Abstract
Computational anthropomorphic phantoms have become an important investigation tool for medical imaging and dosimetry for radiotherapy and radiation protection. The development of computational phantoms with realistic anatomical features contribute significantly to the development of novel methods in medical physics. For many applications, it is desirable that such computational phantoms have a real-world physical counterpart in order to verify the obtained results.
In this work, we report the development of a voxelised phantom, the HIGH_RES_HEAD, modelling a paediatric head based on the commercial phantom 715-HN (CIRS). HIGH_RES_HEAD is unique for its anatomical details and high spatial resolution (0.18 × 0.18 mm2 pixel size). The development of such a phantom was required to investigate the performance of a new proton computed tomography (pCT) system, in terms of detector technology and image reconstruction algorithms.
The HIGH_RES_HEAD was used in an ad-hoc Geant4 simulation modelling the pCT system. The simulation application was previously validated with respect to experimental results. When compared to a standard spatial resolution voxelised phantom of the same paediatric head, it was shown that in pCT reconstruction studies, the use of the HIGH_RES_HEAD translates into a reduction from 2% to 0.7% of the average relative stopping power difference between experimental and simulated results thus improving the overall quality of the head phantom simulation.
The HIGH_RES_HEAD can also be used for other medical physics applications such as treatment planning studies.
A second version of the voxelised phantom was created that contains a prototypic base of skull tumour and surrounding organs at risk.
Keywords: high-resolution, voxelised phantom, DICOM, Geant4
1 Introduction
The development of anthropomorphic phantoms, both physical and computational, is an active field of investigation in medical physics [1]. Anthropomorphic computational phantoms have undergone an evolution from simple stylized phantoms to voxelised phantoms and, more recently, to hybrid phantoms offering a mixture of surface-based and voxelised representations [2–4]. Stylized mathematical phantoms [3], which are based on 3D surface equations for internal organs definition, provide only a rough approximation of the true anatomy of individual patients. Voxelised [4] and hybrid phantoms [2] are usually generated from CT and/or MRI data of patients or volunteers. They provide a better anatomical detail, but are frequently compromised by image noise, partial-volume averaging and imaging artefacts. Despite these drawbacks, it has been well established that voxelised phantoms can be successfully used in a wide range of medical physics applications [5–10]. In this work we describe the development and use of a novel high resolution voxelised head phantom, called here HIGH_RES_HEAD, based on a high resolution CT acquisition of a physical paediatric head phantom (HN715, CIRS).
The HIGH_RES_HEAD was initially created for proton computed tomography (pCT) studies when it became clear that simple geometrical phantoms such as, for example, the Catphan® 600 series (The Phantom Laboratory, Salem, New York, USA) were not sufficient to fully characterize pCT, but that an accurate representation of the human anatomy was necessary. PCT is a promising imaging technique that could add to or substitute for x-ray CT in treatment planning and in-room image guidance applications, as it allows for direct reconstruction of proton relative stopping power (RSP) from proton energy loss measurements [11].
2 Material and methods
2.1 Development of the HIGH_RES_HEAD
The HIGH_RES_HEAD was created from the CT scan of a commercially available tissue-equivalent dosimetry phantom (ATOM®, Model 715 HN, CIRS Inc., Norfolk, VA) (Fig. 1a).
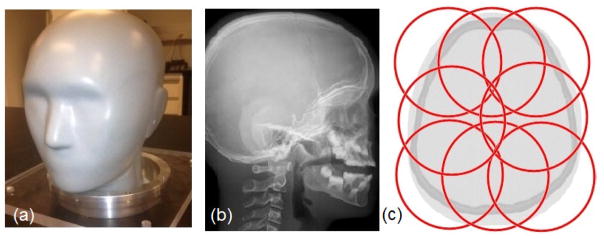
Figure 1.
(a) Head phantom (HN715, CIRS); (b) lateral x-ray radiograph of the head phantom demonstrating its anatomical detail; (c) arrangement of eight partially overlapping FOVs represented by the red circles of 9.6 cm diameter.
The physical phantom provides very realistic anatomical details of the head and spine of a 5-year-old child including skeletal and soft tissue features, intra-cranial and paranasal sinuses, ear canals, and outer head contours (Fig. 1b). The physical phantom is composed of the following seven tissue-equivalent materials (density in g/cm3): soft tissue (1.055), brain (1.07), paediatric spinal disc (1.10), paediatric trabecular bone (1.13), 5-year-old compact bone (1.75), tooth dentine (1.66), and tooth enamel (2.04). All materials of the real phantom are homogeneous in their density and composition; a few minor defects such as small cavities can be present, which were not included in the HIGH_RES_HEAD. The proprietary atomic composition of each material is available from CIRS upon request.
Eight separate helical CT scans of the entire physical head phantom were acquired with a 64-detector-row CT scanner (Lightspeed, GE Healthcare, Waukesha, WI) using an image matrix size of 512×512 pixels and a display FOV of 9.6 cm, corresponding to a pixel size of 0.1875 mm × 0.1875 mm. The slice thickness was 1.25 mm. The display FOVs were partially overlapped so that each part of the phantom was covered by at least one display FOV (Fig. 1c). A single DICOM study with 128 slices and matrix size of 1024×1024 pixels was generated from the CT scan with a segmentation study performed with Matlab (The MathWorks Inc., Natick, MA, USA).
Another voxelised virtual phantom, called here CONVENTIONAL_HEAD, was created scanning the same physical head phantom with the same x-ray CT scanner at Loma Linda University Medical Center using an image matrix size of 512×512 pixels and a display FOV of 37 cm, in order to cover the entire phantom with one scan. Its spatial resolution is lower than the case of the HIGH_RES_HEAD. The pixel size was 0.72 mm × 0.72 mm, the slice thickness was 1.25 mm and 171 slices were collected in a single DICOM study. The CONVENTIONAL_HEAD was not subjected to any image segmentation process but was developed to be used as a term of comparison to quantify the effect of adopting a high spatial resolution and a noiseless virtual phantom, such as the HIGH_RES_HEAD, when characterising a pCT system.
2.2 Segmentation of the anatomy of the HIGH_RES_HEAD
The different tissue regions of the phantom were segmented in each CT slice using ImageJ version 1.46r (http://imagej.nih.gov/ij). The Hounsfield unit (HU) values of the outer air and most of the tissue regions were found to be well described by well-separated Gaussian distributions with mean and standard deviations listed in Table 1.
Table 1.
Mean and standard deviation (SD) of the HU values 7 tissue-equivalent phantom materials in the HN715 phantom, ordered from lowest to highest mean value.
| Material | Mean (HU) | SD (HU) |
|---|---|---|
| Soft tissue | 24 | 9 |
| Brain | 52 | 8 |
| Spinal disc | 92 | 2 |
| Trabecular bone | 197 | 7 |
| Cortical bone | 923 | 107 |
| Tooth dentin | 1280 | 27 |
| Tooth enamel | 2310 | 80 |
The first step of the segmentation process consisted in identifying continuous boundaries between the voxelised phantom and the surrounding air, and between the different tissues of the head itself by means of a thresholding process. To detect entire tissue regions and their boundaries, different windows of HU were selected using a custom thresholding macro in ImageJ. Imperfections in the boundaries were manually edited as guided by anatomical knowledge or by the fact that they were obvious artefacts.
The second step consisted in importing the thresholded ImageJ images in black and white in Microsoft (MS) Paint, where the different tissue regions were assigned to specific colours. Voxels which were found to deviate from their immediate neighbours in the interior of each tissue region were assigned to the tissue of the surrounding medium, thus eliminating single-voxel errors.
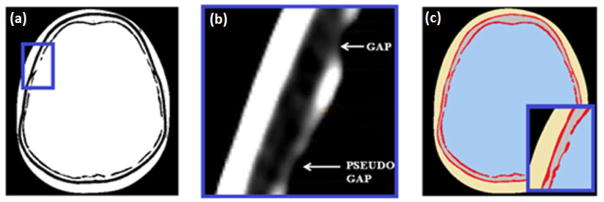
The third step consisted in assigning the corresponding mean HU value, listed in Table 1, to the voxels of each identified tissue region, to eliminate the noise affecting the phantom. The regions with sinus, consisting of lung inhale tissue equivalent material in the physical head phantom, were assigned to the HU value of air in the HIGH_RES_HEAD. Fig. 2 illustrates, for a given slice, the steps from the thresholded image to the final phantom bitmap image.
Figure 2.
(a) Thresholded ImageJ image; (b) close-up view of the inner table with the DicomWorks viewer showing a real gap and pseudo gap; (c) final bitmap image after the segmentation process; the pseudo gap has been eliminated while the real gap has been kept. The image is noiseless. Each tissue is identified with the corresponding mean HU, as listed in Table 1.
After all slices were segmented as described above, the digital head phantom images were combined into a single DICOM study.
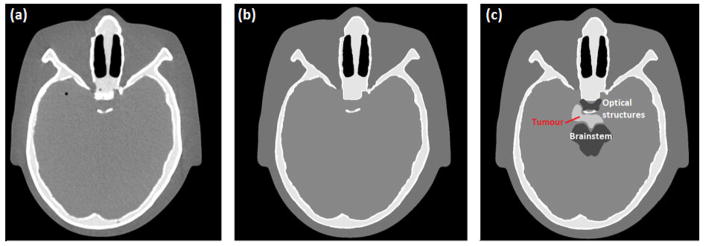
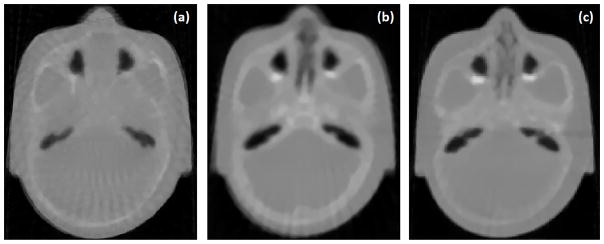
Fig. 3 shows a slice of the x-ray CT scan of the physical head phantom and the corresponding image of the HIGH_RES_HEAD (Fig 3a and b, respectively). A second version of the HIGH_RES_HEAD was created by implementing a base of skull tumour and surrounding organs at risk, shown in Fig. 3c. In order to visualize these regions, HU values of brain +100 were assigned to tumour and brain -100 to OARs, respectively. Position, shape, size and HU value of the tumour were decided under the supervision of one of the authors, Dr. R. W. Schulte, who is a board-certified radiation oncologist.
Figure 3.
(a) x-ray CT scan of the physical head phantom; (b) corresponding image of the HIGH_RES_HEAD; (c) same image of the modified version of HIGH_RES_HEAD with tumour and surrounding OAR.
2.3 Implementation of the HIGH_RES_HEAD in an ad-hoc Geant4 simulation for pCT
The HIGH_RES_HEAD was adopted to characterize a prototype pCT system developed by the pCT collaboration. The design of the prototype pCT scanner is described in [12].
Individual protons are tracked before entering and after exiting the phantom or patient with 2D-sensitive silicon trackers. Each telescope consists of four planes of paired silicon strip sensors with orthogonally arranged strip orientation [13]. The scanned objects are placed at the centre between the telescopes. When performing a full pCT scan, the phantom is rotated on a vertical axis in discrete steps of 4 degrees. In addition to proton coordinates, the residual energy of the protons traversing the scanned object is measured and calibrated in terms of water equivalent path length (WEPL) [14]. The WEPL, required as input in the pCT reconstruction, is measured with a 5-stage scintillating detector made of polystyrene with a sensitive area of 36 cm × 10 cm and a thickness of 5.1 cm per stage [15]. The data acquisition has been described elsewhere [16]. 3D images of the RSP are generated using an image reconstruction software that takes WEPL, position and direction of individual protons as input [17].
The HIGH_RES_HEAD was included in a Geant4 (version 10.1) [18] simulation specifically developed to study the performance of the prototype pCT scanner (Fig. 4). In particular, it was implemented in the Geant4 DICOM extended example [19], which was then integrated in the validated ad-hoc Geant4 simulation, developed to study the novel pCT system [20]. The Geant4 DICOM extended example creates a Geant4 voxelised geometry based on the information of input DICOM files, using the Geant4 Parameterised Volumes geometry functionality (G4VPVParameterisation). The default G4RegularNavigation/G4PhantomParameterization was used, with the default voxel skipping option activated. By default, in the Geant4 example, the HU value of each voxel, information contained in the input DICOM study, is converted into the corresponding density based on the stoichiometric calibration method, described by Schneider et al. [21]. In addition, lower and upper bounds of density intervals must be defined by the user in the DICOM example in order to assign corresponding tissue materials.
Figure 4.
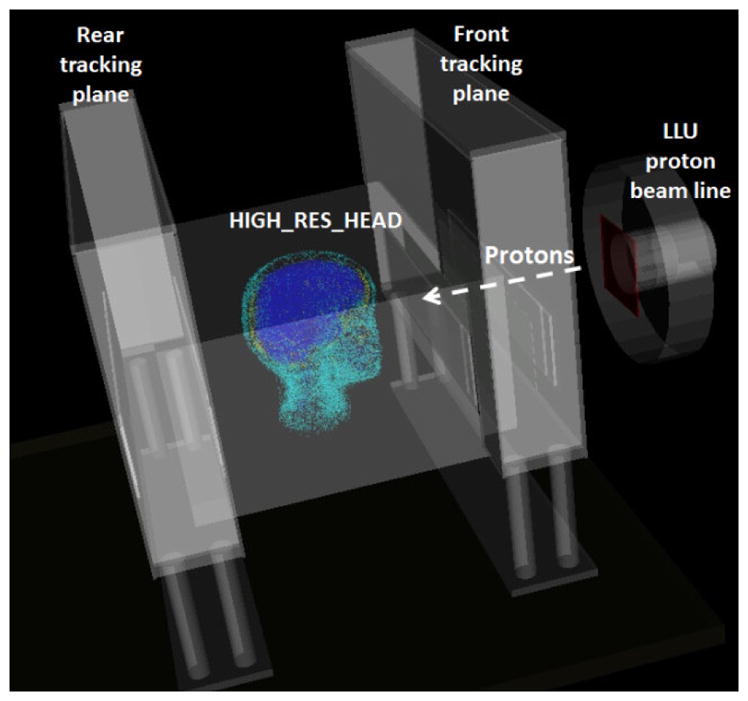
Visualisation of the Geant4 application set-up modelling the novel pCT scanner including the HIGH_RES_HEAD.
The uncertainties introduced by converting HU values into tissue materials using a calibration curve were investigated by Paganetti et al. [22–24] for proton dose calculations and range verification studies. It was shown that when the conversion is based on a proper stoichiometric calibration, the proton beam range uncertainty is about 0.2%. Therefore, the calibration curve based on the characteristic of the LLUMC CT scanner was used to model the CONVENTIONAL_HEAD in the Geant4 DICOM application. Since the HIGH_RES_HEAD contains only 8 HU values, resulting from the segmentation work described in Section 2.2, it was decided to assign each HU value directly to the corresponding material of the physical phantom (see Table 1), bypassing the stoichiometric calibration curve.
In the Geant4 simulation dedicated to the characterization of the novel pCT system, the research proton beam line of the medical proton synchrotron at Loma Linda University Medical Center (LLUMC) [25] was modelled, together with the silicon strip tracking planes and the multi stage scintillator detector (see Fig. 4). The Low Energy Package, based on the Livermore data libraries [26], was selected to model the electromagnetic interactions. The G4HadronPhysicsQGSP_BIC_HP and the G4HadronElasticPhysicsHP were chosen to describe the inelastic and elastic scattering of hadrons, respectively. The neutron High Precision (HP) model was selected to describe neutron interactions up to 20 MeV. Ion hadronic interactions were described by means of the G4IonBinaryCascadePhysics [18].
In this work, all simulated pCT scans of the HIGH_RES_HEAD were obtained with 90 projections (4-degree intervals). The total number of proton histories generated for each projection was 12 × 106. An iterative algebraic reconstruction algorithm [27] was used to calculate the 3D RSP map of the HIGH_RES_HEAD taking WEPL, position and direction of individual protons as input [17].
The reconstructed 3D RSP map consists of a sequence of images (pCT slices) in which each pixel corresponds to one RSP value. For comparative analysis, the RSPs of the different materials were determined using ImageJ. A region of interest was selected within the boundaries of each tissue and mean and standard deviation (SD) of RSP were calculated using standard ImageJ functions. Since some material regions in the head phantom, e.g. enamel and cortical bone, had very limited spatial extension, the RSP was calculated by combining the results typically from 3–5 slices (up to 10) reconstructed pCT slices. RSP values reconstructed from simulated pCT data obtained with the HIGH_RES_HEAD and the CONVENTIONAL_HEAD were compared with those derived from experimental pCT data of the underlying physical head phantom. The experimental RSP values were obtained and reconstructed with the same algorithm and image parameters as the simulated pCT data.
3 Results
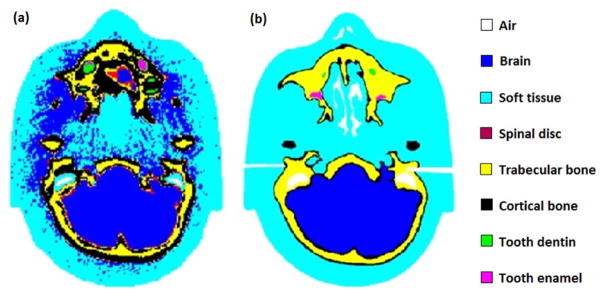
Fig. 5 shows the visualisation of the CONVENTIONAL_HEAD and HIGH_RES_HEAD in Geant4. Fig. 6 shows the pCT images reconstructed from (a) simulated CONVENTIONAL_HEAD data, (b) simulated HIGH_RES_HEAD data, and (c) experimental data with the physical phantom. Approximately the same number of protons per projection (~3·106) were used for each image reconstruction. The mean and standard deviation (SD) of tissue RSP values in the three different images were calculated and are compared in Table 2.
Figure 5.
(a) CONVENTIONAL_HEAD and (b) HIGH_RES_HEAD, visualized in Geant4.
Figure 6.
pCT image reconstruction of (a) simulated CONVENTIONAL_HEAD (b) simulated HIGH_RES_HEAD and (c) experimental real phantom. The visible radial strikes are reconstruction imaging artefacts.
Table 2.
Comparison between mean +/− standard deviation (SD) of RSP values derived from experimental data and simulated data of the anthropomorphic pediatric head phantom.
| Experimental RSP | Simulated RSP CONVENTIONAL_HEAD | Simulated RSP HIGH_RES_HEAD | |||
|---|---|---|---|---|---|
| Material | Mean ± SD | Mean ± SD | Difference1 [%] | Mean ± SD | Difference2 [%] |
| Soft Tissue | 1.032±0.025 | 1.025±0.018 | -0.7 | 1.026±0.014 | -0.6 |
| Brain Tissue | 1.044±0.08 | 1.038±0.012 | -0.6 | 1.043±0.012 | -0.1 |
| Spinal Disc | 1.069±0.017 | 1.089±0.025 | 1.9 | 1.053±0.039 | -1.5 |
| Trabecular Bone | 1.111±0.008 | 1.122±0.028 | 1.0 | 1.110±0.020 | -0.1 |
| Cortical Bone | 1.331±0.032 | 1.288±0.046 | -3.2 | 1.312±0.082 | -1.4 |
| Tooth Dentin | 1.524±0.122 | 1.551±0.089 | 1.8 | 1.521±0.080 | -0.2 |
| Tooth Enamel | 1.651±0.050 | 1.732±0.078 | 4.9 | 1.640±0.064 | -0.7 |
| Average of the absolute value of the RSP difference [%] | 2.0 % | 0.7 % | |||
(simulated RSP CONVENTIONAL_HEAD – experimental RPS)/experimental RSP
(simulated RSP HIGH_RES_HEAD – experimental RSP)/experimental RSP
It is obvious that the different tissues in the CONVENTIONAL_HEAD (Fig. 5(a)) were not as well defined as in the HIGH_RES_HEAD (Fig. 5(b)). For example, in CONVENTIONAL_HEAD, many soft tissue voxels were incorrectly modelled as brain voxels. Also, in CONVENTIONAL_HEAD, the extent of the cortical bone region was overestimated, and spinal disc voxels were found in regions close to cortical bone where they are not present anatomically. The noiseless digital phantom HIGH_RES_HEAD, on the other hand, not only has a higher spatial resolution but also provides a more accurate representation of the physical head phantom in terms of shapes and boundaries of the anatomical structures. This leads to a more accurate reconstruction of RSP values when simulating the pCT scanner as shown in Table 2. The comparison between experimental and simulated RSP of different tissue-equivalent materials shows that the agreement is within 1.5% for the HIGH_RES_HEAD and 4.9% for the CONVENTIONAL_HEAD.
The improvement can vary significantly depending on the specific tissue. In particular, the RSP of spinal disc, cortical bone and tooth enamel are the ones differing the most from the experimental values for both the phantoms. The RSP difference for spinal disc calculated with the HIGH_RES_HEAD and the CONVENTIONAL_HEAD is -1.5% and 1.9%, respectively. This can be explained by the fact that in the reconstructed images the spinal disc is difficult to be distinguished from the brain because their RSPs are very similar.
The tooth enamel and cortical bone are very thin anatomical structures (1–2 mm), thus making it difficult to select homogeneous regions to measure their RSPs, explaining the difference between simulated and experimental measurements for both phantoms. However, it can be observed that the results are significantly better for the HIGH_RES_HEAD. A limited spatial resolution, such the one of the CONVENTIONAL_HEAD, does not allow resolving such small regions, which are strongly compromised by partial-volume averaging effect with the surrounding tissues. This result shows how a phantom with a higher spatial resolution can be a more powerful tool to study the accuracy of novel imaging techniques and novel imaging reconstruction algorithm.
On average, the RSP difference between simulated and experimental results improved from 2% to 0.7% using the HIGH_RES_HEAD.
4 Discussion
A high-resolution CT-scan based digital phantom, HIGH_RES_HEAD, was created as an accurate representation of human head anatomy and was here used in an ad-hoc Geant4 simulation modelling an experimental pCT system. The phantom has a physical counterpart in the real world that was used to generate corresponding experimental data with the pCT system. Eight high resolution CT scans were combined and treated with a segmentation process creating a noise-free digital phantom. This was compared to the CONVENTIONAL_HEAD, which was created with the standard method of converting a CT scan of the underlying physical head phantom into a Geant4 geometry model. Note that the HIGH_RES_HEAD has been validated for imaging purposes [20] but not yet for dosimetry. In particular, when using the HIGH_RES_HEAD for dosimetric studies in Geant4, the G4NestedParametrization is recommended instead of G4RegularNavigation for a proper calculation of the dose in each voxel [28].
Having a counterpart in the real world has the advantage that performance of an experimental CT system can be predicated and optimized in the virtual world before the actual scanner is being built or as part of an iterative process of simulations and obtaining real-time data with stepwise improvement of the scanner hardware. The simulated data are also helpful in analysing and understanding the origin of real-world reconstruction artefacts. Having an accurate representation of the real-world phantom in the Geant4 simulation turned out to be very helpful in this respect. The HIGH_RES_HEAD can also be shared with other investigators who want to develop and optimize a different line of scanning hardware. A modified version of the commercial phantom 715-HN (CIRS) with a film stack insert in the posterior fossa also exists for range error measurements in proton therapy. Range error experiments can be simulated ahead of time in the corresponding modified version of the HIGH_RES_HEAD.
The HIGH_RES_HEAD is one of the highest resolution phantoms currently existing [1,29] that can be implemented in a MC code with feasible computational costs. The voxel size is considered one of the limitations of the tomographic models in modelling small organs [4] and with the HIGH_RES_HEAD this problem is overcome, thanks to the manual segmentation process adopted. Nipper et al. [4] also pointed out that in order to model microstructures such as mucosal layer, skeletal endosteum and trabecular bone, some assumption has to be made, especially when dealing with children imaging. With HIGH_RES_HEAD no assumptions were made to define the trabecular bone but an accurate analysis of each slice was conducted with the supervision of a board-certified radiation oncologist.
MC studies using virtual phantoms were previously used for imaging purposes [30] but this work is the first study that aims at quantifying how a higher spatial resolution digital phantom can impact the final reconstructed image, using experimental data as a benchmark. A similar study was conducted by Songxiang Gu et al. [31]: seven high-resolution heart phantoms for medical imaging and dosimetric purposes were created using triangular meshes for segmenting computed tomography angiograms images. The spatial resolution achieved was 0.35 × 0.35 × 0.40 mm3 but no physical counterpart in the real world is available.
A second version of the HIGH_RES_HEAD phantom that was also created with a prototypic base of skull tumour and surrounding OAR (Fig. 3c) has applications in treatment planning studies. As a recent application example (unpublished), this digital phantom version was imported into the research version of RayStation (RaySearch Laboratories, Stockholm, Sweden) and used for treatment planning studies calculating the dose delivered to the tumour and surrounding OAR by a very high energy electron scanning pencil beam (VHEE) plan with a Monte Carlo simulation assuming treatment delivery with a scanning electron pencil beam.
5 Conclusion
It can be concluded that the use of the HIGH_RES_HEAD in the application of simulating the performance of a pre-clinical pCT scanner led to an improvement of the resolution of the reconstructed RSP. The average RSP difference between simulated and experimental results was reduced from 2% (using the version of the phantom created with standard voxel size) to 0.7% (using the HIGH_RES_HEAD). Thus, the phantom is more suitable representing the real-world phantom in that simulation application. The HIGH_RES_HEAD also has applications in planning range verification and experiments with pCT or other CT modalities in the future. Furthermore, it can be used to simulate new treatment modalities in treatment planning studies as demonstrated in this work. The DICOM version of the HIGH_RES_HEAD can be made available to other investigators.
Development of a novel voxelized head phantom with high (sub-mm) resolution
The phantom is an accurate representation of an existing physical head phantom
The phantom was modelled in Geant4 to characterize a novel proton CT (pCT) system
The study shows the advantages of adopting a high resolution digital phantom in pCT
The digital phantom can be adopted in dosimetric and imaging studies
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zaidi H, Xu XG. Computational anthropomorphic models of the human anatomy: the path to realistic Monte Carlo modeling in radiological sciences. Annu Rev Biomed Eng. 2007;9:471–500. doi: 10.1146/annurev.bioeng.9.060906.151934. [DOI] [PubMed] [Google Scholar]
- 2.Lee C, Lodwick D, Hurtado J, Pafundi D, Williams JL, Bolch WE. The UF family of reference hybrid phantoms for computational radiation dosimetry. Phys Med Biol. 2010;55:339–63. doi: 10.1088/0031-9155/55/2/002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C, Williams JL, Lee C, Bolch WE. The UF series of tomographic computational phantoms of pediatric patients. Med Phys. 2005;32:3537–48. doi: 10.1118/1.2107067. [DOI] [PubMed] [Google Scholar]
- 4.Nipper JC, Williams JL, Bolch WE. Creation of two tomographic voxel models of. Phys Med Biol. 2002;47:3143–64. doi: 10.1088/0031-9155/47/17/307. http://dx.doi.org/10.1088/0031-9155/47/17/307. [DOI] [PubMed] [Google Scholar]
- 5.Caon M, Bibbo G, Pattison J, Staton RJ, Lee C, Lee C, et al. An EGS4-ready tomographic computational model of a 14-year-old female torso for calculating organ doses from CT examinations voxel-based computational models Organ and effective doses in newborn patients during MSCT An EGS4-ready. Phys Med Biol. 1999;44:2213–25. doi: 10.1088/0031-9155/44/9/309. [DOI] [PubMed] [Google Scholar]
- 6.Gualdrini G, Battisti P, Biagini R, De Felice P, Fazio A, Ferrari P. Development and characterisation of a head calibration phantom for in vivo measurements of actinides. Appl Radiat Isot. 2000;53:387–93. doi: 10.1016/S0969-8043(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 7.Milian FM, Attili A, Russo G, Marchetto F, Bourhaleb F, Cirio R. Development of virtual CT DICOM images of patients with tumors. Application for TPS and Monte Carlo dose evaluation. 2013 [Google Scholar]
- 8.Ferreira P, Baptista M, Di Maria S, Vas P. Cancer risk estimation in Digital Breast Tomosynthesis using GEANT4 Monte Carlo simulations and voxel phantoms. Phys Medica. 2016;32:717–23. doi: 10.1016/j.ejmp.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Leni P-E, Laurent R, Salomon M, Gschwind R, Makovicka L, Henriet J. Development of a 4D numerical chest phantom with customizable breathing. Phys Medica. 2016;32:795–800. doi: 10.1016/j.ejmp.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Ioppolo JL, Price RI, Tuchyna T, Buckley CE. Diagnostic x-ray dosimetry using Monte Carlo simulation. Phys Med Biol. 2002;47:1707–20. doi: 10.1088/0031-9155/47/10/307. [DOI] [PubMed] [Google Scholar]
- 11.Hanson KM, Bradbury JN, Cannon TM, Hutson RL, Laubacher DB, Macek RJ, et al. Computed tomography using proton energy loss. Phys Med Biol. 1981;26:965–83. doi: 10.1088/0031-9155/26/6/001. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RP, DeWitt J, Holcomb C, Macafee S, Sadrozinski HFW, Steinberg D. Tracker Readout ASIC for Proton Computed Tomography Data Acquisition. Psychol Sci. 2013;60:3262–9. doi: 10.1109/TNS.2013.2274663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadrozinski HWF, Johnson RP, Macafee S, Plumb A, Steinberg D, Zatserklyaniy A, et al. Development of a Head Scanner for Proton CT. Nucl Inst Methods Phys Res A. 2013;699:205–10. doi: 10.1016/j.nima.2012.04.029.Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley RF, Schulte RW, Bashkirov VA, Wroe AJ, Ghebremedhin A, Sadrozinski HWF, et al. Water-equivalent path length calibration of a prototype proton CT scanner. Med Phys. 2012;39:2438–46. doi: 10.1118/1.3700173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashkirov VA, Schulte RW, Hurley RF, Sadrozinski HWF, Zatserklyaniy A, Plautz TE, et al. Novel scintillation detector design and performance for proton radiography and computed tomography. Med Phys. 2016;43:664–74. doi: 10.1118/1.4939255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RP, Bashkirov VA, Dewitt L, Giacometti V, Hurley RF, Piersimoni P, et al. A Fast Experimental Scanner for Proton CT: Technical Performance and First Experience With Phantom Scans. IEEE Trans Nucl Sci. 2016;63:52–60. doi: 10.1109/TNS.2015.2491918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penfold SN, Schulte RW, Censor Y, Bashkirov VA, Rosenfeld AB. Characteristics of proton CT images reconstructed with filtered backprojection and iterative projection algorithms. IEEE Nucl Sci Symp Conf Rec. 2009:4176–80. doi: 10.1109/NSSMIC.2009.5402282. [DOI] [Google Scholar]
- 18.Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, et al. GEANT4 - A simulation toolkit. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers, Detect Assoc Equip. 2003;506:250–303. doi: 10.1016/S0168-9002(03)01368-8. [DOI] [Google Scholar]
- 19.n.d http://geant4.web.cern.ch/geant4/UserDocumentation/Doxygen/examples_doc/html/DICOM._8cc_source.html.
- 20.Giacometti V, Bashkirov VA, Piersimoni P, Guatelli S, Plautz TE, Sadrozinski HWF, et al. Software Platform for Simulation of a Prototype Proton CT Scanner. Med Phys. 2016 doi: 10.1002/mp.12107. accepted:29/09/2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider U, Pedroni E, Lomax A. The calibration of CT Hounsfield units for radiotherapy treatment planning. Phys Med Biol. 1996;41:111–24. doi: 10.1088/0031-9155/41/1/009. [DOI] [PubMed] [Google Scholar]
- 22.Paganetti H. Range uncertainties in proton therapy and the role of Monte Carlo simulations. Phys Med Biol. 2012;57:99–117. doi: 10.1088/0031-9155/57/11/R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jianga H, Seco J, Paganetti H. Effects of Hounsfield number conversion on CT based proton Monte Carlo dose calculations. Med Phys. 2007;34:1439–49. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paganetti H, Zhu X, España S, Daartz J, Paganetti H. The impact of uncertainties in the CT conversion algorithm when predicting proton beam ranges in patients from dose and PET-activity distributions. n.d doi: 10.1088/0031-9155/55/24/011. [DOI] [PubMed] [Google Scholar]
- 25.Slater JM, Archambeau JO, Miller DW, Notarus MI, Preston W, Slater JD. The proton treatment center at Loma Linda University Medical Center: Rationale for and description of its development. Int J Radiat Oncol Biol Phys. 1992;22:383–9. doi: 10.1016/0360-3016(92)90058-P. [DOI] [PubMed] [Google Scholar]
- 26.Chauvie S, Guatelli S, Ivanchenko V, Longo F, Mantero A, Mascialino B, et al. Geant4 low energy electromagnetic physics. IEEE Symp Conf Rec Nucl Sci 2004. 2004;3:1881–5. doi: 10.1109/NSSMIC.2004.1462612. [DOI] [Google Scholar]
- 27.Penfold SN. Image reconstruction and Monte Carlo simulations in the development of proton computed tomography for applications in proton radiation therapy. University of Wollongong; 2010. [Google Scholar]
- 28.Schümann J, Paganetti H, Shin J, Faddegon B, Perl J. Efficient voxel navigation for proton therapy dose calculation in TOPAS and Geant4. Phys Med Biol. 2012;57:3281–3293. doi: 10.1016/j.pestbp.2011.02.012.Investigations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu XG. An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: a review of the fifty-year history. Phys Med Biol. 2014;59:R233–302. doi: 10.1088/0031-9155/59/18/R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son IY, Winslow M, Yazici B, Xu XG. X-ray imaging optimization using virtual phantoms and computerized observer modelling. Phys Med Biol. 2006;51:4289–310. doi: 10.1088/0031-9155/51/17/011. [DOI] [PubMed] [Google Scholar]
- 31.Gu S, Gupta R, Kyprianou I. Computational high-resolution heart phantoms for medical imaging and dosimetry simulations. Phys Med Biol. 2011;56:5845–64. doi: 10.1088/0031-9155/56/18/005. [DOI] [PubMed] [Google Scholar]