Assembling billions of short sequencing reads into a contiguous genome is a formidable challenge
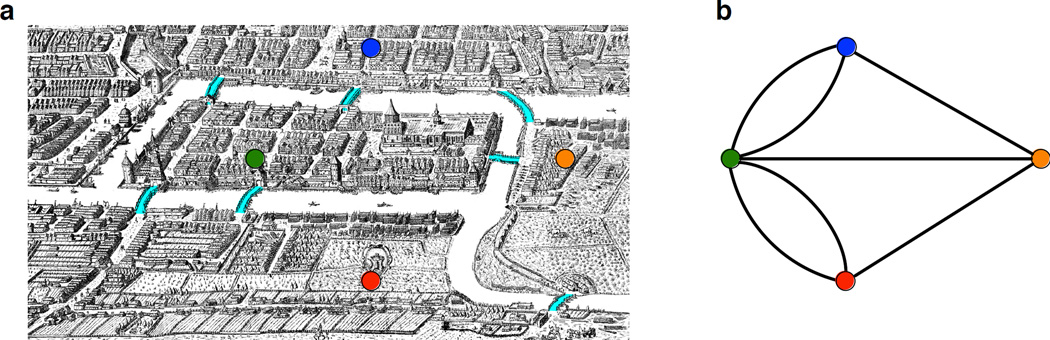
The development of algorithmic ideas for Next-Generation Sequencing (NGS) can be traced back three hundred years to the Prussian city of Königsberg (present-day Kaliningrad, Russia), where seven bridges joined the four parts of the city located on opposing banks of the Pregel River and two river islands (Fig. 1a). Königsberg’s residents enjoyed strolling through the city, and they wondered: is it possible to visit every part of the city by walking across each of the seven bridges exactly once and returning to one’s starting location? Remarkably, the conceptual breakthrough used in 1735 to solve this Bridges of Königsberg Problem by the great mathematician Leonhard Euler1 also enables the assembly of billions of short sequencing reads.
Figure 1. Bridges of Konigsberg problem.
(a) A map of old Königsberg, in which each area of the city is labeled with a differently colored point. (b) The Königsberg Bridge Graph, formed by representing each of four land areas as a node and each of the city’s seven bridges as an edge.
Euler’s first insight was to represent each landmass as a point (called a node) and each bridge as a line segment (called an edge) connecting the appropriate two points. This creates a graph—a network of nodes connected by edges (Fig. 1b). By describing a procedure for determining whether an arbitrary graph contains an Eulerian cycle (a path through the graph that visits every edge exactly once and returns back where it started), Euler not only resolved the Bridges of Königsberg Problem but also effectively launched the entire branch of mathematics known today as graph theory2.
Computational issues arise from alignment-based assembly
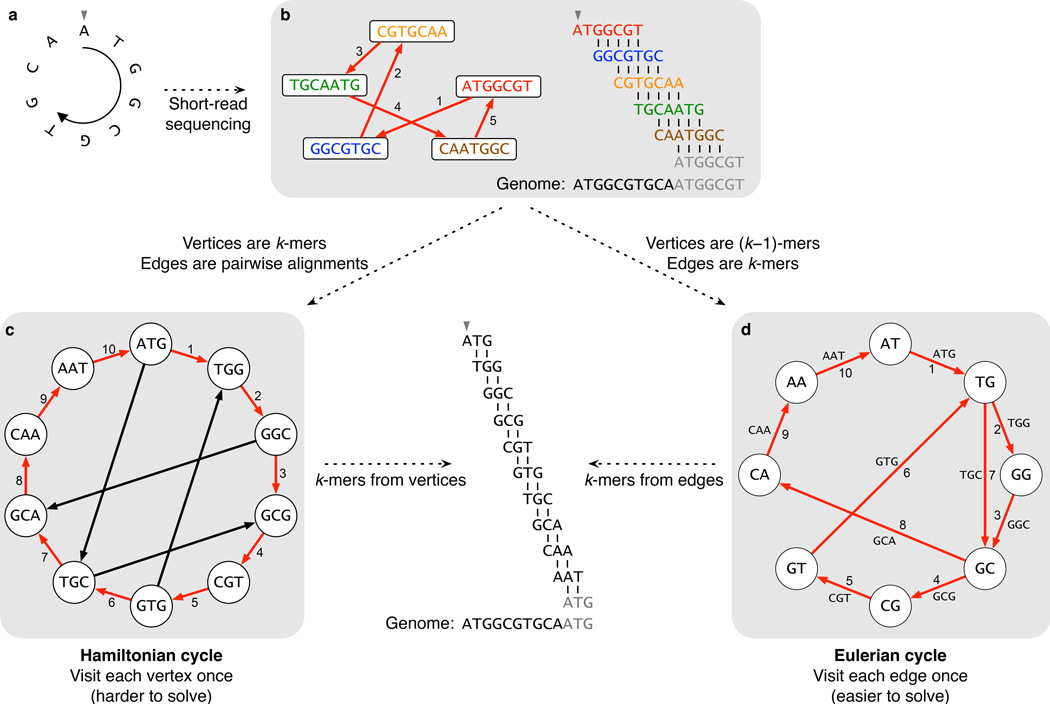
To illustrate why graphs are useful for genome assembly, we will use a simple example with five very short reads (CGTGCAA, ATGGCGT, CAATGGC, GGCGTGC and TGCAATG) sequenced from a small circular genome, ATGGCGTGCA (Fig. 2a). Current NGS methods produce reads that vary in length, but the most popular technology today generates approximately 100-nucleotide reads. A straightforward method for assembling reads into longer contiguous sequences—and the one used for assembling the human genome3,4 in 2001 as well as for all other projects based on Sanger sequencing—uses a graph in which each read is represented by a node and overlap between reads is represented by an arrow (called a directed edge) joining two reads. For instance, two nodes representing reads may be connected with a directed edge if the reads overlap by at least five nucleotides (Fig. 2b).
Figure 2. Two strategies for genome assembly: from Hamiltonian cycles to Eulerian cycles.
(a) A simplified example of a small circular genome. (b) In traditional Sanger sequencing algorithms, reads were represented as nodes in a graph, and edges represented alignments between reads. Walking along a Hamiltonian cycle by following the edges in numerical order allows one to reconstruct the circular genome by combining alignments between successive reads. At the end of the cycle, the sequence wraps around to the start of the genome; the repeated part of the sequence is grayed out in the alignment diagram. (c) An alternative assembly technique first splits reads into all possible k-mers: with k = 3, “ATGGCGT” comprises ATG, TGG, GGC, GCG and CGT. Following a Hamiltonian cycle (indicated by red edges) allows one to reconstruct the genome by forming an alignment in which each successive k-mer (from successive nodes) is shifted by one position. This procedure recovers the genome but does not scale well to large graphs. (d) Modern short-read assembly algorithms construct a de Bruijn graph by representing all k-mer prefixes and suffixes as nodes, then drawing edges that represent k-mers having a particular prefix and suffix. For example, k-mer edge ATG has prefix AT and suffix TG. Finding an Eulerian cycle allows one to reconstruct the genome by forming an alignment in which each successive k-mer (from successive edges) is shifted by one position. This generates the same cyclic genome sequence without the computational strain of finding a Hamiltonian cycle.
Visualizing an ant walking along the edges of this graph provides a useful illustrative aid for understanding a broad class of algorithms used to derive insights from graphs. In the case of genome assembly, the ant’s path traces a series of overlapping reads, and thus represents a candidate assembly. Specifically, if the ant follows the path ATGGCGT → GGCGTGC → CGTGCAA → TGCAATG → CAATGGC → ATGGCGT, its walk induces a Hamiltonian cycle in our graph, which is a cycle that travels to every node exactly once (but closes with the starting node), meaning that each read will be included once in the assembly. The circular “genome” ATGGCGTGCA resulting from a Hamiltonian cycle contains all five reads and thus reconstructs the original genome (although we may have to “wrap around” the genome, for example in order to locate CAATGGC in ATGGCGTGCA).
Modern assemblers usually work with strings of a particular length k (k-mers), which are shorter than entire reads (see Box 2 for an explanation of why researchers prefer k-mers to reads). For example, a 100-nucleotide read may be divided into 46 overlapping 55-mers. We can generalize the Hamiltonian Cycle approach to k-mers by constructing a graph as follows.
Box 2. Practical strategies for applying de Bruijn graphs.
We have noted that in practice, attempting to apply de Bruijn graphs to experimental data is not a straightforward procedure. Below we describe some key computational techniques that have been devised in order to address the practical challenges introduced by errors and quirks in current sequencing technologies, as well as to resolve the complexities created by repeat-rich genomes.
For instance, the astute observer will have noticed that the de Bruijn method for fragment assembly relies upon four hidden assumptions that do not hold for NGS. We took for granted that we can generate all k-mers present in the genome, that all k-mers are error-free, that each k-mer appears at most once in the genome, and that the genome consists of a single circular chromosome. For example, Illumina technology, which generates 100-nucleotide long reads, may miss some 100-mers present in the genome (even if the read coverage is high), and the 100-mers that it does generate typically have errors.
Generating (nearly) all k-mers present in the genome
100-mer reads generated by Illumina technology capture only a small fraction of 100-mers from the genome (even for high-coverage NGS projects), thus violating the key assumption of the de Bruijn graphs. However, if one breaks these reads into shorter k-mers, the resulting k-mers often represent nearly all k-mers from the genome for sufficiently small k. For example, NGS assemblers may break every 100-nucleotide read into 46 overlapping 55-mers and further assemble the resulting 55-mers using de Bruijn graphs. Even if some 100-mers occurring in the genome are not generated as reads, this read breaking procedure17 ensures that nearly all 55-mers appearing in the genome are detected. In our recurring example, our five reads do not account for all 7-mer substrings of the genome, but they do contain all 3-mers present in the genome; we have seen that this is sufficient to reconstruct the genome.
Handling errors in reads
Each error in a read creates a bulge in the de Bruijn graph (Supplementary Fig. 1), complicating assembly. To make matters even worse, in a genome with inexact repeats (such as two regions differing by a single nucleotide or other small variation), reads from the two repeat copies will also generate bulges in the de Bruijn graph. An approach for error-correcting reads, to resolve errors before even beginning assembly, was proposed18 in 2001 and such a phase is now common to do before assembly. An approach for removing bulges from de Bruijn graphs was outlined19 in 2004 and, with some variations, is used in most existing NGS assemblers (EULER-SR20, Velvet21, ALLPATHS22, ABySS23, SOAPdenovo24). These and other recently developed tools introduced many new algorithmic and software engineering ideas in assembly algorithms and paved the way towards assembling large (e.g., mammalian) genomes with NGS data. See refs. 25–26 for a detailed comparison of these and other NGS assemblers.
Handling DNA repeats
Imagine for a moment that our (cyclic) genome is ATGCATGC. Then we would hopefully detect four 3-mers: ATG, TGC, GCA, and CAT; however, the present definition of de Bruijn graphs would lead us to reconstruct the genome as ATGC. As one can see, the problem is that each of the 3-mers detected actually occurs twice in the original genome. Therefore, we will need to adjust genome reconstruction so that we not only find all k-mers occurring in the genome, but we also find how many times each such k-mer appears, which is called its k-mer multiplicity. The good news is that we can still handle fragment assembly in the case when k-mer multiplicities are known.
We simply use the same method to construct the de Bruijn graph E, except that if the multiplicity of a k-mer is m, we will connect its prefix to its suffix via m directed edges (instead of just one). Extending our ongoing example, if during read generation we discover that each of the four 3-mers TGC, GCG, CGT, and GTG has multiplicity 2, and that each of the six 3-mers ATG, TGG, GGC, GCA, CAA, and AAT has multiplicity 1, we create the graph shown in Supplementary Figure 2. Furthermore, it is easy to see that the graph resulting from adding multiplicity edges is Eulerian, as both the indegree and outdegree of a node (representing a (k − 1)-mer) equals the number of times this (k − 1)-mer appears in the genome.
In practice, information about the multiplicities of k-mers in the genome may be difficult to obtain with NGS technologies. However, computer scientists have found ways to reconstruct the genome even when these data are unavailable. One such technique involves paired reads. Reads are typically generated in pairs by sequencing both ends of a long fragment of DNA whose length can be estimated well. If one read maps at or before the entrance to a repeat in the graph, and the other maps at or after the exit, the read pair may be used to determine the correct traversal through the graph.
Handling multiple and linear chromosomes
Our presentation focused on a genome consisting of one circular chromosome. If our chromosome is linear, then we will instead need to search for an Eulerian path, which is not required to end at the node where it begins; if there are multiple linear chromosomes, then we will have one path for each chromosome. Fortunately, Euler’s work can be adapted to handle these complexities.
Handling unsequenced regions
Unsequenced regions and sequencing errors may further break the chromosomes into contigs (a sequenced contiguous region of DNA) and gaps (unsequenced regions), with one path for each contig. Increasing the value of k will tend to reduce the number of bulges and give longer contigs in places with high coverage but some errors. However, it will also tend to break contigs in regions that have low coverage. Successive contigs along a chromosome may have overlaps of fewer than k nucleotides, or may have gaps between them.
The correct order and orientation of the contigs, and the approximate sizes of the gaps, is determined in the scaffolding phase of assembly. This uses additional information, including paired reads, to determine the order of contigs.
First, from a set of reads, form a node for every k-mer appearing in these reads. Second, given a k-mer, define its prefix as the string formed by all its nucleotides except the final one and its suffix as the string formed by all its nucleotides except the first one. Connect one k-mer to another with a directed edge if the suffix of the former equals the prefix of the latter—that is, if the two k-mers completely overlap except for one nucleotide at each end (Fig. 2c). Third, look for a Hamiltonian cycle, which represents a candidate genome because it visits each detected k-mer; moreover, that path will also have minimal length because a Hamiltonian cycle travels to each k-mer exactly once.
However, this method is not as easy to implement as it might seem. Imagine attempting to create a similar graph for a single run of an Illumina sequencer that generates many reads. A million (106) reads will require a trillion (1012) pairwise alignments. A billion (109) reads necessitate a quintillion (1018) alignments. What’s more, there is no known efficient algorithm for finding a Hamiltonian cycle in a large graph with millions (let alone billions) of nodes. The Hamiltonian cycle approach5,6 was feasible for sequencing the first microbial genome7 in 1995 and the human genome in 2001, as well as for all other projects based on Sanger sequencing. However, the computational burden was so large that most NGS sequencing projects have abandoned the Hamiltonian cycle approach.
And here is where genome sequencing faces the limits of modern computer science: the computational problem of finding a Hamiltonian cycle belongs to a class of problems that are collectively called NP-Complete (see ref. 2 for further background). To this day, some of the world’s top computer scientists have worked to find an efficient solution to any NP-Complete problem, with no success. What makes their failure doubly frustrating is that neither has anyone been able to prove that NP-Complete problems are intractable; efficient solutions to these problems may actually exist, but such solutions have not yet been discovered.
Scalable assembly with de Bruijn graphs
We have observed that finding a cycle visiting all nodes of a graph exactly once (called the Hamiltonian cycle problem) is a difficult computational problem; however, as we will soon see, finding a cycle visiting all edges of a graph exactly once is much easier. This algorithmic contrast has motivated computer scientists to cast fragment assembly as such a problem. So instead of assigning each k-mer to a node, we will now assign each k-mer located within a read to an edge. This allows the construction of a de Bruijn graph, which we call E, as follows. First, form a node for every distinct prefix or suffix of a k-mer, meaning that a given sequence of length k − 1 can appear only once as a node of the graph. Then, connect node x to node y with a directed edge if some k-mer has prefix x and suffix y, and label the edge with this k-mer (Fig. 2d). For a discussion on the origin of de Bruijn graphs, see Box 1.
Box 1. Historical notes.
Origin of de Bruijn graphs
In 1946, the Dutch mathematician Nicolaas de Bruijn became interested in the Superstring Problem12: find a shortest circular superstring that contains all possible substrings of length k (k-mers) in a given alphabet. There exist nk k-mers in an alphabet containing n symbols: for example, given the alphabet {A,T,G,C}, there are 43 = 64 trinucleotides. If our alphabet is instead {0,1}, then all possible 3-mers are simply given by all 3-digit binary numbers: 000, 001, 010, 011, 100, 101, 110, 111. The circular superstring “0001110100” not only contains all 3-mers but is also as short as possible, since it contains each 3-mer exactly once. But how can one construct such a superstring for all k-mers in the case of an arbitrary k and an arbitrary alphabet? De Bruijn answered this question by borrowing Euler’s solution of the Bridges of Königsberg Problem.
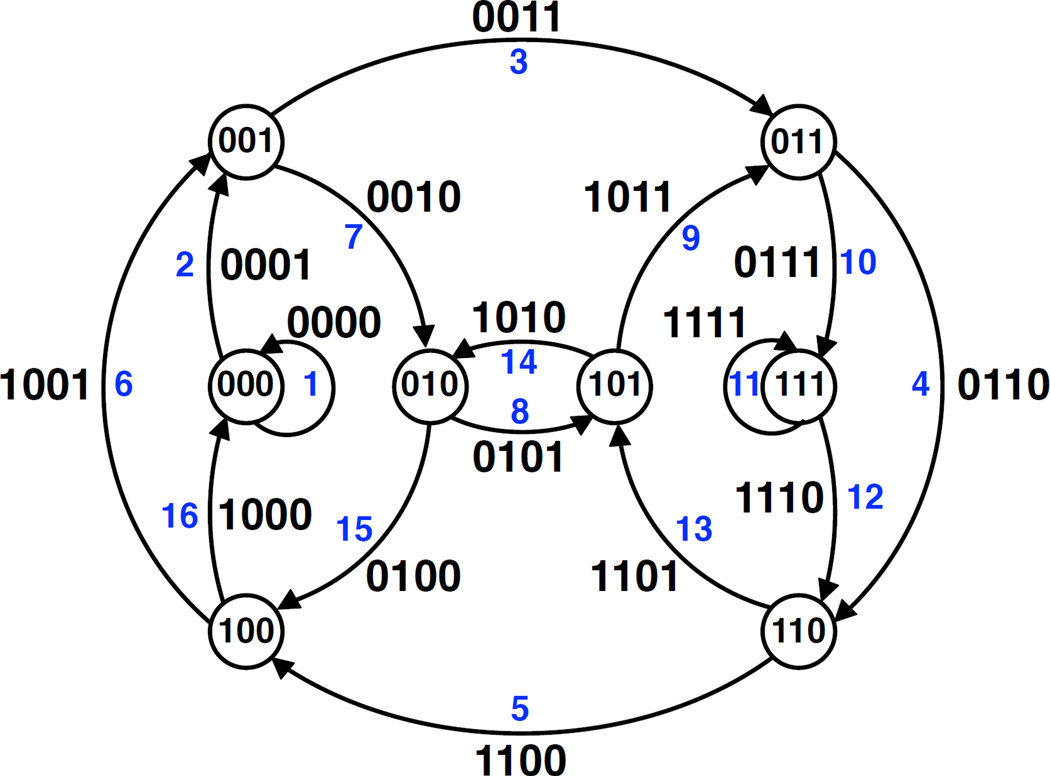
So construct a graph B (the original graph called the de Bruijn graph) for which every possible (k − 1)-mer is assigned to a node; connect one (k − 1)-mer by a directed edge to a second (k − 1)-mer if there is some k-mer whose prefix is the former and whose suffix is the latter (Fig. 3). Edges of the de Bruijn graph represent all possible k-mers, and thus an Eulerian cycle in B represents a shortest (cyclic) superstring that contains each k-mer exactly once. By checking that the indegree and outdegree of every node in B equals the size of the alphabet, we can verify that B contains an Eulerian cycle. In turn, we can construct an Eulerian cycle using Euler’s algorithm, therefore solving the superstring problem. It should now be apparent why the “de Bruijn graph” construction described in the main text, which does not use all possible k-mers as edges but rather only those generated from our reads, is also named in honor of de Bruijn.
Sequencing by Hybridization
Few people remember that the idea of DNA sequencing using short reads (like in NGS) goes back to late 1980s. In fact, the very first short read sequencing technology, called Sequencing by Hybridization (or SBH), aimed at exactly this goal. SBH13–15 (proposed in 1988) is based on building a microarray containing every possible oligonucleotide of length k. After hybridization of such an array with an unknown genome, one would get information about all k-mers present in the genome. De Bruijn graphs were first brought to bioinformatics in 1989 as a method to assemble k-mers generated by SBH16; this method is very similar to the key algorithmic step of NGS assemblers today.
DNA arrays ultimately failed to realize the dream (DNA sequencing) that motivated their inventors because the fidelity of DNA hybridization with the array turned out to be too low and the value of k was too small. Yet the failure of DNA arrays was short-lived: while the original goal (DNA sequencing) was still out of reach, two new unexpected applications emerged: measuring gene expression and analyzing genetic variations. Today these applications have become so ubiquitous that most people have forgotten that the original goal of the inventors of DNA arrays was to sequence the human genome!
Now imagine an ant that follows a different strategy: instead of visiting every node of the graph (as before), it now visits every edge of E exactly once. Sound familiar? This is exactly the kind of path that would solve the Bridges of Königsberg Problem and is called an Eulerian cycle. Since it visits all edges of E, which represent all possible k-mers, this new ant also spells out a candidate genome: for each edge that the ant traverses, one tacks on the first nucleotide of the k-mer assigned to that edge.
Euler considered graphs for which there exists a path between every two nodes (called connected graphs). He proved that a connected graph with undirected edges contains an Eulerian cycle exactly when every node in the graph has an even number of edges touching it. For the Königsberg Bridge Graph, this is not the case because each of the four nodes has an odd number of edges touching it (Fig. 1b), and so the desired stroll through the city does not exist.
The case of directed graphs (i.e. graphs with directed edges) is similar. For any node v in a directed graph, define the indegree of v as the number of edges leading into v and the outdegree of v as the number of edges leaving v. A graph in which indegrees are equal to outdegrees for all nodes is called balanced. Euler’s theorem states that a connected directed graph has an Eulerian cycle if and only if it is balanced. In particular, Euler’s theorem implies that the graph E contains an Eulerian cycle as long as we have located all k-mers present in the genome. Indeed, in this case, for any node, both its indegree and outdegree represent the number of times the (k − 1)-mer assigned to that node occurs in the genome.
It is easy to see that a graph possessing an Eulerian cycle is balanced because every time an ant traversing an Eulerian cycle passes through a particular vertex, it enters on one edge of the cycle and exits on the next edge. This pairs up all the edges touching each vertex, showing that half the edges on the vertex lead into it and half lead out from it.. It is a bit harder to see that every connected balanced graph contains an Eulerian cycle. To prove this fact, Euler sent an ant to randomly explore the graph under a single constraint: the ant cannot traverse a previously traversed edge. Sooner or later, the ant must get stuck at a certain node (with all outgoing edges previously traversed), and Euler noticed that because the graph is balanced, this “no exit” node is exactly the vertex where the ant started, no matter how the ant traveled through the graph. This implies that the ant has completed a cycle; if this cycle happens to traverse all edges, then the ant has found an Eulerian cycle! Otherwise, Euler sent another ant to randomly traverse unexplored edges and thereby to trace a second cycle in the graph. Euler further showed that the two cycles discovered by the two ants can be combined into a single cycle. If this (larger) cycle contains all the edges in the graph, then the two ants have together found an Eulerian cycle! If not, Euler’s method recruits a third (fourth, fifth, etc.) ant, and eventually finds an Eulerian cycle.
On modern computers, this algorithm can efficiently find Eulerian cycles in huge graphs having billions of nodes, thus avoiding the quagmire of NP-Completeness. Therefore, simply recasting our original problem into a slightly different framework has converted fragment assembly into a tractable computational problem; this is a commonly used strategy in computer science.
The run time required by a computer implementation of Euler’s algorithm is roughly proportional to the number of edges in the graph E. In the Hamiltonian approach, the time is potentially a lot larger, due to the large number of pairwise alignments needed to construct the graph, and to the NP-Completeness of finding a Hamiltonian cycle. A more detailed comparison of these approaches is given in ref. 8.
Unfortunately, de Bruijn graphs are not a cure-all. Throughout our exposition, we have made several simplifying assumptions, which require much work to iron out formally. Yet for every apparent complication to sequence assembly, it has proven fruitful to apply some cousin of de Bruijn graphs to transform a question involving Hamiltonian cycles into a different question regarding Eulerian cycles (Box 2). Moreover, analogs of de Bruijn graphs have been useful in many other bioinformatics problems, including antibody sequencing9, synteny block reconstruction10, and RNA assembly11. In each of these applications, the de Bruijn graph represents the experimental data in a manner that leads to a tractable computational problem.
As new sequencing technologies emerge, the best computational strategies for assembling genomes from reads may change. The factors that influence the choice of algorithms include the quantity of data (measured by read length and coverage); quality of data (including error rates); and genome structure (such as number and size of repeated regions, and GC content). Short read sequencing technologies produce very large numbers of reads, which currently favors the use of de Bruijn graphs. De Bruijn graphs are also well suited to representing genomes with repeats, whereas overlap methods need to mask repeats that are longer than the read length. However, if a future sequencing technology produces high quality reads with tens of thousands of bases, a smaller number of reads would be needed, and the pendulum could swing back towards favoring overlap-based approaches for assembly.
Supplementary Material
Figure 3. De Bruijn graph.
The de Bruijn graph B for k = 4 and a 2-character alphabet composed of the digits 0 and 1. This graph has an Eulerian cycle since each node has indegree and outdegree equal to 2. Following the blue numbered edges in order 1, 2, …, 16 gives an Eulerian cycle 0000, 0001, 0011, 0110, 1100, 1001, 0010, 0101, 1011, 0111, 1111, 1110, 1101, 1010, 0100, 1000, which spells the cyclic superstring 0000110010111101.
Acknowledgments
This work was supported by grants from Howard Hughes Medical Institute (HHMI grant 52005726), the National Institutes of Health (NIH grant 3P41RR024851-02S1), and the National Science Foundation (NSF grant DMS-0718810).
Contributor Information
Phillip E. C. Compeau, Department of Mathematics, University of California San Diego, La Jolla, CA, USA
Pavel A. Pevzner, Department of Computer Science and Engineering, University of California San Diego, La Jolla, CA, USA
Glenn Tesler, Department of Mathematics, University of California San Diego, La Jolla, CA, USA.
References
- 1.Euler L. Solutio problematis ad geometriam situs pertinentis. Commentarii Academiae Scientiarum Petropolitanae. 1741;8:128–140. [Google Scholar]
- 2.Skiena S. The Algorithm Design Manual. Berlin: Springer; 2008. [Google Scholar]
- 3.Lander E, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Venter J, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Kececioglu J, Myers E. Combinatorial algorithms for DNA sequence assembly. Algorithmica. 1995;13:7–51. [Google Scholar]
- 6.Adams M, et al. The genome sequence of drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 7.Fleischmann R, et al. Whole-genome random sequencing and assembly of haemophilus influenzae rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Schatz M, Delcher A, Salzberg S. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandeira N, Pham V, Pevzner P, Arnott D, Lill J. Automated de novo protein sequencing of monoclonal antibodies. Nat Biotechnol. 2008;26:1336–1338. doi: 10.1038/nbt1208-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham S, Pevzner P. DRIMM-synteny: decomposing genomes into evolutionary conserved segments. Bioinformatics. 2010;26:2509–2516. doi: 10.1093/bioinformatics/btq465. [DOI] [PubMed] [Google Scholar]
- 11.Grabherr M, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bruijn N. A combinatorial problem. Proc. Nederl. Akad. Wetensch. 1946;49:758–764. [Google Scholar]
- 13.Drmanac R, Labat I, Brukner I, Crkvenjakov R. Sequencing of megabase plus DNA by hybridization: Theory of the method. Genomics. 1989;4:114–128. doi: 10.1016/0888-7543(89)90290-5. [DOI] [PubMed] [Google Scholar]
- 14.Southern, E. United Kingdom patent application gb8810400. 1988 [Google Scholar]
- 15.Lysov Y, et al. DNA sequencing by hybridization with oligonucleotides. Doklady Academy Nauk USSR. 1988;303:1508–1511. [PubMed] [Google Scholar]
- 16.Pevzner P. l-tuple DNA sequencing: Computer analysis. Journal of Biomolecular Structure and Dynamics. 1989;7:63–73. doi: 10.1080/07391102.1989.10507752. [DOI] [PubMed] [Google Scholar]
- 17.Idury R, Waterman M. A new algorithm for DNA sequence assembly. Journal of Computational Biology. 1995;2:291–306. doi: 10.1089/cmb.1995.2.291. [DOI] [PubMed] [Google Scholar]
- 18.Pevzner P, Tang H, Waterman M. An Eulerian path approach to DNA fragment assembly. Proceedings of the National Academy of Science of the United States of America. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pevzner P, Tang H, Tesler G. De novo repeat classification and fragment assembly. Genome Res. 2004;14:1786–1796. doi: 10.1101/gr.2395204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaisson M, Pevzner P. Short read fragment assembly of bacterial genomes. Genome. Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerbino D, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome. Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler J, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson J, et al. ABySS: a parallel assembler for short read sequence data. Genome. Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paszkiewicz K, Studholme D. De novo assembly of short sequence reads. Brief Bioinform. 2010;11:457–472. doi: 10.1093/bib/bbq020. [DOI] [PubMed] [Google Scholar]
- 26.Miller J, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.