Abstract
Castration-resistant prostate cancer (CRPC) remains a major clinical challenge due to the lack of effective targeted therapy for its treatment. The mechanism underlying how CRPC gains resistance towards hormone depletion and other forms of chemotherapy is poorly understood. Research on understanding the factors that drive these processes is desperately needed to generate new therapies to cure the disease. Here, we discovered a fundamental role of S-phase protein kinase 2 (Skp2) in the formation and progression of CRPC. In transgenic adenocarcinoma mouse prostate model, Skp2 depletion leads to a profound repression of prostate tumor growth and distal metastasis and substantially prolonged overall survival. We revealed that Skp2 regulates CRPC through Twist-mediated oncogenic functions including epithelial-mesenchymal transition (EMT) and cancer stem cell (CSC) acquisitions. Mechanistically, Skp2 interacted with Twist and promoted the non-degradative ubiquitination of Twist. Consequently, Skp2 stabilized Twist protein expression by preventing proteasomal degradation of Twist by β-TrCP. We found that Twist overexpression augments CSC self-renewal and population and that Skp2 inhibition reverts Twist’s effects on CSC regulation. Furthermore, genetically depleting or pharmacologically inactivating Skp2 synergistically re-sensitized CRPC cells towards chemotherapies such as paclitaxel or doxorubicin. Together, the current study uncovering Skp2-mediated Twist stabilization and oncogenic functions in CRPC offers new knowledge on how CRPC progresses and acquires chemoresistance during tumor progression. It provides prove-of principle that Skp2 targeting is a promising approach to combat metastatic CRPC by targeting Twist and CSCs.
Keywords: Skp2, Twist, cancer stem cell, castration resistance and chemotherapy
Introduction
Prostate cancer is the second-leading cause of cancer mortality in men. The androgen signaling pathway accounts for the proliferation of prostate cancer cells and thus androgen deprivation therapy (ADT) represents the mainstay therapy for advanced prostate cancer. Prostate tumors initially respond to ADT but ultimately progress after 2–3 years even with continuous hormonal withdrawal.1 This results in a lethal and drug-resistant form of prostate cancer known as castration-resistant prostate cancer (CRPC). Systematic chemotherapy with taxanes is a standard treatment for metastatic CRPC,2 yet the overall benefit on patient survival is modest.3 Mechanisms that underline the progression of androgen-independent prostate cancer includes gene amplification, activating mutations and increased activity of AR and the crosstalk of AR to other growth factor signaling.4,5 In recognition of the sustained AR signaling in the progression of CRPC, several new inhibitors of AR signaling such as MDV3100 (now known as enzalutamide) and abiraterone that act beyond antagonists have been investigated in clinical trials and approved by US Food and Drug Administration. Although these novel cytotoxic agents effectively block AR signaling and display initial success in improving clinical responses of CRPC patients, their prostate tumors eventually progress.6,7 Better understanding of how CRPC become resistant to hormone therapy or chemotherapy is in great demand.
The vital roles of epithelial-mesenchymal transition (EMT) in acquiring cancer stem cell (CSC) feature and drug resistance has been well established. Recent studies revealed that following androgen deprivation, EMT and increased CSCs emerge in prostate tumors developed in prostate-xenograft mouse model.8,9 Twist transcription factor is a key driver bestowing cancer cells with EMT and CSC features. Twist protein is overexpressed in castration-resistant tumors and in prostate cancer cells with either de novo or acquired castration resistance.8 Overexpression of Twist in prostate cancer cells confers resistance to Taxol.10 Inactivation of Twist mitigates the EMT process and inhibits cell growth and migration of prostate cancer.10 These reports together illustrated a critical role of Twist in prostate tumorigenesis and drug resistance. Twist protein is upregulated in majority (90%) of malignant prostate cancer tissues.10 Contrary to the high incidence of overexpression at protein level, genetic alterations including mutation, amplification and upregulation of Twist are less frequently detected in prostate tumors. Current knowledge regarding how Twist protein is upregulated in prostate cancer is limited. Deciphering the regulatory machinery that controls post-translational regulation of Twist is a crucial to develop new cancer therapy against CRPC.
Skp2 (S-phase kinase associated protein-2), an F-Box protein responsible for substrate recognition, is a critical component of the SCF E3 ubiquitin ligase complex.11 We and others have reported that Skp2 interact with multiple signaling pathways including Akt and pRb and genetic silencing of Skp2 restricts the development of tumors driven by these alterations.12,13 Skp2 is overexpressed in prostate cancer and its overexpression is correlated with tumor stage, recurrence and poor patient survival.14,15 Skp2 gene expression is associated with aggressive behavior of prostate cancer, evident by its upregulation in androgen-independent metastatic tumors.16 Animal studies have established Skp2’s essential role in the development of prostate cancer. For instance, overexpression of Skp2 induces proliferation and subsequent hyperplasia, dysplasia and low-grade carcinoma in the prostate gland.17 Loss of Skp2 suppresses the growth of Pten-deficient prostate cancer by triggering cellular senescence through up-regulation of p21, p27 and ATF4 in vivo.18 Skp2 expression is reduced upon castration of androgen-dependent CWR22 xenograft tumors and more importantly, Skp2 is re-expressed when CWR22 xenografts progress to androgen independency post castration,19 implying a critical role of Skp2 in castration resistance. In this study, we revealed a novel link between Skp2 and Twist in castration-resistant prostate cancer cells. We presented several lines of evidence supporting Skp2-Twist axis is a crucial pathway for CRPC development. Skp2 promoted non-degradative Twist ubiquitination and stabilized Twist, thereby inducing EMT and CSC characteristics. Skp2 inhibition by either genetic or pharmacological approaches diminished Twist-mediated CSC and chemoresistance in CRPC cells. Our study collectively provides new knowledge that Skp2 is required for Twist-mediated EMT and stem-like features of CRPC, suggesting Skp2 targeting is a compelling strategy to combat chemoresistant CRPC.
Results
Genetic depletion of Skp2 inhibits the cancer growth and progression in TRAMP mouse
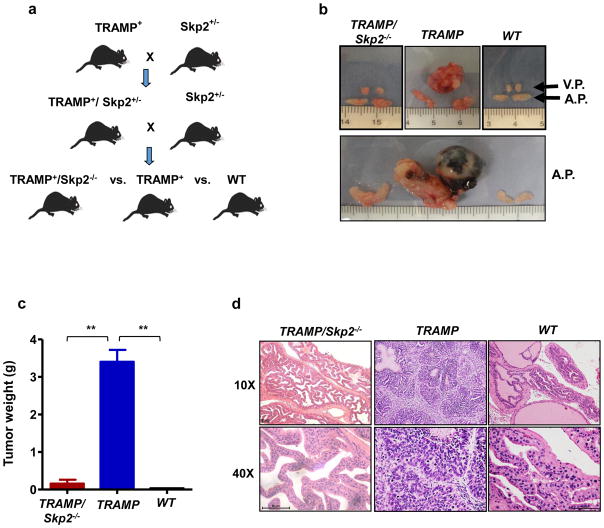
While much has emerged regarding Skp2 and its oncogenic role in prostate cancer, the current knowledge of the downstream targets of Skp2 related to CRPC still remains limited. The TRAMP (transgenic adenocarcinoma mouse prostate) mouse carries the SV40 large T antigen (Tag) under the control of the probasin regulatory element, selectively activated by androgens in the prostate epithelia. TRAMP mice develop spontaneous prostate cancer, wherein tumor development resembles disease progression in humans, from prostatic intraepithelial neoplasia (PIN) to metastatic CRPC.20,21 To determine the role of Skp2 in CRPC and its crosstalk with Twist, we crossed Skp2+/− mice with TRAMP mice to compare the tumorigenesis in the cohort consisting of wild type (WT), TRAMP and TRAMP/Skp2−/− male mice (Figure 1a). As expected, the TRAMP progeny mice in our cohort spontaneously developed tumors in ventral (V.P.) and/or anterior (A.P.) prostate glands (Figure 1b). 16 out of 16 TRAMP mice analyzed at the age of 9–10 months carried huge tumor mass in the prostate (1.8–6.3 g in weight) (Figures 1b and c). There was only one out of 15 age-matched mice analyzed that developed noticeable tumor (1.5g in weight) in TRAMP/Skp2−/− mice (Figures 1b and c). Quantification analysis of tumor weight in the various cohorts of mice demonstrated that Skp2 ablation profoundly inhibits tumor growth (Figure 1c). Histological analysis showed that multifocal hyperplasia/adenocarcinoma lesions appeared in the prostate tissues of TRAMP mice (Figure 1d, middle and Supplementary Figure S1a). Only one TRAMP/Skp2−/− mouse ever progressed beyond the PIN stage in a total of 19 mice examined, 15 mice at the age of 9–10 months and an additional 4 mice at around 14 months. The majority of TRAMP/Skp2−/− mice examined were found to have developed hyperplasia or dysplasia (Figure 1d, left). Enlarged spleen was detected in tumor-bearing TRAMP mice whereas this effect is relieved by Skp2 knockout (Supplementary Figure S1b). Together, these results dictate a crucial role of Skp2 in tumor initiation and formation in TRAMP mice.
Figure 1. Skp2 deficiency in vivo restricts prostate tumor formation in TRAMP mouse model.
(a) Schematic illustration of the generation of TRAMP/Skp2−/− (null) mice. (b) Ventral (V.P.) and anterior prostate (A.P.) glands obtained from TRAMP/Skp2−/−, TRAMP and wild type (WT) mice at the age of 9–10 months. (c) Prostate tumors were collected and weighed for a quantification analysis from a cohort of age-matched TRAMP/Skp2−/− n=15, TRAMP n=16 and WT mice n=10. (d) Histopathological analysis of prostate tumor extracted from TRAMP/Skp2−/−, TRAMP and WT mice at 9–10 months of age. Histopathological images are shown with magnification of 10X and 40X. Quantitative results are presented as means ± SD; **, p < 0.01.
Loss of Skp2 inhibits distant metastasis of prostate cancer in TRAMP mice
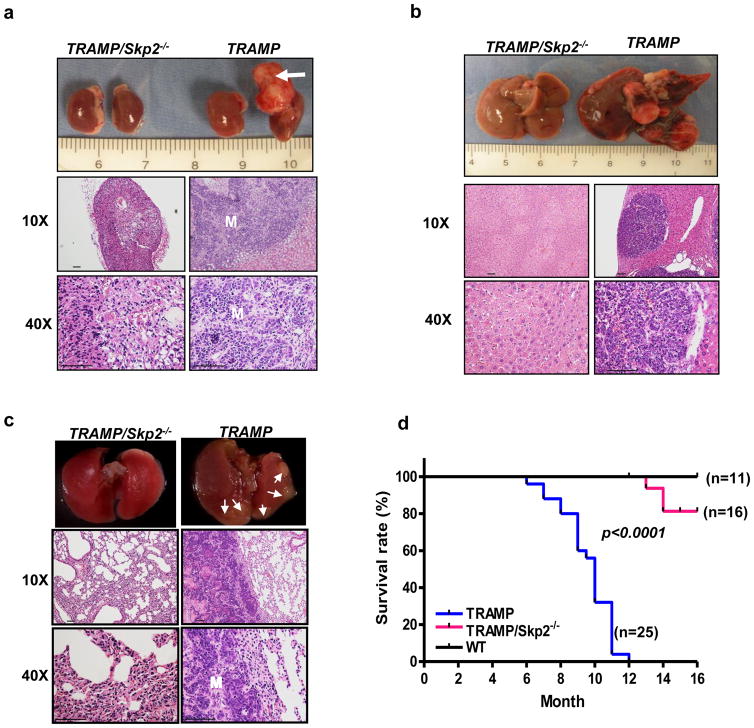
Prostate cancer deaths are largely attributed to the occurrence of distant organ metastasis.22 Following the disease course developed in TRAMP mice in our cohort, we found that 100% of TRAMP mice displayed lymph node metastasis by 8 months, 55% of mice develop distant organ metastasis in adrenal glands and about 30% of mice with lung and/or liver metastasis (Figures 2a–c). These findings are consistent with previous reports dictating that TRAMP mice is a useful tool to study the metastatic process to lymph node and distant organs. Intriguingly, loss of the Skp2 gene, drastically reduced prostate cancer metastasis potential to adrenal glands, livers and lungs (Figures 2a–c). Histopathological analysis performed at the metastatic organs further illustrated that there were no metastatic prostate cancer phenotypes displayed in TRAMP/Skp2−/− mice (Figures 2a–c), underscoring that Skp2 knockout prevents the distal metastatic process of prostate cancer. In addition to the markedly reduction in tumor formation and distant metastasis, Kaplan-Meier plot analysis showed that Skp2 ablation greatly prolonged the survival of TRAMP for more than 6 months (median survival from 10 months [TRAMP] to greater than 16 months [TRAMP/Skp2−/−]) in our cohort (Figure 2d). Of note, three out of the 16 TRAMP/Skp2−/− mice analyzed in our cohort that died by the age of 16 months did not develop detectable distant metastasis. For the three TRAMP/Skp2−/− mice that died before 16 months, one displayed PIN whereas the other two had dysplasia. This immense improvement of survival rate in vivo suggests that targeting Skp2 is a powerful approach to prevent and/or treat CRPC.
Figure 2. Skp2 ablation prevents distal metastasis and prolonged overall survival of TRAMP mice.
(a–c), Representative images and histopathological analysis of adrenal (a), liver (b) and lung (c) organs from TRAMP/Skp2−/− and TRAMP mice of 9–10 months old are presented. Histopathological images are shown with magnification of 10X and 40X. Arrow indicates metastatic adrenal tumor formation and M indicates metastasis. Smaller arrows indicate lung metastasis. (d) Kaplan-Meier plot analysis of cumulative survival rate of TRAMP/Skp2−/−, TRAMP and WT mice.
Skp2 deficiency decreases Twist protein expression by promoting Twist proteasomal degradation
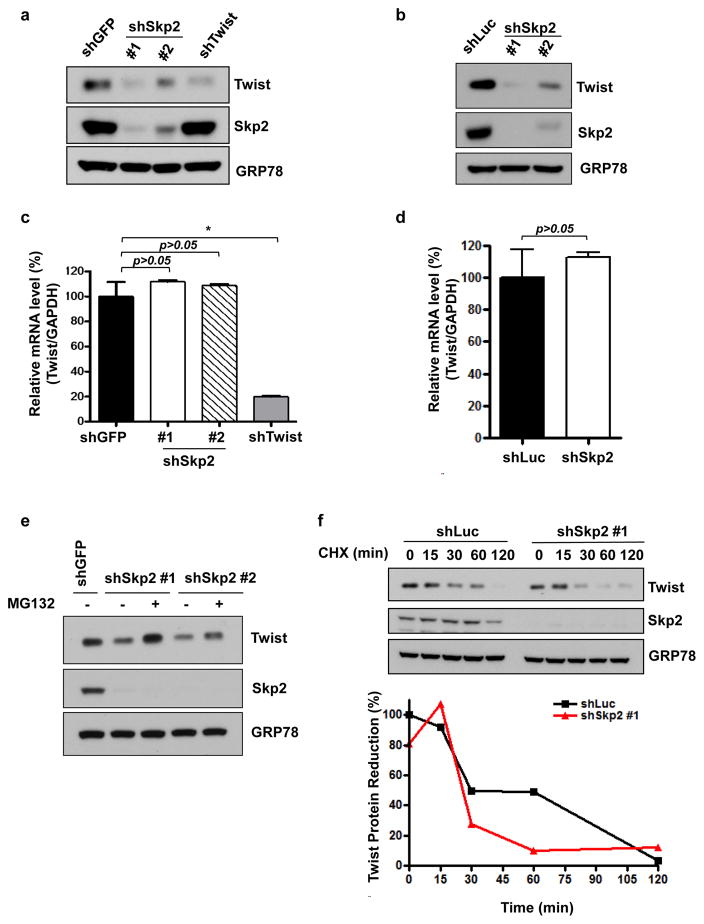
A recent gene expression analysis of CRPC tissues revealed a positive correlation between Twist expression and CRPC metastases.23 Studies with TRAMP mice showed increased Twist expression in prostate tumors and its overexpression is associated with disease progression.20,24 To further study the crosstalk between Skp2 and Twist as well as their requirements in the development and progression of CRPC. Two prostate cancer cell lines were selected to study CRPC biology: PC3 and CWR22Rv1 (termed 22Rv1 hereafter). PC3 cells display de nova resistance to castration due to its intrinsic activation of androgen-independent cell growth mediated by double deficiency of Pten and p53 tumor suppressors. 22Rv1 cells harbor acquired resistance by continual in vivo selection after castration-induced regression and relapse.25 Using two CRPC-relevant cell lines, we depleted Skp2 expression using shRNA (short hair pin RNA). Western blot revealed that upon Skp2 knockdown in PC3 cells, Twist protein expression level was significantly reduced to a level comparable to Twist knockdown (Figure 3a). Likewise, Skp2 deficiency in 22Rv1 cells resulted in Twist downregulation (Figure 3b). Two different shRNAs were used in both CPRC cell models to ensure the reduction of Twist protein expression was attributed to Skp2 gene silencing. While Skp2 has been shown to regulate a myriad of protein substrates post-translationally, our previous report illustrated that Skp2 can work in concert with Myc for regulating gene transcription.26 To study if Skp2 regulates Twist expression on a transcriptional level, we performed real time (RT)-PCR to examine Skp2’s effects on Twist gene expression. We found that Twist mRNA level remains intact upon genetic silencing of Skp2 in two CRPC cells (Figure 3c and d). These observations indicate that Skp2 is not involved in the gene transcription of Twist. Furthermore, decreased Twist expression level by Skp2 knockdown can be rescued upon treatment of the proteasome inhibitor MG132 (Figure 3e and Supplementary Figure S2). Additionally, Skp2 deficiency resulted in a more rapid degradation of Twist protein upon cycloheximide (CHX) treatment when compared to control (Figure 3f). Together, these results indicate that Skp2 orchestrates Twist expression on a post-translational level in both intrinsic and acquired CRPC cells and this regulation acts through proteasome-mediated proteolysis.
Figure 3. Skp2 deficiency in CRPC cells reduces Twist expression level post-translationally through proteasomal degradation.
(a, b) Twist expression level in PC3 (a) or 22Rv1 (b) cells upon GFP, luciferase, Skp2 or Twist knockdown were analyzed by western blot. Two different Skp2 shRNAs were used in this assay. (c, d) Real-time PCR analysis of Twist in PC3 (c) or 22Rv1 (d) cells with GFP, luciferase, Skp2 or Twist knockdown. Quantified results are presented as means ± SD; *, p < 0.05. (e) Western blot assay for Twist expression level with GFP or Skp2 knockdown in PC3 cells with the absence or presence of proteasome inhibitor, MG132. (f) Western blot assay for Twist expression level in 22Rv1 cells with luciferase or Skp2 knockdown upon treatment of 50 μg/mL of cycloheximide (CHX) at various time points.
Skp2 mediates ubiquitination of Twist and prevents Twist degradation by β-TrCP
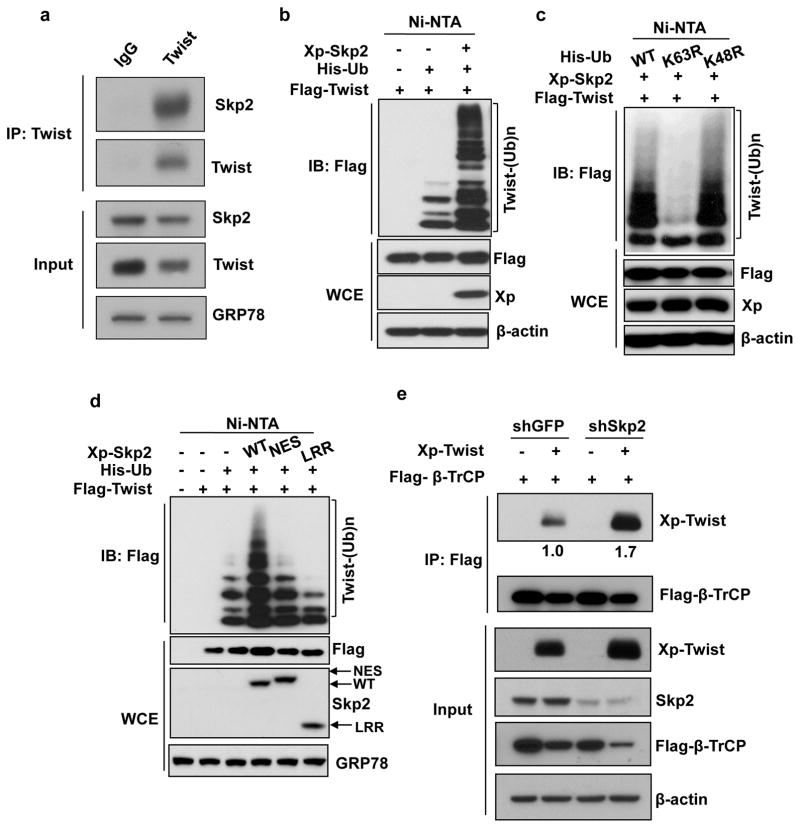
Ubiquitination is a post-translational modification that is important for regulating proteasome-dependent degradation of protein substrates and is known to regulate a variety of biological processes including cell proliferation, apoptosis, autophagy, etc.27 Skp2, as a substrate recognition protein of SCF E3 ligase complex, regulates downstream protein substrates primarily through promoting ubiquitination.11 In view of Skp2’s ability to regulate Twist post-transcriptionally, we questioned whether Skp2 does so by triggering ubiquitination of Twist. We examined the interaction between Skp2 and Twist through co-immunoprecipitation. Our data revealed that immunoprecipitation of Twist successfully pulled down Skp2 endogenously (Figure 4a and Supplementary Figure S3a), indicating that Skp2 and Twist endogenously interact with each other. We next explored whether Twist is a substrate of Skp2-mediated ubiquitination. Indeed, in vivo ubiquitination assay demonstrated that Skp2 robustly promoted ubiquitination of Twist (Figure 4b). Skp2-promoted Twist ubiquitination was found to stabilize Twist protein expression (Figure 4b), consistent with our finding that Skp2 prevents proteasome-mediated Twist degradation as shown in Figures 3a and b. Moreover, Skp2 promoted K63-but not K48-linked ubiquitination of Twist (Figure 4c). To explore if Skp2’s effect on Twist is attributed to its E3 ligase activity, two Skp2 SCF E3 ligase dead mutants, Skp2-NES (nuclear export signal) and Skp2-LRR (leucine-rich repeat),12 were included for in vivo ubiquitination assay. Compared to WT, both E3 ligase mutants of Skp2 exhibited profound impairment in promoting Twist ubiquitination (Figure 4d), concluding the importance of E3 ligase activity of Skp2 in regulating Twist ubiquitination. These findings suggest that Skp2 is a new mechanism for activating Twist protein functions and Twist-mediated cellular processes by promoting Twist ubiquitination and stabilization.
Figure 4. Skp2 interacts with and promotes non-degradative ubiquitination of Twist.
(a) PC3 cells were used for immunoprecipitation (IP) with Twist antibody followed by western blot analysis. (b) In vivo ubiquitination assay in 293T cells transfected with Xpress (Xp) tagged Skp2, Histidine-tagged ubiquitin (His-Ub) and Flag-tagged Twist. Ni-NTA indicates nickel nitrilotriacetic acid bead precipitate; WCE indicates whole cell extract. (c) In vivo ubiquitination assay in 293T cells transfected with various plasmids. (d) In vivo ubiquitination assay in 293T cells transfected with Xp-Skp2 and Flag-Twist along with His-Ub WT, His-Ub K63R or His-Ub K48R constructs. (e) 293 cells with GFP or Skp2 knockdown transfected with Flag-tagged β-TrCP and Xp-tagged Twist were subjected to IP after 48 hours with Flag antibody followed by western blot analysis. The relative intensities of immunoprecipitated Twist were quantified with Image J software and normalized with Twist in the input.
Twist is a transcription factor that exerts its biological functions through transcriptional events in the nucleus. We recently discovered that Twist ubiquitination by RNF8 E3 ligase in breast cancer accounts for Twist localization to the nucleus.28 We therefore examined if Skp2 deficiency in CRPC cells would affect Twist localization to the nucleus. To answer this question, we performed biochemical fractionation assay in control and Skp2-knockdown PC3 cells. Our data showed that although Twist expression in the nuclear extract was reduced upon Skp2 knockdown, its level of reduction is proportional to the reduction detected in whole cell extract (Supplementary Figure S3b). Moreover, the Twist reduction in nucleus was not accompanied by Twist accumulation in the cytoplasm (Supplementary Figure S3b). Together, these results suggest that Skp2 does not affect nuclear localization of Twist and indicate that an alternative mechanism is involved in Skp2’s regulation of Twist.
A previous study showed that β-TrCP E3 ligase has the ability to ubiquitinate Twist, leading to its proteasomal degradation.29 Consequently, others have shown that Skp2 degrades β-TrCP and that Skp2 knockdown leads to the accumulation of β-TrCP.30 These findings sparked us to determine whether Skp2 stabilizes Twist through downregulating β-TrCP. Western blot analyses on PC3 and 22Rv1 cells with control and Skp2 knockdown showed that Skp2 deficiency did not result in β-TrCP accumulation (Supplementary Figures S4a and b). We then investigated whether Skp2 downregulates Twist by impairing the interaction between β-TrCP and Twist using a co-immunoprecipitation assay (Figure 4e). In order to visualize the binding of Twist to β-TrCP, we added MG132 to prevent Twist degradation. In agreement with previous reports, the interaction between Twist and β-TrCP was detected in control cells (Figure 4e). Importantly, we found that Skp2 knockdown enhanced Twist binding to β-TrCP (Figure 4e), supporting the notion that that Skp2 stabilizes Twist through antagonizing β-TrCP-mediated Twist proteolysis.
Skp2 regulates EMT in CRPC cells
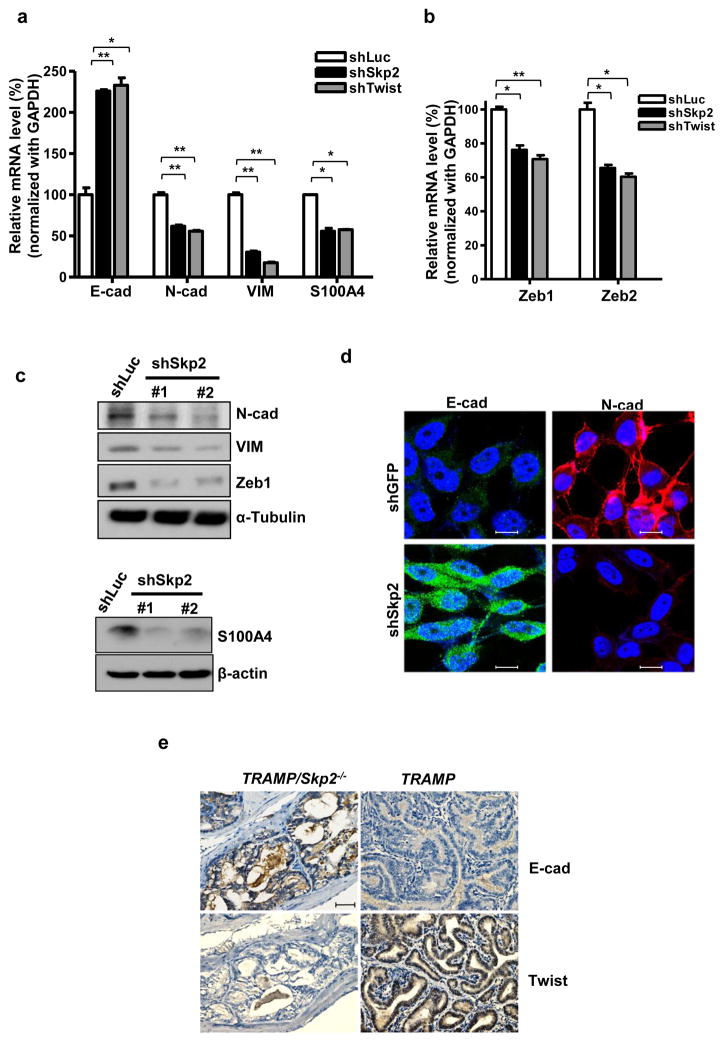
The EMT process is a prerequisite for cancer cell metastasis which is associated with the loss of expression in epithelial markers and gain of expression in mesenchymal markers.31 Twist, an EMT inducing transcription factor, has been known to play a key role in the EMT process and in CSC acquisition.32 Thus, we sought to explore the transcription levels of several EMT markers. Upon Skp2 knockdown in PC3 cells, we discovered that the epithelial marker E-cadherin mRNA level was increased while several mesenchymal markers such as N-cadherin, Vimentin, S100A4, were significantly reduced (Figure 5a). Notably, Skp2 genetic silencing also downregulates gene transcription of Zeb1 and Zeb2, EMT-inducing transcription factors (Figure 5b). This observation supports a previous report showing Twist’s essential role in promoting Zeb1 gene expression.33 We examined the gene expression of stem cell markers BMI1 and Nanog and found reduced BMI1 and Nanog mRNA levels in response to deficiency of Skp2, similar to Twist deficiency (Supplementary Figures S5a and b). Besides transcriptional levels, we also examined the protein expression level of EMT markers and found that Skp2 ablation reduces protein expression of multiple mesenchymal markers including N-cadherin, Vimentin and Zeb1 (Figure 5c). We used immunofluorescent staining to examine the E-cadherin and N-cadherin protein expression and distribution in response to Skp2 deficiency. Skp2 depletion resulted in increased E-cadherin and decreased N-cadherin in the plasma membrane compared to control PC3 cells (Figure 5d). Of note, Skp2 deficiency consistently reduced the protein expression of Snail but not Slug in CRPC cells (Supplementary Figure S6). Importantly, IHC analysis demonstrated that Skp2 depletion led to reduced Twist protein expression and upregulated E-cadherin in prostate tumors/tissues of our mouse cohort (Figure 5e), establishing Skp2’s critical role in Twist expression and EMT in vivo. Together, our data suggests that Skp2 recapitulates Twist’s effects on EMT and CSC, two biological processes important for CRPC progression.
Figure 5. Skp2 recapitulates Twist’s effect on EMT in CRPC cells.
(a, b) Real-time PCR analysis of epithelial marker E-cadherin (E-cad), mesenchymal markers N-cadherin (N-cad), Vimentin (VIM), S100A4 (a) and EMT regulators Zeb1 and Zeb2 (b) in PC3 cells with luciferase, Skp2 or Twist silencing. Quantified results are presented as means ± SD; *, p < 0.05 and **, p < 0.01. (c) Western blot analysis of EMT markers in PC3 cells with luciferase or Skp2 knockdown. (d) Immunofluorescence (IF) staining for E-cadherin and N-cadherin in PC3 cells with GFP or Skp2 knockdown. Scale bar represents 10 μm. (e) Representative images of histological analyses of Twist and E-cadherin protein expression in prostate tumors/tissues isolated from TRAMP/Skp2−/− and TRAMP mice. Scale bar, 100 μm.
Skp2 regulates CSCs and chemosensitivity in CRPC cells
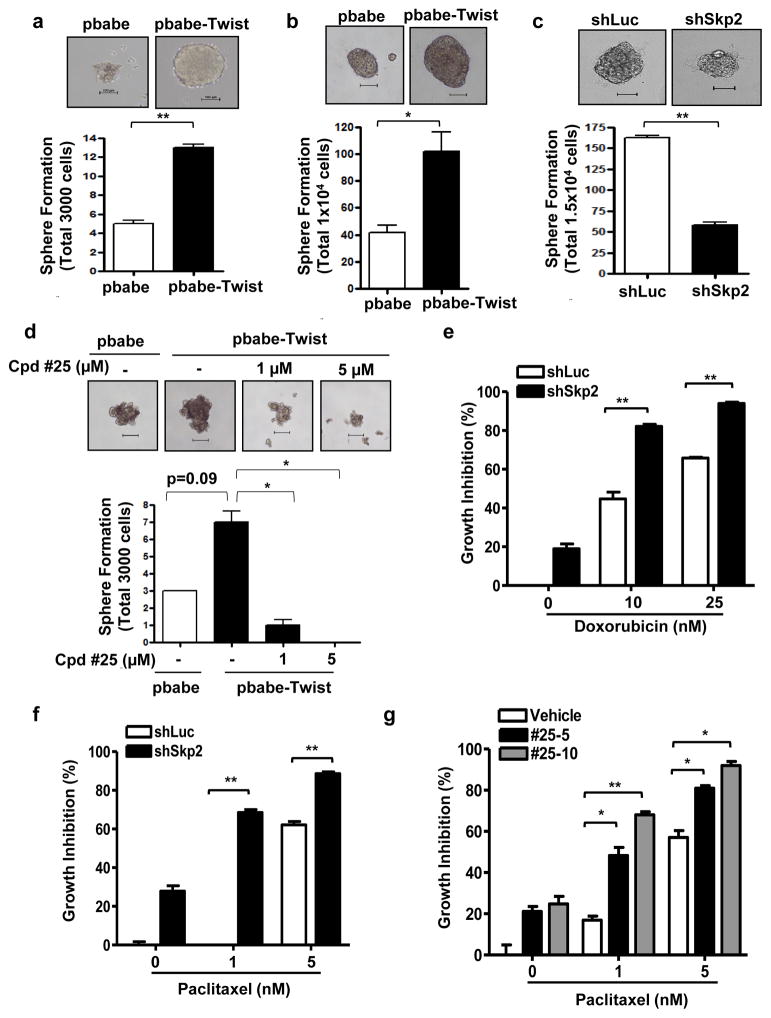
Systemic chemotherapy with taxanes is the frontline treatment for CRPC patients with metastasis. Although docetaxel indeed prolonged survival of CRPC patients, most patients would experience disease progression in around 7 months. Many studies have documented the existence of CSCs in prostate cancer. CSCs which possess unlimited self-renewal ability and can regenerate tumorigenic progenies, have been demonstrated to play a pivotal role in tumor initiation, progression and therapy resistance. Recent evidence by Puhr et al. documented reduced E-cadherin expression in docetaxel-resistant cells.34 Marín-Aguilera et al. showed that docetaxel-resistant cells harbored increased expression of EMT and stem-like cell markers.35 These reports together implicate EMT as a mechanism conferring chemoresistance and endowing CSC phenotype in prostate cancer. Tumor sphere formation assay is a gold standard assay used to determine the self-renewal ability and population CSCs.36 To determine whether Twist is indeed the driver for CSC generation and expansion in CRPC, we induced Twist expression in PC3 and 22Rv1 cells and confirmed successful Twist overexpression using Western blot analysis (Supplementary Figures S7a and b). We then analyzed Twist’s impact on the tumor sphere forming ability of PC3 and 22Rv1 cells. Our data showed that prostate tumor spheres appear to be larger in size as well as in number upon Twist overexpression, depicting that cancer stemness corresponds to the Twist expression level (Figures 6a and b). Having proven Skp2 as a new upstream regulator of Twist, we next determined if Skp2 also affects tumor sphere formation of CRPC cells. Skp2 deficiency in 22Rv1 cells reduced both the size and the total number of tumor spheres (Figure 6c), signifying the importance of Skp2 in regulating CSCs of CRPC. In our previous work, we identified a specific small molecule Skp2 inhibitor denoted as compound #25.37 Pharmacological inhibition of Skp2 by compound #25 elicited a dose-dependent Twist protein reduction in both 22Rv1 and PC3 cells (Supplementary Figures S7c and d), recapitulating the effects mediated by Skp2 genetic silencing. Twist-increased self-renewal of CSCs was greatly abolished upon administration of compound #25 (Figure 6d), underscoring the requirement of Skp2 in Twist-mediated CSC expansion.
Figure 6. Skp2 is essential for Twist-mediated CSC self-renewal and chemotherapy resistance.
(a, b) Tumor sphere formation assay of PC3 (a) or 22Rv1 (b) cells overexpressed with vector control and Twist. Scale bar represents 100 μm. (c) Tumor sphere formation assay of luciferase or Skp2-depleted 22Rv1 cells. Scale bar represents 100 μm. (d) Tumor sphere formation assay of PC3 cells overexpressed with vector control and Twist in the absence or presence of Skp2 inhibitor, compound #25. Scale bar represents 100 μm. (e, f) Cell growth inhibition assay of 22Rv1 cells with luciferase or Skp2 silencing treated with doxorubicin (e) or paclitaxel (f). (g) Cell growth inhibition assay of 22Rv1 cells treated with a combination of paclitaxel and compound #25. Quantified results are presented as means ± SD; *, p < 0.05 and **, p < 0.01.
We next determined whether Skp2 targeting sensitizes CRPC cell responses towards chemotherapeutics. Indeed, genetic silencing of Skp2 synergistically induced cell responses to doxorubicin in two CRPC cells (Figure 6e and Supplementary Figure S8a). Likewise, Skp2 knockdown augmented 22Rv1 CRPC cell sensitivity to paclitaxel treatment (Figure 6f). Combination therapy is often used for optimal effectiveness of chemotherapy therefore, we sought to test whether compound #25 can be used as a therapeutic regimen to improve chemosensitivity of CRPC. Intriguingly, doxorubicin or paclitaxel in conjunction with compound #25 substantially heightened cytotoxicity of CRPC cells (Figure 6g and Supplementary Figure S8b). To explore the mechanistic insight of how compound #25 inhibits Twist activity on CSCs, we performed Twist ubiquitination assay and found that Skp2-promoted Twist ubiquitination and protein expression were impaired by compound #25 (Supplementary Figure S9). Our data collectively suggest that Skp2 targeting is a promising approach for preventing the development and chemoresistance of CRPC by destabilizing Twist.
Discussion
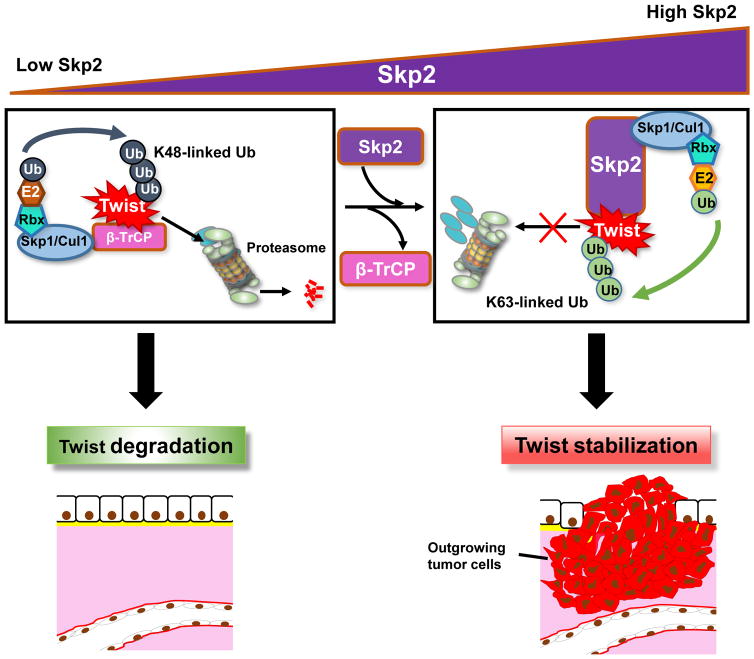
Skp2 and Twist separately have been associated with androgen-independent growth and malignancy of prostate cancer but the crosstalk between Skp2 and Twist and the causal role of Skp2/Twist pathway in CRPC remains elusive. In this study, we reveal that Skp2 is essential for CRPC progression and chemoresistance by stabilizing Twist and Twist-mediated EMT and CSC self-renewal (Figure 7).
Figure 7. Schematic illustration of Skp2’s interaction with Twist to induce progression and chemoresistance of CPRC.
In conditions of low Skp2 levels, β-TrCP ubiquitinates Twist leading to its proteasomal degradation, wherein with high levels of Skp2, Twist is stabilized and activated through ubiquitination by Skp2 allowing for CRPC progression.
Studies in TRAMP mice have shown that they produce neuroendocrine (NE) differentiation feature.38 While NE differentiation is not a frequent phenotype associated with human prostate cancer, it occurs as prostate cancer progresses to CRPC and some CRPC patients indeed develop NE tumors.25,39,40 TRAMP model is a useful tool to study the complex pathogenesis for CRPC. Previous reports have shown that the TRAMP tumors overexpress Twist and conversely, inhibiting Twist suppresses tumor progression in TRAMP mice.20,24 Establishing TRAMP/Skp2−/− mice allows us to visualize the effects of depleting Skp2 in a CRPC environment with Twist upregulation. We showed that Skp2 depletion greatly represses tumorigenesis and metastasis in TRAMP mice. We further show that Skp2 inactivation inhibits Twist-increased CSCs in CRPC cells. Results from this work indicate that Skp2-mediated Twist upregulation is an important mechanism for CRPC progression and chemoresistance. Aside from Twist, a number of genetic alternations, such as co-inactivation of tumor suppressors, p53 with pRb and p53 with Pten, have been implicated in the development of CRPC.41,42 Double knockout of p53 and pRb in murine prostate epithelial cells leads to invasive prostate carcinoma which can be repressed upon genetic loss of Skp2, has been shown to repress the development of invasive prostate carcinoma in p53/pRb double knockout mice, although it is unclear whether the analyzed prostate cancer have progressed into CRPC.13 Skp2 inactivation restricts tumorigenesis driven by singe deficiency of p53 or Pten.18,43 While it remains undetermined whether Skp2 is required for the development of CRPC driven by Pten/p53 double deficiency, a recent study reported Skp2 upregulation in such CRPC tissues.44 Consistently, our study showed that Skp2 is essential for regulating CSCs and chemosensitivity in Pten/p53 double-null PC3 cells. Future animal studies are needed to determine whether Skp2 inhibition can counteract the CRPC progression by Pten/p53 co-inactivation and or other oncogenic insults. Our study together with previous reports45–47 suggest that Skp2 inhibitors can be promising therapeutics not only for treatment-naïve but also CRPC.
Twist protein is overexpressed is in 90% of prostate tumors and is correlated with prostate cancer metastasis.10 Twist promotes AR transcription and thus accounts for AR overexpression, a primary mechanism for CRPC.48 The knowledge of how Twist is upregulated post-transcriptionally in CRPC is very limited. Zhu’s group has demonstrated that Skp2 expression closely follows the prostate tumor responses to castration. Skp2 is shown to be downregulated upon castration and re-expressed when prostate cancer becomes castration-resistant.19 In the current study, we showed that Skp2 encompasses the ability to stabilize Twist protein expression through preventing Twist degradation by β-TrCP. Therefore, our work reveals that Skp2 overexpression is a novel upstream signal leading to Twist overexpression in CRPC. These findings together functionally connect Skp2 to Twist and add Twist into a growing list of Skp2 oncogenic substrates.
Skp2 promotes tumorigenesis through interplay with a variety of cancer-regulating substrates.49 Skp2 triggers K48-linked ubiquitination and degradation of cell cycle inhibitors, p27 and p21.50,51 Skp2 activates EGFR-mediated Akt signaling pathway through inducing K63-linked ubiquitination and membrane recruitment of Akt.12 Skp2 blocks p53-mediated apoptosis by antagonizing the binding to p300.52 These reports sparked the development of inhibitors that target Skp2 at different mechanisms of action. We previously identified a small-molecule compound (cpd) #25 that selectively inhibits Skp2 through disruption of Skp2-Skp1 interaction but not other SCF E3 ligase activities.37 Consequently, cpd #25 enable concomitant suppression of multiple Skp2 oncogenic pathway including Akt activation and p27 and p21 downregulation. In this report, administration of cpd #25 in CRPC cells was found to reduce Twist protein expression as Skp2 genetic silencing does. Moreover, cpd #25 re-sensitizes the 22Rv1 CRPC cells towards frontline chemotherapy paclitaxel supporting the idea to disrupt Skp2-Skp1 interface as potential strategy for developing new cancer therapeutics. Several other Skp2 inhibitors have also been identified, such as cpd C1, C2/C16/C20, A and M1. Small molecular cpds C1, C2, C16 and C20 identified through in silico screens were shown to bind the Skp2/Cks1 pocket in order to prevent Skp2 interaction with p27 which results in p27 accumulation and p27-mediated cell cycle arrest.53 Among these inhibitors, cpd C1 was further characterized for its efficacy in targeting the growth of CRPC organoids.45 Cpd A was identified through an in vitro Skp2-mediated p27 ubiquitination assay. Cpd A blocks the recruitment of Skp2 to the SCF ligase complex for inhibiting Skp2 functions.54 Cpd M1, on the other hand, inhibits Skp2’s non-proteolytic action. Cpd M1 binds to Skp2/p300 interface, thereby releasing p300 for p53 acetylation and activation but does not affect Skp2 proteolytic activities.55 Since Skp2 stability is regulated by p300-mediated acetylation, cpd M1 also downregulates Skp2.55,56 However, in vivo efficacy of cpd C2/C16/ C20, cpd A and cpd M1 remains unclear. Further studies are required to characterize their effects on tumor formation. Our discovery that Skp2 regulates Twist stabilization and functions in CRPC offers new knowledge about how CRPC acquires chemoresistance during tumor progression and shed light on the application of Skp2 inhibitors for CRPC
Materials and Methods
Cell culture and reagents
PC3 and 22Rv1 cells (from ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI 1640, respectively supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The cell lines used were authenticated by STR profiling and routinely verified to be free of mycoplasma contamination. (His)6-ubiquitin construct and Skp2-LRR mutant was from Drs. D. Bohmann and W. Tansey, respectively. Flag-Twist was a kind gift from Dr. Mien-Chie Hung. MG132 was from Calbiochem while compound #25 was from ChemBridge. Doxorubicin and Paclitaxel were from Sigma.
Mouse model
TRAMP/Skp2−/− mouse models were generated from breeding TRAMP with Skp2+/− mice. Female TRAMP/Skp2+/− mice were used to breed with male Skp2+/− mice. TRAMP mouse (C57BL/6) was purchased from the Jackson Laboratory. At 8 months TRAMP/Skp2−/−, TRAMP, and wild type mice were dissected and the adrenal glands, prostate tumors, spleen, adrenal glands, lung and liver were collected, examined, weighed and subjected to histological analysis. All animal experiments were performed under IACUC approved protocol.
Viral Infection, Western Blot, Immunoprecipitation, Immunofluorescent staining, RT-PCR, In vivo ubiquitination and Nuclear Fractionation assays
Viral infection, Western blot, immunoprecipitation, immunofluorescent staining, RT-PCR, in vivo ubiquitination and nuclear fractionation assays were performed as previously described.57 Target sequence, antibody and primer sequence details are provided in Tables 1, 2 and 3, respectively.
Table 1.
Target sequences used in this study
| Target | shRNA sequence |
|---|---|
| Skp2 (#1) | 5′-GATAGTGTCATGCTAAAGAAT-3′ |
| Skp2 (#2) | 5′-GCCTAAGCTAAATCGAGAGAA-3′ |
| Twist | 5′-CCTGAGCAACAGCGAGGAAGA-3′ |
| GFP | 5′-GCAAGCTGACCCTGAAGTTC-3′ |
Abbreviation: shRNA, short hairpin RNA
Table 2.
Antibodies used in this study
| Primary Antibody | Source | Dilution |
|---|---|---|
| Twist – IP | Santa Cruz | 1:750 |
| Twist – WB | Abcam | 1:1000 |
| Twist – IHC | Biorbyt | 1:100 |
| Flag | Sigma | 1:2000 |
| Skp2 | Invitrogen | 1:2000 |
| Xpress | Invitrogen | 1:10,000 |
| GRP78 | BD Biosciences | 1:10,000 |
| β-actin | Sigma | 1:20,000 |
| N-cadherin | Cell Signaling | 1:3000 |
| E-cadherin-WB | Cell Signaling | 1:3000 |
| E-cadherin-IHC | Cell Signaling | 1:200 |
| Vimentin | Cell Signaling | 1:8000 |
| TCF8/ZEB1 | Cell Signaling | 1:2000 |
| S100A4 | Cell Signaling | 1:500 |
| β-TrCP – PC3 | Genetex | 1:1000 |
| β-TrCP – 22Rv1 | Abcam | 1:1000 |
| α-Tubulin | Sigma | 1:10,000 |
| Lamin B | Santa Cruz | 1:5000 |
Table 3.
Primer sequences used in this study
| Target (F/R) | Primer sequence (5′ to 3′) |
|---|---|
| Twist-F | 5′-TCCATGTCCGCGTCCCACTA-3′ |
| Twist-R | 5′-ATTCAAAGAAACAGGGCGTG-3′ |
| GAPDH-F | 5′-GATTCCACCCATGGCAAATTC-3′ |
| GAPDH-R | 5′-CTTCTCCATGGTGGTGAAGAC-3 |
| E-cad-F | 5′-CAATGCCGCCATCGCTTACACCAT-3′ |
| E-cad-R | 5′-TCAGCAGCTTGAACCACCAGGGTA-3′ |
| N-cad-F | 5′-AGCCTGACACTGTGGAGCCT-3′ |
| N-cad-R | 5′-GGAGTCATATGGTGGAGCTGT-3′ |
| Vimentin-F | 5′-AAAGTGTGGCTGCCAAGAACCTGC-3′ |
| Vimentin-R | 5′-ACTCAGTGGACTCCTGCTTTGCCT-3′ |
| S100A4-F | 5′-TGATGGTGTCCACCTTCCACAAGT-3′ |
| S100A4-R | 5′-CCTGTTGCTGTCCAAGTTGCTCAT-3′ |
| Zeb1-F | 5′-GGGCCTGAAGCTCAGGCAGATGA-3′ |
| Zeb1-R | 5′-CTCTGGTCCTCTTCAGGTGCCTC-3′ |
| Zeb2-F | 5′-AGAAGCCACGATCCAGACCGCAATTA-3′ |
| Zeb2-R | 5′-GGTAAATAATGGCTGTGTCACTGCGC-3′ |
Tumor sphere formation assay
PC3 and 22Rv1 cells were seeded in 6-well ultra-low attachment plates (Corning, Corning, NY) in plating medium (MEGM) at a density of 3000 cells/well for PC3 cells and 1.0×104 cells/well for 22Rv1 cells. The appearance of tumor spheres for PC3 and 22Rv1 cells were evaluated after 14 and 12 days, respectively. Tumor spheres with diameters of ≥100 μm were manually counted under a microscope.
Cell growth inhibition assay
PC3 and 22Rv1 cells were seeded at 3000 cells/well in a 12-well plate in triplicates. 24 hours later, cells were treated with specified drugs. Three days after treatment, cells were washed with PBS, trypsinized and re-suspended with complete media. Trypan blue exclusion were used to stain the dead cells and live cells were counted using a hemocytometer.
Statistical Analysis
All data are shown as mean ± SEM for at least three independent experiments unless otherwise indicated. All statistical significance was determined by un-paired two-tailed Student’s t-tests, and p < 0.05 was considered statistically significant. For in vivo mouse experiment, the mouse number was determined by power analysis based on the fact that, with a sample size of six with a two-sided type I error rate of 0.05, the study will have 90% power to detect a 30% difference in cancer metastasis.
Supplementary Material
Acknowledgments
We sincerely appreciate Dr. Keiichi I. Nakayama for providing Skp2+/− mice. This work was supported by the Stony Brook School of Medicine and Cancer Center Fund, the Feldstein Medical Foundation Award, a NYS grant (DOH01-ROWLEY-2015-00011) and a NIH grant (5 K22 CA181412) to C.H.C, and a MOHW grant (MOHW103-TDU-M-221-123017) and a NHRI grant (CA-106-PP-36) to C.F.L, as well as NIH grants (R01 CA182424 and R01CA193813) to H.K.L.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Rescigno P, Buonerba C, Bellmunt J, Sonpavde G, De Placido S, Di Lorenzo G. New perspectives in the therapy of castration resistant prostate cancer. Curr Drug Targets. 2012;13:1676–1686. doi: 10.2174/138945012803529956. [DOI] [PubMed] [Google Scholar]
- 2.Dayyani F, Gallick GE, Logothetis CJ, Corn PG. Novel therapies for metastatic castrate-resistant prostate cancer. J Natl Cancer Inst. 2011;103:1665–1675. doi: 10.1093/jnci/djr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plymate SR, Bhatt RS, Balk SP. Taxane resistance in prostate cancer mediated by AR-independent GATA2 regulation of IGF2. Cancer Cell. 2015;27:158–159. doi: 10.1016/j.ccell.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–992. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 9.Yun EJ, Zhou J, Lin CJ, Hernandez E, Fazli L, Gleave M, et al. Targeting Cancer Stem Cells in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2016;22:670–679. doi: 10.1158/1078-0432.CCR-15-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 11.Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. Scientific World Journal. 2010;10:1001–1015. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Bauzon F, Fu H, Lu Z, Cui J, Nakayama K, et al. Skp2 deletion unmasks a p27 safeguard that blocks tumorigenesis in the absence of pRb and p53 tumor suppressors. Cancer Cell. 2013;24:645–659. doi: 10.1016/j.ccr.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang G, Ayala G, De Marzo A, Tian W, Frolov A, Wheeler TM, et al. Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin Cancer Res. 2002;8:3419–3426. [PubMed] [Google Scholar]
- 15.Nguyen PL, Lin DI, Lei J, Fiorentino M, Mueller E, Weinstein MH, et al. The impact of Skp2 overexpression on recurrence-free survival following radical prostatectomy. Urol Oncol. 2011;29:302–308. doi: 10.1016/j.urolonc.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 17.Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, et al. Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate. Cancer Res. 2003;63:1583–1588. [PubMed] [Google Scholar]
- 18.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Sun D, Ji P, Mohler J, Zhu L. An AR-Skp2 pathway for proliferation of androgen-dependent prostate-cancer cells. J Cell Sci. 2008;121:2578–2587. doi: 10.1242/jcs.030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardiani A, Farsaci B, Rogers CJ, Protter A, Guo Z, King TH, et al. Combination therapy with a second-generation androgen receptor antagonist and a metastasis vaccine improves survival in a spontaneous prostate cancer model. Clin Cancer Res. 2013;19:6205–6218. doi: 10.1158/1078-0432.CCR-13-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingrich JR, Barrios RJ, Kattan MW, Nahm HS, Finegold MJ, Greenberg NM. Androgen-independent prostate cancer progression in the TRAMP model. Cancer Res. 1997;57:4687–4691. [PubMed] [Google Scholar]
- 22.Cookson MS, Roth BJ, Dahm P, Engstrom C, Freedland SJ, Hussain M, et al. Castration-resistant prostate cancer: AUA Guideline. The Journal of urology. 2013;190:429–438. doi: 10.1016/j.juro.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Haider M, Zhang X, Coleman I, Ericson N, True LD, Lam HM, et al. Epithelial mesenchymal-like transition occurs in a subset of cells in castration resistant prostate cancer bone metastases. Clinical & experimental metastasis. 2016;33:239–248. doi: 10.1007/s10585-015-9773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwilas AR, Ardiani A, Dirmeier U, Wottawah C, Schlom J, Hodge JW. A poxviral-based cancer vaccine the transcription factor twist inhibits primary tumor growth and metastases in a model of metastatic breast cancer and improves survival in a spontaneous prostate cancer model. Oncotarget. 2015;6:28194–28210. doi: 10.18632/oncotarget.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchiani S, Tamburrino L, Nesi G, Paglierani M, Gelmini S, Orlando C, et al. Androgen-responsive and -unresponsive prostate cancer cell lines respond differently to stimuli inducing neuroendocrine differentiation. Int J Androl. 2010;33:784–793. doi: 10.1111/j.1365-2605.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- 26.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nature cell biology. 2010;12:457–467. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HJ, Li CF, Ruan D, Powers S, Thompson PA, Frohman MA, et al. The DNA Damage Transducer RNF8 Facilitates Cancer Chemoresistance and Progression through Twist Activation. Molecular cell. 2016;63:1021–1033. doi: 10.1016/j.molcel.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong J, Ogura K, Wang Z, Inuzuka H. Degradation of the Transcription Factor Twist, an Oncoprotein that Promotes Cancer Metastasis. Discovery Medicine. 2013;15:7–15. [PMC free article] [PubMed] [Google Scholar]
- 30.Wei S, Chu P, Chuang H, Hung W, Kulp S, Chen C. Targeting the Oncogenic E3 Ligase Skp2 in Prostate and Breast Cancer Cells with a Novel Energy Restriction- Mimetic Agent. PLOS ONE. 2012;7:e47298. doi: 10.1371/journal.pone.0047298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 33.Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. The Journal of biological chemistry. 2011;286:12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puhr M, Hoefer J, Schafer G, Erb HH, Oh SJ, Klocker H, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. The American journal of pathology. 2012;181:2188–2201. doi: 10.1016/j.ajpath.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Marin-Aguilera M, Codony-Servat J, Reig O, Lozano JJ, Fernandez PL, Pereira MV, et al. Epithelial-to-mesenchymal transition mediates docetaxel resistance and high risk of relapse in prostate cancer. Molecular cancer therapeutics. 2014;13:1270–1284. doi: 10.1158/1535-7163.MCT-13-0775. [DOI] [PubMed] [Google Scholar]
- 36.Johnson S, Chen H, Lo P. In vitro Tumorsphere Formation Assays. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiaverotti T, Couto SS, Donjacour A, Mao JH, Nagase H, Cardiff RD, et al. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. The American journal of pathology. 2008;172:236–246. doi: 10.2353/ajpath.2008.070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Epstein JI. Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. The American journal of surgical pathology. 2008;32:65–71. doi: 10.1097/PAS.0b013e318058a96b. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:1–15. doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan CH, Gao Y, Moten A, Lin HK. Novel ARF/p53-independent senescence pathways in cancer repression. J Mol Med (Berl) 2011;89:857–867. doi: 10.1007/s00109-011-0766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu W, Liu S, Li B, Xie Y, Izban M, Ballard B, et al. SKP2 loss destabilizes EZH2 by promoting TRAF6-mediated ubiquitination to suppress prostate cancer. Oncogene. 2016 doi: 10.1038/onc.2016.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, Lu Z, Bauzon F, Fu H, Cui J, Locker J, et al. P27T187A knockin identifies Skp2/Cks1 pocket inhibitors for advanced prostate cancer. Oncogene. 2016:1–11. doi: 10.1038/onc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: a novel potential therapeutic target for prostate cancer. Biochim Biophys Acta. 2012;1825:11–17. doi: 10.1016/j.bbcan.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Lu Y, Wang L, Mizokami A, Keller ET, Zhang J, et al. Skp2 is associated with paclitaxel resistance in prostate cancer cells. Oncology Reports. 2016;36:559–566. doi: 10.3892/or.2016.4809. [DOI] [PubMed] [Google Scholar]
- 48.Shiota M, Yokomizo A, Tada Y, Inokuchi J, Kashiwagi E, Masubuchi D, et al. Castration resistance of prostate cancer cells caused by castration-induced oxidative stress through Twist1 and androgen receptor overexpression. Oncogene. 2010;29:237–250. doi: 10.1038/onc.2009.322. [DOI] [PubMed] [Google Scholar]
- 49.Chan CH, Morrow JK, Zhang S, Lin HK. Skp2: a dream target in the coming age of cancer therapy. Cell Cycle. 2014;13:679–680. doi: 10.4161/cc.27853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrano A, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biology. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 51.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitagawa M, Lee SH, McCormick F. Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Mol Cell. 2008;29:217–231. doi: 10.1016/j.molcel.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 53.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Q, Xie W, Kuhn D, Voorhees P, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCFSkp2 results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood Journal. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh M, Lee JH, Moon H, Hyun YJ, Lim HS. A Chemical Inhibitor of the Skp2/p300 Interaction that Promotes p53-Mediated Apoptosis. Angew Chem Int Ed Engl. 2016;55:602–606. doi: 10.1002/anie.201508716. [DOI] [PubMed] [Google Scholar]
- 56.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee H, Li C, Ruan D, Powers S, Thompson P, Frohman M, et al. The DNA Damage Transducer RNF8 Facilitates Cancer Chemoresistance and Progression through Twist Activation. Molecular Cell. 2016;63:1021–1033. doi: 10.1016/j.molcel.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.