ABSTRACT
The centromere is a critical genomic region that enables faithful chromosome segregation during mitosis, and must be distinguishable from other genomic regions to facilitate establishment of the kinetochore. The centromere-specific histone H3-variant CENP-A forms a special nucleosome that functions as a marker for centromere specification. In addition to the CENP-A nucleosomes, there are additional H3 nucleosomes that have been identified in centromeres, both of which are predicted to exhibit specific features. It is likely that the composite organization of CENP-A and H3 nucleosomes contributes to the formation of centromere-specific chromatin, termed ‘centrochromatin’. Recent studies suggest that centrochromatin has specific histone modifications that mediate centromere specification and kinetochore assembly. We use chicken non-repetitive centromeres as a model of centromeric activities to characterize functional features of centrochromatin. This review discusses our recent progress, and that of various other research groups, in elucidating the functional roles of histone modifications in centrochromatin.
KEYWORDS: centromere, CENP-A, histone modification, kinetochore
Introduction
The centromere is a critical genomic region that enables faithful chromosome segregation during mitosis. A large, multimeric protein structure called the kinetochore that is assembled on the centromere, binds microtubules during mitosis. The centromere region is not specified by a particular DNA sequence, but rather by sequence-independent epigenetic mechanisms. The centromere-specific histone H3-variant CENP-A forms a specific nucleosome, and is thought to be a critical epigenetic marker for centromere specification. This hypothesis is supported by previous studies that showed that most active centromeres include CENP-A-containing nucleosomes,1 and that localization of CENP-A to a non-centromeric region induces centromere formation under specific conditions in some organisms.2-5 In addition to CENP-A-containing nucleosomes, canonical histone H3-containing nucleosomes also exist in centromere regions (Fig. 1).6-8 CENP-A nucleosomes are interspersed with chromatin-containing canonical histone H3 in the centromere region, and this composite organization may contribute to the formation of centromere-specific chromatin called ‘centrochromatin’ (Fig. 1).6,9,10
Figure 1.
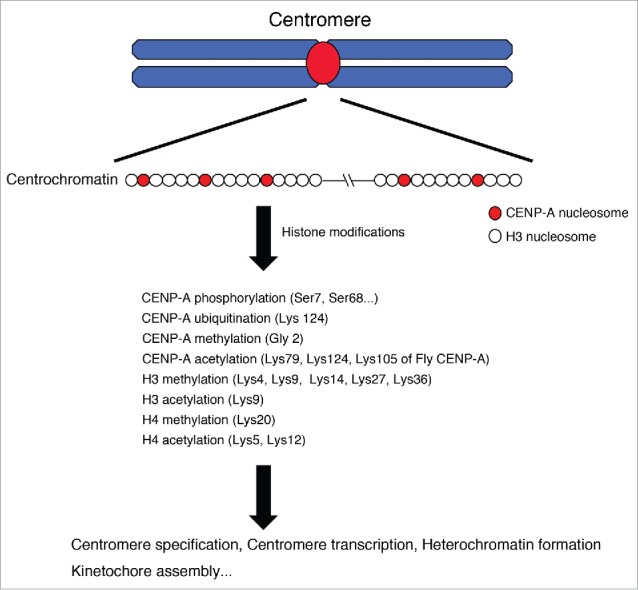
Post-transcriptional histone modifications in centrochromatin. In centromere regions, CENP-A nucleosomes are interspersed with chromatin-containing canonical histone H3 and this composite organization must be critical for the formation of centromere-specific chromatin called ‘centrochromatin’. Various histones are modified in centromeres and these modifications contribute to centromere functions including centromere specification, centromere transcription, heterochromatin formation, kinetochore assembly.
To facilitate the assembly of kinetochore proteins, centrochromatin is characterized by a unique pattern of post-translational histone modifications that contribute to an overall status that is neither heterochromatic nor euchromatic (Fig. 1). In general, histone modifications are associated with the genome status (i.e. an ‘open’ versus a ‘closed‘ chromatin conformation) as ascertained by the assessment of genome-wide distribution patterns of various histone modifications using specific antibodies.11 Transcriptionally-active genomic regions are usually associated with an open chromatin status and acetylated histones, while heterochromatic regions are instead associated with closed chromatin and methylated histones. Although centrochromatin is believed to include specific histone modifications, these are difficult to investigate because of the massively repetitive nature of underlying centromeric and peri-centromeric DNA sequences.
Sullivan and colleagues used an extended-chromatin-fiber technique, combined with antibodies against histone modifications and/or CENP-A, to reveal the organization of centrochromatin.6,12 Earnshaw and coworkers used an artificial chromosome that harbored a centromere sequence that was able to be manipulated to examine centromere histone modifications in artificial chromosomes.13 In addition, they altered the centromere histone modification status by tethering a histone demethylase onto the artificial chromosomes, and thus examined the biological significance of such histone modifications.13-15 Furthermore, there are various studies on CENP-A modifications, including phosphorylation, methylation, acetylation, and ubiquitination, which may be critical for centromere functions (Fig. 1).16-21 Our group focuses on non-repetitive centromeres as a model in chicken cells.8 Although in most organisms, centromeres generally contain highly repetitive sequences, chicken chromosomes 5, 27, and Z do not contain such sequences,22,23 (notably, other chicken chromosomes do exhibit repetitive centromeres). This is also true for the equine chromosome 11 centromere.24 By analyzing these non-repetitive centromeres with specific antibodies directed against various histone modifications, we identified centromere-specific histone modifications.25,26 In this review, we focus on modifications of canonical histones (other than for CENP-A) in centromeres and, in particular, discuss our recent findings combined with other reports, as well as the biological significance of histone modifications for centromere specification and/or kinetochore assembly.
Modifications of canonical histones in centrochromatin
Since the concept of ‘centrochromatin’ was first proposed, the histone modification status of centrochromatin in different organisms has been examined using various methodologies (Fig. 1). H3K4me2 has been shown to be highly enriched in centrochromatin in humans and flies.6,9 Conversely, maize centromeres have been found to contain significant amounts of H3K9me2 and H3K9me3, but only low levels of H3K4me2.27 Notably, this inconsistency may be the result of the difficulty of accurately mapping histone modifications on repetitive sequences using ChIP analysis.28 To exclude this issue, we used non-repetitive centromeres found in chicken chromosomes 5, 27, and Z.22,23 Although human neo-centromeres also lack repetitive sequences,29 they vary significantly in size and chromatin features.30 In addition, human chromosomes that contain neo-centromeres are less stable than endogenous chromosomes with normal centromeres.31 Given that chicken non-repetitive centromeres naturally exist in chromosomes, they are excellent models for examining centromeric chromatin features. Furthermore, centromere sizes among these chromosomes are similar,22,23 as are their kinetochore protein contents, compared with those of repetitive centromeres in other chicken chromosomes.32
Thus, we extensively analyzed various histone H3 modifications in non-repetitive centromeres using ChIP-seq analysis with appropriate antibodies, but did not detect specific enrichment of any of the tested H3 modifications25, (including H3K4me2, H3K9me3, and H3K36me2). H3K9me3 is associated with, and considered to mark, heterochromatin, and has previously been shown to be predominantly enriched in the peri-centromeric region. Our results were consistent with these previous findings, such that H3K9me3 was found to be enriched in the repetitive centromeres in chicken cells; however, it was not detected in association with non-repetitive centromeres.25 This may either reflect the fact that the heterochromatic structure is not established in non-repetitive centromeres, or instead indicate that non-repetitive centromeres may form their heterochromatin structure via different mechanisms. For example, non-repetitive centromeres may associate with heterochromatic structures established at a distant point to the centromere region via complex 3-dimensional chromosomal organization. To ensure faithful chromosomal segregation, as well as the correct attachment of microtubules to kinetochores, sister-chromatid cohesion must be established in centromere regions. This implies that centromere regions must be heterochromatic to some extent, and hence it is likely that non-repetitive centromeres are too. The recently-developed chromosome-conformation-capture method, including the ‘Hi-C’ technique, allows 3-dimensional organization studies of centrochromatin structures; hence, we must set out to use this technique to evaluate our hypotheses.
Although our analyses to date have not identified specific enrichment of H3 modifications, we predict centromere-specific H3 modifications since there are sufficient H3 nucleosomes in centromeres to establish centrochromatin. Thus, identifying centromere-specific H3 modifications will be a challenging, but vital, topic for future research.
In addition to histone H3 modifications, we analyzed histone H4 modifications in non-repetitive centromeres, and found H4K20me1, H4K5ac, and H4K12ac modifications to be enriched in centromeric regions via ChIP-seq analysis using appropriate antibodies.25,26 Of these 3 modifications, H4K20me1 mainly occurs at CENP-A nucleosomes in centrochromatin (Fig. 2).25 This methylation is mediated by PR-Set7,33 and our preliminary experiments have suggested H4K20me1 in centrochromatin to be diminished in PR-Set7-deficient DT40 cells (unpublished results). Furthermore, we have demonstrated that H4K20me1 does not occur in the H4-CENP-A pre-deposition complex. Considering these observations, we conclude that H4K20me1 is present in centrochromatin through PR-Set7 activity (Fig. 2). We next sought to elucidate the underlying mechanisms by which PR-Set7 selectively methylates H4K20 in centrochromatin. While it is possible that PR-Set7 may transiently bind particular centromere proteins, to date we have been unable to detect centromeric proteins by co-immunoprecipitation with PR-Set7. Alternatively, the H4 tail position in the CENP-A nucleosome may be different from its position in the H3 nucleosome, rendering the H4 tail more readily accessible to PR-Set7 in the former context. Although the H4 tail is flexible, and it is difficult to compare the structures of the CENP-A and H3 nucleosomes, it is in fact possible - indeed necessary - to test the accessibility of the H4 tail to PR-Set7 in each context in vitro.
Figure 2.
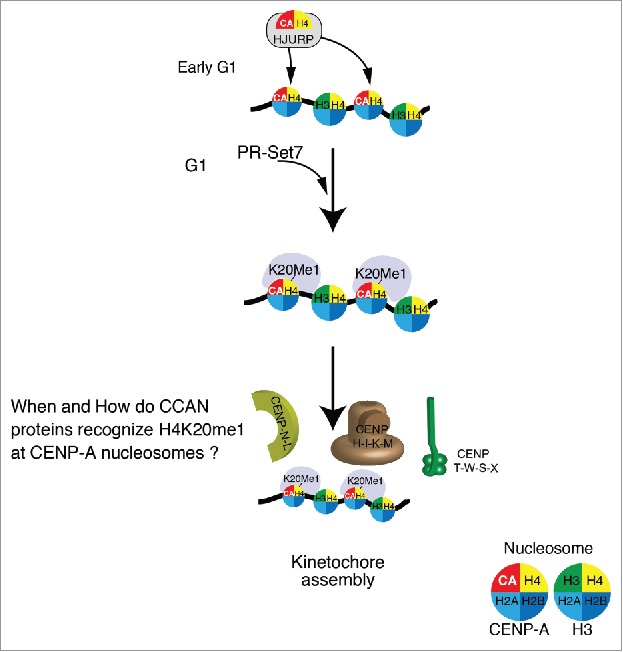
Schematic model for the functional role of H4K20me1 in centrochromatin. The CENP-A-H4 complex is associated with the CENP-A chaperone HJURP, and has been shown to be incorporated into centromeres during early G1 phase of the cell cycle. Given that H4 subunits in CENP-A nucleosomes have been found to be consistently methylated at K20, it is likely that this modification occurs just after CENP-A incorporation by PR-Set7. This would allow various Constitutive-Centromere-Associated-Network (CCAN) proteins to recognize H4K20me1 in centrochromatin, and thereby assemble kinetochores. It is vital that future research elucidates the timing and mechanisms by which CCAN proteins mediate kinetochore assembly.
In contrast to H4K20me1, H4K5ac and H4K12ac mainly occur in the H4-CENP-A pre-deposition complex, which is predominantly associated with the CENP-A chaperone HJURP.26 Since the pre-deposition complex is transiently associated with the RbAp46/48-Hat1 complex, H4K5ac and H4K12ac mainly occur before CENP-A deposition. Consistent with this observation, we previously showed that once CENP-A-H4 is incorporated into chromatin, the level of these modifications was reduced.26 Therefore, we propose that these modifications are involved in CENP-A deposition, rather than with a centrochromatin-specific function (see below and Fig. 3).
Figure 3.
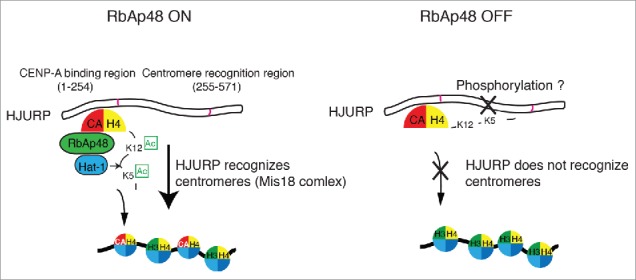
Role for the RbAp48-Hat1 complex-mediated acetylation of the H4 tail in CENP-Adeposition. The H4 tail in the CENP-A-H4 complex is acetylated at K5 and K12 by the RbAp48-Hat1 complex before H4-CENP-A centromere deposition. The CENP-A-H4 complex binds the N-terminus of HJURP, and the middle region of HJURP recognizes centromeres (via the Mis18 complex) to facilitate centromere deposition. Acetylation of the H4 tail normally facilitates this process; however, if acetylation does not occur correctly (as in RbAp48-deficient cells), the non-acetylated tail interferes with HJURP centromere recognition, preventing CENP-A from being incorporated into centromere domains. It is unclear how H4 acetylation facilitates CENP-A deposition. Given that HJURP is a highly phosphorylated protein, it may be that a combination of HJURP phosphorylation and H4 acetylation is critical for correct H4-CENP-A deposition.
Histone modifications contribute to kinetochore assembly and centromere specification
We have demonstrated that centromeres contain specific histone H4 modifications, and have illustrated the importance of elucidating their biological significance. By conditionally reducing the level of H4K20me1 in centromeres in cells,25 we revealed that a reduced level of centromeric H4K20me1 induced both cell cycle arrest at mitosis and the loss of the kinetochore proteins CENP-H and CENP-T.25 Taken together, these data suggest that H4K20me1 is required for kinetochore assembly (Fig. 2). Data from other current studies indicate that other kinetochore proteins, CENP-N and CENP-C directly bind the CENP-A nucleosome.34,35 Similarly, CENP-C and CENP-T have been shown to be associated with the H3 nucleosome in centrochromatin.7 Given that CENP-C levels were not altered in centromeres with reduced H4K20me1, CENP-T or CENP-N (and CENP-N-associated proteins) are promising candidates for interaction with H4K20me1. Nevertheless, H4K20me1 also occurs in non-centromere regions, (for example, it is particularly enriched in transcriptionally active regions), and thus, the mechanisms by which kinetochore proteins recognize H4K20me1 in centrochromatin remain to be elucidated (Fig. 2). The simplest possibility is that some kinetochore proteins specifically bind to H4K20me1. However, we reconstituted CENP-A nucleosomes with H4K20me1 and tested whether centromere proteins bind to the reconstituted CENP-A nucleosomes. So far, we have not observed clear binding of centromeric proteins with reconstituted CENP-A nucleosomes in vitro (unpublished data). Additional histone modifications may be required for CENP-T- or CENP-N-H4K20me1 interactions. An alternative possibility is that modification of CENP-T- or CENP-N may be necessary for interaction with H4K20me1. In either case, it will be essential to address molecular mechanisms of how H4K20me1 recognizes centromere proteins.
Given that recombinant CENP-N and CENP-N-associated proteins have been shown to be capable of binding in vitro-reconstituted CENP-A nucleosomes (which do not contain H4K20me1 modifications),36,37 H4K20me1 may not be necessary to enable the interaction of kinetochore proteins with the CENP-A nucleosome. Notably however, this result was limited to an in vitro analysis, and, therefore, H4K20me1 may in fact facilitate the stable association of kinetochore proteins with centrochromatin in vivo. Continued research is required to clarify the molecular basis for the functional role of H4K20me1 in kinetochore assembly.
As previously discussed, H4K5ac and H4K12ac predominantly occur in the H4-CENP-A pre-deposition complex, and thus, we believe that they may mediate the recruitment of H4-CENP-A to centromeres. Indeed, cells expressing H4K5R and H4K12R mutants have been previously shown to exhibit improper centromere deposition of H4-CENP-A,26 and, furthermore, we observed that the CENP-A chaperone HJURP does not recognize centromeres in H4K5ac- or K12ac-deficient cells26 (Fig. 3). This led us to develop a model suggesting that the acetylation-deficient H4 tail may interfere with centromere recognition by HJURP (Fig. 3), although the molecular mechanism by which H4K5ac and H4K12ac facilitate centromere deposition of H4-CENP-A remains unknown. HJURP is a highly phosphorylated protein, and some reports suggest that such phosphorylation may regulate its functions.38,39 Thus, it is possible that phosphorylated HJURP may not be capable of recognizing centromeres and we speculate that histone H4 acetylation in the H4-CENP-A complex may facilitate the recognition of centromeres by phosphorylated-HJURP. It is certainly likely that a combination of HJURP phosphorylation, H4 acetylation, and/or CENP-A modification is critical for correct H4-CENP-A deposition, but this will require confirmation through future studies.
Epigenetic engineering reveals the significance of histone modifications in centrochromatin
Although we did not detect specific accumulation of H3K4me2 or H3K36me2 in our chicken non-repetitive centromeres,25 these modifications have previously been demonstrated to be centromerically-enriched on a human artificial chromosome (HAC).13 These modifications are predominantly associated with transcriptionally active genomic regions, suggesting that the centromere region of HACs may undergo transcription. To examine the functional significance of these modifications, a unique epigenetic engineering method was used, whereby the H3K4me2 demethylase LSD1 was specifically targeted to the HAC centromere region.13 As expected, centromeric H3K4me2 on the HAC was lost completely. Interestingly, CENP-A and CENP-C levels on the HAC were gradually reduced, centromere H3K36me2 and transcripts from alphoid DNA on the HAC centromere were decreased, and the HAC subsequently became unstable in these cells.13
Bergmann et al. further investigated why a decrease in H3K4me2 and centromeric transcripts resulted in incorrect CENP-A and CENP-C localization.13 They found that newly synthesized CENP-A was not properly targeted to the centromere region, because HJURP did not recognize H3K4me2-deficient chromatin.13 These observations suggest that an open chromatin structure mediated by centromeric transcripts may be critical for the recruitment of HJURP, and for the consequent incorporation of CENP-A into centromeres. However, given that a high level of transcripts was also observed to disrupt centromere formation, they proposed “moderate” transcription to be critical for centromere formation.13 Earnshaw and colleagues continue to analyze centromeric chromatin based on epigenetic engineering methods.14,15 They recently combined a decrease of H3K4me2 with an elevation of histone acetylation, whereby induction of CENP-28/Eaf6 and p65 was used to induce H4K12 and H3K9 acetylation, respectively. Their results demonstrated the defects of CENP-A incorporation observed in H3K4me2-deficient chromatin to be rescued by induction of H3K9 acetylation by p65, but not by Eaf6-mediated H4K12 acetylation.14,15 From these data, they proposed that open chromatin, mediated by H3K4me2 and/or centromeric transcripts, is essential for new CENP-A incorporation, and that a barrier formed by H3K9ac may prevent both the spread of heterochromatin and kinetochore inactivation. Further studies are needed to reach a comprehensive understating of the effects of these histone modifications on centromere formation.
In addition to the HAC reports, recent studies also suggest that centromere transcripts may be important for centromere specification and kinetochore assembly in various other organisms.40-45 Consolidation of data from these various studies is problematic; hence understanding of the manner in which centromere transcripts are involved in centromere formation and function is still unclear, and requires further elucidation.
Concluding perspectives on histone modifications in centrochromatin
The centromere is a specialized region of the chromosome and, as such, has distinct features compared with other chromatin regions. The centromere-specific histone H3 CENP-A forms a nucleosome that may have different features compared with canonical H3 nucleosomes.1 Although recent studies suggest that CENP-A forms an octameric nucleosome with other histones (as observed for the canonical H3 nucleosome1,46) throughout the cell cycle, specific details surrounding CENP-A nucleosome structure are somewhat different from those of the H3 nucleosome.47 Nevertheless, the structure of H3 nucleosomes in centromere regions is similar to that involving non-centromeric domains. Therefore, H3 nucleosomes that are present in centromere regions must exhibit features distinct from other H3 nucleosomes. We have thus attempted to identify and characterize centromere-specific histone modifications using non-repetitive chicken centromeres. To date, we have demonstrated that H4K20me1, H4K5ac, and H4K12ac are enriched in centromeric regions.25,26 However, given the well-established complexity of histone modifications in general, we expect additional modifications to be present in these regions. Recently, Foltz and colleagues analyzed various post-translational modifications involving centromeric chromatin using proteomic approaches.48 As in our studies, they identified H4K20me1, H4K5ac, and H4K12ac in centrochromatin, and additional modifications. Not only the various modifications alone, but their combinations also might be important for correct centrochromatin structure and function. Furthermore, while modifications including methylation, acetylation, and phosphorylation have been thus far characterized in centrochromatin, other modifications such as ubiquitination, farnesylation, sumoylation, and crotonylation might also be possible. In fact, CENP-A has been shown to ubiquitinated at K124 in human cells,18 although the significance of this observation is controversial.49,50 In this case, H3 or H4 ubiquitination in centromeres may be involved in centromere function. Continued research is required to comprehensively investigate histone modifications in centromere specification and function, and in kinetochore assembly.
Although this review focused on modifications to canonical histones in centromeres, CENP-A modifications must be critical for centromere specification and kinetochore assembly.16-21 For example, tri-methylation of the human CENP-A N-terminus is essential for recruitment of other centromeric proteins.20 This suggests that while CENP-A is an important epigenetic marker for centromere specification, CENP-A modifications may be also involved in downstream mechanisms of kinetochore assembly. Detailed characterizations of CENP-A modifications also represent an important topic in this field.
Whereas the effects of histone modifications on centromere functions are important, it is also essential to elucidate the mechanisms by which centrochromatin is modified. One hypothesis is that enzymes responsible for these modifications may directly associate with centromere proteins. We recently demonstrated that RbAp46/48-Hat1 directly associates with CENP-A to produce H4K5ac and H4K12ac modifications,26 which is also observed in Drosophila cells.19 However, it is still unclear how PR-Set7 (responsible for H4K20me1) is targeted to centromeres. Recently, Ohzeki et al. showed that KAT7 (the histone acetyl transferase for H3K14ac), specifically localizes to centromeres.51 Although the significance of H3K14ac in centromere function is still unclear, the observed centromere-specific localization of KAT7 is notable.
Studies on histone modifications in centrochromatin represent a new field of research, and thus many hypotheses are as yet unresolved. It is, however, clear that a complex and coordinated combination of various histone modifications, including CENP-A modifications, is likely to mediate centromere specification and function, as well as kinetochore assembly. The importance of these cellular functions emphasizes the need for continued research on mechanisms by which they are regulated by epigenetic processes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The author is very grateful to Tetsuya Hori and Wei-Hao Shang, who contribute to analysis of histone modifications in chicken centromeres. The author also thanks other members of the Fukagawa Lab for useful discussion.
Funding
This work in the Fukagawa Lab was supported by MEXT KAKENHI (Grant Numbers 25221106 and 15H05972).
References
- [1].Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 2011; 144:471-9; PMID:21335232; https://doi.org/ 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 2011; 477:354-8; PMID:21874020; https://doi.org/ 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science 2011; 334:686-90; PMID:22053052; https://doi.org/ 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- [4].Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 2011; 194:229-43; PMID:21768289; https://doi.org/ 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 2013; 200:45-60; PMID:23277427; https://doi.org/ 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 2004; 11:1076-83; https://doi.org/ 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al.. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008; 135:1039-52; PMID:19070575; https://doi.org/ 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- [8].Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 2014; 30:496-508; PMID:25203206; https://doi.org/ 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2002; 2:319-30; PMID:11879637; https://doi.org/ 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A 2010; 107:10484-9; PMID:20483991; https://doi.org/ 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet 2008; 9:179-91; PMID:18250624; https://doi.org/ 10.1038/nrg2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A 2006; 103:4186-91; PMID:16537506; https://doi.org/ 10.1073/pnas.0507947103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 2011; 30:328-40; PMID:21157429; https://doi.org/ 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, et al.. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci 2012; 125:411-21; PMID:22331359; https://doi.org/ 10.1242/jcs.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 2016; 7:13334; PMID:27841270; https://doi.org/ 10.1038/ncomms13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell 2003; 5:853-64; PMID:14667408; https://doi.org/ 10.1016/S1534-5807(03)00364-2 [DOI] [PubMed] [Google Scholar]
- [17].Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al.. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell 2015; 32:68-81; PMID:25556658; https://doi.org/ 10.1016/j.devcel.2014.11.030 [DOI] [PubMed] [Google Scholar]
- [18].Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell 2015; 32:589-603; PMID:25727006; https://doi.org/ 10.1016/j.devcel.2015.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Boltengagen M, Huang A, Boltengagen A, Trixl L, Lindner H, Kremser L, Offterdinger M, Lusser A. A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. Nucleic Acids Res 2016; 44:2145-59; PMID:26586808; https://doi.org/ 10.1093/nar/gkv1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sathyan KM, Fachinetti D, Foltz DR. alpha-amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat Commun 2017; 8:14678; PMID:28266506; https://doi.org/ 10.1038/ncomms14678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bui M, Pitman M, Nuccio A, Roque S, Donlin-Asp PG, Nita-Lazar A, Papoian GA, Dalal Y. Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenetics Chromatin 2017; 10:17; https://doi.org/ 10.1186/s13072-017-0124-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 2010; 20:1219-28; PMID:20534883; https://doi.org/ 10.1101/gr.106245.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shang WH, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, et al.. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 2013; 24:635-48; PMID:23499358; https://doi.org/ 10.1016/j.devcel.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al.. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009; 326:865-7; PMID:19892987; https://doi.org/ 10.1126/science.1178158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al.. Histone H4 Lys 20 Monomethylation of the CENP-A Nucleosome Is Essential for Kinetochore Assembly. Dev Cell 2014; 29:740-9; PMID:24960696; https://doi.org/ 10.1016/j.devcel.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, et al.. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun 2016; 7:13465; PMID:27811920; https://doi.org/ 10.1038/ncomms13465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 2006; 173:1571-83; PMID:16624902; https://doi.org/ 10.1534/genetics.106.056853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stimpson KM, Sullivan BA. Epigenomics of centromere assembly and function. Curr Opin Cell Biol 2010; 22:772-80; PMID:20675111; https://doi.org/ 10.1016/j.ceb.2010.07.002 [DOI] [PubMed] [Google Scholar]
- [29].Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet 2003; 12:2711-21; PMID:12928482; https://doi.org/ 10.1093/hmg/ddg282 [DOI] [PubMed] [Google Scholar]
- [30].Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin 2010; 3:6; https://doi.org/ 10.1186/1756-8935-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol 2010; 190:177-85; PMID:20643881; https://doi.org/ 10.1083/jcb.201001035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol 2010; 189:937-43; PMID:20548100; https://doi.org/ 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 2012; 26:325-37; https://doi.org/ 10.1101/gad.177444.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 2009; 11:896-902; PMID:19543270; https://doi.org/ 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 2013; 340:1110-3; PMID:23723239; https://doi.org/ 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM. The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol Cell 2015; 60:886-98; PMID:26698661; https://doi.org/ 10.1016/j.molcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischbock J, Pentakota S, Keller J, Pesenti ME, Pan D, et al.. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 2016; 537:249-53; PMID:27580032; https://doi.org/ 10.1038/nature19333 [DOI] [PubMed] [Google Scholar]
- [38].Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell reports 2014; 8:190-203; PMID:25001279; https://doi.org/ 10.1016/j.celrep.2014.06.002 [DOI] [PubMed] [Google Scholar]
- [39].Stankovic A, Guo LY, Mata JF, Bodor DL, Cao XJ, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE, et al.. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol Cell 2017; 65:231-46; PMID:28017591; https://doi.org/ 10.1016/j.molcel.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chan FL, Wong LH. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res 2012; 40:11178-88; PMID:23066104; https://doi.org/ 10.1093/nar/gks921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Quenet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 2014; 3:e03254; PMID:25117489; https://doi.org/ 10.7554/eLife.03254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rosic S, Kohler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 2014; 207:335-49; PMID:25365994; https://doi.org/ 10.1083/jcb.201404097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Catania S, Pidoux AL, Allshire RC. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet 2015; 11:e1004986; PMID:25738810; https://doi.org/ 10.1371/journal.pgen.1004986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Grenfell AW, Heald R, Strzelecka M. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol 2016; 214:133-41; PMID:27402954; https://doi.org/ 10.1083/jcb.201604029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Blower MD. Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep 2016; 15:1624-33; PMID:27184843; https://doi.org/ 10.1016/j.celrep.2016.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AY, Nguyen K, Dekker C, et al.. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J Cell Biol 2017; 216:607-21; PMID:28235947; https://doi.org/ 10.1083/jcb.201608083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al.. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 2011; 476:232-5; PMID:21743476; https://doi.org/ 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- [48].Bailey AO, Panchenko T, Shabanowitz J, Lehman SM, Bai DL, Hunt DF, Black BE, Foltz DR. Identification of the posttranslational modifications present in centromeric chromatin. Mol Cell Proteomics 2015; 15(3):918-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, Black BE. CENP-A modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev Cell 2017; 40:104-13; PMID:28073008; https://doi.org/ 10.1016/j.devcel.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Niikura Y, Kitagawa R, Kitagawa K. CENP-A Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev Cell 2017; 40:7-8; PMID:28073011; https://doi.org/ 10.1016/j.devcel.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H. KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Dev Cell 2016; 37:413-27; PMID:27270040; https://doi.org/ 10.1016/j.devcel.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]


