Abstract
Domestication and plant breeding are ongoing, 10,000-year evolutionary experiments that have radically altered wild species to meet human needs. Maize has undergone a particularly striking transformation. Researchers have sought for decades to identify the genes underlying maize evolution1,2, but these efforts have been limited in scope. Here, we report a comprehensive assessment of the evolution of modern maize based on the genome-wide resequencing of 75 wild, landrace, and improved maize lines3. We find evidence of recovery of diversity post-domestication, likely introgression from wild relatives, and evidence for stronger selection during domestication than improvement. We identify a number of genes with stronger signals of selection than those previously shown to underlie major morphological changes4,5. Finally, through transcriptome-wide analysis of gene expression, we find evidence consistent with removal of cis-acting variation during maize domestication and improvement and suggestive of modern breeding having increased dominance in expression while targeting highly expressed genes.
Archaeological6 and genetic7,8 evidence indicate that maize (Zea mays ssp. mays) was domesticated approximately 10,000 B.P. in the Balsas River Basin of southwestern Mexico. Domestication involved a radical phenotypic transformation from the wild progenitor, Zea mays ssp. parviglumis (hereafter parviglumis; Fig. 1), resulting in an unbranched plant with seed attached to a cob, making maize entirely dependent on humans for propagation. Subsequent to domestication, maize has been subject to intensive improvement efforts, culminating in the development of hybrid maize lines highly adapted to modern agricultural practices. We present a population genomic analysis of maize evolution based on resequencing of 75 genomes of maize and its wild relatives (Fig. 1, Supplementary Fig. 1, and Supplementary Table 1). We generated 781 gigabases of sequence from 35 improved maize lines, 23 traditional landraces, and 17 wild relatives (14 parviglumis; two Zea mays ssp. mexicana, hereafter mexicana; one Tripsacum dactyloides var. meridionale) using short-read technology, sequencing each line to an average depth of more than 5× (Supplementary Table 1; ref. 3). Reads were mapped to the maize reference genome (release 4a.53) and analyses are based on a final set of 21,141,953 high quality single nucleotide polymorphisms (SNPs).
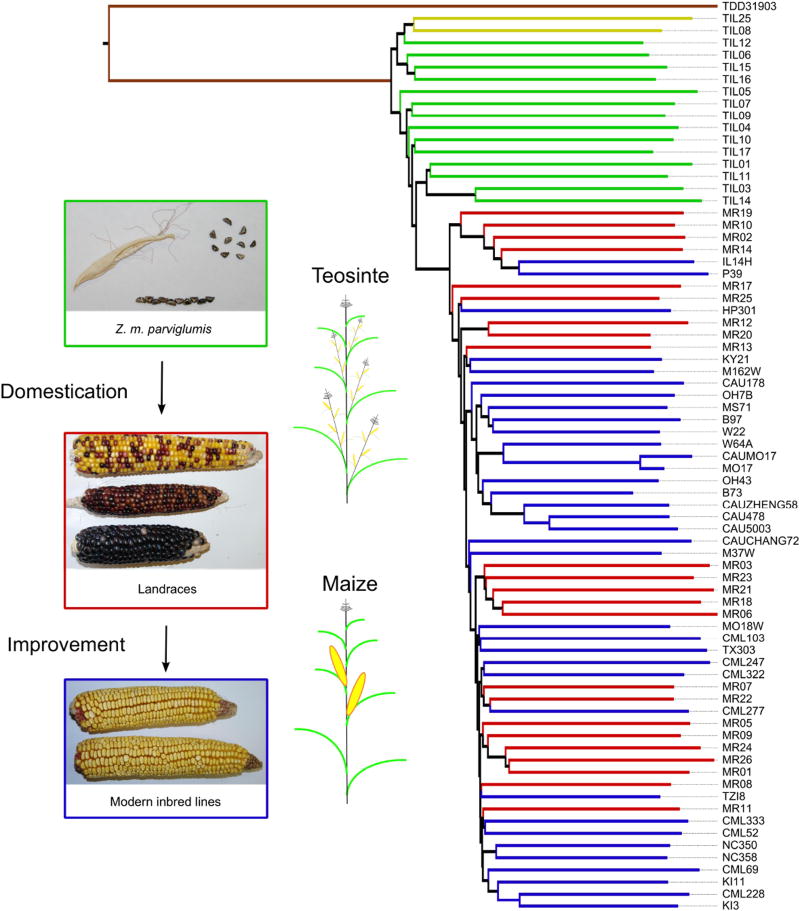
Figure 1. Neighbor-joining tree and changing morphology of domesticated maize and its wild relatives.
Taxa in the neighbor-joining tree (right) are represented by different colors: parviglumis (green), landraces (red), improved lines (blue), mexicana (yellow), and Tripsacum (brown). Morphological changes (left) are shown for female inflorescences and plant architecture during domestication and improvement.
Maize landraces retain more nucleotide diversity (83%; Fig. 2a) and show lower genetic differentiation from their wild progenitor (FST=0.11) than other crop species9,10. This is likely due to large census size and an outcrossing mating system in maize landraces. Linkage disequilibrium has increased dramatically as a result of domestication, with genome-wide estimates of the population recombination rate ρ in landraces estimated to be 25% of the rate in parviglumis and average haplotype length increasing from 22 to 30kb (Supplementary Fig. 2). These results are consistent with the effects of a domestication bottleneck, but an excess of rare SNPs (Supplementary Table 2 and Supplementary Fig. 3) suggests that variation has begun to recover across most of the genome. Gene-rich regions, however, exhibit fewer SNPs unique to landraces (t-test, p<0.001) and no excess of rare SNPs (Supplementary Tables 2 and 3), a difference likely due to the effects of background selection against deleterious mutations slowing the post-domestication recovery of variation at linked sites. Modern breeding appears to have had negligible effects on genome-wide diversity or mean haplotype lengths in our broad sample of modern lines (Fig. 2a, Supplementary Fig. 2, Supplementary Table 2). While our estimates of nucleotide diversity in improved lines may be inflated by the diverse inbreds chosen, the relationships between inbred and landrace lines (Fig. 1) suggest a weaker genome-wide bottleneck during improvement. Finally, comparison of maize landraces to our two mexicana genomes identifies several extended regions of high genetic similarity (Supplementary Fig. 4), consistent with previous observations of admixture between these taxa8,11 and suggestive of the possibility that mexicana may have contributed alleles important for maize evolution.
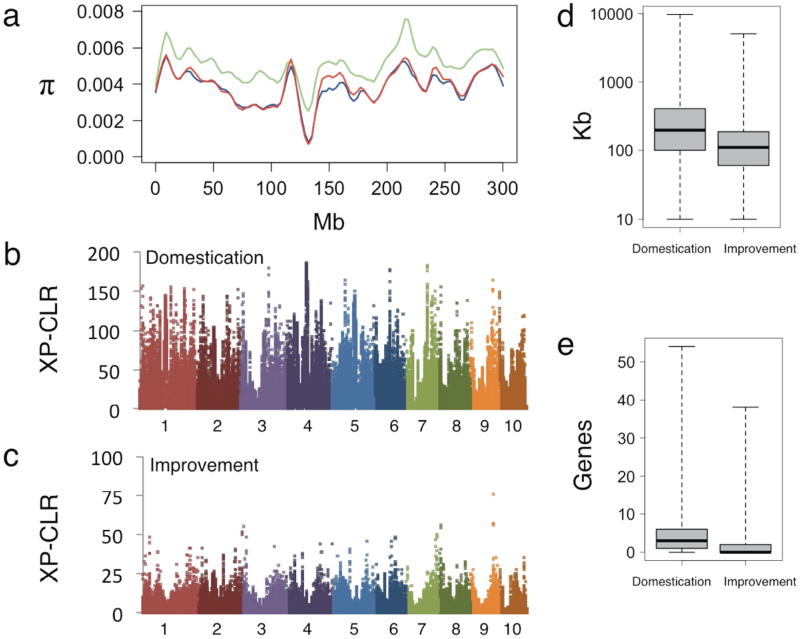
Figure 2. Genome-wide analysis of nucleotide diversity and selection.
(a) LOWESS curves of nucleotide diversity (π) along chromosome 1 in parviglumis (green), landraces (red), and improved lines (blue). Genome-wide likelihood values (XP-CLR) for selection during domestication (b) and improvement (c) with chromosome number indicated along the x-axis. Distributions of feature size (d) and gene counts within features (e) in domestication and improvement scans.
To identify regions of the genome most affected by selection during maize evolution, we used a likelihood method (XP-CLR; ref. 12) to scan for extreme allele frequency differentiation over extended linked regions (Fig. 2b, c). Adjacent windows of high XP-CLR were grouped into “features,” each likely representing the effect of a single selective sweep. Features in multiple centromeres show high XP-CLR values (Fig. 2b, c, and Supplementary Fig. 5). Combined with evidence for change in abundance of centromeric retroelements (Supplementary Table 4 and Supplementary Fig. 6; ref. 3), this finding suggests rapid centromere evolution. However, because centromeres harbor few genes and our genetic map may underestimate the extended LD in these regions, we masked centromeres from further analysis. We also masked a newly discovered ~50-Mb inversion polymorphism on chromosome 1 (Supplementary Fig. 7; further characterized in submitted publication by Z. Fang, T.P., A.L. Weber, R.K. Dawe, J.C.G., J. Sánchez-González, C. Ross-Ibarra, J.D., P.L.M., J.R.I.).
We focused analyses on the 484 domestication and 695 improvement features in the highest 10% of XP-CLR values (Fig. 2b, c). Domestication features contain an average of 3.4 genes and have a mean size of 322 kb (Fig. 2d, e), cover approximately 7.6% of the maize genome, and show multiple signatures of selection, including elevated differentiation, low nucleotide diversity, and an excess of high-frequency derived SNPs (Supplementary Table 5 and Supplementary Fig. 8). We estimate the mean strength of selection in these features as s = 0.015, which is within the range of estimates based on archaeological data from other domesticates13 and more than an order of magnitude higher than the mean value of 0.0011 across the rest of the genome.
While selection during maize improvement can be strong14,15, XP-CLR values and estimated selection coefficients (mean s = 0.003) from our improvement scan were substantially lower than observed for domestication (Fig. 2b, c). Consistent with this finding, improvement features have smaller average size (Fig. 2d) and contain fewer genes (Fig. 2e) than domestication features. One explanation for these results may be that our diverse tropical and temperate lines derive from distinct landrace founders (Fig. 1) and have been subject to different selective pressures16. Indeed, independent scans of temperate and tropical lines find stronger evidence of selection and little overlap of selected features (Supplementary Fig. 9). However, previous estimates of effect size for loci involved in domestication and improvement traits provide some independent evidence of stronger selection during domestication17. Twenty-three percent (107) of domestication features show additional evidence of selection during improvement, indicating that a subset of domestication loci may contribute to phenotypes of continued agronomic importance.
Individual features likely result from a single selective event, and we assigned the gene closest to the 10-kb window with the maximum XP-CLR score in each feature as the most likely candidate (Supplementary Tables 6 and 7). Our domestication and improvement candidate lists, each including 1–2% of the maize filtered gene set (FGS), represent our best estimate of the direct targets of selection within features, but linked genes have also been affected by selection, limiting the diversity available for modern improvement for many of the 3,040 genes found within features. Thus, while our candidates are of most interest for understanding genes directly related to maize evolution, breeding programs would likely benefit from efforts to incorporate diversity from exotic germplasm in these genomic regions.
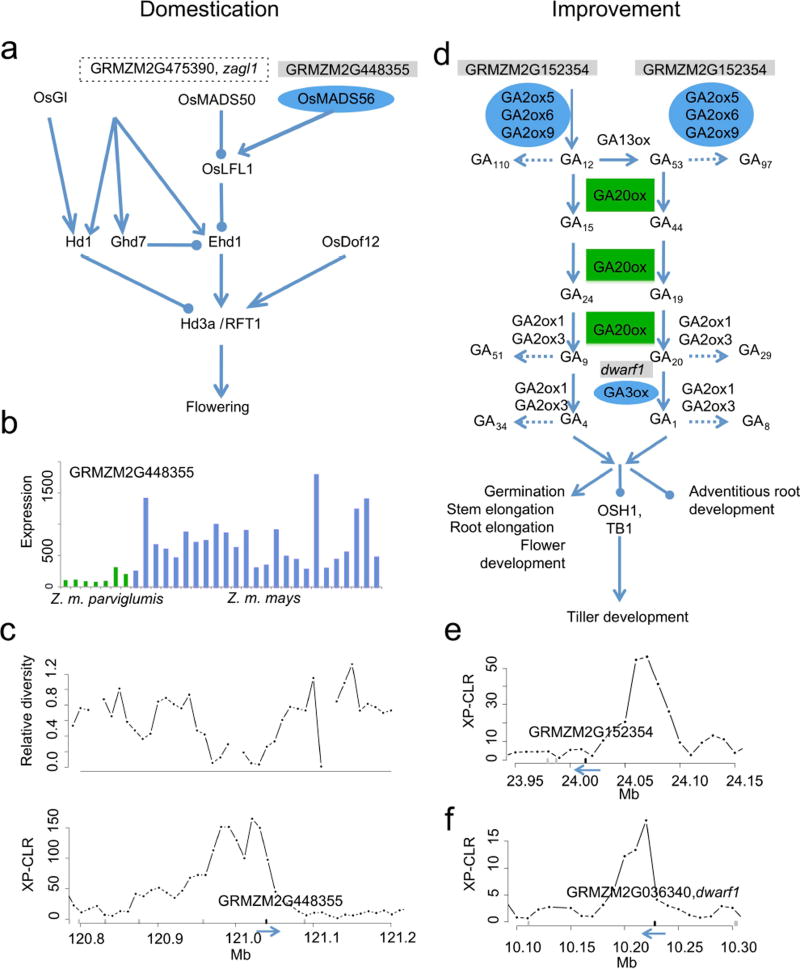
A sizeable fraction of our domestication and improvement features contain no annotated sequence (6% and 11%), a result that could implicate regulatory variants in the process of maize evolution. However, the majority of our features contain genes in the high-confidence FGS, and should prove useful both in dissecting existing QTLs and identifying novel candidate genes. For example, the domestication candidate GRMZM2G448355, an ortholog of the rice gene OsMADS56 which delays flowering under long-day conditions (Fig. 3 a–c), is found within a flowering-time QTL on chromosome 918 and two improvement candidates implicated in nitrogen metabolism, GRMZM2G036464 (glutamine synthetase) and GRMZM2G428027 (nitrate reductase), both reside in a QTL for multiple traits including thousand kernel weight and nitrogen mobilization19. Only a fraction of our novel candidate genes are functionally characterized in maize; one example is the domestication candidate abph1 (GRMZM2G035688) that is known to affect phyllotaxy20. However, function can often be inferred from orthology; the domestication candidate GRMZM2G010290 has no known function, but shows close sequence identity to Arabidopsis DAG1 and DAG2 proteins that affect seed germination21. Two improvement candidates, gibberellin 2-oxidase (GRMZM2G152354) and gibberellin 3-oxidase (GRMZM2G036340, dwarf1) are found in the plant growth hormone gibberellin biosynthesis pathway (Fig. 3 d–f) upstream and downstream of the “green revolution gene,” gibberellin 20-oxidase22. Other notable improvement candidates include GRMZM2G082468, a homolog of Arabidopsis farnesyltransferase that has been engineered as a drought tolerance transgene in canola23, and GRMZM2G087612, whose Arabidopsis ortholog, SDP1, initiates storage oil breakdown in seed24.
Figure 3. Domestication (a–c) and improvement (d–f) candidate genes in relation to two pathways in rice.
Zea mays genes are on a grey background (candidates) or boxed (in selected regions) above their rice orthologs. (a) The flowering-time pathway29, including GRMZM2G448355 and zagl1. (b) Seedling expression pattern of GRMZM2G448355 in parviglumis and maize inbreds. (c) XP-CLR and relative diversity near GRMZM2G448355; gene orientation is indicated with arrows. (d) The gibberellin biosynthesis pathway30. (e) and (f) XP-CLR near the improvement candidates GRMZM2G152354 and dwarf1. The high-yielding rice variety IR8 has a mutation in GA20ox22.
To further characterize the genomic impact of domestication, we used long-oligo array hybridization to survey expression of 18,242 genes in the FGS in seedling tissue of a subset of 25 improved maize and seven parviglumis lines (Supplementary Table 1). Compared to non-candidates, our domestication candidates show greater absolute change in expression between parviglumis and maize (29% versus 22% in non-candidates, p=0.004, Supplementary Fig. 10), upregulation in maize relative to parviglumis (11.4% of candidates upregulated versus 6.5% of non-candidates, p=0.001, Supplementary Fig. 10) and a 10% lower coefficient of variation in expression among maize lines (p=0.006). Reduced variation in expression is observed throughout candidate features, suggesting removal of cis-variation at sites linked to the target of selection. Improvement candidates also show decreased variation in expression in maize relative to parviglumis (8% reduction in CoV, p=0.019), but do not show a significant change in the magnitude of expression. While the reduction in variation in expression could be due to selection on linked sites, the directional change seen in domestication candidates suggests the action of selection on cis-acting regulation.
Although changes in expression during domestication are unlikely to be limited to seedling tissue, our domestication candidates show no tissue-specific patterns of expression (Supplementary Fig. 11a; ref. 25). Improvement candidates also show no tissue specificity, but are more highly expressed than non-candidates in all but one of the tissue groups evaluated (p=0.025–0.044; Supplementary Fig. 11b). Because improvement candidates show no significant change in expression between teosinte and modern inbreds, this latter result suggests that modern maize improvement may have targeted loci that were already highly expressed. Comparison to the full FGS finds no evidence for an overall bias towards constitutive expression in our candidates, in contrast to previous resequencing scans2,26 (Supplemental Table 8).
Finally, we took advantage of expression data from crosses between inbred lines to evaluate levels of dominance in our candidate genes27. Domestication candidates show elevated dominance (p=0.001), but no significant difference in dominance (t-test, p=0.74) between crosses from the same or different genetic (heterotic) groups, a result that can be explained simply by the loss of additive cis-regulatory variation. Improvement candidates, in contrast, show higher dominance of expression than non-candidates (p=0.007) mostly due to higher dominance in crosses between heterotic groups (p=0.001), likely reflecting the important role that complementation between heterotic groups has played in maize improvement.
Our comparative genomic analysis of wild, landrace, and modern maize sheds light on the complexities of crop evolution and offers guidance to modern breeding. George Beadle28 and others4,5 have shown that a few genes (e.g., tb1 and tga1) radically altered some aspects of morphology during domestication. The majority of domestication features we identify show stronger evidence of selection than these canonical domestication genes, implying domestication targeted hundreds of genes of diverse biological function that likely involved unstudied aspects of phenotype. The loss of diversity at sites linked to selection and the observed enrichment of improvement candidates for highly expressed genes suggests that modern breeding has mostly worked with the “low-hanging fruit” of the genome and that much could be gained by focusing breeding efforts on the effective incorporation of diversity at other loci.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Methods
Sequencing and read mapping
As part of the maize Hapmap II project, sequence was generated and paired-end libraries were prepared in a subset of 75 of the 103 lines described in Chia et al.3 Sampled accessions include 35 improved maize lines, 23 traditional landraces, and 17 wild relatives but exclude 28 tropical inbreds from the International Maize and Wheat Improvement Center (CIMMYT) sampled by Chia et al.3 Sequence depth for each line is detailed in Supplementary Table 1. Lines were sequenced to a mean depth of 5.05X (median 4.58X) with mean coverage in analyzed regions of 3.74X (median 3.70X). Paired-end sequence was aligned to the AGPv1 B73 reference genome31, and SNPs called following algorithms described in Chia et al.3 We further filtered SNPs by retaining only non-singleton, biallelic SNPs with ≤ 50% missing data across all three groups of interest (parviglumis, landraces, and improved lines) resulting in a final dataset of 21,141,953 SNPs.
Error rate estimation
Per-nucleotide error rates for the HapMap II data set were estimated to be ~0.1% when compared to Sanger-sequenced BACs, on par with ~0.12% error across loci genome-wide for Sanger resequencing data3. To estimate the genotypic error rate in our subset of typed SNPs, SNP calls from the maize Hapmap II and maize Illumina Infinium 55K chip datasets were compared for a subset of 66 lines (SNP data for the 55K lines are available at www.panzea.org). The Infinium 55K data set is comprised of 51,584 SNPs, of which 37,279 were in common with the subset of Hapmap II SNPs used in our study. Out of 2.2 million comparable genotypes, 2.45% differed between the SNP calls from the two datasets. Most (88%) of the genotypic discrepancies consisted of a heterozygous versus homozygous call; hence, if we assume no errors in the Infinium 55K data set, the mean and median homozygous error rates in our data set per line (0.31% and 0.20% respectively) were much lower than the overall genotypic error rates and the actual perallele error rate was ~1%. Most (59.1%) SNPs had a genotypic error rate of 2% or less, 30.2% of SNPs had zero errors, and only 1.5% of SNPs had an error rate higher than 10%. Both the genotypic and homozygous error rates followed a unimodal distribution with a mode of zero, and there was no obvious second class of SNPs with poor performance with respect to error rates. B73 had the lowest genotypic error rate (0.87%) and the third lowest homozygous error rate (0.12%); this suggests that most of the genotyping errors in the Hapmap II data resulted from alignment issues rather than raw sequencing errors. The four teosinte lines with the highest homozygous error rates (TIL01, TIL07, TIL08, and TIL09) were represented in the two datasets by individuals of different selfing generations (e.g., S5 vs. S8 for TIL08).
Genome scan for selection
We performed a genome scan using the composite likelihood approach (XP-CLR) of Chen et al.12, modified to incorporate missing data (code available on request). Evidence for selection across the genome during domestication and improvement was evaluated in two contrasts: landraces vs. parviglumis for domestication and improved lines versus landraces for improvement. Our scan used a 0.05-cM sliding window, stepping every 100 bp across the genome. Individual SNPs were assigned a position along the genetic map32 by assuming uniform recombination between mapped markers. To ensure comparability of the composite likelihood score in each window, we fixed the number of SNPs assayed in each window at 50. Following Chen et al.12, we down-weighted pairs of SNPs in high LD (R2 > 0.70) to minimize the effect of dependence on the composite likelihood score. Final estimates were tabulated in non-overlapping 10-kb windows across the genome, assigning each 10-kb window the mean likelihood score (XP-CLR) and selection coefficient (s) estimated by the method of Chen et al.12
To partially account for the non-independence of XP-CLR scores along the physical map, we grouped regions into putatively selected “features”. Features were defined as groups of 10-kb windows with XP-CLR values above the genome-wide 80th percentile uninterrupted by more than one window below this threshold. Features falling within 0.05 cM of functional centromeres33 and an inversion on chromosome 1 (Supplementary Note) were masked from subsequent analyses. Our analyses of regions selected during domestication and improvement focused on features in the highest 10th percentile of mean feature-wise XP-CLR. However, we applied a more stringent criterion for identifying candidate genes, drawing only from features in which the observed reduction in nucleotide diversity during domestication or improvement was lower than the median observed from 1000 random windows of similar width and nucleotide diversity. We assigned the maize FGS gene closest to the window with the maximum XP-CLR value as the most likely candidate.
Population genetic analyses
Individual SNPs for each gene were classified as noncoding, synonymous coding, or nonsynonymous coding based on annotations of the first transcript in the FGS. Standard population genetic summary statistics (π, ρ, FST, Tajima’s D, Fay and Wu’s H′) were calculated for non-overlapping, 10-kb windows across the genome and separately for individual genes in the FGS using a combination of custom scripts, programs written using the libsequence C++ library34, and SAMtools35. In addition to statistics within groups, we calculated a weighted FST36 between groups and net pair-wise divergence between parviglumis and the outgroup Tripsacum. We tested for outliers of our summary statistics in candidate regions by comparing average values to a distribution calculated for randomly sampled, non-overlapping, genomic regions of identical width. Site frequency spectra were rescaled to address missing data and differing sample sizes (Supplementary Note).
To estimate mean haplotype lengths in each group, we used a custom perl script to choose 1 million starting points uniformly across the genome. At each point we chose two random lines from within a group (parviglumis, landraces, improved lines). We compared the two lines’ genotypes at the focal point, extending outward in both directions until we found different genotypes. Missing data and heterozygous SNPs were ignored.
Historical recombination rates (ρ=4Ne) were estimated using the composite likelihood approach of Hudson37. For estimating recombination, we treated the data as haploid, coding all heterozygous sites as missing data. We subsequently removed all SNPs with minor allele counts of ≤ 2. For windows with ≥ 10 remaining SNPs, values of ρ per bp were estimated across a grid of values from 10−4 to 0.2, assuming no homologous gene conversion
Expression analyses
Three separate datasets were used to assess patterns of gene expression in maize and parviglumis transcriptomes. First, using a custom long oligonucleotide microarray38 designed by NimbleGen (GPL10846, Roche NimbleGen, Madison, WI), we characterized variation in gene expression in a subset of our 25 improved maize and 7 parviglumis inbred lines (Supplementary Table 1). Multiple replications of the maize inbred lines B73 and Mo17 were included to assess consistency. Plants were grown and seedling leaf tissue was harvested 8 days after germination. RNAs were isolated using the commercial TRIzol (Invitrogen, Carlsbad, CA; cat# 15596026) from above-ground tissue and purified by Lithium Chloride treatment followed by 3 M sodium acetate (0.1 vol) and 95% ethanol (2.5 vol) precipitation. Purified RNAs (10 ug) were reverse transcribed and labeled according to the array manufacturer protocol. Per sample, ~20 µg of Cy3- or Cy5-labeled RNAs were hybridized for 16–20 hours at 42 °C using the NimbleGen Hybridization System. Post hybridization, slides were washed (NimbleGen Wash Buffer Kit) and dried for two minutes by centrifugation. Slides were immediately scanned using the GenePix 4000B Scanner (Molecular Devices, Sunnyvale, CA) according to the array manufacturer protocol.
Array images and data were processed using NimbleScan software. Briefly, images from each slide were separated into 12 subarrays and aligned to a grid to extract signal intensity for each feature on the array. Experimental integrity was verified by evaluation of the signal intensities of the sample tracking control features for each subarray. In addition, metrics reports were produced for each array to describe the signal uniformity across the array and the intensity of known empty features, random probes and experimental probes. Signal-to-noise ratios were estimated by dividing the average signal intensity of experimental gene probes by that of the control probes. Only slides with a signal-to-noise ratio ≤ 2 were retained. NimbleScan was used to generate RMA-normalized39 gene expression values from the spatially-corrected probe signal intensities on a per probe and per gene basis. Normalized gene expression values across multiple replications (technical or biological) of the same genotype were averaged, when possible. Comparisons of the distributions of signal intensity for control and experimental probes (Supplementary Fig. 12) were used to determine a reasonable signal threshold for positive expression across all slides. Genes with average probe log2 signals of > 10 in at least three arrays were retained as expressed (N=19,792 expressed genes).
Nucleotide polymorphism may contribute to differences in hybridization between transcripts from divergent genotypes, though previous results suggest as many as 4–5 SNPs are needed to strongly affect probe hybridization40. To minimize the impact of polymorphism on hybridization, we further filtered the probeset based on a previous comparative genomic hybridization dataset developed using many of the same genotypes and the same array platform38. Probes that exhibited substantially reduced CGH values for at least three genotypes (26,937 probes) were removed resulting in a set of 46,167 probes that detected expression of 18,242 genes with one to four probes per gene. This subset of the data was used for all subsequent analyses. Finally, a Spearman’s rank correlation showed a weakly positive correlation between parviglumis expression and FST parviglumis/maize (rho = 0.043) providing no evidence for hybridization bias.
Our second data set comprised expression estimates in 60 different tissues of the inbred line B73 from a NimbleGen array of 23,740 genes in the FGS25. These data were curated and categorized following conventions described in Sekhon et al.25.
The third dataset consisted of expression from 5 improved lines and their F1 hybrids characterized using an Affymetrix GeneChip® Maize Genome Array (GMGA)27. Probesets with fewer than two biological replicates were dropped, and remaining probesets were annotated by comparing the AGPv1 physical map positions of array probes to the FGS41. Probesets were included in the analysis only if they mapped to a single gene. When multiple probesets mapped to a single gene, expression data from all probesets were averaged for further analysis.
Significant differences between candidate and non-candidate expression values were determined by bootstrap resampling of log2-transformed, RMA-normalized data from non-candidate genes. The validity of a bootstrap approach was assessed by plotting the mean of each bootstrap sample against its variance in order to confirm calculated test statistics were pivots42. For tissue-specific expression, bootstrap significance values were adjusted for multiple tests with a Benjamini-Hochberg False Discovery Rate correction at the 0.05-level. Dominance was assessed as:
Finally, in order to ensure that the relatively low coefficient of variation in expression observed in candidates did not result in an inflation of estimates of dominance, an analysis of covariance was conducted with the coefficient of variation included as a covariate.
Supplementary Material
Acknowledgments
The authors would like to thank T. Kono, S. Watson and Captain M. Watson for photographs of inflorescences, P. Brown for help with QTL delineation, and B.S. Gaut and A.M. Gonzales for comments on an earlier version of the manuscript. This work was supported by funding from the maize diversity project - NSF IOS-0820619 (E.S.B., J.D., M.D.M.) and USDA-ARS (E.S.B., M.D.M., D.W.), USDA Hatch Funds (P.T. and N.M.S.), the Chinese 973 program 2007CB815701 (J.W.), the Chinese Ministry of Agriculture 984 program no. 2010-Z13 (G.Z.), the Shenzhen Municipal Government Basic Research Program (J.W.), the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494), Contract No. DE-AC02-05CH11231 from the Office of Science of the U.S. Department of Energy to the U.S. Department of Energy Joint Genome Institute, and by grants NSF IOS-0922703 and USDA-NIFA 2009-01864 (J.R.-I.).
Footnotes
Data availability. Expression data are available from the Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo/ under the series accession number GSE30036.
Note: Supplementary information is available on the Nature Genetics website.
Author Contributions
M.B.H., X.X., J.v.H., and T.P. contributed equally to this work. J.D., M.D.M., E.S.B., D.W., and J.R.-I. designed the project; M.B.H., J.v.H., T.P., and J.R.-I. performed most data analyses; J.D. developed wild and landrace inbred lines; E.S.B., S.K., J.L., M.D.M., and D.W. contributed sequence data for inbred maize and parviglumis; K.E.G. and R.J.E. developed libraries and managed sequencing for inbred maize and parviglumis; X.X., S.Y., J.W., and G.Z. directed sequencing for landrace maize, mexicana, and Tripsacum; E.S.B., J.R.-I., D.W., and X.X. directed bioinformatics; J.-M.C. and C.S. performed read mapping, SNP calling and annotation, and analysis of coding sequence; E.S.B., J.-M.C., and J.C.G. performed quality control filtering of SNPs; N.M.S., R.A.S.-W., and P.T. generated Nimblegen expression data for maize and parviglumis; S.K. provided early access expression data; L.S. reanalyzed QTL data for domestication traits; R.A.C. analyzed site frequency spectra; M.B.H., J.v.H., T.P., P.L.M., and J.R.-I. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Briggs WH, McMullen MD, Gaut BS, Doebley J. Linkage mapping of domestication loci in a large maize-teosinte backcross resource. Genetics. 2007;177:1915–1928. doi: 10.1534/genetics.107.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright SI, et al. The effects of artificial selection of the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 3.Chia J-M, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. doi: 10.1038/ng.2313. (Accept in Principle) [DOI] [PubMed] [Google Scholar]
- 4.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium BP maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci U S A. 2009;106:5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci U S A. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci U S A. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 11.Wilkes HG. Teosinte: The Closest Relative of Maize. The Bussey Institute of Harvard University; Cambridge, Massachusetts: 1967. [Google Scholar]
- 12.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purugganan MD, Fuller DQ. Archaeological data reveal slow rates of evolution during plant domestication. Evolution. 2011;65:171–183. doi: 10.1111/j.1558-5646.2010.01093.x. [DOI] [PubMed] [Google Scholar]
- 14.Olsen KM, et al. Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palaisa K, Morgante M, Tingey S, Rafalski A. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci U S A. 2004;101:9885–9890. doi: 10.1073/pnas.0307839101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camus-Kulandaivelu L, et al. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics. 2006;172:2449–2463. doi: 10.1534/genetics.105.048603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown PJ, et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011;7:e1002383. doi: 10.1371/journal.pgen.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 19.Gallais A, Hirel B. An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot. 2004;55:295–306. doi: 10.1093/jxb/erh006. [DOI] [PubMed] [Google Scholar]
- 20.Jackson D, Hake S. Control of phyllotaxy in maize by the abphyl1 gene. Development. 1999;126:315–323. doi: 10.1242/dev.126.2.315. [DOI] [PubMed] [Google Scholar]
- 21.Gualberti G, et al. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell. 2002;14:1253. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki A, et al. Green revolution: A mutant gibberellin-synthesis gene in rice - New insight into the rice variant that helped to avert famine over thirty years ago. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005;43:413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- 24.Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekhon RS, et al. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 26.Hufford KM, Canaran P, Ware DH, McMullen MD, Gaut BS. Patterns of selection and tissue-specific expression among maize domestication and crop improvement loci. Plant Physiol. 2007;144:1642–1653. doi: 10.1104/pp.107.098988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stupar RM, et al. Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol. 2008;8 doi: 10.1186/1471-2229-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beadle GW. Teosinte and the origin of maize. J Hered. 1939;30:245–247. [Google Scholar]
- 29.Ryu CH, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- 30.Lo SF, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 32.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 33.Wolfgruber TK, et al. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 2009;5:e1000743. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornton K. libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- 35.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 37.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson-Wagner RA, et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010;20:1689–1699. doi: 10.1101/gr.109165.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 40.Springer NM, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5:e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence CJ, Dong OF, Polacco ML, Seigfried TE, Brendel V. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 2004;32:D393–D397. doi: 10.1093/nar/gkh011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge University Press; New York, New York: 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.