6B) [22]. On both continuous and corralled SLBs, integrin-RGD cluster growth coincided with focal adhesion kinase, talin, and paxillin recruitment and promoted actin polymerization over NAs, which caused clusters to initially move inwards in pairs [22]. Several clusters associated with FHOD1, which was activated by Src family kinases to promote actin polymerization driving lamellipodia spreading and cluster outwards translocation [22, 157]. This was the first observation of outwards cluster motion in adhesion, which is not visible on glass or polymer substrates. On glass, FHOD1 signaling was required for polarized actin polymerization, traction force organization, and NA maturation [157]. On SLBS, cells retracted following spreading, and clusters were again translocated inwards, driven by Myosin II. Vinculin associated with clusters, suggesting talin unfolding during retraction [3, 22]. Cells on gridlines aggregated integrins on the outside of barriers and formed stable adhesions, and cells on continuous substrates clustered integrins in tight rings and became round. Further studies are needed to measure forces generated by Myosin II on an SLB and to investigate whether these forces are sufficient to drive integrin tension, but measurement of very weak and highly cooperative Myo1c forces on an SLB coated bead suggest that this behavior likely also requires many engaged myosins [84].
Figure 6.
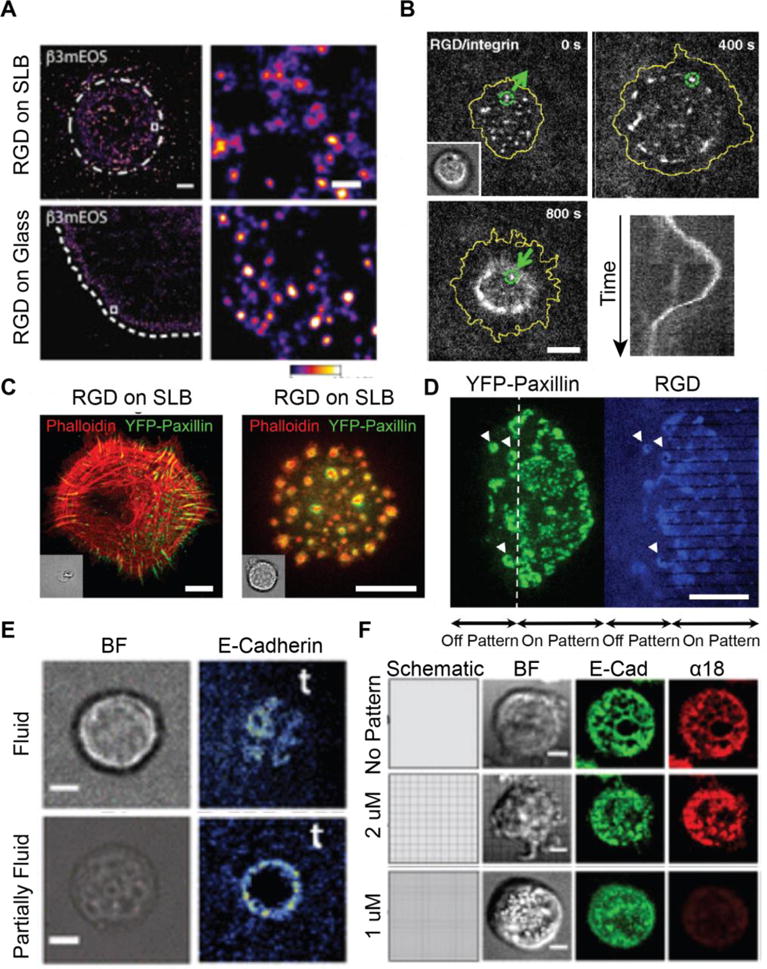
Mechanotransduction in integrin and cadherin-mediated adhesion assembly. (A) Integrins form uniform-sized clusters on both glass and SLBs, indicating early cluster formation regardless of substrate mechanics. Scale bar 5 μm. (Reprinted from [24] with permission from publisher). (B) Timelapse and kymograph (bottom right) of integrin cluster translocation on an SLB. Clusters are transported radially outwards before forming a tight contractile ring. Scale bar 5 μm. Reprinted from [22]. Copyright 2011, National Academy of Sciences). (C) Focal adhesion (left) and podosome (right) formation in fibroblasts on glass and SLBs, respectively. Scale bar 10 μm. (D) Podosomes form in the absence of traction forces on continuous SLBs but not on SLBs patterned with resistive chromium barriers. Scale bar 10 μm. (C,D reprinted from [23] with permission from publisher). (E) Hybrid adherens junctions form on partially fluid bilayers containing NBD-PC but fail to form on fluid bilayers demonstrating the importance of viscous drag. Scale bar 5 μm. (Reprinted from [26]. Copyright 2015, National Academy of Sciences). (F) Resistive barriers restrict cadherin transport and serve as sites of mechanotransduction, causing altered α-catenin activation (marked by a18). Scale bar 5 μm. (Reprinted from [27] with permission from publisher).
NA formation and migration required talin. Expression of the talin head domain rescued NA formation and motility, but NAs were slightly smaller than WT. Expression of the rod domain rescued NA formation but with less mobility. Talin rod domains can dimerize, which aided in clustering, but the full protein was required for full and motile NAs [24]. These results fit closely with Elosegui-Artola, et. al’s recent results that talin expression is required for stiffness-dependent NA maturation on rigid substrates [148]. TFM revealed that on low stiffness substrates, fibroblast traction forces increased with rigidity independent of talin expression. On rigid substrates, traction forces increased with rigidity in talin-expressing cells and decrease with rigidity in talin-depleted cells [148]. Talin head domain expression activated integrins in talin-depleted cells, but could not recover high traction forces on rigid substrates. The tail-domain alone also failed to rescue the stiffness response, which could only be recovered by expression of the entire protein. Thus, talin binding and unfolding are critical decision-making steps in mechanotransduction. On fluid substrates, talin regulates NA motility, whereas on nonfluid substrates talin regulates the loading-dependent stiffness response.
Traction forces also impact the formation of podosome-like adhesions, mature FAs, and receptor internalization (Figure 6C) [23, 80]. NAs in macrophages and fibroblasts on continuous SLBs ultimately transition into podosome-like adhesions in the absence of strong traction forces. NAs initially formed as described above, but adhesion proteins later segregated to a ring surrounding a core of polymerizing actin. These adhesions closely mimicked monocytic podosomes and transformed fibroblast’s invadopodia, which are protrusive structures. When fibroblasts spread on 1 μm line pitch corralled bilayers which could support traction forces, mature adhesions like those observed on glass substrates were recovered [23, 24, 80]]. Yu, et. al. proposed a mechanotransduction pathway in which traction forces serve as a checkpoint in forming stable FAs and failure to mount traction forces leads to Class 1a phosphatidylinositide 3-kinase (PI3K) recruitment, which initiates a biochemical cascade resulting in podosome formation [23]. Notably these studies suggest a model of local rather than global integrin mechanotransduction. The introduction of barriers on SLBs altered local force generation and signaling [22, 35]. When cells spanned the boundary of a continuous and corralled bilayer, they formed traction force stabilized FAs in the patterned region and podosomes in the continuous, fluid region (Figure 6C) [23].
A subset of integrins were internalized on SLBs, but not on glass. After initial NA formation, integrin-β3 clusters colocalized with Dab2, an adaptor protein in clathrin-mediated endocytosis. These NAs anti-colocalized with talin, indicating that recruitment of endocytosis machinery occurred downstream of mechanosensing. When actomyosin contractility was inhibited on glass, NA also colocalized with Dab2. Thus, failure to generate traction forces and stable FA can lead to integrin internalization [61].
5.3. Mechanobiology of Cadherin-Mediated Adhesion
Cadherin mediated adherens junctions form at the cell-cell interface and are the main junctions responsible for tissue integrity. In adherens junctions, cadherins cluster in cis on the cell surface and bind to cadherins on the opposing cell surface through trans interactions. Several lines of evidence suggest that cadherins both transmit and respond to mechanical forces. AFM studies suggest cadherins can form both slip and catch bonds depending on binding configuration [40]. Micropillar arrays deflected by cadherin-mediated forces demonstrated that cadherins apply traction forces to their substrate, but the unnatural spatial arrangement of cadherins in these studies obscures the physiological relevance of the results [158]. Borghi, et. al. inserted genetically encoded spider silk tension sensors into the cytoplasmic tails of cadherin, allowing the first measurement of mechanical forces across adherens junctions. Their results revealed that membrane associated E-cadherin is constitutively under tension and that tension is transmitted across adherens junctions through cadherins [17]. However, as with FA, AJ formation has been challenging to probe. It was unclear how cadherins associated to form AJs, and how mechanical forces contributed to their assembly. Recently, SLBs have provided a platform to spatiomechanically resolve AJ and cadherin cluster formation, while also offering improved physiological relevance [26, 61, 110].
Membrane technologies have been used for more than a decade to study cadherin-mediated adhesion, but only recent works have successfully mimicked AJ formation. The earliest studies characterizing the mechanics of cadherin-mediated adhesion at a fluid interface employed simple cell-free systems [159, 160]. Giant unilamellar vesicles (GUVs) were decorated with E-cadherin and allowed interaction with bilayers containing E-cadherin. Adhesion was observed by puckering in reflection interference contrast microscopy. Because cadherins bind weakly, vesicle adhesion to the bilayer required high concentrations of surface-presented E-cadherin [159]. Adhesions withstood thermal fluctuations but ruptured under shear force, suggesting weak clustering at adhesion sites [160].
Inspired by SLB studies using a bilayer to mimic an antigen presenting cell, Perez, et. al. published the first model of cadherin-mediated adhesion at the living-nonliving interface. MCF-7 cells were adhered to an SLB containing glycosylphosphatidyl inositol (GPI)-linked cadherins (hEFG) [161]. A small percentage of cells loosely attached and clustered hEFG, but the majority of cells could not spread without anchors. The incorporation of immobile 5 μm fibronectin islands within the bilayers permitted cell spreading and hEGF enhancement under the cell.
In 2015, Biswas, et. al. used phase tuning to develop the first SLB platform to support artificial AJ formation [26]. 1% cells clustered cadherins into an AJs on highly fluid bilayers, but cells on partially fluid bilayers containing NBD-PC readily enriched E-cadherins into AJs (Figure 6D). FCS revealed that cadherins on SLB diffused as monomers, suggesting they only associated during AJ formation. AJ formation was achieved by cluster coalescence during filopodial retraction. In both fluid and partially fluid bilayers, AJs formed in an all-or-nothing fashion; partial junctions were not observed. This observation together with the enhanced AJ formation on low fluidity SLBs suggest that adherens junctions require mechanical forces and kinetic nucleation to form. Very viscous SLBs generated resisting forces that could possibly support catch bonds across bound cadherins which would elongate bond lifetime. Low diffusivity promoted clustering and active nucleation, allowing junction formation to proceed. Within junctions, FRAP revealed that cadherins had low turnover and instead were immobilized within stable junctions [27]. Surprisingly, mechanical resistance alone was insufficient to form adherens junctions. Although corralled bilayer gridlines slowed transport and served as sites of force generation, cells on fluid corralled bilayers failed to form AJs. Thus, long range lateral transport of receptors is required (Figure 5E) [26]. This mechanism is in contrast with integrin adhesion, where forces at barriers locally determined the adhesion pathway.
SLBs with reduced fluidity by the addition of high density poly-His E-cadherin ectodomain also supported AJ formation and were used to study α-catenin mechanobiology in cadherin-mediated adhesions [27]. Cells spread on these SLBs exhibited two populations of cadherins that clustered during filapodial retraction: AJs at the cell periphery and cadherins loosely clustered in “central adhesions” underneath the cell. AJs and central adhesions both contained α- and β-catenin, but only AJs colocalized with actin, vinculin, and phosphorylated myosin light chain. Interestingly, the vinculin head domain and α18 could bind both populations of α-catenin; this indicated that α-catenin was active both AJ and central adhesions, which was unexpected because activated α-catenin usually is bound to actin. Cells spread on SLBs with widely spaced chromium grids exhibited normal cluster formation, but spreading and cluster formation by filapodia, along with α18 binding, was reduced on narrow grids (Figure 6E). Thus, α-catenin activation required mechanotransduction during cell spreading and retraction to activate, but sustained forces were not necessary for it to maintain its open conformation in cadherin clusters [27].
6. Conclusions and Future Directions
Major breakthroughs in SLB technologies include the ability to precisely pattern fluid and anchored ligands, to incorporate properly oriented and fluid transmembrane proteins, to generate multiple stacked bilayers, and to measure mechanical forces at the cell-SLB interface using ratiometric tension probes [25, 109, 110, 123, 136, 137]. Current studies of integrin and cadherin mediated adhesion using SLBs offer new insight into NA and AJ formation and demonstrate the power of spatiomechanical mutation using SLBs. By combining the techniques described in this review, we envision mechanically tunable cell substrates and sensors to probe specific signaling events in mechanotransduction. Hybrid adhesions consisting of immobilized ligands on STONAs and mobile ligands into the surrounding SLB will reveal the role of ligand mobility and transport in adhesion and cluster formation. Adjusting fluidity and stiffness while maintaining ligand density will deepen or understanding of how cells respond to ligand mobility versus substrate rigidity.
An open topic in the literature remains the dynamics of molecular mechanics: How do forces evolve in time and space across individual proteins, adhesion complexes, and the cell? SLBs provide a useful platform to approach this question, particularly relating to early events in adhesion formation. Ratiometric MTFM will track tension density evolution in spreading and retracting NAs and podosomes. Moving forward, fluorescence lifetime imaging may provide a simpler, concentration independent method to measure forces on an SLB.
SLBs are notable for their reductionist approach to biological interfaces, however further advancements in SLB engineering will focus on the fabrication of more sophisticated and physiologically relevant mechanical niches. Very recently, Vafaei, et. al. introduced ECM functionalized SLBs to mimic very soft neural tissue. This method will allow the probing of mechanotransduction and gene expression in previously inaccessible regimes [91]. The role of the glycocalyx remains poorly understood; decorating SLBs with glycolipids could offer a novel approach to explore its mechanical influence in juxtacrine signaling. Recent literature suggests that the glycocalyx is important regulating receptor clustering and FA assembly in cancer metastasis and in cell recognition in the immune system [162–165]. Incorporating ratiometric tension probes into SLBs mimicking the glycocalyx could provide direct evidence of how the glycocalyx influences mechanotransduction independent of cytoskeletal forces [162].
Although Afensenkau, et. al. succeeded in culturing neurons on an SLB for nearly 3 weeks, SLBs exhibited degradation during this timeframe that would prevent high quality molecular imaging [166]. Therefore, SLBs can used to study initial adhesions, but they cannot yet serve as dual culture and imaging platforms for longer processes such as synaptogenesis, stem cell differentiation, and embryo development. Further work is needed to optimize SLB stability for long-term studies. One approach has been to chelate calcium following SLB formation, but given the calcium dependence of many adhesion receptors, this unlikely to be a viable strategy for live cell studies [167]. In addition, while SLBs capture the planar interface at the cell-cell or cell-ECM junction, cells behave differently in 2D and 3D. Although 3D SLB-coated wells have been developed, these can only contain a single cell, not a cluster of cells that more accurately models tissue. More stable and 3D platforms are needed to understand mechanotransduction during development.
Despite these challenges, SLBs can still offer improved physiological relevance to model cell-cell and cell-matrix interactions. We envisage that SLBs will be widely used as mechanically tunable substrates to spatiomechanically mutate and probe events in adhesion. Beyond adhesion, these techniques will also be useful for studying mechanotransduction pathways in immune cell activation and viral entry. SLB technologies provide a sensitive and controllable toolset to study the link between physics and biology.
Highlights.
SLBs functionalized with adhesion proteins form artificial junctions with cells.
Chromium barriers serve as diffusion gates and sites of mechanical resistance.
Molecular tension probes report pN forces on SLBs.
Cells adhered to stacked or patterned SLBs exhibit distinct phenotypes.
Acknowledgments
This work was supported by the NIH (R01-GM097399) and the NSF CAREER Award (1350829) for financial support. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-444932.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimers: Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References
- 1.Bell GI. Models for the specific adhesion of cells to cells. Science (New York, NY) 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 2.Minajeva A, Kulke M, Fernandez JM, Linke WA. Unfolding of titin domains explains the viscoelastic behavior of skeletal myofibrils. Biophysical journal. 2001;80:1442–1451. doi: 10.1016/S0006-3495(01)76116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science (New York, NY) 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Montell C. Forcing open TRP channels: Mechanical gating as a unifying activation mechanism. Biochemical and biophysical research communications. 2015;460:22–25. doi: 10.1016/j.bbrc.2015.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillespie PG, Muller U. Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shawky JH, Davidson LA. Tissue mechanics and adhesion during embryo development. Developmental biology. 2015;401:152–164. doi: 10.1016/j.ydbio.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steward AJ, Kelly DJ. Mechanical regulation of mesenchymal stem cell differentiation. Journal of anatomy. 2015;227:717–731. doi: 10.1111/joa.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Kakimoto Y, Toda K, Naruse K. Mechanobiology in cardiac physiology and diseases. Journal of cellular and molecular medicine. 2013;17:225–232. doi: 10.1111/jcmm.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer metastasis reviews. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Z, Chen X, Chen H, Ji Q, Chen Y, Wang J, Cao Y, Wang F, Lou J, Tang Z, Liu W. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. eLife. 2015;4:e06925. doi: 10.7554/eLife.06925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:5610–5615. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. The Journal of cell biology. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo BH, Springer TA. Integrin structures and conformational signaling. Current opinion in cell biology. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nature communications. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingber D. Integrins as mechanochemical transducers. Current opinion in cell biology. 1991;3:841–848. doi: 10.1016/0955-0674(91)90058-7. [DOI] [PubMed] [Google Scholar]
- 17.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annual review of cell and developmental biology. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 19.Murrell M, Oakes PW, Lenz M, Gardel ML. Forcing cells into shape: the mechanics of actomyosin contractility, Nature reviews. Molecular cell biology. 2015;16:486–498. doi: 10.1038/nrm4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophysical journal. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sackmann E. How actin/myosin crosstalks guide the adhesion, locomotion and polarization of cells. Biochimica et biophysica acta. 2015;1853:3132–3142. doi: 10.1016/j.bbamcr.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Yu C-h, Law JBK, Suryana M, Low HY, Sheetz MP. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proceedings of the National Academy of Sciences. 2011;108:20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu CH, Rafiq NB, Krishnasamy A, Hartman KL, Jones GE, Bershadsky AD, Sheetz MP. Integrin-matrix clusters form podosome-like adhesions in the absence of traction forces. Cell reports. 2013;5:1456–1468. doi: 10.1016/j.celrep.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Changede R, Xu X, Margadant F, Sheetz MP. Nascent Integrin Adhesions Form on All Matrix Rigidities after Integrin Activation. Developmental cell. 2015;35:614–621. doi: 10.1016/j.devcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Lautscham LA, Lin CY, Auernheimer V, Naumann CA, Goldmann WH, Fabry B. Biomembrane-mimicking lipid bilayer system as a mechanically tunable cell substrate. Biomaterials. 2014;35:3198–3207. doi: 10.1016/j.biomaterials.2013.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas KH, Hartman KL, Yu CH, Harrison OJ, Song H, Smith AW, Huang WY, Lin WC, Guo Z, Padmanabhan A, Troyanovsky SM, Dustin ML, Shapiro L, Honig B, Zaidel-Bar R, Groves JT. E-cadherin junction formation involves an active kinetic nucleation process. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:10932–10937. doi: 10.1073/pnas.1513775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas KH, Hartman KL, Zaidel-Bar R, Groves JT. Sustained alpha-catenin Activation at E-cadherin Junctions in the Absence of Mechanical Force. Biophysical journal. 2016;111:1044–1052. doi: 10.1016/j.bpj.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dustin ML. Insights into function of the immunological synapse from studies with supported planar bilayers. Current topics in microbiology and immunology. 2010;340:1–24. doi: 10.1007/978-3-642-03858-7_1. [DOI] [PubMed] [Google Scholar]
- 29.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science (New York, NY) 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 30.Brian AA, McConnell HM. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalb E, Frey S, Tamm LK. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochimica et biophysica acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- 32.Tamm LK, McConnell HM. Supported phospholipid bilayers. Biophysical journal. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwang TJ, Fletcher WR, Lane TJ, Johal MS. Quantification of the layer of hydration of a supported lipid bilayer. Langmuir. 2010;26:4598–4601. doi: 10.1021/la100275v. [DOI] [PubMed] [Google Scholar]
- 34.Anderson TH, Min Y, Weirich KL, Zeng H, Fygenson D, Israelachvili JN. Formation of Supported Bilayers on Silica Substrates. Langmuir. 2009;25:6997–7005. doi: 10.1021/la900181c. [DOI] [PubMed] [Google Scholar]
- 35.Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of Receptor Movement Alters Cellular Response: Physical Force Sensing by EphA2. Science (New York, NY) 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science (New York, NY) 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 37.Yu CH, Groves JT. Engineering supported membranes for cell biology. Medical & biological engineering & computing. 2010;48:955–963. doi: 10.1007/s11517-010-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Weerd J, Karperien M, Jonkheijm P. Supported Lipid Bilayers for the Generation of Dynamic Cell-Material Interfaces. Advanced healthcare materials. 2015;4:2743–2779. doi: 10.1002/adhm.201500398. [DOI] [PubMed] [Google Scholar]
- 39.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. The Journal of cell biology. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proceedings of the National Academy of Sciences. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narui Y, Salaita K. Membrane tethered delta activates notch and reveals a role for spatio-mechanical regulation of the signaling pathway. Biophysical journal. 2013;105:2655–2665. doi: 10.1016/j.bpj.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez MF, Levi V, Weidemann T, Carrer DC. Agonist mobility on supported lipid bilayers affects Fas mediated death response. FEBS letters. 2015;589:3527–3533. doi: 10.1016/j.febslet.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Care BR, Soula HA. Receptor clustering affects signal transduction at the membrane level in the reaction-limited regime. Physical review E, Statistical, nonlinear, and soft matter physics. 2013;87:012720. doi: 10.1103/PhysRevE.87.012720. [DOI] [PubMed] [Google Scholar]
- 45.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villard V, Kalyuzhniy O, Riccio O, Potekhin S, Melnik TN, Kajava AV, Ruegg C, Corradin G. Synthetic RGD-containing alpha-helical coiled coil peptides promote integrin-dependent cell adhesion. Journal of peptide science: an official publication of the European Peptide Society. 2006;12:206–212. doi: 10.1002/psc.707. [DOI] [PubMed] [Google Scholar]
- 47.Edwards LJ, Zarnitsyna VI, Hood JD, Evavold BD, Zhu C. Insights into T cell recognition of antigen: significance of two-dimensional kinetic parameters. Frontiers in immunology. 2012;3:86. doi: 10.3389/fimmu.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Parajo MF, Cambi A, Torreno-Pina JA, Thompson N, Jacobson K. Nanoclustering as a dominant feature of plasma membrane organization. Journal of cell science. 2014;127:4995–5005. doi: 10.1242/jcs.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duke T, Graham I. Equilibrium mechanisms of receptor clustering. Progress in biophysics and molecular biology. 2009;100:18–24. doi: 10.1016/j.pbiomolbio.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Dustin ML, Groves JT. Receptor signaling clusters in the immune synapse. Annual review of biophysics. 2012;41:543–556. doi: 10.1146/annurev-biophys-042910-155238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selhuber-Unkel C, Lopez-Garcia M, Kessler H, Spatz JP. Cooperativity in adhesion cluster formation during initial cell adhesion. Biophysical journal. 2008;95:5424–5431. doi: 10.1529/biophysj.108.139584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartman NC, Nye JA, Groves JT. Cluster size regulates protein sorting in the immunological synapse. Proceedings of the National Academy of Sciences. 2009;106:12729–12734. doi: 10.1073/pnas.0902621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattheyses AL, Simon SM, Rappoport JZ. Imaging with total internal reflection fluorescence microscopy for the cell biologist. Journal of cell science. 2010;123:3621–3628. doi: 10.1242/jcs.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das C, Sheikh KH, Olmsted PD, Connell SD. Nanoscale mechanical probing of supported lipid bilayers with atomic force microscopy. Physical review E, Statistical, nonlinear, and soft matter physics. 2010;82:041920. doi: 10.1103/PhysRevE.82.041920. [DOI] [PubMed] [Google Scholar]
- 55.Portet T, Dimova R. A New Method for Measuring Edge Tensions and Stability of Lipid Bilayers: Effect of Membrane Composition. Biophysical journal. 2010;99:3264–3273. doi: 10.1016/j.bpj.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steltenkamp S, Müller MM, Deserno M, Hennesthal C, Steinem C, Janshoff A. Mechanical Properties of Pore-Spanning Lipid Bilayers Probed by Atomic Force Microscopy. Biophysical journal. 2006;91:217–226. doi: 10.1529/biophysj.106.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picas L, Rico F, Scheuring S. Direct Measurement of the Mechanical Properties of Lipid Phases in Supported Bilayers. Biophysical journal. 2012;102:L01–L03. doi: 10.1016/j.bpj.2011.11.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seitz M, Park CK, Wong JY, Israelachvili JN. Long-Range Interaction Forces between Polymer-Supported Lipid Bilayer Membranes. Langmuir: the ACS journal of surfaces and colloids. 2001;17:4616–4626. doi: 10.1021/la0103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leckband D, Muller W, Schmitt FJ, Ringsdorf H. Molecular mechanisms determining the strength of receptor-mediated intermembrane adhesion. Biophysical journal. 1995;69:1162–1169. doi: 10.1016/S0006-3495(95)79990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S. Direct measurement of a tethered ligand-receptor interaction potential. Science (New York, NY) 1997;275:820–822. doi: 10.1126/science.275.5301.820. [DOI] [PubMed] [Google Scholar]
- 61.Yu C-h, Rafiq NBM, Cao F, Zhou Y, Krishnasamy A, Biswas KH, Ravasio A, Chen Z, Wang Y-H, Kawauchi K, Jones GE, Sheetz MP. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nature communications. 2015;6:8672. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science (New York, NY) 2013;340:1587–1590. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaz WLC, Goodsaid-Zalduondo F, Jacobson K. Lateral diffusion of lipids and proteins in bilayer membranes. FEBS letters. 1984;174:199–207. [Google Scholar]
- 64.Saffman PG. Brownian motion in thin sheets of viscous fluid. Journal of Fluid Mechanics. 2006;73:593–602. [Google Scholar]
- 65.Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophysical journal. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knight JD, Lerner MG, Marcano-Velazquez JG, Pastor RW, Falke JJ. Single molecule diffusion of membrane-bound proteins: window into lipid contacts and bilayer dynamics. Biophysical journal. 2010;99:2879–2887. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziemba BP, Falke JJ. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chemistry and physics of lipids. 2013;172–173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machan R, Hof M. Lipid diffusion in planar membranes investigated by fluorescence correlation spectroscopy. Biochimica et Biophysica Acta (BBA) – Biomembranes. 2010;1798:1377–1391. doi: 10.1016/j.bbamem.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 69.Forstner MB, Yee CK, Parikh AN, Groves JT. Lipid lateral mobility and membrane phase structure modulation by protein binding. Journal of the American Chemical Society. 2006;128:15221–15227. doi: 10.1021/ja064093h. [DOI] [PubMed] [Google Scholar]
- 70.Niemela PS, Miettinen MS, Monticelli L, Hammaren H, Bjelkmar P, Murtola T, Lindahl E, Vattulainen I. Membrane Proteins Diffuse as Dynamic Complexes with Lipids. Journal of the American Chemical Society. 2010;132:7574–7575. doi: 10.1021/ja101481b. [DOI] [PubMed] [Google Scholar]
- 71.Tamm LK. Lateral diffusion and fluorescence microscope studies on a monoclonal antibody specifically bound to supported phospholipid bilayers. Biochemistry. 1988;27:1450–1457. doi: 10.1021/bi00405a009. [DOI] [PubMed] [Google Scholar]
- 72.Nguyen TT, Sly KL, Conboy JC. Comparison of the Energetics of Avidin, Streptavidin, NeutrAvidin, and Anti-Biotin Antibody Binding to Biotinylated Lipid Bilayer Examined by Second-Harmonic Generation. Analytical Chemistry. 2012;84:201–208. doi: 10.1021/ac202375n. [DOI] [PubMed] [Google Scholar]
- 73.Sako Y, Kusumi A. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. The Journal of cell biology. 1994;125:1251–1264. doi: 10.1083/jcb.125.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sako Y, Kusumi A. Barriers for lateral diffusion of transferrin receptor in the plasma membrane as characterized by receptor dragging by laser tweezers: fence versus tether. The Journal of cell biology. 1995;129:1559–1574. doi: 10.1083/jcb.129.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edidin M, Kuo SC, Sheetz MP. Lateral movements of membrane glycoproteins restricted by dynamic cytoplasmic barriers. Science (New York, NY) 1991;254:1379–1382. doi: 10.1126/science.1835798. [DOI] [PubMed] [Google Scholar]
- 76.Smith AS, Sengupta K, Goennenwein S, Seifert U, Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proceedings of the National Academy of Sciences. 2008;105:6906–6911. doi: 10.1073/pnas.0801706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delanoe-Ayari H, Al Kurdi R, Vallade M, Gulino-Debrac D, Riveline D. Membrane and acto-myosin tension promote clustering of adhesion proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2229–2234. doi: 10.1073/pnas.0304297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marcy Y, Prost J, Carlier MF, Sykes C. Forces generated during actin-based propulsion: A direct measurement by micromanipulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5992–5997. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Footer MJ, Kerssemakers JWJ, Theriot JA, Dogterom M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proceedings of the National Academy of Sciences. 2007;104:2181–2186. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Labernadie A, Bouissou A, Delobelle P, Balor S, Voituriez R, Proag A, Fourquaux I, Thibault C, Vieu C, Poincloux R, Charrière GM, Maridonneau-Parini I. Protrusion force microscopy reveals oscillatory force generation and mechanosensing activity of human macrophage podosomes. Nature communications. 2014;5:5343. doi: 10.1038/ncomms6343. [DOI] [PubMed] [Google Scholar]
- 81.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 82.Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tabdanov E, Gondarenko S, Kumari S, Liapis A, Dustin ML, Sheetz MP, Kam LC, Iskratsch T. Micropatterning of TCR and LFA-1 ligands reveals complementary effects on cytoskeleton mechanics in T cells. Integrative biology: quantitative biosciences from nano to macro. 2015;7:1272–1284. doi: 10.1039/c5ib00032g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pyrpassopoulos S, Arpag G, Feeser EA, Shuman H, Tuzel E, Ostap EM. Force Generation by Membrane-Associated Myosin-I. Scientific reports. 2016;6:25524. doi: 10.1038/srep25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kahya N, Schwille P. How Phospholipid-Cholesterol Interactions Modulate Lipid Lateral Diffusion, as Revealed by Fluorescence Correlation Spectroscopy. Journal of Fluorescence. 2006;16:671–678. doi: 10.1007/s10895-006-0108-6. [DOI] [PubMed] [Google Scholar]
- 86.Hjort Ipsen J, Karlstrom G, Mourtisen OG, Wennerström H, Zuckermann MJ. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 87.Redondo-Morata L, Giannotti MI, Sanz F. Influence of cholesterol on the phase transition of lipid bilayers: a temperature-controlled force spectroscopy study. Langmuir. 2012;28:12851–12860. doi: 10.1021/la302620t. [DOI] [PubMed] [Google Scholar]
- 88.Loura LMS, Fernandes F, Fernandes AC, Ramalho JPP. Effects of fluorescent probe NBD-PC on the structure, dynamics and phase transition of DPPC. A molecular dynamics and differential scanning calorimetry study. Biochimica et Biophysica Acta (BBA) – Biomembranes. 2008;1778:491–501. doi: 10.1016/j.bbamem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 89.Smith KA, Gale BK, Conboy JC. Micropatterned Fluid Lipid Bilayer Arrays Created Using a Continuous Flow Microspotter. Analytical Chemistry. 2008;80:7980–7987. doi: 10.1021/ac800860u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uhl E, Donati A, Reviakine I. Platelet Immobilization on Supported Phospholipid Bilayers for Single Platelet Studies. Langmuir. 2016;32:8516–8524. doi: 10.1021/acs.langmuir.6b01852. [DOI] [PubMed] [Google Scholar]
- 91.Vafaei S, Tabaei SR, Biswas KH, Groves JT, Cho NJ. Dynamic Cellular Interactions with Extracellular Matrix Triggered by Biomechanical Tuning of Low-Rigidity, Supported Lipid Membranes. Advanced healthcare materials. 2017:1700243–n/a. doi: 10.1002/adhm.201700243. [DOI] [PubMed] [Google Scholar]
- 92.Andreasson-Ochsner M, Romano G, Hakanson M, Smith ML, Leckband DE, Textor M, Reimhult E. Single cell 3-D platform to study ligand mobility in cell-cell contact. Lab on a chip. 2011;11:2876–2883. doi: 10.1039/c1lc20067d. [DOI] [PubMed] [Google Scholar]
- 93.Oliver AE, Parikh AN. Templating membrane assembly, structure, and dynamics using engineered interfaces. Biochimica et Biophysica Acta (BBA) – Biomembranes. 2010;1798:839–850. doi: 10.1016/j.bbamem.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 94.Scomparin C, Lecuyer S, Ferreira M, Charitat T, Tinland B. Diffusion in supported lipid bilayers: influence of substrate and preparation technique on the internal dynamics. The European physical journal E, Soft matter. 2009;28:211–220. doi: 10.1140/epje/i2008-10407-3. [DOI] [PubMed] [Google Scholar]
- 95.Merkel R, Sackmann E, Evans E. Molecular friction and epitactic coupling between monolayers in supported bilayers. Journal de Physique. 1989;50:1535–1555. [Google Scholar]
- 96.Evans E, Sackmann E. Translational and rotational drag coefficients for a disk moving in a liquid membrane associated with a rigid substrate. Journal of Fluid Mechanics. 1988;194:553–561. [Google Scholar]
- 97.Eghiaian F, Rigato A, Scheuring S. Structural, Mechanical, and Dynamical Variability of the Actin Cortex in Living Cells. Biophysical journal. 2015;108:1330–1340. doi: 10.1016/j.bpj.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han X, Achalkumar AS, Cheetham MR, Connell SD, Johnson BR, Bushby RJ, Evans SD. A self-assembly route for double bilayer lipid membrane formation. Chemphyschem: a European journal of chemical physics and physical chemistry. 2010;11:569–574. doi: 10.1002/cphc.200900798. [DOI] [PubMed] [Google Scholar]
- 99.Murray DH, Tamm LK, Kiessling V. Supported double membranes. Journal of structural biology. 2009;168:183–189. doi: 10.1016/j.jsb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabaei SR, Jonsson P, Branden M, Hook F. Self-assembly formation of multiple DNA-tethered lipid bilayers. Journal of structural biology. 2009;168:200–206. doi: 10.1016/j.jsb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 101.Zhu Y, Negmi A, Moran-Mirabal J. Multi-Stacked Supported Lipid Bilayer Micropatterning through Polymer Stencil Lift-Off. Membranes. 2015;5:385–398. doi: 10.3390/membranes5030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaizuka Y, Groves JT. Structure and dynamics of supported intermembrane junctions. Biophysical journal. 2004;86:905–912. doi: 10.1016/S0006-3495(04)74166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 104.Goennenwein S, Tanaka M, Hu B, Moroder L, Sackmann E. Functional Incorporation of Integrins into Solid Supported Membranes on Ultrathin Films of Cellulose: Impact on Adhesion. Biophysical journal. 2003;85:646–655. doi: 10.1016/S0006-3495(03)74508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wagner ML, Tamm LK. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophysical journal. 2000;79:1400–1414. doi: 10.1016/S0006-3495(00)76392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Diaz AJ, Albertorio F, Daniel S, Cremer PS. Double cushions preserve transmembrane protein mobility in supported bilayer systems. Langmuir. 2008;24:6820–6826. doi: 10.1021/la800018d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sterling Sarah M, Dawes R, Allgeyer Edward S, Ashworth Sharon L, Neivandt David J. Comparison Actin- and Glass-Supported Phospholipid Bilayer Diffusion Coefficients. Biophysical journal. 2015;108:1946–1953. doi: 10.1016/j.bpj.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Richards MJ, Hsia CY, Singh RR, Haider H, Kumpf J, Kawate T, Daniel S. Membrane Protein Mobility and Orientation Preserved in Supported Bilayers Created Directly from Cell Plasma Membrane Blebs. Langmuir. 2016;32:2963–2974. doi: 10.1021/acs.langmuir.5b03415. [DOI] [PubMed] [Google Scholar]
- 109.Minner DE, Herring VL, Siegel AP, Kimble-Hill A, Johnson MA, Naumann CA. Iterative layer-by-layer assembly of polymer-tethered multi-bilayers using maleimide-thiol coupling chemistry. Soft matter. 2013;9:9643–9650. doi: 10.1039/c3sm51446c. [DOI] [PubMed] [Google Scholar]
- 110.Ge Y, Lin YH, Lautscham LA, Goldmann WH, Fabry B, Naumann CA. N-cadherin-functionalized polymer-tethered multi-bilayer: a cell surface-mimicking substrate to probe cellular mechanosensitivity. Soft matter. 2016;12:8274–8284. doi: 10.1039/c6sm01673a. [DOI] [PubMed] [Google Scholar]
- 111.Lohmüller T, Aydin D, Schwieder M, Morhard C, Louban I, Pacholski C, Spatz JP. Nanopatterning by block copolymer micelle nanolithography and bioinspired applications. Biointerphases. 2011;6:MR1–MR12. doi: 10.1116/1.3536839. [DOI] [PubMed] [Google Scholar]
- 112.Deeg J, Axmann M, Matic J, Liapis A, Depoil D, Afrose J, Curado S, Dustin ML, Spatz JP. T Cell Activation is Determined by the Number of Presented Antigens. Nano Letters. 2013;13:5619–5626. doi: 10.1021/nl403266t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jackson BL, Groves JT. Scanning probe lithography on fluid lipid membranes. Journal of the American Chemical Society. 2004;126:13878–13879. doi: 10.1021/ja046040a. [DOI] [PubMed] [Google Scholar]
- 114.Yee CK, Amweg ML, Parikh AN. Membrane Photolithography: Direct Micropatterning and Manipulation of Fluid Phospholipid Membranes in the Aqueous Phase Using Deep-UV Light. Advanced Materials. 2004;16:1184–1189. [Google Scholar]
- 115.Hovis JS, Boxer SG. Patterning and Composition Arrays of Supported Lipid Bilayers by Microcontact Printing. Langmuir. 2001;17:3400–3405. [Google Scholar]
- 116.Groves JT, Mahal LK, Bertozzi CR. Control of cell adhesion and growth with micropatterned supported lipid membranes. Langmuir. 2001;17:5129–5133. [Google Scholar]
- 117.Huang RT. Cell adhesion mediated by glycolipids. Nature. 1978;276:624–626. doi: 10.1038/276624a0. [DOI] [PubMed] [Google Scholar]
- 118.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural Photoreceptors as a Source of Fluorescent Proteins, Biosensors, and Optogenetic Tools. Annual review of biochemistry. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DeMond AL, Starr T, Dustin ML, Groves JT. Control of Antigen Presentation with a Photoreleasable Agonist Peptide. Journal of the American Chemical Society. 2006;128:15354–15355. doi: 10.1021/ja065304l. [DOI] [PubMed] [Google Scholar]
- 120.Nakayama K, Tachikawa T, Majima T. Spatial Control of Protein Binding on Lipid Bimembrane Using Photoeliminative Linker. Langmuir. 2008;24:6425–6428. doi: 10.1021/la801028m. [DOI] [PubMed] [Google Scholar]
- 121.Kung LA, Groves JT, Ulman N, Boxer SG. Printing via photolithography on micropartitioned fluid lipid membranes. Advanced Materials. 2000;12:731–+. [Google Scholar]
- 122.Lohmüller T, Triffo S, O’Donoghue GP, Xu Q, Coyle MP, Groves JT. Supported Membranes Embedded with Fixed Arrays of Gold Nanoparticles. Nano Letters. 2011;11:4912–4918. doi: 10.1021/nl202847t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pi F, Dillard P, Alameddine R, Benard E, Wahl A, Ozerov I, Charrier A, Limozin L, Sengupta K. Size-Tunable Organic Nanodot Arrays: A Versatile Platform for Manipulating and Imaging Cells. Nano Lett. 2015;15:5178–5184. doi: 10.1021/acs.nanolett.5b01400. [DOI] [PubMed] [Google Scholar]
- 124.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science (New York, NY) 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 125.Nair PM, Salaita K, Petit RS, Groves JT. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nature protocols. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Groves JT, Boxer SG. Electric field-induced concentration gradients in planar supported bilayers. Biophysical journal. 1995;69:1972–1975. doi: 10.1016/S0006-3495(95)80067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. T Cell Receptor Microcluster Transport through Molecular Mazes Reveals Mechanism of Translocation. Biophysical journal. 2008;94:3286–3292. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cremer PS, Boxer SG. Formation and Spreading of Lipid Bilayers on Planar Glass Supports. The Journal of Physical Chemistry B. 1999;103:2554–2559. [Google Scholar]
- 129.Kung LA, Kam L, Hovis JS, Boxer SG. Patterning Hybrid Surfaces of Proteins and Supported Lipid Bilayers. Langmuir. 2000;16:6773–6776. [Google Scholar]
- 130.Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA. Mast Cell Activation on Patterned Lipid Bilayers of Subcellular Dimensions. Langmuir. 2003;19:1599–1605. [Google Scholar]
- 131.Jurchenko C, Salaita KS. Lighting Up the Force: Investigating Mechanisms of Mechanotransduction Using Fluorescent Tension Probes. Molecular and cellular biology. 2015;35:2570–2582. doi: 10.1128/MCB.00195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Meth. 2012;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 133.Liu Y, Medda R, Liu Z, Galior K, Yehl K, Spatz JP, Cavalcanti-Adam EA, Salaita K. Nanoparticle tension probes patterned at the nanoscale: impact of integrin clustering on force transmission. Nano Lett. 2014;14:5539–5546. doi: 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chang Y, Liu Z, Zhang Y, Galior K, Yang J, Salaita K. A General Approach for Generating Fluorescent Probes to Visualize Piconewton Forces at the Cell Surface. Journal of the American Chemical Society. 2016;138:2901–2904. doi: 10.1021/jacs.5b11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Galior K, Liu Y, Yehl K, Vivek S, Salaita K. Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions. Nano Lett. 2016;16:341–348. doi: 10.1021/acs.nanolett.5b03888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma VPY, Liu Y, Blanchfield L, Su H, Evavold BD, Salaita K. Ratiometric Tension Probes for Mapping Receptor Forces and Clustering at Intermembrane Junctions. Nano Letters. 2016;16:4552–4559. doi: 10.1021/acs.nanolett.6b01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nowosad CR, Spillane KM, Tolar P. Germinal center B cells recognize antigen through a specialized immune synapse architecture. Nature immunology. 2016;17:870–877. doi: 10.1038/ni.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular Tension Sensors Report Forces Generated by Single Integrin Molecules in Living Cells. Nano Letters. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. The FEBS journal. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nature cell biology. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 142.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li F, Redick SD, Erickson HP, Moy VT. Force measurements of the alpha5beta1 integrin-fibronectin interaction. Biophysical journal. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Developmental cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 147.Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Agarose gel stiffness determines rate of DRG neurite extension in 3D cultures. Biomaterials. 2001;22:1077–1084. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- 148.Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Perez-Gonzalez C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nature cell biology. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- 149.Kourouklis AP, Lerum RV, Bermudez H. Cell adhesion mechanisms on laterally mobile polymer films. Biomaterials. 2014;35:4827–4834. doi: 10.1016/j.biomaterials.2014.02.052. [DOI] [PubMed] [Google Scholar]
- 150.Garcia AS, Dellatore SM, Messersmith PB, Miller WM. Effects of supported lipid monolayer fluidity on the adhesion of hematopoietic progenitor cell lines to fibronectin-derived peptide ligands for alpha5beta1 and alpha4beta1 integrins. Langmuir. 2009;25:2994–3002. doi: 10.1021/la802772y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Koçer G, Jonkheijm P. Guiding hMSC Adhesion and Differentiation on Supported Lipid Bilayers. Advanced healthcare materials. 2017;6:1600862–n/a. doi: 10.1002/adhm.201600862. [DOI] [PubMed] [Google Scholar]
- 152.Wong DSH, Li J, Yan X, Wang B, Li R, Zhang L, Bian L. Magnetically Tuning Tether Mobility of Integrin Ligand Regulates Adhesion, Spreading, and Differentiation of Stem Cells. Nano Letters. 2017;17:1685–1695. doi: 10.1021/acs.nanolett.6b04958. [DOI] [PubMed] [Google Scholar]
- 153.Attwood SJ, Cortes E, Haining AWM, Robinson B, Li D, Gautrot J, del Río Hernández A. Adhesive ligand tether length affects the size and length of focal adhesions and influences cell spreading and attachment. Scientific reports. 2016;6:34334. doi: 10.1038/srep34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pompe T, Kaufmann M, Kasimir M, Johne S, Glorius S, Renner L, Bobeth M, Pompe W, Werner C. Friction-Controlled Traction Force in Cell Adhesion. Biophysical journal. 101:1863–1870. doi: 10.1016/j.bpj.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nature cell biology. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iskratsch T, Yu C-H, Mathur A, Liu S, Stévenin V, Dwyer J, Hone J, Ehler E, Sheetz M. FHOD1 is needed for directed Forces and Adhesion Maturation during Cell Spreading and Migration. Developmental cell. 2013;27:545–559. doi: 10.1016/j.devcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ganz A, Lambert M, Saez A, Silberzan P, Buguin A, Mege RM, Ladoux B. Traction forces exerted through N-cadherin contacts. Biology of the cell / under the auspices of the European Cell Biology Organization. 2006;98:721–730. doi: 10.1042/BC20060039. [DOI] [PubMed] [Google Scholar]
- 159.Fenz SF, Merkel R, Sengupta K. Diffusion and intermembrane distance: case study of avidin and E-cadherin mediated adhesion. Langmuir. 2008;25:1074–1085. doi: 10.1021/la803227s. [DOI] [PubMed] [Google Scholar]
- 160.Puech PH, Feracci H, Brochard-Wyart F. Adhesion between Giant Vesicles and Supported Bilayers Decorated with Chelated E-Cadherin Fragments. Langmuir. 2004;20:9763–9768. doi: 10.1021/la048682h. [DOI] [PubMed] [Google Scholar]
- 161.Perez TD, Nelson WJ, Boxer SG, Kam L. E-Cadherin Tethered to Micropatterned Supported Lipid Bilayers as a Model for Cell Adhesion. Langmuir. 2005;21:11963–11968. doi: 10.1021/la052264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, Rubashkin MG, Magbanua MJ, Thorn KS, Davidson MW, Rugo HS, Park JW, Hammer DA, Giannone G, Bertozzi CR, Weaver VM. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Paszek MJ, Boettiger D, Weaver VM, Hammer DA. Integrin Clustering Is Driven by Mechanical Resistance from the Glycocalyx and the Substrate. PLOS Computational Biology. 2009;5:e1000604. doi: 10.1371/journal.pcbi.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu GK, Qian J, Hu J. The glycocalyx promotes cooperative binding and clustering of adhesion receptors. Soft matter. 2016;12:4572–4583. doi: 10.1039/c5sm03139g. [DOI] [PubMed] [Google Scholar]
- 165.Dustin ML, Cooper JA. The immunological synapse and the actin cytoskeleton: molecular hardware for T cell signaling. Nature immunology. 2000;1:23–29. doi: 10.1038/76877. [DOI] [PubMed] [Google Scholar]
- 166.Afanasenkau D, Offenhausser A. Positively Charged Supported Lipid Bilayers as a Biomimetic Platform for Neuronal Cell Culture. Langmuir. 2012;28:13387–13394. doi: 10.1021/la302500r. [DOI] [PubMed] [Google Scholar]
- 167.Moller I, Seeger S. Solid supported lipid bilayers from artificial and natural lipid mixtures - long-term stable, homogeneous and reproducible. Journal of Materials Chemistry B. 2015;3:6046–6056. doi: 10.1039/c5tb00437c. [DOI] [PubMed] [Google Scholar]


