The δ-opioid receptor (δR) is retained in intracellular structures in neurons, but the mechanisms of retention and regulated export are not known. The atypical phosphoinositide-3 kinase C2A is required and sufficient for NGF-regulated δR export from the trans-Golgi network and surface transport.
Abstract
The interplay between signaling and trafficking by G protein–coupled receptors (GPCRs) has focused mainly on endocytic trafficking. Whether and how surface delivery of newly synthesized GPCRs is regulated by extracellular signals is less understood. Here we define a signaling-regulated checkpoint at the trans-Golgi network (TGN) that controls the surface delivery of the delta opioid receptor (δR). In PC12 cells, inhibition of phosphoinositide-3 kinase (PI3K) activity blocked export of newly synthesized δR from the Golgi and delivery to the cell surface, similar to treatment with nerve growth factor (NGF). Depletion of class II phosphoinositide-3 kinase α (PI3K C2A), but not inhibition of class I PI3K, blocked δR export to comparable levels and attenuated δR-mediated cAMP inhibition. NGF treatment displaced PI3K C2A from the Golgi and optogenetic recruitment of the PI3K C2A kinase domain to the TGN-induced δR export downstream of NGF. Of importance, PI3K C2A expression promotes export of endogenous δR in primary trigeminal ganglion neurons. Taken together, our results identify PI3K C2A as being required and sufficient for δR export and surface delivery in neuronal cells and suggest that it could be a key modulator of a novel Golgi export checkpoint that coordinates GPCR delivery to the surface.
INTRODUCTION
G protein–coupled receptors (GPCRs), the largest family of signaling receptors, are highly regulated by membrane trafficking (Pierce et al., 2002). The interplay between signaling and trafficking has been explored mainly using endocytic and postendocytic trafficking, which modify the number of receptors on the cell surface (Hanyaloglu and von Zastrow, 2008; Marchese et al., 2008). Activated GPCRs are internalized and trafficked to the endosome, causing a decrease in cellular responsiveness to the signal. From the endosome, receptors are returned to the surface by recycling or are degraded in the lysosome (Hanyaloglu and von Zastrow, 2008; Marchese et al., 2008; Bowman and Puthenveedu, 2015). In the latter case, new receptors need to be made and inserted at the cell surface in order for cells to recover signaling.
Compared to endocytic trafficking, relatively little is known about whether and how the delivery of newly synthesized GPCRs is regulated by signaling pathways. A physiologically relevant prototype for this is the delta opioid receptor (δR), for which regulated delivery of newly synthesized receptors has clear physiological and clinical significance (Vanderah, 2010; Gendron et al., 2016). The δR is an inhibitory Gi protein–coupled receptor and a promising alternative target for pain management (Cahill et al., 2007; Vanderah, 2010; Pradhan et al., 2011; Gendron et al., 2016). Several studies have demonstrated that increased surface expression of δR in neurons is correlated with changes in δR physiology. Factors such as nerve growth factor (NGF), bradykinin, and δR activation itself can alter the sensitivity of neurons to δR agonists (Patwardhan et al., 2005; Cahill et al., 2007; Bie et al., 2010; Mittal et al., 2013; Pettinger et al., 2013). Further, studies evaluating expression and localization of δR in cellular systems found that δR is localized to intracellular pools in neuronal cells, raising the exciting idea that the basal surface expression of δR is tightly controlled by physiological signals that regulate the surface delivery of δR (Roth et al., 1981; Zhang et al., 1998; Petaja-Repo et al., 2000; Wang et al., 2001; Cahill et al., 2001a, b; Bao et al., 2003; Kim and von Zastrow, 2003; Gendron et al., 2015; Shiwarski et al., 2017; St-Louis et al., 2017). The cellular mechanisms that regulate δR surface delivery downstream of extracellular pathways, however, are relatively unknown. In this study, we elucidate the physiological checkpoint at the trans-Golgi network (TGN) that limits δR export from the Golgi and surface expression in neuroendocrine cells.
RESULTS
Phosphoinositide-3 kinase activity is required for δR export from the Golgi
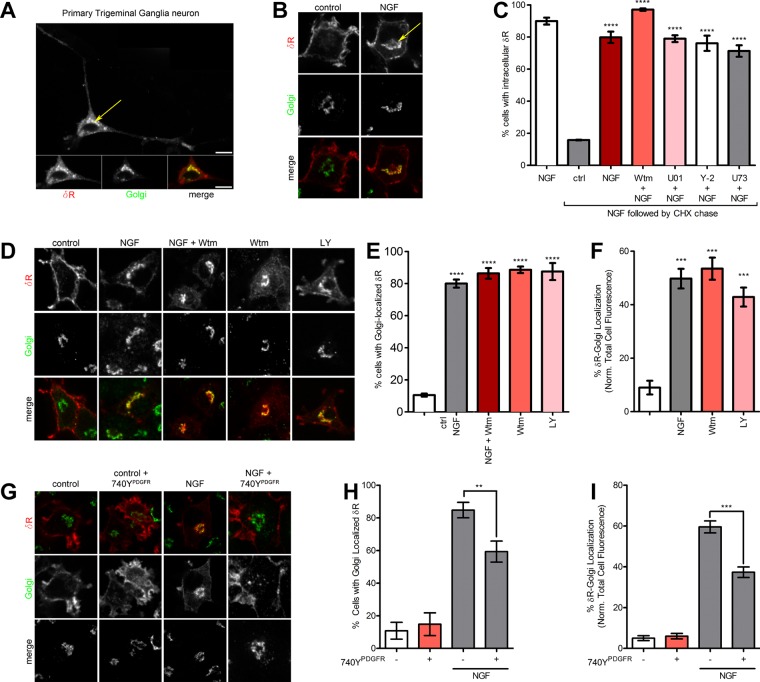
To visualize intracellular δR retention, we used the expression of an N-terminally FLAG-tagged δR in cultured neuroendocrine cells. This expression system has been highly useful to study many GPCRs, including δR, and we and others have extensively confirmed that tagged receptors are functional (Guan et al., 1992; Kim and von Zastrow, 2003; Arttamangkul et al., 2008; Puthenveedu et al., 2010; Soohoo and Puthenveedu, 2013; Vistein and Puthenveedu, 2013; Bowman et al., 2015; Shiwarski et al., 2017). To confirm proper localization of this construct, we expressed FLAG-tagged δR in cultured adult mouse trigeminal ganglia (TG) neurons. In TG neurons, after fixed cell immunofluorescence, FLAG-tagged δR was localized predominantly in intracellular structures that overlapped with the Golgi apparatus, recapitulating earlier studies showing δR localization to intracellular pools (Figure 1A). As an experimental system to start addressing the mechanistic regulation of δR retention, we used neuroendocrine pheochromocytoma-12 (PC12) cells expressing N-terminally FLAG-tagged δR in a fixed-cell immunofluorescence assay. Consistent with previous observations, δR expressed in PC12 cells showed predominantly surface expression (Figure 1B). After NGF treatment for 1 h, δR is retained in an intracellular compartment broadly overlapping with the TGN marker TGN-38 (Figure 1B; Kim and von Zastrow, 2003). We next directly visualized the NGF-mediated accumulation of δR retention by expressing an N-terminally green fluorescent protein (GFP)–tagged δR and imaging δR using live-cell confocal fluorescence microscopy. When exposed to NGF and imaged live, NGF caused robust intracellular δR accumulation within 1 h (Supplemental Movie S1). Taken together, our results suggest that NGF signaling in PC12 cells actively reduces δR export from the Golgi and recapitulates the intracellular δR retention seen in neurons.
FIGURE 1:
PI3K inhibition is required and sufficient for intracellular δR retention. (A) Representative confocal image from a fixed primary cultured TG neuron expressing a FLAG-tagged δR. Internal δR (red in merge) colocalizes with the Golgi marker GPP130 (green in merge). (B) Example image (of three independent experiments) of a PC12 cell showing surface δR. NGF treatment (60 min at 100 ng/μl) causes internal δR accumulation (arrow), colocalizing with TGN-38 (Golgi, green). (C) NGF-treated PC12 cells, chased with cycloheximide (CHX) for 1 h to prevent additional δR delivery, in the presence of inhibitors of MEK (10 µM U0126, denoted as U01), ROCK (5 µM Y-27632, denoted as Y-2), and PLC (10 µM U73122, denoted as U73). Percentage of cells with Golgi-localized δR >100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. ctrl). None of the inhibitors prevented NGF-mediated δR retention. (D) Representative images (of three independent experiments) showing that inhibition of PI3K with 10 μM Wtm or LY is sufficient to cause δR retention. (E) Quantitation of percentage of cells with δR Golgi localization showing that PI3K inhibition with Wtm or LY is sufficient for δR Golgi localization (>100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. NT). (F) Quantitation of the percentage of δR localization within the Golgi normalized to the total cell δR fluorescence, showing that PI3K inhibition with Wtm or LY is sufficient for δR retention (>100 cells each; mean ± SEM; ***p < 0.001). (G) Example images (of three independent experiments) for PC12 cells expressing FLAG-δR, untreated (left two columns) or treated with NGF (100 ng/ml) for 1 h. NGF treatment induces intracellular retention of δR (red), which colocalizes with the Golgi (green). Activation of PI3K by the p85 subunit–binding peptide 740YPDGFR (50 µg/ml) decreased NGF-induced Golgi localization of δR. Images without and with 740YPDGFR. (H) Quantitation of percentage of cells with δR Golgi localization, showing significant reduction in percentage of cells with Golgi-localized δR in the NGF condition after addition of the PI3K-activating peptide 740YPDGFR (>100 cells each; mean ± SEM; **p < 0.01 by one-way ANOVA with Dunn’s multiple comparison test). (I) Image analysis and quantification shows a significant reduction in percentage of total δR fluorescence that overlaps with the Golgi in the NGF condition after addition of PI3K-activating peptide 740YPDGFR. 740YPDGFR had no effect on Golgi localization of δR on its own (>100 cells each; mean ± SEM; ***p < 0.001 by one-way ANOVA with Dunn’s multiple comparison test).
To establish that this accumulation represented a change in export from the Golgi and not a transient pool of newly synthesized receptors, we first accumulated δR in the TGN by treating cells with NGF for 1 h to induce Golgi retention. After NGF, an increase in the percentage of cells containing an intracellular pool of δR dramatically increases (Figure 1C). We then chased this accumulated pool by blocking the synthesis of new δR with cycloheximide, thereby preventing new proteins from entering the Golgi. This chase was performed either in the presence of continued NGF or after NGF was removed. The intracellular pool was rapidly lost in the absence of NGF, suggesting that NGF induced a block in export. In the continued presence of NGF, the intracellular pool persisted even when synthesis of new δR was blocked (Figure 1C).
Considering that δR is retained in neurons potentially in the absence of NGF (Zhang et al., 1998; Cahill et al., 2001a, b; Bao et al., 2003; Kim and von Zastrow, 2003; Gendron et al., 2015), we next attempted to identify additional signaling factors that could cause retention under physiological conditions in neurons. We focused on factors downstream of the TrkA receptor, the primary target of NGF in PC12 cells, because it is required for the retention of δR in this system (Kim and von Zastrow, 2003). Inhibitors of the main TrkA effectors Rho-associated protein kinase (ROCK; Y-27632), MEK (U0126), Akt (Akt1/2 kinase inhibitor), phospholipase C (PLC; U73122), protein kinase C (PKC; chelerythrine), and c-Src (PP2) did not reduce the NGF-induced retention of δR in our cycloheximide assay. However, inhibition of phosphoinositide-3 kinase (PI3K) by wortmannin (Wtm) increased NGF-mediated δR retention (Figure 1C). Strikingly, PI3K inhibition by either Wtm or LY294002 (LY) was sufficient to cause δR retention in the absence of NGF, as measured by the percentage of cells exhibiting intracellular δR (Figure 1, D and E; Vlahos et al., 1994; Wymann et al., 1996). To better quantitate the fraction of total cellular δR that was retained, we used immunofluorescence of a TGN marker, TGN-38, as a mask to measure the δR that colocalized with the Golgi (Supplemental Figure S1). NGF treatment or PI3K inhibition significantly increased the fraction of Golgi-localized δR (Figure 1F), indicating that PI3K activity is required for δR export. We next directly visualized δR retention via PI3K inhibition by imaging retention of GFP-δR in live cells as before. When cells are exposed to the PI3K inhibitor Wtm and imaged live, there is robust intracellular δR accumulation within 1 h (Supplemental Movie S2).
To determine whether PI3K inhibition required activation of its common downstream kinase targets, we tested whether inhibition of the main PI3K targets Akt, PKC, and cSrc was sufficient to induce δR retention. In PC12 cells expressing FLAG-tagged δR, treatment with the Akt inhibitor (Akt 1/2 inhibitor), PKC inhibitor (chelerythrine), or cSrc inhibitor (PP2) was not sufficient to induce intracellular retention of δR (Supplemental Figure S2A). On quantitation, neither the percentage of cells with Golgi-localized δR nor the percentage of δR localized to the Golgi increased after inhibition of PKC, Akt, or cSrc (Supplemental Figure S2, B and C), indicating that these kinases were not required for δR export.
PI3K activation is sufficient to reduce the NGF-induced retention of δR
We showed that biosynthetic trafficking of δR and its retention in the Golgi can be controlled via NGF signaling and PI3K inhibition; however, the exact relationship between PI3K inhibition and NGF-induced Golgi retention of δR was still unknown. Inhibition of PI3K with either LY or Wtm was sufficient to induce Golgi retention of δR (Figure 1D). We next wanted to determine whether PI3K activation was sufficient to prevent δR Golgi retention after NGF treatment. To activate PI3K in the presence of NGF, we used a PI3K-activating peptide, 740YPDGFR (Derossi et al., 1998). Activation of PI3K by the p85 subunit–binding peptide 740YPDGFR decreased the Golgi retention phenotype after NGF. Activation of PI3K by 740YPDGFR had no effect on δR Golgi retention on its own (Figure 1G). Quantification of the immunofluorescence confirmed that NGF was sufficient to increase the percentage of δR colocalized with the Golgi and the percentage of cells exhibiting Golgi-localized δR (Figure 1, H and I). The PI3K-activating peptide 740YPDGFR significantly decreased the percentage of δR colocalized with the Golgi and the percentage of cells exhibiting Golgi-localized δR after NGF treatment (Figure 1, H and I). These data taken together suggest that activation of PI3K is sufficient to reduce the NGF-mediated Golgi retention of δR.
PI3K inhibition does not induce retention of μR
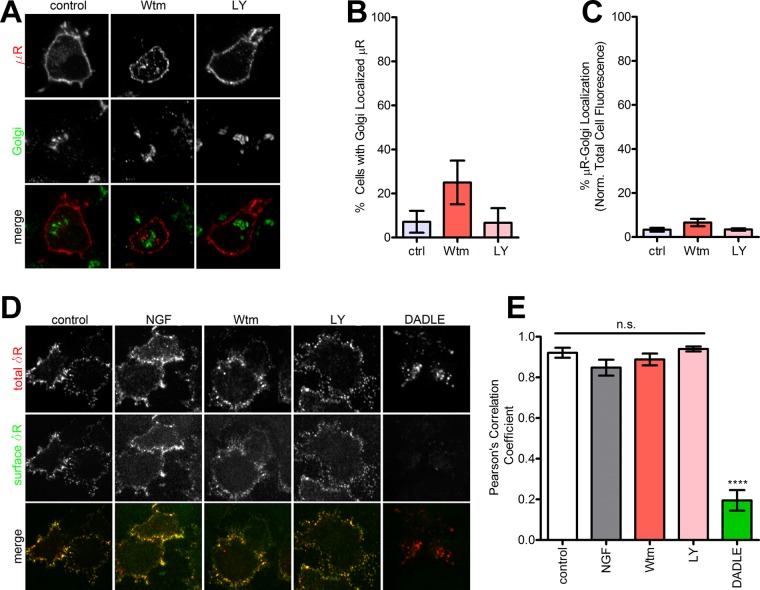
We next tested whether the δR retention induced by NGF and PI3K inhibition represented a general block in surface cargo trafficking or a regulated process controlling δR export. To differentiate these, we asked whether NGF treatment or PI3K inhibition induced retention of the related GPCR, the mu opioid receptor (μR). In PC12 cells expressing FLAG-tagged μR, neither NGF treatment nor inhibition of PI3K by Wtm or LY altered the localization of μR (Figure 2A). On quantitation, the percentage of cells showing intracellular retention of μR did not change with these treatments (Figure 2B). The fraction of μR in the Golgi also did not change with these treatments (Figure 2C). This suggests that the NGF-PI3K–regulated retention of δR is not due to a general block in receptor surface trafficking but represents a regulated exocytic pathway.
FIGURE 2:
PI3K inhibition does not induce intracellular retention of μR. (A) Representative images (of three independent experiments) of PC12 cells expressing FLAG-μR as a control do not show retention after PI3K inhibition. (B) Quantitation of percentage of cells showing μR retention after PI3K inhibition (>100 cells each; mean ± SEM; not significant [p > 0.05] by two-sided t test vs. control). (C) Quantitation of percentage of total μR fluorescence that overlaps with the Golgi after PI3K inhibition (>100 cells each; mean ± SEM; not significant [p > 0.05] by two-sided t test vs. control). (D) Representative images (of three independent experiments) for δR endocytosis estimated by selectively labeling the surface vs. total pool of δR as described in Materials and Methods. PI3K inhibition did not cause endocytosis. The δR agonist DADLE was used as a control for endocytosis. (E) Pearson’s r of colocalization of the primary and secondary antibodies. High correlation denotes minimal endocytosis. DADLE significantly reduced the correlation, consistent with endocytosis (three representative fields; mean ± SEM; ****p < 0.0001 by two-sided t test vs. control).
The NGF and PI3K inhibition–induced retention of δR is not due to surface receptor internalization
To ensure that the intracellular pool of δR was not derived from receptors internalized from the cell surface, PC12 cells expressing the N-terminally FLAG-tagged δR were prelabeled live with Alexa 647–conjugated anti-FLAG antibodies to isolate and follow the surface pool after NGF, Wtm, or LY addition. None of these treatments redistributed surface δR to intracellular compartments (Figure 2D). As a positive control, the δR agonist [D-Ala2, D-Leu5]-enkephalin (DADLE) caused robust internalization and redistribution of receptors to endosomes (Figure 2D). To quantitate the amount of internalization, we incubated the cells with Alexa 488–conjugated secondary antibodies at the end of the treatment. This allowed us to specifically detect the remaining surface pool of labeled δR and quantitatively estimate the fraction of the surface pool that colocalized with the total pool of δR. The surface and the total pools of δR showed robust colocalization in cells treated with NGF, Wtm, or LY, comparable to control, indicating that these treatments do not induce any noticeable redistribution of surface δR to intracellular compartments (Figure 2, D and E). DADLE, as expected, caused low surface fluorescence and a significant loss of colocalization, consistent with internalization. These data reveal that the intracellular δR pool induced by NGF or PI3K inhibition is not due to receptor endocytosis. Blocking new protein synthesis before NGF treatment, in contrast, abolished the intracellular pool of δR (Figure 1C). Together our results suggest that PI3K activity is required for δR export from the Golgi and that NGF inhibits this export, causes δR retention, and limits surface δR delivery.
PI3K activity is required for δR export from the Golgi in human embryonic kidney 293 cells
In PC12 cells, we showed specificity of PI3K inhibition on the Golgi retention of δR by demonstrating that PI3K inhibition does not result in Golgi retention of the related μR. In addition, Kim and von Zastrow (2003) showed that TrkA receptor expression was required for NGF-induced δR retention in PC12 cells. Therefore we sought to determine whether changes in PI3K activity could control the biosynthetic trafficking of δR in cell lines not expressing the TrkA receptor, such as human embryonic kidney 293 (HEK 293) cells. FLAG-tagged δR and μR were transfected into HEK 293 cells, grown under antibiotic selection to obtain a stable pool of expressing cells, and evaluated for receptor localization via fixed-cell immunofluorescence detection and confocal microscopy. In HEK 293 cells expressing FLAG-δR under control conditions, δR was predominantly localized to the cell surface and did not colocalize with the Golgi marker GPP130 (Figure 3A). When cells were treated with the PI3K inhibitor Wtm, there was a dramatic increase in the amount of Golgi-localized δR (Figure 3A). Quantification of the δR immunofluorescence confirmed that treatment with Wtm increased both the percentage of δR localized to the Golgi and the percentage of cells exhibiting Golgi-localized δR (Figure 3, B and C). Identical experiments were performed with the FLAG-μR HEK 293 cells. Like δR, μR was basally localized to the cell surface; however, upon treatment with the PI3K inhibitor Wtm or LY, there was no change in μR localization (Figure 3D). Quantification of the μR immunofluorescence confirmed that treatment with neither Wtm nor LY altered the percentage of μR localized to the Golgi or the percentage of cells exhibiting Golgi-localized μR (Figure 3, E and F). These data demonstrate that PI3K inhibition is sufficient to induce the Golgi retention of δR independently of TrkA signaling in a receptor-specific manner.
FIGURE 3:
PI3K inhibition delays export of δR from the Golgi in HEK 293 cells. (A) Example images of HEK 293 cells expressing FLAG-δR (δR). In control conditions (ctrl), δR is present on the cell surface; however, in HEK 293 cells, PI3K inhibition by Wtm (10 µM) increases the Golgi localization of δR. (B) Image analysis and quantification show a significant increase in percentage of cells with the Golgi-localized δR after PI3K inhibition by 10 µM Wtm (>100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. ctrl) (C) Image analysis and quantification reveal a significant increase in percentage of total δR fluorescence that overlaps with the Golgi after PI3K inhibition by 10 µM Wtm (>100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. ctrl). (D) Example images of HEK 293 cells expressing FLAG-μR. In control conditions (ctrl), μR is present on the cell surface; however, unlike for δR, PI3K inhibition by 10 µM Wtm or LY does not increase the Golgi localization of μR. (E) Image analysis and quantification resulted in a nonsignificant change in percentage of cells with the Golgi-localized μR after PI3K inhibition by Wtm or LY (ctrl, n = 45; Wtm, n = 80; LY, n = 88; mean ± SEM; not significant by two-sided t test vs. ctrl). (F) Image analysis and quantification show a nonsignificant change in percentage of total μR fluorescence that overlaps with the Golgi after PI3K inhibition by Wtm or LY (ctrl, n = 45; Wtm, n = 80; LY, n = 88; mean ± SEM; not significant by two-sided t test vs. ctrl). (G) Example time-lapse images (of four independent experiments) for HEK 293 cells expressing VSVG-δRtail-GFP imaged at the permissive temperature of 32°C. PI3K inhibition by 10 µM Wtm and LY resulted in sustained intracellular Golgi signal compared with control. (H) Quantification of the Golgi-associated fluorescence signal revealed a kinetic slowing of VSVG-δRtail-GFP trafficking from the Golgi after treatment with PI3K inhibitors Wtm and LY (control, 12 cells; Wtm, 14 cells; LY, 12 cells; mean ± SEM). (I) Quantification of the Golgi-associated VSVG fluorescence signal after 30 min, normalized to total, reveals that VSVG-δRtail-GFP requires the δRtail to show PI3K-dependent trafficking from the Golgi (control VSVG, 9 cells; VSVG LY294002, 9 cells; VSVG-δRtail control, 11 cells; VSVG-δRtail LY294002, 11 cells; mean ± SEM; ***p < 0.001).
PI3K activity controls the Golgi export of GFP–vesicular stomatitis virus glycoprotein–δRtail
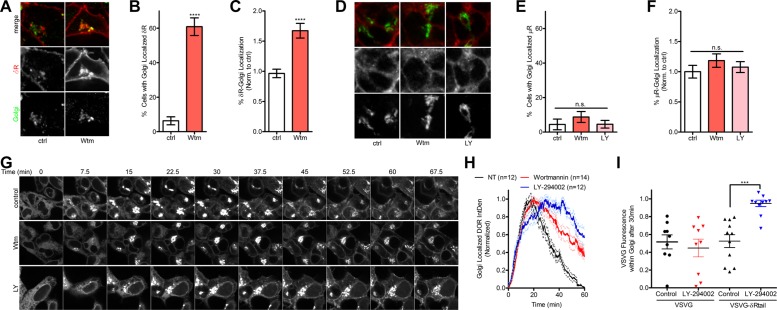
To test whether PI3K activity was directly affecting the biosynthetic trafficking of δR in a sequence-specific manner and evaluate the kinetics of δR retention and release, we used the temperature-sensitive mutant (ts045) of the vesicular stomatitis virus glycoprotein (VSVG) to visualize dynamic protein trafficking through the secretory pathway. The temperature-sensitive VSVG allows for a temperature-controlled release of VSVG from the endoplasmic reticulum (ER) to the Golgi upon transition from 40 to 32°C and has widely been used for studying protein trafficking through the secretory pathway (Kreis, 1986; Nakamura et al., 1995; Scales et al., 1997). For our experiments, we created a chimeric protein consisting of the temperature-sensitive mutant (ts045) VSVG linked to the last 27 amino acids of the C-terminal tail domain of δR because the δR C-terminal tail has been shown to be sufficient for NGF-induced Golgi retention in PC12 cells (Kim and von Zastrow, 2003). To visualize the trafficking in real-time, we C-terminally tagged our chimeric protein with GFP for live-cell fluorescence imaging capabilities (VSVG-δRtail-GFP). HEK 293 cells were transfected with VSVG-δRtail-GFP and grown under antibiotic selection for stable expression. To inhibit exit of the VSVG-δRtail-GFP from the ER, the cells were grown at 40°C. At the time of imaging, the cells were transitioned to 32°C and visualized by live-cell fluorescence microscopy.
At time zero, all HEK 293 cells expressing the VSVG-δRtail-GFP exhibited a GFP signal resembling ER localization. Under control conditions, trafficking of the VSVG-δRtail-GFP appeared normal, exiting the ER, accumulating within the Golgi, and budding off into vesicular structures trafficking toward the membrane. After 1 h at 32°C, most of the Golgi-accumulated VSVG-δRtail-GFP signal had dissipated, and the cell membrane showed increased surface fluorescence (Figure 3G and Supplemental Movie S3). When cells were treated with the PI3K inhibitor Wtm, trafficking of the VSVG-δRtail-GFP from the ER to the Golgi appeared normal; however, a significant amount of Golgi-accumulated fluorescence remained after 1 h at 32°C (Figure 3G and Supplemental Movie S3). Similarly, inhibition of PI3K with LY demonstrated strong intracellular and Golgi retention of VSVG-δRtail-GFP (Figure 3G). To evaluate quantitatively the kinetics of VSVG-δRtail-GFP trafficking, we measured the Golgi fluorescence signal accumulation and decay over the course of the experiment. Inhibition of PI3K by Wtm or LY delayed the trafficking of the VSVG-δRtail-GFP from the Golgi and resulted in a prolonged Golgi-localized fluorescence intensity compared with control (Figure 3H). Quantification of the total Golgi-localized fluorescence for VSVG-δRtail-GFP over time was performed by calculating the area under the curve (AUC) for each experimental condition. Inhibition of PI3K by Wtm or LY significantly increased the total Golgi-localized δR (Supplemental Figure S3). As a control, HEK 293 cells expressing VSVG without the δRtail were imaged and treated with LY. Without the last 27 amino acids of the C-terminal tail domain of δR, treatment with LY did not delay Golgi export (Figure 3I). These data suggest that inhibiting PI3K decreases Golgi export of VSVG-δRtail-GFP in HEK 293 cells and that the last 27 amino acids of δR are sufficient to confer PI3K sensitivity.
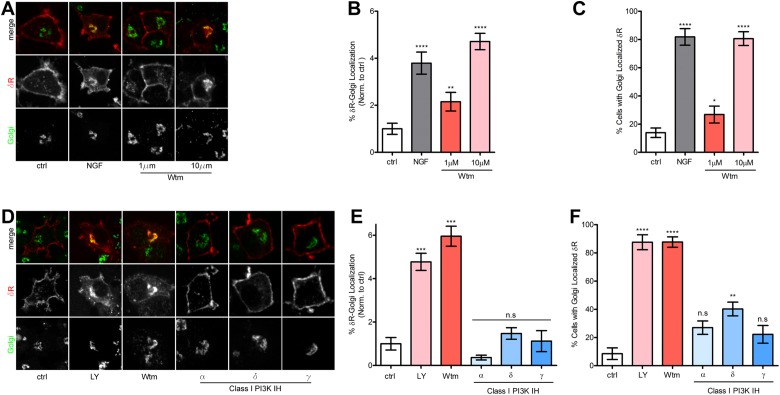
Inhibition of class I PI3Ks is not sufficient to retain δR in the Golgi
There are several classes of PI3Ks: I–III. The best-known and commonly targeted are the class I PI3Ks, which are made up of a p85 regulatory subunit and a p110 catalytic subunit. These PI3Ks are primarily responsible for the production of phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3). Within class I PI3Ks, there are several subtypes of the p110 catalytic subunit: α, β, δ, and γ. At nanomolar and low micromolar concentrations, the PI3K inhibitor Wtm preferentially inhibits class I PI3Ks; however, at concentrations ≥1 µM, Wtm begins to inhibit the class II PI3Ks (Domin et al., 1997). To test whether the Golgi retention of δR after treatment with Wtm was specific to class I PI3K inhibition, we administered 1 or 10 µM Wtm to PC12 cells expressing FLAG-δR. Treatment with 1 µM Wtm had a small but statistically significant effect on inducing a Golgi-localized pool of δR, whereas 10 µM Wtm demonstrated a large and significant increase in the Golgi-localized pool of δR (Figure 4A). Quantification and image analysis of these results showed a twofold increase in the percentage of δR fluorescence localized to the Golgi after 1 µM Wtm, whereas NGF and 10 µM Wtm resulted in an increase in the percentage of δR fluorescence localized to the Golgi of approximately fourfold (Figure 4B). Similarly, the percentage of cells exhibiting visual intracellular δR minimally increased after 1 µM Wtm, whereas NGF and 10 µM Wtm resulted in 80% of cells exhibiting intracellularly localized δR (Figure 4C).
FIGURE 4:
Class I PI3K is not required for Golgi retention of δR. (A) Example images (of three independent experiments) for PC12 cells expressing FLAG-δR, untreated (ctrl), or treated with NGF (100 ng/ml) for 1 h. NGF treatment induces intracellular retention of δR (red), which colocalizes with the Golgi (green). Inhibition of PI3K via 1 µM Wtm leads to partial accumulation of δR in the Golgi. Inhibition of PI3K via 10 µM Wtm leads to large accumulation of δR in the Golgi. (B) Image analysis and quantification show significant increase in percentage of cells with Golgi-localized δR after NGF treatment and PI3K inhibition by 1 and 10 µM Wtm (ctrl, 105 cells; NGF, 64 cells; 1 µM Wtm, 56 cells; 10 µM Wtm, 67 cells; mean ± SEM; **p < 0.01, ****p < 0.0001 by two-sided t test vs. ctrl). (C) Further quantification demonstrated a significant increase in percentage of total δR fluorescence that overlaps with the Golgi in the NGF condition and a dose-dependent effect on percentage of δR-Golgi localization for 1 and 10 µM Wtm (ctrl, 105 cells; NGF, 64 cells; 1 µM Wtm, 56 cells; 10 µM Wtm, 67 cells; mean ± SEM; *p < 0.05, ****p < 0.0001 by two-sided t test vs. ctrl). (D) Example images (of three independent experiments) for PC12 cells expressing FLAG-δR, untreated (ctrl), or treated with NGF (100 ng/ml), LY (10 µM), or Wtm (10 µM) for 1 h induces intracellular retention of δR (red), which colocalizes with the Golgi (green). Class I–specific PI3K inhibitors PI-103 (PI3K C1 α, 50 nM), IC87114 (PI3K C1 δ, 5 µM), and AS605240 (PI3K C1 γ, 25 nM) were not sufficient to cause Golgi retention of δR. (E) Quantification demonstrated a significant increase in percentage of cells with Golgi-localized δR after NGF treatment and PI3K inhibition by LY (10 µM) or Wtm (10 µM) but not after class I–specific PI3K inhibition by PI3K inhibitors PI-103 (PI3K C1 α, 50 nM), IC87114 (PI3K C1 δ, 5 µM), and AS605240 (PI3K C1 γ, 25 nM; ctrl, 42 cells; NGF, 73 cells; LY, 77 cells; Wtm, 45 cells; α, 82 cells; δ, 99 cells; γ, 28 cells; mean ± SEM; ***p < 0.001 by one-way ANOVA with Dunn’s multiple comparison test). (F) Image analysis and quantification demonstrated a significant increase in percentage of cells with the Golgi-localized δR after NGF treatment and PI3K inhibition by 10 µM LY or Wtm but not after class I–specific PI3K inhibition by PI3K inhibitors PI-103 (PI3K C1 α, 50 nM), IC87114 (PI3K C1 δ, 5 µM), and AS605240 (PI3K C1 γ, 25 nM; ctrl, 42 cells; NGF, 73 cells; LY, 77 cells; Wtm, 45 cells; α, 82 cells; δ, 99 cells; γ, 28 cells; mean ± SEM; **p < 0.01, ****p < 0.0001 by one-way ANOVA with Dunn’s multiple comparison test).
To rule out inhibition of class I PI3Ks as the driver of δR Golgi retention, we used the class I isoform–specific PI3K inhibitors PI-103 (PI3K C1, α), IC87114 (PI3K C1, δ), and AS605240 (PI3K C1, γ; Sadhu et al., 2003; Camps et al., 2005; Park et al., 2008). Using our fixed-cell immunofluorescence assay in PC12 cells expressing FLAG-δR, Wtm (10 µM) and LY (10 µM), which are both pan–class I and II PI3K inhibitors at these concentrations, resulted in significant Golgi-localized δR fluorescence; however, the PI3K class I–specific inhibitors did not cause any change in δR localization pattern (Figure 4D). Quantification and image analysis of these data revealed a significant increase in the percentage of δR fluorescence localized to the Golgi and the percentage of cells with Golgi-localized δR after 10 µM Wtm or LY treatment but no significant change for either measurement after treatment with the PI3K class I–specific inhibitors (Figure 4, E and F). Therefore inhibition of class I PI3Ks is not sufficient to induce Golgi retention of δR, and a concentration of >1 µM Wtm is required to stimulate a large increase in δR Golgi localization.
Class II PI3K C2A is required for complete surface trafficking of δR
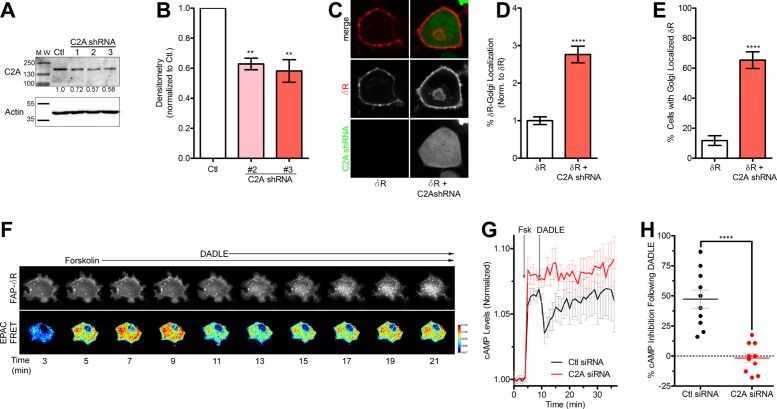
Our results suggest that inhibition of class I PI3Ks is not sufficient to cause the Golgi retention of δR; however, concentrations of Wtm and LY of 10 µM, which can inhibit class II PI3Ks, are sufficient to induce significant retention of δR. In addition, whereas class I PI3Ks are traditionally believed to be catalytically active at the plasma membrane, class II PI3Ks are active at the TGN and involved in intracellular trafficking and clathrin recruitment (Domin et al., 2000; Gaidarov et al., 2001, 2005). For these reasons, we chose to determine the role of the class II PI3K α (PI3K C2A) in δR Golgi retention by knocking down the endogenous PI3K C2A in PC12 cells. Rat species–specific PI3K C2A short hairpin RNAs (shRNAs) were designed using the RNAi Central shRNA psm2 Design tool from the Hannon laboratory and two previously published shRNAs targeting PI3K C2A (Silva et al., 2005; Wang et al., 2006; Bridges et al., 2012). All PI3K C2A shRNAs were cloned into the pENTR-pSM2(CMV)GFP vector and transiently transfected into PC12 cells for initial evaluation of knockdown (Supplemental Table S1). Three of the five sequences tested (1–3) showed promising knockdown of PI3K C2A and were transferred into the pLenti X1 Puro Dest vector using Gateway cloning, and lentiviral particles were produced following a modified protocol from Campeau et al. (2009). Stably transduced PC12 cells expressing the PI3K C2A shRNAs were evaluated for PI3K C2A expression via immunoblotting (Figure 5A). The shRNA sequences 2 and 3 produced ∼50% knockdown of PI3K C2A (Figure 5B), which we observed were optimal for these studies, as higher levels of knockdown caused higher cell death, consistent with an essential role for PI3K C2A. The PC12 cells expressing PI3K C2A shRNA 3 (PI3K C2A shRNA) was chosen for use in our further studies.
FIGURE 5:
Class II PI3K C2A is required for surface trafficking of δR. (A) Immunoblotting for PI3K C2A (C2A) confirmed knockdown of C2A in PC12 cells stably expressing C2A lentiviral shRNA. Three different shRNA sequences were used to determine the most efficient knockdown. Representative immunoblot, with actin as a loading control. Blot densitometry was performed to quantitate the percentage of knockdown for C2A shRNA sequences 1–3 compared with control (Ctl) cells. (B) Densitometry (of three independent experiments) revealed C2A shRNA sequences 2 and 3 as providing significant reduction in C2A protein expression, with shRNA 3 providing the best knockdown at 49% (three independent experiments; mean ± SEM; **p < 0.01 by two-sided t test vs. Ctl). (C) Example images (of three independent experiments) for PC12 cells expressing FAP-δR (δR) with and without lentiviral PI3K C2A shRNA 3 (C2A shRNA) with a GFP reporter. Cells stably expressing the PI3K C2A shRNA were identified via GFP expression and had increased intracellular δR. (D) Image analysis and quantification revealed a significant increase in percentage of total δR fluorescence that overlaps with the Golgi in cells stably expressing the PI3K C2A shRNA compared with δR-only cells (δR, 102 cells; δR + C2A shRNA, 75 cells; mean ± SEM; ****p < 0.0001 by two-sided t test vs. δR). (E) Further quantification confirmed a significant increase in percentage of cells with Golgi-localized δR in cells stably expressing the PI3K C2A shRNA compared with δR-only cells (δR, 102 cells; δR + C2A shRNA, 75 cells; mean ± SEM; ****p < 0.0001 by two-sided t test vs. δR). (F) Example images from PC12 FAP- δR cells expressing EPAC cAMP biosensor and control nontargeted siRNA. Images reveal an increase in cAMP EPAC FRET ratio over time after 5 µM forskolin addition and subsequent decrease in cAMP and receptor internalization after δR agonist DADLE (10µM) addition. (G) Quantitative analysis of the EPAC FRET ratio, showing an increase in cAMP after forskolin addition and decrease after δR agonist DADLE addition in control (Ctl) siRNA cells. Similar experiments performed in cells transfected with PI3K C2A siRNA exhibited no decrease in cAMP after δR agonist DADLE (Ctl siRNA, 12 cells; C2A siRNA, 10 cells; mean ± SEM). (H) Analysis of percentage of cAMP inhibition after δR agonist addition demonstrates PI3K C2A siRNA ability to significantly reduce cAMP inhibition compared with control siRNA cells (Ctl siRNA, 12 cells; C2A siRNA, 10 cells; mean ± SEM; ****p < 0.0001 by two-sided t test Ctl vs. C2A siRNA).
The PC12 cells expressing PI3K C2A shRNA were transfected with a fusion construct expressing a previously characterized fluorogen-activated peptide (FAP) N-terminally tagged to δR (FAP-δR) and imaged live via fluorescence confocal microscopy. The FAP-δR was chosen for its far-red emission spectra, which results from a malachite green (MG)–based fluorogen (Szent-Gyorgyi et al., 2008, 2013; Wu et al., 2014; Pratt et al., 2015). In control cells not expressing the PI3K C2A shRNA, FAP-tagged δR was localized to the cell surface; however, in the cells stably expressing the PI3K C2A shRNA, a prominent Golgi-localized pool of δR was observed (Figure 5C). Image analysis and quantification demonstrated a significant increase in the percentage of δR fluorescence localized to the Golgi and in the percentage of cells that had Golgi-localized δR in PI3K C2A shRNA–expressing cells compared with cells that only expressed the FAP-tagged δR (Figure 5, D and E). To evaluate the functional consequence of PI3K C2A knockdown on δR function, we measured cAMP inhibition, as a readout for δR activation and Gi coupling, using the exchange protein directly activated by cAMP (EPAC)–based Förster resonance energy transfer (FRET) sensor (DiPilato et al., 2004; Shiwarski et al., 2017). PC12 cells stably expressing FAP-δR were transiently transfected with the EPAC FRET sensor and siRNA against PI3K C2A. Activation of δR was quantified based on inhibition of forskolin-stimulated cAMP using live-cell fluorescence imaging as previously described (Shiwarski et al., 2017). Cells transfected with control siRNA showed an expected increase in cAMP after 5 µM forskolin, followed by inhibition after addition of the δR agonist DADLE (10 µM) and subsequent receptor internalization (Figure 5F). Quantification of the change in cAMP over time in these cells revealed ∼50% inhibition of the forskolin-stimulated cAMP after δR agonist addition. In cells expressing PI3K C2A siRNA, in contrast, there was no significant reduction in the cAMP levels after δR agonist addition (Figure 5, G and H). This suggests that PI3K C2A expression is required for surface trafficking of functionally competent δR.
Exogenous expression of class II PI3K C2A is sufficient to decrease NGF-induced retention of δR
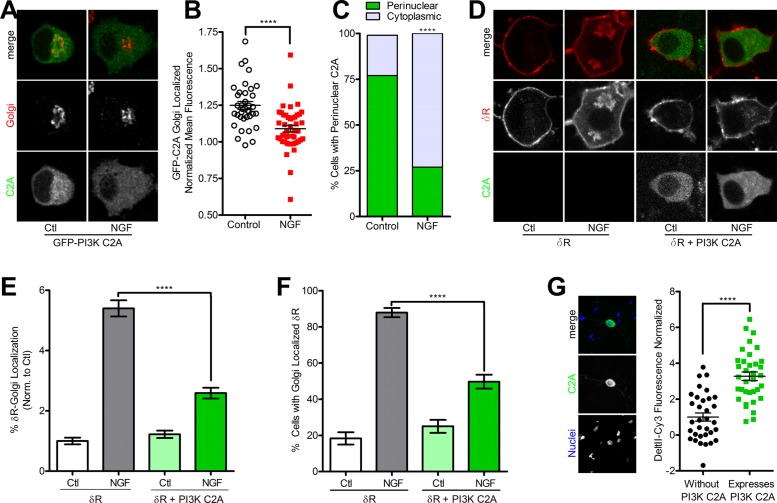
Because PI3K C2A was required for proper trafficking and function of δR to the surface in PC12 cells, we tested whether PI3K C2A localization was altered after NGF treatment. PC12 cells were transfected with GFP-PI3K C2A, treated with or without NGF for 1 h, and imaged via fixed-cell fluorescence confocal microscopy. GFP-PI3K C2A displayed an expected cytoplasmic and Golgi localization in control untreated conditions. After NGF, GFP-PI3K C2A appeared much more diffusely localized within the cytoplasm and less concentrated around the Golgi (Figure 6A). Quantification and image analysis of these data revealed a significant decrease in the percentage of GFP-PI3K C2A fluorescence localized to the Golgi and the percentage of cells with perinuclear-localized GFP-PI3K C2A after NGF (Figure 6, B and C).
FIGURE 6:
Expression of class II PI3K C2A is sufficient to induce surface trafficking of δR. (A) Example images (of three independent experiments) for PC12 cells expressing GFP-PI3K C2A (green) demonstrate a pool of GFP-PI3K C2A enriched around the Golgi (TGN-38, red) in control conditions (Ctl) that is reduced after 1 h of treatment with NGF (100 ng/ml). (B) Quantification of GFP-PI3K C2A mean fluorescence intensity revealed a significant decrease in Golgi localization after NGF treatment (control, 36 cells; NGF, 45 cells; mean ± SEM; ****p < 0.0001 by two-sided t test vs. control). (C) Additional quantification of the observed shift in GFP-PI3K C2A distribution shows a shift from perinuclear localization to diffuse cytoplasmic localization after NGF treatment (control, 28 of 36 perinuclear; NGF, 12 of 33 perinuclear; mean ± SEM; ****p < 0.0001 by two-sided Fisher’s exact test vs. control). (D) Example images (of three independent experiments) for PC12 cells expressing FAP-δR (δR, red) alone and in combination with overexpression of GFP-PI3K C2A (PI3K C2A, green) untreated (Ctl) or treated with NGF (100 ng/ml) for 1 h. Overexpression of GFP-PI3K C2A was sufficient to prevent the NGF-induced Golgi retention of δR. (E) Quantification and image analysis show a significant decrease in percentage of total δR fluorescence that overlaps with the Golgi in cells treated with NGF when overexpressing GFP-PI3K C2A compared with δR-only cells (>100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. δR Ctl). (F) Further quantification demonstrated a significant decrease in percentage of cells with Golgi-localized δR in cells treated with NGF when overexpressing GFP-PI3K C2A compared with δR-only cells (>100 cells each; mean ± SEM; ****p < 0.0001 by two-sided t test vs. δR Ctl). (G) Example image showing GFP-PI3K C2A (green) expression in primary TG neuron counterstained live with Hoechst DNA stain (blue). TG neurons expressing GFP-PI3K C2A have ∼3.27-fold higher deltorphin II–Cy3 binding compared with nonexpressing (without PI3K-C2A) TG neurons (without PI3K-C2A, 33 cells; expresses PI3K-C2A, 35 cells; mean ± SEM; ****p < 0.0001 by two-sided t test vs. without PI3K-C2A).
We next asked whether exogenous expression of PI3K C2A was sufficient to reduce the NGF-induced retention of δR. PC12 cells stably expressing FAP- δR were transiently transfected with GFP-PI3K C2A and imaged via live-cell fluorescence confocal microscopy. In cells expressing only the FAP-δR, control treated cells expressed predominantly surface-localized δR, whereas 1 h of NGF treatment resulted in the expected Golgi localization of δR (Figure 6D). For the cells expressing FAP-δR and GFP-PI3K C2A, δR was localized to the cell surface, with minimal detectable Golgi localization in both control and NGF-treated conditions (Figure 6D). Quantification of these experiments demonstrated an increase in the percentage of δR fluorescence localized to the Golgi and in the percentage of cells that had Golgi-localized δR after NGF treatment for cells that only expressed the FAP-δR. In cells expressing FAP-δR and GFP-PI3K C2A, a significant decrease in the NGF-induced retention of δR, measured via the percentage of δR fluorescence localized to the Golgi and in the percentage of cells that had Golgi-localized δR, was observed (Figure 6, E and F).
To test whether PI3K C2A induces the trafficking of endogenous δR in neurons, we used the δR-specific ligand deltorphin II, C-terminally conjugated with a Cy3 fluorescent dye (Shiwarski et al., 2017), to directly visualize endogenous receptors on the cell surface. Primary TG neurons were isolated and dissociated from adult mice, grown in culture, and microinjected with plasmid DNA for GFP-PI3K C2A. After 72 h in culture, we measured surface deltorphin II–Cy3 binding in neurons, with and without the expression of GFP-PI3K C2A, by live-cell confocal microscopy. In TG neurons expressing GFP-PI3K C2A, there was a 3.27-fold increase in deltorphin II–Cy3 fluorescence compared with control cells on the same coverslips that were not expressing GFP-PI3K C2A (Figure 6G). Taken together, these data suggest that increased PI3K C2A expression is sufficient to reduce NGF-induced retention of δR and increase the functional pool of endogenous surface receptors in TG neurons.
Optogenetic recruitment of the PI3K C2A kinase domain to the TGN is sufficient to promote δR surface trafficking and exocytosis
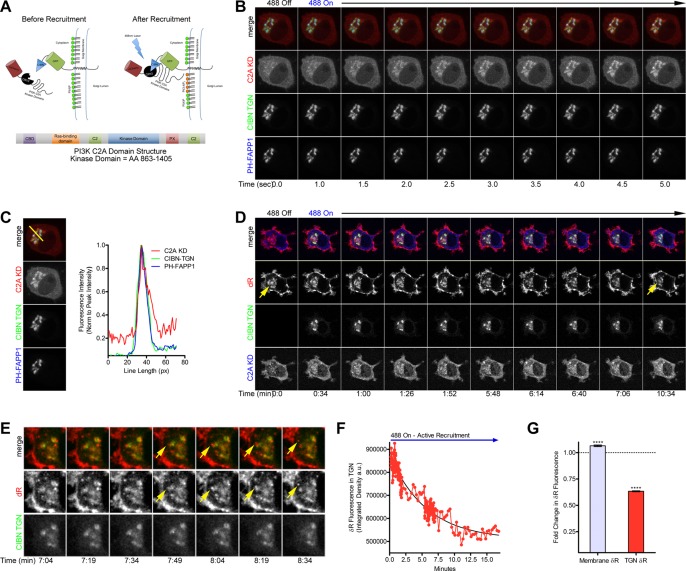
To determine whether the kinase domain of PI3K C2A at the TGN is sufficient to acutely induce trafficking of δR to the cell surface after NGF-induced retention of δR, we developed a TGN-targeted optogenetic recruitment strategy. It was previously shown that the CRY2 and CIBN optical dimerization system developed by the Tucker laboratory could be used to control phosphoinositide metabolism at the plasma membrane (Kennedy et al., 2010; Idevall-Hagren et al., 2012). To use this strategy for targeting of PI3K C2A to the TGN, we exchanged the plasma membrane–targeting domain on the CIBN-GFP for a TGN-targeting domain from the N-terminus of 2,6-sialyltransferase (CIBN-GFP-TGN). The PI3K C2A kinase domain (KD; amino acids 863–1405) was cloned onto the C-terminus of an mCherry-CRY2 chimeric construct to make mCherry-CRY2-PI3K-C2A-KD (Figure 7A). The GFP-CIBN-TGN has a basal TGN localization, and the mCherry-CRY2-PI3K-C2A-KD is mostly cytoplasmic before recruitment. Optogenetic recruitment and dimerization between the CRY2 and CIBN is activated by blue light stimulation at 488 nm, driving the mCherry-CRY2-PI3K-C2A-KD to the TGN (Figure 7A).
FIGURE 7:
Optogenetic recruitment of the PI3K C2A kinase domain to the TGN is sufficient to induce δR surface trafficking. (A) Schematic showing the optogenetic recruitment strategy for targeting the PI3K C2A kinase domain to the TGN upon stimulation with blue light. The CRY2-CIBN optical recruitment system was adapted by cloning the PI3K C2A kinase domain (amino acids 863–1405, KD) onto the C-terminus of an mCherry-CRY2 chimeric construct to make mCherry-CRY2-PI3K-C2A-KD. In addition, the GFP-CIBN was adapted for TGN-specific recruitment by cloning the N-term of 2,6-sialyltransferase onto the C-terminus of GFP-CIBN to make GFP-CIBN-SialyT. The GFP-CIBN-SialyT has a basal TGN localization and the mCherry-CRY2-PI3K-C2A-KD is mostly cytoplasmic before recruitment. Optorecruitment is activated by blue light stimulation at 488 nm driving the mCherry-CRY2-PI3K-C2A-KD to the TGN. (B) PC12 cells were transiently transfected with the mCherry-CRY2-PI3K-C2A-KD (C2A KD, red), GFP-CIBN-SialyT (CIBN-TGN, green), and the TGN-localized PI(4)P sensor PH-FAPP1-iRFP (PH-FAPP1, blue) for live imaging of the optogenetic recruitment. During the live stimulation with 488-nm light, recruitment of the C2A KD to the CIBN-TGN and PH-FAPP1 can be observed within 5 s. (C) Analysis of the recruitment and colocalization of the C2A KD to the CIBN-TGN and PH-FAPP1 region was performed via a line-scan plot (example line in yellow) to identify relative spatiotemporal fluorescence intensity over a pixel area. Over time, the peak fluorescence intensity and the fluorescence intensity distribution for C2A KD (red), CIBN-TGN (green), and PH-FAPP1 (blue) are all highly overlapping after 5 s. (D) PC12 cells were transiently transfected with the mCherry-CRY2-PI3K-C2A-KD (C2A KD, blue), GFP-CIBN-SialyT (CIBN-TGN, green), and FAP-δR (δR, red) for live imaging of the optogenetic recruitment and induced surface trafficking after NGF-induced retention of δR. NGF (100 ng/ml) was added for 1 h to cause Golgi retention of δR (yellow arrow). Optical recruitment of the mCherry-CRY2-PI3K-C2A-KD to the TGN was performed to stimulate PI3K C2A and induce surface trafficking of the retained pool of δR. After recruitment, the Golgi pool of δR decreases and appears to vesiculate. (E) Analysis of live-imaging frames reveals the formation of vesicles containing δR beginning in the TGN (yellow arrow), bud off, and appear to be trafficking to the cell surface. (F) Image analysis and quantification of the complete recruitment movie demonstrate a decrease in the δR fluorescence intensity within the TGN over time after active and sustained recruitment of the mCherry-CRY2-PI3K-C2A-KD to the TGN. (G) Quantification and analysis (of eight independent experiments) show the fold change in δR membrane and TGN-localized fluorescence normalized to their starting values (dotted blue line) after recruitment of mCherry-CRY2-PI3K-C2A-KD to the TGN. There is a significant decrease in the fluorescence intensity of δR within the TGN after recruitment (TGN δR, 37%), as well as a significant increase in total δR membrane fluorescence (membrane δR, 7%; membrane δR, eight cells; TGN δR, eight cells; mean ± SEM; ****p < 0.0001 by two-sided t test vs. respective Ctl).
To confirm functionality and correct localization of the TGN-targeted CIBN-GFP-TGN, and verify proper recruitment of the mCherry-CRY2-PI3K-C2A-KD upon blue light activation, we transfected PC12 cells with CIBN-GFP-TGN, mCherry-CRY2-PI3K-C2A-KD, and the TGN marker PH-FAPP1-GFP, which binds to phosphatidylinositol 4-phosphate (PI(4)P). The cells were then imaged via live-cell fluorescence confocal microscopy. Before recruitment, the mCherry-CRY2-PI3K-C2A-KD was localized to the cytoplasm and the CIBN-GFP-TGN to the region overlapping with the PH-FAPP1-GFP. Upon stimulation with 488-nm blue light, the mCherry-CRY2-PI3K-C2A-KD was rapidly recruited to the CIBN-GFP-TGN within 5 s (Figure 7B and Supplemental Movie S4). To evaluate the colocalization between CIBN-GFP-TGN, mCherry-CRY2-PI3K-C2A-KD, and PH-FAPP1-GFP, we performed a line-scan analysis for the images over time. Over the course of 5 s after 488-nm excitation and recruitment, the fluorescence intensity for the mCherry-CRY2-PI3K-C2A-KD increases within a localized region (Supplemental Figure S4). Five seconds after recruitment is initiated, the peak fluorescence intensity of the CIBN-GFP-TGN, mCherry-CRY2-PI3K-C2A-KD, and PH-FAPP1-GFP are superimposed with each other, suggesting colocalization and effective recruitment of the mCherry-CRY2-PI3K-C2A-KD to the TGN (Figure 7C).
With the ability to spatially and temporally control the recruitment of the kinase domain of PI3K C2A to the TGN, we tested whether PI3K C2A recruitment would rescue δR Golgi retention. For these experiments, we transfected PC12 cells with FAP-δR, CIBN-GFP-TGN, and mCherry-CRY2-PI3K-C2A-KD. This allowed for the simultaneous imaging of δR trafficking and optogenetic recruitment of mCherry-CRY2-PI3K-C2A-KD to the TGN. Before PI3K C2A recruitment, the cells were pretreated with NGF for 1 h to induce Golgi retention of δR. Once retention of δR was confirmed, optical recruitment was initiated by activation with 488-nm blue light. Over the course of 10 min, the internal Golgi pool of δR decreases in both size and intensity in response to the recruitment of mCherry-CRY2-PI3K-C2A-KD to the TGN (Figure 7D; full imaging series showing decrease in Supplemental Movie S5). Further examination of the time-lapse images reveals de novo δR-positive formation of vesicles from the TGN, which appear to traffic toward the cell surface (Figure 7E). Quantification of the δR fluorescence intensity within the TGN region defined by the CIBN-GFP-TGN over time shows a dramatic decrease in Golgi-localized δR fluorescence intensity after mCherry-CRY2-PI3K-C2A-KD recruitment (Figure 7F). In addition, analysis of the initial versus postrecruitment membrane and Golgi-localized δR fluorescence demonstrates a small but significant increase in δR membrane fluorescence and a significant decrease in the Golgi-localized δR fluorescence (Figure 7G). These data demonstrate that the PI3K C2A kinase domain is sufficient to acutely release the Golgi-retained pool of δR and stimulate its trafficking to the cell surface.
DISCUSSION
Our results implicate the atypical PI3K C2A in regulating surface trafficking of newly synthesized δRs downstream of extracellular signals mediated by NGF. Inhibition of PI3K C2A, either pharmacologically or by genetic methods, induced retention of δR in intracellular compartments independently of extracellular NGF. After NGF-induced retention of δR in the Golgi, recruitment of the kinase domain of PI3K C2A to the TGN was sufficient to induce export and delivery of δR to the surface. These data suggest that the localization of PI3K controls the phosphoinositide checkpoint, preventing newly synthesized δR from reaching the cell surface.
PI3Ks, of which there are several classes, control the generation of 3′-phosphoinositides within lipid membranes and specify compartmentalization of proteins within membranes (De Matteis and Godi, 2004). Class I PI3Ks, consisting of a p85 regulatory subunit and a p110 catalytic subunit, have been implicated in trafficking. Inhibition of the p110δ subunit of class I PI3K can impair the trafficking of tumor necrosis factor α (TNFα) from the Golgi by preventing tubule scission in macrophages (Low et al., 2010). Active PI3K converts phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) into PI(3,4,5)P3, creating a docking site for proteins containing the pleckstrin homology (PH) domain to regulate trafficking. The class III PI3K Vps34 is similar to class I PI3K but primarily synthesizes phosphatidylinositol 3-phosphate (PI(3)P) and regulates endosomal recycling (Mazza and Maffucci, 2011; Carpentier et al., 2013). Class II PI3Ks, in contrast, are single-subunit kinases comprising α, β, and γ isoforms and containing a unique C2 domain and a PX domain (Domin et al., 1997; Backer, 2008; Mazza and Maffucci, 2011). These mainly use PI and PI(4)P as substrates to produce PI(3)P and phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2). Class II PI3K α (PI3K C2A) has been shown to localize to the TGN, where it can produce PI(3,4,5)P3 from PI(4,5)P2 and recruit clathrin-coated vesicles via the AP-1 adaptor protein (Domin et al., 2000).
To the best of our knowledge, this study provides the first evidence that PI3K C2A mediates Golgi export and surface delivery of GPCRs in the biosynthetic pathway. This is consistent with our recent data that inhibition of phosphatase and tensin homologue (PTEN), a phosphatase that counters the activity of PI3K C2A, functionally increases the surface numbers of δR in TG neurons and improves the efficacy of δR agonists in a mouse model of chronic pain (Shiwarski et al., 2017). PI3K C2A has been implicated in changing signaling characteristics of receptors, including GPCRs, but its role in trafficking has been studied largely in the context of the endocytic pathway (Biswas et al., 2013; Campa et al., 2015). After clathrin-coated pit formation, PI3K C2A can regulate endocytic dynamics by generating a local population of PI(3,4)P2 and recruiting the BAR domain–containing protein SNX9 (Posor et al., 2013). Consistent with this, depletion of PI3K C2A reduced the amount of VEGFR2 localized to FYVE-containing endosomes (Yoshioka et al., 2012). PI3K C2A can localize to clathrin-coated vesicles with the AP-1 adaptor protein and also directly associate with the AP-2 adaptor protein (Domin et al., 2000; Gaidarov et al., 2001). In addition, the N-terminal region of PI3K C2A can directly bind and recruit clathrin. This interaction activates the enzyme and increases PI(3,4)P2 and PI(3,4,5)P3 generation (Gaidarov et al., 2005; Zhao et al., 2007). Because PI3K C2A localizes primarily to the TGN, our data that PI3K C2A at the TGN is required and sufficient for regulated release of δR might reflect analogous roles for PI3K C2A in endocytosis and TGN export.
This PI3K C2A–mediated pathway likely represents an export pathway for subsets of cargo molecules requiring on-demand release from intracellular storage compartments. The canonical pathway for cargo export from the TGN acts through activation of PI4KIII and PI4KII, which stimulate PI(4)P synthesis in the Golgi and recruits trafficking components like AP1 and GGA to the TGN (Van Lint et al., 2002; Weixel et al., 2005; Di Paolo and De Camilli, 2006; Wang et al., 2007; De Matteis and Luini, 2008; Szentpetery et al., 2010; Balla et al., 2012; Cruz-Garcia et al., 2013; Guo et al., 2013; Bonifacino, 2014). This process is regulated by protein kinase D (PKD) activity at the TGN, which controls synthesis of PI(4)P and PI(4,5)P2 and regulates constitutive VSVG export (Van Lint et al., 2002). Although GGA adapter proteins can mediate Golgi export of some members of the GPCR family (Zhang et al., 2016a, b), it is unlikely that PI3K C2A–mediated export of δR reflects this constitutive export. Neither NGF nor PI3K inhibition caused retention of µR, whereas PI3K C2A inhibition caused retention of δR and impaired GLUT4 translocation and hormone secretion (Meunier et al., 2005; Falasca et al., 2007). Further, using a fluorescent PI(4)P reporter, GFP-PH-FAPP1, we observed no apparent change in PI(4)P levels in the TGN after NGF treatment, and Wtm at 150 nM, which inhibits PI4K (Meyers and Cantley, 1997), does not cause δR retention (unpublished data; Figure 3B). Therefore the PI3K C2A–dependent pathway might act as receptor-selective alternative trafficking route to the more traditional PI(4)P-mediated constitutive export pathway.
Regulated surface delivery of δR was first proposed many years ago, but the exact pathway that δR takes to the surface is still under debate. Unlike the more conventionally surface localized µR, the δR appeared to display cell type–specific localization patterns. Of interest, in dorsal root ganglion neurons and cells overexpressing chromogranin-A and pre–protachykinin-A (PPT-A), δR was immunolocalized to large dense-core vesicles (Zhang et al., 1998; Guan et al., 2005). Because this localization was lost in mice lacking PPT-A, it was suggested that interactions of the lumenal domain of δR with the substance P (SP) domain of protachykinin caused δR retention (Guan et al., 2005). Further, in a knock-in mouse expressing δR that was C-terminally tagged with GFP, the localization of δR in intracellular and surface pools was not dependent on SP (Scherrer et al., 2009). One potential caveat about this mouse, however, is that tagging δR on the C-terminus with enhanced GFP (eGFP) could prevent normal interactions required for regulated trafficking because the C-terminal 27 amino acids of δR mediate intracellular δR retention (Kim and von Zastrow, 2003). Supporting this, in cultured cells, a C-terminally tagged δR showed predominant surface localization compared with an N-terminally tagged δR (Wang et al., 2008).
Several studies attempted to evaluate how extracellular signaling from δR agonists and inflammatory molecules can modulate δR function at the surface. One example is that δR activation itself might increase δR coupling to voltage-dependent calcium channels on the surface via a ROCK-LIMK pathway (Mittal et al., 2013). Similarly, the proinflammatory peptide bradykinin increases surface levels and δR responses, although it is not clear whether this is a result of increased total expression, changes in GRK2 binding, or increased coupling to channels (Patwardhan et al., 2005; Pettinger et al., 2013; Brackley et al., 2016). Our results show a direct role for PI3K C2A in regulating δR export from the TGN and surface delivery, independent of expression and receptor activation. NGF, on acute application, can increase brainstem responses to δR agonists in a PI3K-dependent manner (Bie et al., 2010). In our experiments, we see minimal changes in surface δR levels at early time points after NGF, suggesting that the increased response after acute NGF is likely due to changes in coupling of δR to channels. Of interest, acute NGF has been shown to stimulate PI3K activity at the plasma membrane, whereas persistent NGF can decrease PI3K activity (Chen et al., 2012). Although this is consistent with the retention we observe after long-term NGF, which phenocopies the retention observed with PI3K-C2A inhibition, it is possible that the PI3K C2A–regulated exocytic pathway that we define here plays redundant or overlapping roles with the foregoing mechanisms in modulating δR function on the cell surface.
Independent of the diversity in mechanisms and signals that mediate δR surface delivery, a cargo export checkpoint in the Golgi is intriguing at a fundamental level. The prevalent model for cargo transport through the Golgi postulates that compartments containing cargo mature into the next, and that resident enzymes are constantly retrieved by vesicles that travel backward (Papanikou and Glick, 2014). When receptor cargo reaches the TGN, the expected outcome is that they are packaged into export vesicles, unlike resident proteins, which are retrieved to earlier compartments. The retention we observe likely reflects a switch in the sorting of δR between regulated export and retrieval pathways. A recent study shows that δR has sequence elements on its intracellular loops that can interact with COPI, a main mediator of retrieval, and that these interactions prevent constitutive δR surface expression (St-Louis et al., 2017). Because the C-terminal tail of δR is sufficient for NGF-regulated transport, it is possible that additional biochemical mechanisms exist for signal-regulated δR transport in neurons. Of interest, activity of AKT, the best-known kinase activated downstream of PI3Ks, had no effect on modulating the NGF-regulated retention of δR. This suggests that the switch between regulated export and retrieval pathways for δR is driven by the 3′-phosphorylation of phosphoinositides (PIs) at the TGN. Thus δR TGN export and surface expression could be dynamically tuned by altering the balance of 3′-PI phosphorylation at the TGN via extracellular signaling events such as NGF addition. Through precise activation and localization of phosphoinositide kinases in response to physiological cues and environmental signals, receptor surface expression could be controlled independently of transcription.
Our data support a model in which δR export is mediated by a specialized pathway dependent on PI3K C2A–mediated 3′-phosphorylation of PIs at the TGN, distinct from the previously described PI4K-dependent export pathway. We believe that PI(3,4)P2 is the main PI driving δR export at the TGN, as it can be directly generated from phosphorylation of PI(4)P, the predominant phospholipid species in the TGN. This model is further supported by the lack of involvement of class I PI3K and Akt in δR retention. This provides a potential control point for extracellular signals such as NGF to regulate export of receptors that use this pathway. Of interest, δR retention is normally observed only in neuronal cells, raising the possibility that neuronal differentiation induces a new export pathway that is absent from nondifferentiated cells. Our data indicate that this is unlikely because we can recapitulate the neuronal retention phenotype by inhibiting PI3K C2A in either epithelial HEK 293 cells or neuroendocrine PC12 cells. This suggests that δR export is inherently mediated by a general export pathway, dependent on 3′-phosphorylation of PIs, that is poised for regulation by neuron-specific signals.
At a fundamental level, it is established that the number of surface receptors controls the strength and specificity of signals. There are many ways that a cell can control cell surface receptor expression at the transcriptional level. However, how cells acutely change receptor surface expression is less understood, and research has been mostly limited to post-agonist-stimulated conditions. Studies on the modulation of receptor trafficking and resensitization via biased agonism and heterologous signaling pathways have largely focused on receptor endocytosis and recycling (Hanyaloglu and von Zastrow, 2008; Jean-Alphonse and Hanyaloglu, 2011; Vistein and Puthenveedu, 2013, 2014; Bowman et al., 2015; Grimsey et al., 2015). Here we investigated a mechanism of cell surface receptor regulation, independent of agonists, that controls the delivery of newly synthesized δR. For GPCRs such as δR that are predominantly degraded after agonist-mediated internalization, resensitization to δR agonists depends on new receptor synthesis and delivery to the surface. We believe that for cells such as neurons, the presence of a δR storage pool in the TGN might represent ready and waiting receptors that can respond to physiological changes in the extracellular environment through modulation of a Golgi trafficking checkpoint. Therefore manipulation of the PI3K C2A–dependent regulatory pathway controlling signal-mediated GPCR export from the TGN highlights a novel mechanism to drive receptor surface and resensitization of a nonrecycling receptor, similar to the more traditional regulatory mechanisms elucidated for many recycling GPCRs (Hanyaloglu and Zastrow, 2008).
MATERIALS AND METHODS
Cell lines and cell culture
The cell lines used for experimentation were PC12 (CRL-1721), HEK 293 (CRL-1573), and HEK 293T (CRL-11268) cells. The PC12 cell line is a neuroendocrine cell line isolated from rat adrenal medulla tissue. These cells were grown at 37°C with 5% CO2 and cultured in F12K medium (21127-022; Life Technologies) supplemented with 10% horse serum and 5% fetal bovine serum (FBS). This medium was changed every 3 d to maintain proper pH and nutrient supply. Plastic culture vessels were coated with collagen IV (C5533-5MG; Sigma-Aldrich) to allow PC12 cells to adhere. The standard passage ratio for PC12 cells was 1:4 to ensure that sufficient cell–cell contacts were made, facilitating growth. HEK 293 cells were cultured in DMEM supplemented with 10% FBS at 37°C with 5% CO2. Standard passage ratio was 1:10. The HEK 293T cell line is a derivative of HEK 293 cells that contains the SV40 T-antigen to increase replication efficiency of vectors that have an SV40 replication origin. These cells were specifically used for virus production and maintained in a similar manner to the HEK 293 cells.
Primary TG neuron isolation
Dissection and dissociation largely followed publish protocol (Malin et al., 2007). Before the day of dissection, glass coverslips (Corning) were precoated with poly-d-lysine and laminin to enhance cell adhesion and promote cell survival. Mice (p9-15) were obtained from the institutional animal facility and killed via CO2 asphyxiation. The mouse’s head was removed with large scissors. The skin covering the skull was then removed to make cutting away the top of the skull and accessing the brain easier. Once the skull was removed and the brain was exposed, the brain was removed by lifting toward the posterior with forceps. With the brain removed, the TGs were apparent as longitudinal bundles of tissue left behind in the skull. The TGs were cut into small pieces and placed into ice-cold Lebovitz medium. The Lebovitz medium was then removed, replaced with a sterile filtered papain solution, and incubated for 15 min at 37°C. Next the papain solution was aspirated, and a sterile filtered collagenase solution was applied for 15–20 min at 37°C. After completion of digestion and removal of the collagenase solution, 2 ml of Neurobasal A medium (Invitrogen) was added to the TG tissue. The TGs were then triturated with a fire-polished Pasteur pipette until the neurons appeared visually dissociated. The TG cells were then plated onto the poly-d-lysine– and laminin-coated coverslips within a cloning ring to create a highly dense cell population. After 5 h, the cloning ring was removed and fresh Neurobasal A medium added to each well. All animals were housed in Association for Assessment and Accreditation of Laboratory Animal Care–approved facilities, and studies used protocols that received institutional review and approval from the Institutional Animal Care and Use Committees at the respective institutions.
DNA transfection of cultured cells
PC12 cells were plated in collagen IV–coated, six-well plates and grown in F12K medium containing 10% horse serum and 5% FBS for 24 h before transfection. Lipofectamine 2000 lipofection reagent (11668-019; Invitrogen) was used to transiently transfect PC12 cells with the desired plasmid constructs, following the manufacturer’s recommendations. Briefly, Opti-MEM (100 μl) was added to two 1.7-ml microcentrifuge tubes. Lipofectamine 2000 (7.5 μl) and the appropriate plasmid DNA (1.5 μg) were added to the Opti-MEM-containing 1.7-ml microcentrifuge tubes and incubated at room temperature for 5 min. The entire DNA–Opti-MEM mixture was then added to the microcentrifuge tube containing Opti-MEM and Lipofectamine 2000, mixed well, and incubated for 20 min at room temperature. The growth medium was removed from the cells to be transfected, and 1 ml of Opti-MEM was added to each well. After incubation, the entire transfection mixture was added dropwise to one well. Cells were then incubated with the transfection mixture at 37°C for 5 h. Subsequently the transfection solution was aspirated from the wells and replaced with 2 ml of F12K medium containing 10% horse serum and 5% FBS. Experiments were conducted 48–72 h after transfection. Similar protocols were used for HEK 293 cells and TG neurons, substituting appropriate cell-specific media.
Fixed-cell immunofluorescence
HEK 293 and PC12 cells were transfected with either FLAG-δR or FLAG-μR, and stable cells were obtained via antibiotic selection. Stably expressing cells were plated on coverslips (Corning) coated with poly-d-lysine (P7280; Sigma-Aldrich) and grown at 37°C for 24–48 h. After treatment, cells were fixed in 4% paraformaldehyde, pH 7.4. The cells were blocked with phosphate-buffered saline (PBS) containing calcium and magnesium, with 5% FBS, 5% 1 M glycine, and 0.75% Triton X-100. The FLAG-tagged receptors, Golgi, and TGN were labeled for 1 h in blocking buffer with anti–FLAG M1 antibody (1:1000; F3040, Sigma-Aldrich) conjugated with Alexa 647 (A20186; Molecular Probes), anti-GPP130 (a gift from Adam Linstedt, Carnegie Mellon University, Pittsburgh, PA) and anti–TGN-38 rabbit polyclonal antibody (1:2000; T9826; Sigma-Aldrich), respectively. Coverslips were washed three times in calcium/magnesium PBS and then labeled with Alexa 568 goat anti-rabbit secondary antibody (1:1000; A11011) in blocking buffer for 1 h. Coverslips were washed three more times in calcium/magnesium PBS and mounted onto glass slides using Prolong Diamond Reagent (P36962; Molecular Probes). Confocal imaging of the mounted cells was performed using a confocal imaging system (XDi spinning disk; Andor) at 60× magnification (Nikon CFI APO TIRF) on a Nikon TE-2000 inverted microscope. In addition, the imaging setup contained temperature- and humidity-controlled imaging capabilities, a mechanical Piezo XYZ-stage (Nikon), iXon 897 Ultra backilluminated camera (Andor), a laser combiner (Andor) containing 405-, 488-, 515-, 568-, and 647-nm excitation capabilities, a Dell 5400 Workstation optimized for IQ2 imaging software (Andor), and an active isolation air table (TMC). Images of representative fields were taken. Quantification of the fluorescence ratio was taken for a minimum of 50 cells and averaged to ensure that results were representative of the population. A minimum of three biological replicates were performed to confirm the results.
Fixed-cell image quantification
To determine the percentage of FLAG-δR within the Golgi compared with the total cell fluorescence, we obtained confocal images of fluorescently labeled δR and the Golgi marker (TGN-38) on the same confocal imaging system previously described. After image acquisition, the background fluorescence was subtracted from the Golgi channel. The Golgi channel was thresholded to construct a mask of the Golgi region to use as a pass-through filter by converting the Golgi mask to a binary image and multiplying it by the δR image. This allowed for the measurement of the amount of δR fluorescence within the Golgi region. The fluorescence signals from δR within the Golgi and total δR images were thresholded to remove averaging bias from background zeroed pixels. Regions of interest were drawn for the Golgi region and the total δR signal in ImageJ to calculate the total and average fluorescence within these two regions. The fluorescence signal for δR within the Golgi was then divided by the total δR fluorescence signal to get a ratio of the percentage of δR within the Golgi compared with the total expression of δR. An identical procedure was followed to determine the percentage of FLAG-μR within the Golgi compared with the total cell fluorescence. To quantitate the percentage of cells with intracellular δR or μR, we assigned a binary value to cells visibly displaying intracellular receptors (1) and cells without an intracellular pool (0) and took the percentage of the total population exhibiting a clearly visible intracellular pool.
Live-cell imaging of δr retention
To visualize δR in live cells, eGFP was N-terminally tagged to δR in place of the FLAG-tag previously described. PC12 cells were transfected with eGFP-δR using Lipofectamine 2000 as described previously. Before imaging, cells were transferred to poly-d-lysine–coated coverslips. Cells were imaged at 60× magnification using a Nikon TE-2000 inverted microscope with a 37°C heated enclosure and 5% CO2 environment in Leibovitz’s L-15 medium plus 1% FBS. Images were acquired every minute for 1.5–2 h. After 5 min of baseline imaging, NGF (100 ng/ml) was added to the coverslip. The images were analyzed and quantified using ImageJ analysis software. Identical protocol was followed for Wtm (10 μM)–induced retention.
Cycloheximide assay
PC12 FLAG-δR cells were plated onto poly-d-lysine–coated coverslips in a 24-well plate and incubated overnight at 37°C. NGF (100 ng/ml) was added to all appropriate samples for 2 h to achieve complete intracellular retention of δR. The medium was changed to Opti-MEM plus GlutaMAX (Invitrogen) with cycloheximide (3 μg/ml) for an additional 1 h with NGF with or without various pharmacological inhibitors and activators. The cells were then fixed in 4% paraformaldehyde (PFA) and blocked in the immunofluorescence-blocking buffer. Immunofluorescence of FLAG-δR and intracellular compartmental markers were performed as described previously.
δR internalization surface control assay
FLAG-δR–expressing PC12 cells were incubated with Alexa 647 M1 antibody for 15 min at room temperature (1:500). The antibody was removed, and the cells were washed with Ca2+/Mg2+ PBS to remove residual antibody. Pharmacological treatments were added for 1 h at concentrations used during the pharmacological experiments (see Table 1). After 1 h, a secondary Alexa 568 antibody to the Flag-M1 was applied at 4°C for 15 min (1:1000) to reduce the amount of potential internalization from that point on. Cells were washed twice with Ca2+/Mg2+ PBS and fixed in 4% PFA pH 7.4 for 30 min. Cells were again washed three times with Ca2+/Mg2+PBS and mounted to coverslips for imaging. Confocal stacks were taken of the cells for no treatment or treatment with NGF (100 ng/ml), Wtm (10 μM), LY (10 μM, or DADLE (10 μM).
Table 1:
Pharmacological compounds used throughout the article.
| Compound name | Protein target | Concentration | Supplier | Catalog number |
|---|---|---|---|---|
| Nerve growth factor | TrkA | 100 ng/µl | BD Biosciences | 356004 |
| 740YPDGFR | PI3K activator | 50 µg/ml | Tocris Bioscience | 1983 |
| LY294002 | PI3K inhibitor | 10 µM | Tocris Bioscience | 1130 |
| Wortmannin | PI3K inhibitor | 10 µM | Enzo Life Sciences | BML-ST415 |
| PI-103 | PI3K C1 α inhibitor | 50 nM | Echelon Biosciences | B0303 |
| IC87114 | PI3K C1 δ inhibitor | 5 µM | Echelon Biosciences | B0305 |
| AS605240 | PI3K C1 γ | 25 nM | Echelon Biosciences | B0301 |
| Y-27632 | ROCK inhibitor | 5 μM | Cayman Chemical | 10005583 |
| Akt1/2 kinase IH | Akt 1/2 inhibitor | 500 nM | Sigma-Aldrich | A6730-5MG |
| U0126 | MEK inhibitor | 10 μM | Cayman Chemical | 70970 |
| U73122 | PLC inhibitor | 10 μM | Enzo Life Sciences | BML-ST391 |
| Chelerythrine | PKC inhibitor | 10 μM | Sigma-Aldrich | 0C020953 2-1 MG |
| PP2 | cSrc inhibitor | 500 nM | EMD Millipore | 529573-1 MG |
| DADLE | δR-agonist | 10 μM | Tocris | 3790 |
VSVG trafficking assay
HEK 293 cells were transfected with either VSVG-GFP or VSVG-GFP-δRtail construct, grown under antibiotic selection, and maintained at the restrictive temperature of 40°C in standard culture medium. Cells were plated on 25-mm coverslips and imaged live via confocal fluorescence microscopy using a Nikon TE-2000 inverted microscope as described previously. To initiate ER exit and trafficking of the VSVG proteins, live-cell imaging was performed at 32°C, and confocal images were acquired every 10 s for 2 h. Cells were treated with vehicle controls or with the PI3 kinase inhibitor Wtm or LY with (+) or without (–) the PI3K-activating peptide 740YPDGFR. The images were compiled to form a time-lapse movie showing the cellular trafficking of the VSVG-GFP– or VSVG-GFP-δRtail–expressing cells. GFP fluorescence intensity was quantified using ImageJ software.
Inhibitors and activators
For most pharmacological experiments, the treatments were conducted for 1 h, followed by fixation and confocal immunofluorescence microscopy as detailed in individual methods. The concentrations and usage specifications are listed in Table 1.
Lentiviral shRNA knockdown of PI3K C2A
Knockdown of PI3K C2A was performed using shRNA constructs targeting the rat Pik3c2a. The lentiviral vectors pLenti X1 Puro DEST (694-6) and pENTR/pSM2(CMV) GFP (w513-1) were gifts from Eric Campeau (Addgene plasmids19170 and 17297; Campeau et al., 2009), and the shRNA sequences were designed by following an established protocol from the Hannon lab and entering the target sequence into the appropriate site (http://cancan.cshl.edu/RNAi_central/RNAi.cgi?type=shRNA; Silva et al., 2005). The site outputs a 22-mer sequence, as well as a 97–base pair sequence. Restriction sites are added such that the sequences can be inserted into the pSM2 plasmid: an XhoI site at the 5′ end and an EcoR1 site at the 3′ end. After addition of the restriction sites, the sequences were ordered as adapter pairs for cost efficiency. These sequences are given in Supplemental Table S1. Once the sequences were obtained, a double digest of the pSM2 entry vector was performed with EcoR1 (New England Biolabs) and Xho1 (New England Biolabs). The digested product was run on a 1% agarose gel and purified using a Qiagen Gel Extraction Kit (28704; Qiagen). The adapters (purchased from IDT) were annealed and then ligated into the linearized entry vector, pENTR/pSM2(CMV) GFP (w513-1). The ligated constructs were transformed into DH5α cells, and multiple colonies from each transformation were chosen to screen for the correct insert. Correct insertion was confirmed via restriction digest and DNA sequencing. Transient transfection of PC12 cells using Lipofectamine 2000 followed by Western blotting was used to assess the knockdown efficiency of the shRNA constructs. The shRNA construct that produced the most efficient knockdown, shRNA 3 in Figure 3A, was inserted into the pLenti-X1 destination vector via the Gateway Cloning Method using the LR reaction (11791-019; Life Technologies) to produce the pLenti-X1-puro-PI3K C2A shRNA construct.
Lentiviral particles were produced using this construct to stably infect and knock down PI3K C2A in PC12 cells. HEK 293T cells were transfected with 15 μg of pLenti-X1-puro-PI3K C2A shRNA, 15 μg of pMDLg/pRRE (Addgene plasmid 12251), 6 μg of pRSV-Rev (Addgene plasmid 12253), and 3 μg of pMD2.G (Addgene plasmid 12259) as per Campeau et al. (2009). These plasmids were a gift from Didier Trono (EPFL, Lausanne, Switzerland). The medium was changed 24 h posttransfection. A GFP marker in the expression construct was used to determine transfection efficiency. Exactly 64 h posttransfection, the virus-containing medium was removed and centrifuged at 3000 × g to pellet nonadherent cells and debris. The virus-containing medium was further sterile filtered through a 0.45-μm sterile filter to remove any remaining cells debris. To achieve maximum infection efficiency, 1.5 ml of the filtered virus solution was added to each well of a six-well plate containing PC12 cells at 50% confluency. GFP expression was used to assess proper functional viral production and infection efficiency. Approximately 48 h postinfection, puromycin (A11138-03; Life Technologies) selection was added to the growth medium at 1 μg/ml. Stable lines expressing the PI3K C2A shRNA were obtained within 1 wk from initial infection, and knockdown was confirmed via immunoblotting.
Immunoblotting and densitometry
The knockdown of PI3K C2A was assessed by immunoblotting. The six-well plates were placed on ice, growth medium was removed, and each well was rinsed with 1 ml of PBS. A 500-μl amount of PBS was added, and cells were scrape collected and transferred to 1.7-ml tubes. Cells were pelleted at 1200 × g for 5 min. Supernatant was aspirated, and the pellet was resuspended in 30–75 μl of lysis buffer containing 2% SDS and 60 mM Tris-HCl, pH 6.8, with Complete Mini EDTA-free (T00004; Roche) and PhosStop Tab (A00173; Roche). Cells were lysed by vortexing for 15 s, followed by 2 min in a 95°C heat block and then pipetting up and down three to five times. This lysis process was performed twice. Bicinchoninic acid protein estimation (Pierce BCA Assay KIT, 23225; Thermo Fisher) was performed. For each condition, 40-μl samples were prepared from the lysates such that protein content and volume were standardized. A 10-μl amount of 4× loading dye and fresh β-mercaptoethanol and 1 μl of 1 M dithiothreitol were added to the samples. To denature the proteins, samples were heated to 95°C for 5 min and ran on a 4–15% Mini-PROTEAN TGX Stain-Free Protein Gel (4568083; Bio-Rad). An overnight transfer at 4°C to a nitrocellulose membrane was performed. After transfer, the blot was blocked with 5% Tris-buffered saline/Tween 20 (TBST) for 1 h on a shaker. Primary antibody for PI3K C2A (ab154583; Abcam) was prepared at 1:1000 in 5% milk-TBST and added to the blot. The blot was incubated at room temperature on a shaker for 1 h or overnight in cold room. The primary antibody solution was removed, and the blot was washed three times for 5 min with 5% milk-TBST. The secondary antibody goat anti-rabbit (1706515; Bio-Rad) was prepared at 1:3000 in 5% milk-TBST and added to the blot, which was incubated on a shaker at room temperature for 1 h. The secondary antibody solution was removed, and the blot was washed twice for 5 min with TBST and once for 5 min with TBS. The blot was developed with Clarity Western ECL Substrate (1705061; Bio-Rad) and imaged using the ChemiDoc Touch imaging system (Bio-Rad). Densitometry was performed using the built-in ImageJ plug-in to quantify the band intensity in each lane. Values were then normalized and plotted.
Live-cell imaging with a fluorogen-activated peptide
PC12 cells and PC12 cells expressing PI3K C2A shRNA were plated in a six-well plate. Once confluency reached 50%, these cells were transfected with an N-terminally tagged FAP-δR construct using Lipofectamine 2000. At 2 d posttransfection, cells were plated on 25-mm poly-d-lysine–coated coverslips (Micro Coverglass #1½, 72225-01; Electron Microscopy Sciences). The FAP-δR has a far-red emission spectrum, resulting from an MG-based fluorogen (Szent-Gyorgyi et al., 2008; Pratt et al., 2015), which can be visualized via fluorescent microscopy. At 15 min before imaging, the cell-permeable MG ester–based fluorogen (100 nM) was added to the cells. Live confocal imaging was performed at 37°C using a Nikon TE-2000 inverted microscope. FAP-δR localization was quantified using ImageJ software to determine the ratio of internal δR fluorescence to the total fluorescence signal. An identical protocol was followed for studies involving the overexpression of GFP-tagged PI3K C2A. The GFP-PI3K C2A was transfected along with the FAP-δR and imaged in a similar manner (PI3K C2A cDNA was a gift from Jim Keen, Thomas Jefferson University, Philadelphia, PA).
Live-cell imaging of EPAC cAMP biosensor
To quantify the effects of PI3K C2A knockdown on changes in cAMP and visualize δR agonist-mediated internalization, PC12 cells expressing FAP-δR were Lipofectamine 2000 transfected with 1 µg of EPAC cAMP FRET sensor and 125 µM control nontargeting siRNA or PI3K C2A siRNA (Silencer Select Pre-designed, 4390771, GGAGAUAGCAAACUCGAAAtt; Life Technologies) following the manufacturer’s recommended protocol. Experiments followed a validated paradigm for determining GPCR inhibition of cAMP (Shiwarski et al., 2017). Baseline cAMP was stimulated with forskolin (5 µM), and subsequent inhibition after δR agonist addition (10 µM DADLE) was calculated via the EPAC CFP/FRET ratio. At 15 min before imaging, the cell-permeable MG ester–based fluorogen (100 nM) was added to the cells to visualize FAP-δR localization and trafficking. Live wide-field fluorescence imaging was performed at 37°C using a Nikon TE-2000 inverted microscope. The images were analyzed and quantified using custom ImageJ macro analysis software.
TG microinjection and deltorphin II labeling
For microinjection of GFP-PI3K C2A DNA into the cultured TG neurons, we followed previously developed methods from our lab (Puthenveedu and Linstedt, 2001). Briefly, borosilicate with filament (outer diameter, 1.2 mm; inner diameter, 0.94 mm; length; 10 cm; BF120-94-10) micropipettes were pulled using a Sutter Instrument P-87 with a modified program based on the initial RAMP test (Heat = 768, Pull = 70, Velocity = 50, Time = 250, Pressure = 450). GFP-PI3K C2A (2 µg) was diluted into KH buffer (20 µl) containing 125 mM potassium acetate and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid. The diluted GFP-PI3K C2A DNA was backloaded into the micropipettes. Neurons were injected at room temperature using an Eppendorf FemtoJet (pi = 35 hPa, ti(s) = 0.2, pc(hPa) = 20) attached to a Zeiss inverted microscope and then returned to the incubator.
Deltorphin II–Cy3 (100 nM) was added to the cultured TGs expressing GFP-PI3K C2A. Hoechst DNA stain was added to the coverslips to visualize non-GFP-PI3K C2A–expressing neurons. Imaging was performed live at 37°C via confocal fluorescence microscopy 48–72 h after microinjection as previously described. Images were acquired for baseline GFP-PI3K C2A expression and background deltorphin II–Cy3 signal and after 5 min of incubation with the deltorphin II–Cy3. Images were analyzed for GFP-PI3K C2A expression and deltorphin II-Cy3 fluorescence. Comparative analysis was performed in ImageJ to determine the change in fluorescence after deltorphin II–Cy3 addition between GFP-PI3K C2A–expressing and –nonexpressing neurons (Shiwarski et al., 2017).
Optogenetic recruitment of PI3K C2A to the TGN and release of δR
The CRY2-CIBN optical recruitment system was adapted by cloning the PI3K C2A kinase domain (amino acids 863–1405, KD) onto the C-terminus of an mCherry-CRY2 chimeric construct (a gift from Pietro De Camilli, Yale University, New Haven, CT) to make mCherry-CRY2-PI3K-C2A-KD (Idevall-Hagren et al., 2012). GFP-CIBN, a gift from Chandra Tucker (University of Colorado School of Medicine, Aurora, CO; Addgene plasmid 26867), was adapted for TGN-specific recruitment by cloning the N-term of 2,6-sialyltransferase onto the C-terminus of GFP-CIBN to make GFP-CIBN-SialyT (Kennedy et al., 2010). PC12 cells were transiently transfected with FAP-δR (1 µg), mCherry-CRY2-PI3K-C2A-KD (C2A KD, 2.5 µg), GFP-CIBN-SialyT (CIBN-TGN, 1 µg), and the TGN-localized PI(4)P sensor PH-FAPP1-iRFP (PH-FAPP1; PH-FAPP1-GFP was a gift from Tamas Balla, National Institutes of Health, Bethesda, MD; 0.5 µg) for live imaging of the optogenetic recruitment (Balla and Várnai, 2002). The GFP-CIBN-SialyT has a basal TGN localization, and mCherry-CRY2-PI3K-C2A-KD is cytoplasmic before recruitment. Optorecruitment is activated by blue light stimulation at 488 nm driving the mCherry-CRY2-PI3K-C2A-KD to the TGN.
For experiments involving the release of Golgi-retained δR followed by C2A KD recruitment to the TGN, PC12 cells expressing the optogenetic recruitment system were plated onto 25-mm poly-d-lysine–coated coverslips and grown for 48 h. On the day of the experiment, the coverslips were transferred to imaging chambers and treated with NGF (100 ng/ml) for 1 h. The chambers were then moved to the microscope for live-cell confocal imaging performed at 37°C using a Nikon TE-2000 inverted microscope. FAP-δR was labeled with the MG-ester dye (100 nM), and baseline images for δR (640-nm excitation, 700/75-nm emission) and C2A-KD (561-nm excitation, 620/60-nm emission) expression and localization were acquired three times at 5-s intervals. Initial live recruitment of C2A-KD to the CIBN-TGN was performed by dual excitation of 561 nm (35% power) and 488 nm (10% power) for 1 min with a 620/60-nm emission filter to visualize in real time the recruitment of the C2A-KD. To maintain C2A-KD recruitment and visualize δR trafficking over time, subsequent images were acquired for CIBN-TGN (488-nm excitation, 525/50-nm emission), C2A-KD (561-nm excitation, 620/60-nm emission), and FAP-δR (640-nm excitation, 700/75-nm emission) every 30 s for 15 min. Quantification of the live-imaging recruitment movies was performed using ImageJ software. Fluorescence intensity of TGN-localized δR was compared with the total fluorescence for each cell and expressed as a ratio. When δR is released from the TGN, the signal within the TGN decreases as the surface signal increases. Thus the ratio of TGN to total fluorescence decreases as δR is released from the Golgi.
Line-scan analysis
A line-scan analysis was used to confirm recruitment of the C2A-KD to the CIBN-TGN and the PH-FAPP1. Using ImageJ software, the fluorescence intensity was measured over a line that passed through the region of CIBN-TGN fluorescence, and the relative spatiotemporal fluorescence intensity over a pixel area was compared for each of the three channels for C2A-KD, CIBN-TGN, and PH-FAPP1. Colocalization and recruitment was confirmed by visualizing overlapping peaks of fluorescence along the length of this line normalized to the maximal fluorescence intensity in each channel over the line area. During recruitment, the C2A-KD peak increases and localizes to the peaks defined by CIBN-TGN and PH-FAPP1. The data were plotted using GraphPad Prism software.
Image analysis and quantification
All imaging data were quantified using ImageJ, which was used to measure the total δR fluorescence for each cell and the fluorescence intensity of δR within the Golgi for fixed- and live-cell analysis. The values measured using ImageJ were transferred to Microsoft Excel to calculate the ratio of the Golgi fluorescence to the total cell fluorescence. This ratio was calculated for each cell, and all cell ratios were averaged. The average represents the fraction of δR present in the Golgi compared with the total cell fluorescence. In addition, a manual binary quantification was performed to determine the percentage of cells that visually displayed Golgi-localized δR. Based on visual inspection, each cell that had at least half of its δR present in the Golgi was scored as a 1, and cells with little or no δR in the Golgi was scored as a 0. The average of the binary quantification was used to determine percentage of cells with Golgi-localized δR.
Statistics and data analysis
Statistical analyses were performed using GraphPad Prism 5 software, and the appropriate statistical tests were chosen based on sample size, distribution, and experimental conditions. A minimum of three independent experiments were performed for each set of data. For statistical analysis of the fixed- and live-cell immunofluorescence imaging data, two-tailed unpaired t tests were performed between the different experimental conditions and controls. p < 0.05 was considered statistically significant. For experiments in which multiple comparisons were made, one-way analysis of variance (ANOVA) was performed, followed by Dunn’s multiple comparison test in GraphPad Prism 5. In addition, GraphPad Prism 5 software was used to measure area under the curve for the VSVG assay data. All images were quantified using macros constructed from the functions within the ImageJ software package. Figures were constructed with ImageJ and Adobe Photoshop CS6.
Supplementary Material
Acknowledgments
We thank Stephanie Crilly, Zara Weinberg, Elena Shuvaeva, Shanna Bowman, and Cary Shiwarski for technical help and comments. We thank Nathan Urban, Adam Linstedt, Tina Lee, Peter Friedman, Guillermo Romero, Jean-Pierre Vilardaga, and Alessandro Bisello for reagents, comments, and suggestions. We also thank Mark von Zastrow, Jim Keen, and Pietro De Camilli for helpful discussions and reagents. M.P.B. and C.A.T. were supported by National Institutes of Health grant U54GM103529. M.A.P. was supported by National Institutes of Health Grants DA024698 and DA036086.
Abbreviations used:
- AUC
area under the curve
- cAMP
cyclic-AMP
- CHX
cycloheximide
- DADLE
[d-Ala2, d-Leu5]-enkephalin
- EPAC
exchange protein directly activated by cAMP
- ER
endoplasmic reticulum
- FAP
fluorogen-activated peptide
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- GPCR
G protein–coupled receptor
- HEK 293
embryonic kidney 293 cells
- LY
LY294002
- MG
malachite green
- µR
mu opioid receptor
- NGF
nerve growth factor
- PC12
pheochromocytoma-12 cells
- PI(3,4)P2
phosphatidylinositol (3,4)-bisphosphate
- PI(3,4,5)P3
phosphatidylinositol (3,4,5)-trisphosphate
- PI3K
phosphoinositide-3 kinase
- PI3K C2A
class II phosphoinositide-3 kinase α
- PI3K C2A KD
PI3K C2A kinase domain
- PKC
protein kinase C
- PKD
protein kinase D
- PLC
phospholipase C
- PPT-A
pre–protachykinin-A
- PTEN
phosphatase and tensin homologue
- δR
delta opioid receptor
- ROCK
Rho-associated protein kinase
- shRNAs
short hairpin RNAs
- SP
substance P
- TG neurons
trigeminal ganglion neurons
- TGN
trans-Golgi network
- VSVG
vesicular stomatitis virus glycoprotein
- Wtm
wortmannin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0030) on May 31, 2017.
REFERENCES
- Arttamangkul S, Quillinan N, Low MJ, von Zastrow M, Pintar J, Williams JT. Differential activation and trafficking of micro-opioid receptors in brain slices. Mol Pharmacol. 2008;74:972–979. doi: 10.1124/mol.108.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Balla T, Várnai P. Visualizing cellular phosphoinositide pools with GFP-fused protein-modules. Sci STKE. 2002;2002:l3. doi: 10.1126/stke.2002.125.pl3. [DOI] [PubMed] [Google Scholar]
- Balla T, Wymann M, York JD. Dordrecht, Netherlands: Springer; 2012. Phosphoinositides I: Enzymes of Synthesis and Degradation. [Google Scholar]
- Bao L, Jin SX, Zhang C, Wang LH, Xu ZZ, Zhang FX, Wang LC, Ning FS, Cai HJ, Guan JS, et al. Activation of delta opioid receptors induces receptor insertion and neuropeptide secretion. Neuron. 2003;37:121–133. doi: 10.1016/s0896-6273(02)01103-0. [DOI] [PubMed] [Google Scholar]
- Bie B, Zhang Z, Cai Y-Q, Zhu W, Zhang Y, Dai J, Lowenstein CJ, Weinman EJ, Pan ZZ. Nerve growth factor-regulated emergence of functional delta-opioid receptors. J Neurosci. 2010;30:5617–5628. doi: 10.1523/JNEUROSCI.5296-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Yoshioka K, Asanuma K, Okamoto Y, Takuwa N, Sasaki T, Takuwa Y. Essential role of class II phosphatidylinositol-3-kinase-C2α in sphingosine 1-phosphate receptor-1-mediated signaling and migration in endothelial cells. J Biol Chem. 2013;288:2325–2339. doi: 10.1074/jbc.M112.409656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS. Adaptor proteins involved in polarized sorting. J Cell Biol. 2014;204:7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SL, Puthenveedu MA. Postendocytic sorting of adrenergic and opioid receptors: new mechanisms and functions. Prog Mol Biol Transl Sci. 2015;132:189–206. doi: 10.1016/bs.pmbts.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SL, Soohoo AL, Shiwarski DJ, Schulz S, Pradhan AA, Puthenveedu MA. Cell-autonomous regulation of Mu-opioid receptor recycling by substance P. Cell Rep. 2015;10:1925–1936. doi: 10.1016/j.celrep.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackley AD, Gomez R, Akopian AN, Henry MA, Jeske NA. GRK2 constitutively governs peripheral delta opioid receptor activity. Cell Rep. 2016;16:2686–2698. doi: 10.1016/j.celrep.2016.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Ma J-T, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill CM, Holdridge SV, Morinville A. Trafficking of delta-opioid receptors and other G-protein-coupled receptors: implications for pain and analgesia. Trends Pharmacol Sci. 2007;28:23–31. doi: 10.1016/j.tips.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Cahill CM, McClellan KA, Morinville A, Hoffert C, Hubatsch D, O’Donnell D, Beaudet A. Immunohistochemical distribution of delta opioid receptors in the rat central nervous system: evidence for somatodendritic labeling and antigen-specific cellular compartmentalization. J Comp Neurol. 2001a;440:65–84. doi: 10.1002/cne.1370. [DOI] [PubMed] [Google Scholar]
- Cahill CM, Morinville A, Lee MC, Vincent JP, Collier B, Beaudet A. Prolonged morphine treatment targets delta opioid receptors to neuronal plasma membranes and enhances delta-mediated antinociception. J Neurosci. 2001b;21:7598–7607. doi: 10.1523/JNEUROSCI.21-19-07598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa CC, Franco I, Hirsch E. PI3K-C2α: one enzyme for two products coupling vesicle trafficking and signal transduction. FEBS Lett. 2015;589:1552–1558. doi: 10.1016/j.febslet.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Rückle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Françon B, et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- Carpentier S, N’Kuli F, Grieco G, Van Der Smissen P, Janssens V, Emonard H, Bilanges B, Vanhaesebroeck B, Gaide Chevronnay HP, Pierreux CE, et al. Class III phosphoinositide 3-kinase/VPS34 and dynamin are critical for apical endocytic recycling. Traffic. 2013;14:933–948. doi: 10.1111/tra.12079. [DOI] [PubMed] [Google Scholar]
- Chen J-Y, Lin J-R, Cimprich KA, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol Cell. 2012;45:196–209. doi: 10.1016/j.molcel.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia D, Ortega-Bellido M, Scarpa M, Villeneuve J, Jovic M, Porzner M, Balla T, Seufferlein T, Malhotra V. Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. 2013;32:1717–1729. doi: 10.1038/emboj.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Derossi DD, Williams EJE, Green PJP, Dunican DJD, Doherty PP. Stimulation of mitogenesis by a cell-permeable PI 3-Kinase binding peptide. Biochem Biophys Res Commun. 1998;251:148–152. doi: 10.1006/bbrc.1998.9444. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci USA. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domin J, Gaidarov I, Smith ME, Keen JH, Waterfield MD. The class II phosphoinositide 3-kinase PI3K-C2alpha is concentrated in the trans-Golgi network and present in clathrin-coated vesicles. J Biol Chem. 2000;275:11943–11950. doi: 10.1074/jbc.275.16.11943. [DOI] [PubMed] [Google Scholar]
- Domin J, Pages F, Volinia S, Rittenhouse SE, Zvelebil MJ, Stein RC, Waterfield MD. Cloning of a human phosphoinositide 3-kinase with a C2 domain that displays reduced sensitivity to the inhibitor wortmannin. Biochem J. 1997;326:139–147. doi: 10.1042/bj3260139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca M, Hughes WE, Dominguez V, Sala G, Fostira F, Fang MQ, Cazzolli R, Shepherd PR, James DE, Maffucci T. The role of phosphoinositide 3-kinase C2alpha in insulin signaling. J Biol Chem. 2007;282:28226–28236. doi: 10.1074/jbc.M704357200. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Smith ME, Domin J, Keen JH. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol Cell. 2001;7:443–449. doi: 10.1016/s1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Zhao Y, Keen JH. Individual phosphoinositide 3-kinase C2alpha domain activities independently regulate clathrin function. J Biol Chem. 2005;280:40766–40772. doi: 10.1074/jbc.M507731200. [DOI] [PubMed] [Google Scholar]
- Gendron L, Cahill CM, von Zastrow M, Schiller PW, Piñeyro G. Molecular pharmacology of δ-opioid receptors. Pharmacol Rev. 2016;68:631–700. doi: 10.1124/pr.114.008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron L, Mittal N, Beaudry H, Walwyn W. Recent advances on the δ opioid receptor: from trafficking to function. Br J Pharmacol. 2015;172:403–419. doi: 10.1111/bph.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsey NJ, Aguilar B, Smith TH, Le P, Soohoo AL, Puthenveedu MA, Nizet V, Trejo J. Ubiquitin plays an atypical role in GPCR-induced p38 MAP kinase activation on endosomes. J Cell Biol. 2015;210:1117–1131. doi: 10.1083/jcb.201504007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Xu ZZ, Gao H, He SQ, Ma GQ, Sun T, Wang LH, Zhang ZN, Lena I, Kitchen I, et al. Interaction with vesicle luminal protachykinin regulates surface expression of delta-opioid receptors and opioid analgesia. Cell. 2005;122:619–631. doi: 10.1016/j.cell.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Guan XM, Kobilka TS, Kobilka BK. Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J Biol Chem. 1992;267:21995–21998. [PubMed] [Google Scholar]
- Guo Y, Zanetti G, Schekman R. A novel GTP-binding protein-adaptor protein complex responsible for export of Vangl2 from the trans Golgi network. Elife. 2013;2:e00160. doi: 10.7554/eLife.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Alphonse F, Hanyaloglu AC. Regulation of GPCR signal networks via membrane trafficking. Mol Cell Endocrinol. 2011;331:205–214. doi: 10.1016/j.mce.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-A, von Zastrow M. Neurotrophin-regulated sorting of opioid receptors in the biosynthetic pathway of neurosecretory cells. J Neurosci. 2003;23:2075–2085. doi: 10.1523/JNEUROSCI.23-06-02075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low PC, Misaki R, Schroder K, Stanley AC, Sweet MJ, Teasdale RD, Vanhaesebroeck B, Meunier FA, Taguchi T, Stow JL. Phosphoinositide 3-kinase δ regulates membrane fission of Golgi carriers for selective cytokine secretion. J Cell Biol. 2010;190:1053–1065. doi: 10.1083/jcb.201001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Davis BM, Molliver DC. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc. 2007;2:152–160. doi: 10.1038/nprot.2006.461. [DOI] [PubMed] [Google Scholar]
- Marchese A, Paing MM, Temple BRS, Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annu Rev Pharmacol Toxicol. 2008;48:601–629. doi: 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza S, Maffucci T. Class II phosphoinositide 3-kinase C2alpha: what we learned so far. Int J Biochem Mol Biol. 2011;2:168–182. [PMC free article] [PubMed] [Google Scholar]
- Meunier FA, Osborne SL, Hammond GRV, Cooke FT, Parker PJ, Domin J, Schiavo G. Phosphatidylinositol 3-kinase C2alpha is essential for ATP-dependent priming of neurosecretory granule exocytosis. Mol Biol Cell. 2005;16:4841–4851. doi: 10.1091/mbc.E05-02-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers R, Cantley LC. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J Biol Chem. 1997;272:4384–4390. doi: 10.1074/jbc.272.7.4384. [DOI] [PubMed] [Google Scholar]
- Mittal N, Roberts K, Pal K, Bentolila LA, Fultz E, Minasyan A, Cahill C, Pradhan A, Conner D, DeFea K, et al. Select G-protein-coupled receptors modulate agonist-induced signaling via a ROCK, LIMK, and β-arrestin 1 pathway. Cell Rep. 2013;5:1010–1021. doi: 10.1016/j.celrep.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikou E, Glick BS. Golgi compartmentation and identity. Curr Opin Cell Biol. 2014;29:74–81. doi: 10.1016/j.ceb.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Chapuis N, Bardet V, Tamburini J, Gallay N, Willems L, Knight ZA, Shokat KM, Azar N, Viguié F, et al. PI-103, a dual inhibitor of Class IA phosphatidylinositide 3-kinase and mTOR, has antileukemic activity in AML. Leukemia. 2008;22:1698–1706. doi: 10.1038/leu.2008.144. [DOI] [PubMed] [Google Scholar]
- Patwardhan AM, Berg KA, Akopain AN, Jeske NA, Gamper N, Clarke WP, Hargreaves KM. Bradykinin-induced functional competence and trafficking of the delta-opioid receptor in trigeminal nociceptors. J Neurosci. 2005;25:8825–8832. doi: 10.1523/JNEUROSCI.0160-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petaja-Repo UE, Hogue M, Laperriere A, Walker P, Bouvier M. Export from the endoplasmic reticulum represents the limiting step in the maturation and cell surface expression of the human delta opioid receptor. J Biol Chem. 2000;275:13727–13736. doi: 10.1074/jbc.275.18.13727. [DOI] [PubMed] [Google Scholar]
- Pettinger L, Gigout S, Linley JE, Gamper N. Bradykinin controls pool size of sensory neurons expressing functional δ-opioid receptors. J Neurosci. 2013;33:10762–10771. doi: 10.1523/JNEUROSCI.0123-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Signalling: seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- Pradhan AA, Befort K, Nozaki C, Gavériaux-Ruff C, Kieffer BL. The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol Sci. 2011;32:581–590. doi: 10.1016/j.tips.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt CP, He J, Wang Y, Barth AL, Bruchez MP. Fluorogenic green-inside red-outside (GIRO) labeling approach reveals adenylyl cyclase-dependent control of BKα surface expression. Bioconjug Chem. 2015;26:1963–1971. doi: 10.1021/acs.bioconjchem.5b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143:761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Laskowski MB, Coscia CJ. Evidence for distinct subcellular sites of opiate receptors. Demonstration of opiate receptors in smooth microsomal fractions isolated from rat brain. J Biol Chem. 1981;256:10017–10023. [PubMed] [Google Scholar]
- Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol. 2003;170:2647–2654. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Scales SJ, Pepperkok R, Pepperkok R, Kreis TE, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Scherrer G, Imamachi N, Cao Y-Q, Contet C, Mennicken F, O’Donnell D, Kieffer BL, Basbaum AI. Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell. 2009;137:1148–1159. doi: 10.1016/j.cell.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiwarski DJ, Tipton A, Giraldo MD, Schmidt BF, Gold MS, Pradhan AA, Puthenveedu MA. A PTEN-regulated checkpoint controls surface delivery of δ opioid receptors. J Neurosci. 2017;37:3741–3752. doi: 10.1523/JNEUROSCI.2923-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Soohoo AL, Puthenveedu MA. Divergent modes for cargo-mediated control of clathrin-coated pit dynamics. Mol Biol Cell. 2013;24:1725–1734. doi: 10.1091/mbc.E12-07-0550. S1–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Louis É, Degrandmaison J, Grastilleur S, Génier S, Blais V, Lavoie C, Parent JL, Gendron L. Involvement of the coatomer protein complex I in the intracellular traffic of the delta opioid receptor. Mol Cell Neurosci. 2017;79:53–63. doi: 10.1016/j.mcn.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JA, Woolford CA, Yan Q, Vasilev KV, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Stanfield RL, Andreko S, Dempsey A, Ahmed M, Capek S, Waggoner AS, Wilson IA, Bruchez MP. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. J Mol Biol. 2013;425:4595–4613. doi: 10.1016/j.jmb.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Várnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderah TW. Delta and kappa opioid receptors as suitable drug targets for pain. Clin J Pain. 2010;26(Suppl 10):S10–S15. doi: 10.1097/AJP.0b013e3181c49e3a. [DOI] [PubMed] [Google Scholar]
- Van Lint J, Rykx A, Maeda Y, Vantus T, Sturany S, Malhotra V, Vandenheede JR, Seufferlein T. Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]
- Vistein R, Puthenveedu MA. Reprogramming of G protein-coupled receptor recycling and signaling by a kinase switch. Proc Natl Acad Sci USA. 2013;110:15289–15294. doi: 10.1073/pnas.1306340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistein R, Puthenveedu MA. Src regulates sequence-dependent beta-2 adrenergic receptor recycling via cortactin phosphorylation. Traffic. 2014;15:1195–1205. doi: 10.1111/tra.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- Wang H, Wang H, Pickel VM, Pickel VM. Preferential cytoplasmic localization of delta-opioid receptors in rat striatal patches: comparison with plasmalemmal mu-opioid receptors. J Neurosci. 2001;21:3242–3250. doi: 10.1523/JNEUROSCI.21-09-03242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-B, Guan J-S, Bao L, Zhang X. Distinct subcellular distribution of delta-opioid receptor fused with various tags in PC12 cells. Neurochem Res. 2008;33:2028–2034. doi: 10.1007/s11064-008-9678-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun H-Q, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yoshioka K, Azam MA, Takuwa N, Sakurada S, Kayaba Y, Sugimoto N, Inoki I, Kimura T, Kuwaki T, et al. Class II phosphoinositide 3-kinase alpha-isoform regulates Rho, myosin phosphatase and contraction in vascular smooth muscle. Biochem J. 2006;394:581–592. doi: 10.1042/BJ20051471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tapia PH, Jarvik J, Waggoner AS, Sklar LA. Real-time detection of protein trafficking with high-throughput flow cytometry (HTFC) and fluorogen-activating protein (FAP) base biosensor. Curr Protoc Cytom. 2014;67:Unit 9.43. doi: 10.1002/0471142956.cy0943s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Yoshida K, Cui H, Wakayama T, Takuwa N, Okamoto Y, Du W, Qi X, Asanuma K, Sugihara K, et al. Endothelial PI3K-C2α, a class II PI3K, has an essential role in angiogenesis and vascular barrier function. Nat Med. 2012;18:1560–1569. doi: 10.1038/nm.2928. [DOI] [PubMed] [Google Scholar]
- Zhang M, Davis JE, Li C, Gao J, Huang W, Lambert NA, Terry AV, Wu G. GGA3 Interacts with a G protein-coupled receptor and modulates its cell surface export. Mol Cell Biol. 2016a;36:1152–1163. doi: 10.1128/MCB.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Huang W, Gao J, Terry AV, Wu G. Regulation of α2B-adrenergic receptor cell surface transport by GGA1 and GGA2. Sci Rep. 2016b;6:37921. doi: 10.1038/srep37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bao L, Arvidsson U, Elde R, Hökfelt T. Localization and regulation of the delta-opioid receptor in dorsal root ganglia and spinal cord of the rat and monkey: evidence for association with the membrane of large dense-core vesicles. Neuroscience. 1998;82:1225–1242. doi: 10.1016/s0306-4522(97)00341-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Gaidarov I, Keen JH. Phosphoinositide 3-kinase C2alpha links clathrin to microtubule-dependent movement. J Biol Chem. 2007;282:1249–1256. doi: 10.1074/jbc.M606998200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.