Abstract
The HS3ST1 gene controls endothelial cell production of HSAT+ – a form of heparan sulfate containing a specific pentasaccharide motif that binds the anticoagulant protein antithrombin (AT). HSAT+ has long been thought to act as an endogenous anticoagulant; however, coagulation was normal in Hs3st1−/− mice that have greatly reduced HSAT+ (HajMohammadi et al., 2003). This finding indicates that HSAT+ is not essential for AT’s anticoagulant activity. To determine if HSAT+ is involved in AT’s poorly understood inflammomodulatory activities, Hs3st1−/− and Hs3st1+/+ mice were subjected to a model of acute septic shock. Compared with Hs3st1+/+ mice, Hs3st1−/− mice were more susceptible to LPS-induced death due to an increased sensitivity to TNF. For Hs3st1+/+ mice, AT treatment reduced LPS-lethality, reduced leukocyte firm adhesion to endothelial cells, and dilated isolated coronary arterioles. Conversely, for Hs3st1−/− mice, AT induced the opposite effects. Thus, in the context of acute inflammation, HSAT+ selectively mediates AT’s anti-inflammatory activity; in the absence of HSAT+, AT’s pro-inflammatory effects predominate. To explore if the anti-inflammatory action of HSAT+ also protects against a chronic vascular-inflammatory disease, atherosclerosis, we conducted a human candidate-gene association study on >2000 coronary catheterization patients. Bioinformatic analysis of the HS3ST1 gene identified an intronic SNP, rs16881446, in a putative transcriptional regulatory region. The rs16881446G/G genotype independently associated with the severity of coronary artery disease and atherosclerotic cardiovascular events. In primary endothelial cells, the rs16881446G allele associated with reduced HS3ST1 expression. Together with the mouse data, this leads us to conclude that the HS3ST1 gene is required for AT’s anti-inflammatory activity that appears to protect against acute and chronic inflammatory disorders.
Keywords: 3-O-sulfotransferase, HS3ST1, inflammation, sepsis, heparan sulfate, atherosclerosis
1 Introduction
Inflammation contributes to the morbidity and mortality of most diseases, regardless of their position on the acute to chronic continuum. The impact of inflammation is highlighted by both severe sepsis/septic shock (acute) and atherosclerotic cardiovascular disease (chronic) [1, 2]. Annually in the United States, there are more than 750,000 episodes of severe sepsis, resulting in over 220,000 deaths and $16.6B in medical costs [3]. Moreover, each year, the predominant forms of atherosclerotic cardiovascular disease (coronary artery disease (CAD) and stroke) afflict over 15 million people, kill more than 250,000 individuals, and impose about $250B in costs [4].
Both sepsis and cardiovascular disease involve NF-κB activation through TLR4 and members of the TNF receptor-superfamily. This process leads to endothelial cell activation, as well as leukocytes rolling on, adhering to, and migrating across the endothelium into tissues [5, 6]. In Gram-negative sepsis, LPS binding to endothelial TLR4 triggers an initial round of NF-κB activation that is amplified through TNF-mediated positive feedback [7] (Fig. 1). In atherosclerosis, NF-κB activation also involves ligand engagement of both TLR4 (which binds oxidized LDL) and the TNF receptor-superfamily members OX40 and CD137 [11]. The elucidation of novel molecules that regulate these shared pathways and processes will provide new targets for the development of therapeutics to combat the devastating impact of acute and chronic inflammatory disorders.
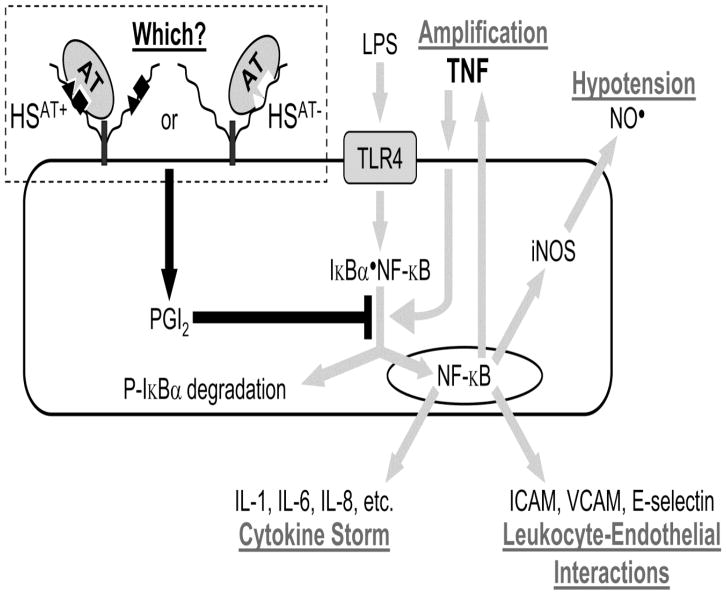
Fig. 1. Mechanism of antithrombin’s anti-inflammatory activity in sepsis.
Gram-negative sepsis is initiated by LPS activation of TLR4 on endothelial cells. This leads to phosphorylation of cytosolic Iκ-Bα•NF-κB complex, which then dissociates. The free NF-κB translocates to the nucleus and activates transcription of numerous pro-inflammatory genes. These include cytokines and chemokines, which lead to cytokine storm; cell adhesion molecules, which lead to leukocyte-endothelial interactions; iNOS that synthesizes vasodilatory nitric oxide (NO•), which contributes to hypotension; and TNF, which stimulates the TNF receptor to further activate NF-κB via phosphorylation of the Iκ-Bα•NF-κB complex [7]. AT’s anti-inflammatory activity involves binding to cell surface HSPGs, which leads to synthesis of prostacyclin (PGI2) that blocks phosphorylation of the Iκ-Bα•NF-κB complex [8–10]. Thus, AT treatment prevents activation of proinflammatory NF-κB. Our study evaluates whether this anti-inflammatory activity involves HSAT+ or HSAT−, which respectively bear or lack the pentasaccharide with high AT affinity.
One pathway that might protect against inflammation involves HSAT+, a form of heparan sulfate (HS). HS is the unbranched polysaccharide of heparan sulfate proteoglycans (HSPGs), which are multifunctional molecules that modulate numerous biologic processes, including inflammation [12–19]. The HS component exhibits tremendous structural complexity that provides an array of motifs for binding a diverse range of effector proteins [20–22]. Although HSAT+ was discovered in the early 1980’s [23], its biologic function remains a mystery [24]. HSAT+ is produced by endothelial cells and is displayed on both lumenal and ablumenal surfaces [25]; thus, HSAT+ is ideally localized to regulate physical interactions/communication between leukocytes and the endothelium [26, 27]. Endothelial synthesis of HSAT+ is almost exclusively controlled by the rate-limiting enzyme HS 3-O-sulfotransferase-1 that is encoded by the HS3ST1 gene [28–30], making this gene an interesting target for investigation of inflammatory processes.
The biologic role of HSAT+ may involve antithrombin (AT), a multifunctional plasma protein that is best known as the body’s most important natural anticoagulant. AT forms 1:1 complexes with, and thereby neutralizes, proteases of the coagulation cascade. This process is dramatically accelerated by the binding of AT to heparin, a subtype of HS containing a specific pentasaccharide motif. The structure-function relationships between the pentasaccharide and AT are the best characterized of all HS•protein interactions. In particular, the central glucosamine residue contains a 3-O-sulfate group that is essential for binding and activating AT [31, 32]. This moiety is attached to HS by the product of the HS3ST1 gene [28].
The pentasaccharide motif also occurs in HSAT+. In vitro, HSAT+ behaves like heparin; it catalyzes AT neutralization of coagulation proteases. Thus, HSAT+ was initially anticipated to convey anticoagulant tone to the blood vessel wall. To test this possibility, we disrupted the mouse Hs3st1 gene, which reduced HSAT+ levels by ~90%. However, coagulation status was completely normal in Hs3st1−/− mice [33]. Since a role in coagulation could not be detected, we hypothesize that HSAT+ modulates other activities of AT
AT possesses context-dependent inflammomodulatory properties [34]. In vitro studies of endothelial cells/leukocytes show that AT can elicit anti-inflammatory effects (such as stimulating prostacyclin synthesis and blocking NF-κB activation) [10, 35] (Fig. 1), or pro-inflammatory effects (such as upregulating iNOS expression and stimulating leukocyte migration), depending on the experimental context [36, 37]. In vivo, AT’s anti-inflammatory effects predominate; AT therapy reduces morbidity and/or mortality in numerous animal and non-human primate models of sepsis [9, 38–40]. However, little is known of the mechanisms that regulate AT’s opposite inflammomodulatory activities.
In vivo and in vitro studies reveal that AT’s beneficial effects in sepsis require the binding of AT to cell surface HSPGs, which triggers anti-inflammatory signaling events [9, 35, 39, 41–45] (Fig. 1). HS exhibits two distinct binding sites for AT – a low-affinity site (HSAT−, KD≈10 μM) and the high-affinity pentasaccharide (KD≈15 nM) [46, 47]. It is unknown which binding site is necessary for AT’s anti-inflammatory activity; however, this activity requires supraphysiologic levels of AT (~7 μM) [48] that would logically implicate the low-affinity site. Thus, it is unclear if the pentasaccharide motif of HSAT+ is required for AT’s anti-inflammatory activity.
The objectives of our study are to determine if AT’s anti-inflammatory activity requires HSAT+ and to determine if HSAT+ has a potential protective role in atherosclerosis. Through analysis of an acute sepsis model we show that HSAT+ selectively regulates the anti-inflammatory activity of AT. This pathway may also serve to protect against chronic vascular inflammation, such as atherosclerosis. Consistent with this role, we show that an HS3ST1 SNP associates with reduced gene expression and increased atherosclerotic cardiovascular disease.
2 Results
2.1 Hs3st1−/− mice exhibit a pro-inflammatory phenotype
2.1.1 HSAT+-deficient mice show enhanced susceptibility to LPS-lethality
We predict that Hs3st1−/− mice, which have greatly reduced levels of HSAT+, should exhibit a pro-inflammatory phenotype. This possibility was evaluated by challenging Hs3st1+/+ and Hs3st1−/− mice with a dose of LPS that induced acute lethal septic shock. Fig. 2 shows that Hs3st1−/− mice, compared with Hs3st1+/+ littermates, succumbed earlier to LPS-induced death, with a sex-adjusted hazard ratio (95% CI) of 4.42 (2.21–9.05). We also found that female mice were more resistant to LPS-lethality than male mice, with a genotype-adjusted hazard ratio of 0.17 (0.08–0.36). Indeed, it is well appreciated that females are relatively resistant to LPS-induced septic shock [49, 50]. Therefore, all subsequent experiments employed exclusively male mice. Overall, these data demonstrate that Hs3st1−/− mice, compared with Hs3st1+/+, are more sensitive to LPS-induced lethal septic shock.
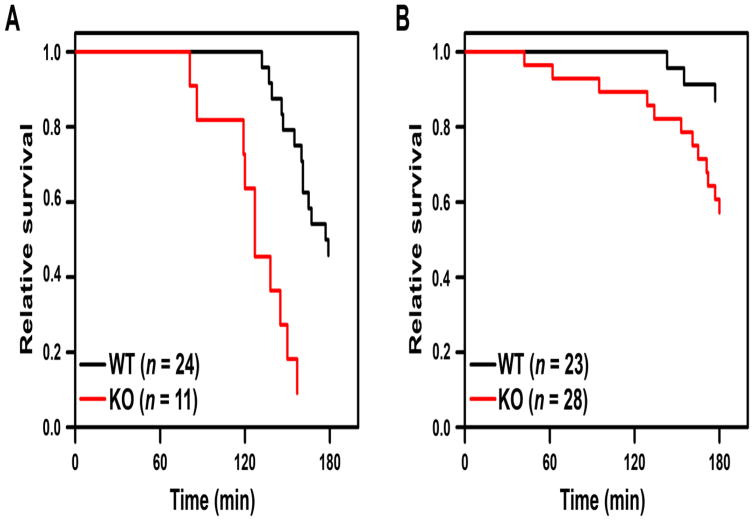
Fig. 2. Hs3st1−/− mice show enhanced LPS-lethality.
Kaplan-Meier survival analysis for LPS-lethality in Hs3st1+/+ (WT) and Hs3st1−/− (KO) mice, (A) males and (B) females. Anesthetized mice were administered 50 mg/kg LPS at T = 0. Time of death was established when the maximum systolic pressure dropped below 0.5 mm Hg. All groups were composed of littermates matched for age and weight. Data were analyzed by Cox proportional hazard analysis using sex and genotype as independent variables, which reveals the degree that each variable (independent of the other) increases the rate of death (adjusted hazard ratio). The adjusted significance for genotype was P < 0.00005 and for sex was P < 0.00001. As a control, five mice of each genotype were administered the LPS vehicle; all maintained normal hemodynamic parameters for the full 3 h monitoring period.
2.1.2 LPS induces cardiogenic shock in both Hs3st1+/+ and Hs3st1−/− mice
Left ventricle hemodynamics were examined to determine if LPS induces a similar mechanism of death in both Hs3st1+/+ and Hs3st1−/− mice. At baseline (T = 0), there were no significant differences in any of the hemodynamic parameters between genotypes, indicating that Hs3st1−/− and Hs3st1+/+ hearts are structurally and functionally indistinguishable. We next examined hemodynamic parameters at the mid-point of cardiovascular collapse, T = Pmax50%, where the maximum systolic pressure fell to 50% of the starting value at T = 0 (Supplementary Fig. 1). Comparison of these two time points (Table 1) reveals that LPS induces lethal cardiogenic shock (Appendix A). For each of the parameters, Hs3st1+/+ and Hs3st1−/− mice exhibited equivalent hemodynamic responses to LPS (Table 1); there were no significant differences between genotypes, and no significant interactions between genotype and time. Combined, these results demonstrate that for both genotypes LPS-induced death occurs by the same mechanism — cardiogenic shock. Thus, Hs3st1−/− mice are more sensitive to LPS than Hs3st1+/+ mice, indicating a pro-inflammatory phenotype.
Table 1.
LPS induces cardiogenic shock in Hs3st1+/+ and Hs3st1−/− mice.
| Parameters | T = 0a | T = Pmax50% | P- valueb | |||
|---|---|---|---|---|---|---|
| Hs3st1+/+ | Hs3st1−/− | Hs3st1+/+ | Hs3st1−/− | |||
| n | 12 | 10 | 12 | 10 | ||
| Maximum pressure | (mmHg) | 118 ± 4 | 121 ± 5 | 61 ± 3 | 60 ± 3 | <0.0001 |
| Heart rate | (bpm) | 578 ± 18 | 574 ± 22 | 565 ± 45 | 564 ± 17 | NS |
| Arterial elastance | (mmHg/μl ) | 5.09 ± 1.93 | 4.55 ± 0.47 | 6.45 ± 0.72 | 8.23 ± 2.17 | 0.034 |
| Cardiac output | (μl/min) | 14976 ± 1690 | 16357 ± 1836 | 5792 ± 674 | 7069 ± 1908 | <0.0001 |
| Cardiac index | (μl/min/g) | 440 ± 66 | 494 ± 52 | 182 ± 20 | 235 ± 52 | <0.0001 |
| End diastolic volume | (μl) | 76.8 ± 3.6 | 78.8 ± 3.3 | 79.1 ± 4.2 | 93.6 ± 8.6 | 0.044 |
| Stroke volume | (μl) | 25.5 ± 2.4 | 28.6 ± 2.9 | 11.0 ± 1.6 | 12.6 ± 3.2 | <0.0001 |
| Ejection fraction | (%) | 32.1 ± 2.4 | 34.7 ± 2.8 | 13.6 ± 1.7 | 12.8 ± 3.0 | <0.0001 |
| Stroke work | (mmHg•μl ) | 2300 ± 183 | 2762 ± 366 | 275 ± 61 | 318 ± 142 | <0.0001 |
| dP/dtmax | (mmHg/s) | 9851 ± 468 | 10732 ± 742 | 3868 ± 288 | 3831 ± 342 | <0.0001 |
| dP/dtmin | (mmHg/s) | −9894 ± 589 | −9838 ± 854 | −2943 ± 337 | −2811 ± 414 | <0.0001 |
| TauWeiss | (ms) | 5.49 ± 0.44 | 5.63 ± 0.50 | 12.46 ±2.47 | 17.27 ± 4.87 | 0.001 |
mean ± SEM
P-value for time effect by repeated measures ANOVA
2.1.3 Left ventricle HSAT+ is dramatically reduced in Hs3st1−/− mice
To test if this cardiac phenotype is associated with a regional reduction of endothelial HSAT+, we examined HSAT+ localization and levels in the left ventricle. In Hs3st1+/+ hearts, all blood vessels including capillaries exhibited intense HSAT+ staining that colocalized with an endothelial marker (Fig. 3A). Such localization is consistent with the well-documented endothelial expression of HSAT+ [28, 48]. Conversely, in Hs3st1−/− hearts, endothelial HSAT+ staining was reduced to undetectable levels (Fig. 3A). These results were confirmed by analyzing HS extracted from mouse left ventricles. Although total HS levels were unaltered, HSAT+ levels in Hs3st1−/− left ventricles were reduced by 80% relative to that of Hs3st1+/+ mice (Fig. 3B). The large decrease in endothelially expressed HSAT+ in the Hs3st1−/−, compared with Hs3st1+/+, left ventricle suggests that endothelial HSAT+ may convey a protective activity against LPS-induced cardiogenic shock.
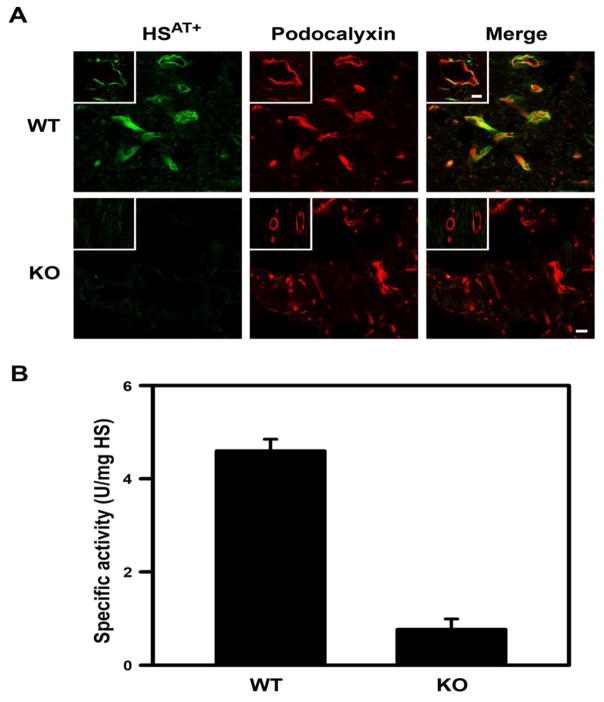
Fig. 3. Endothelial HSAT+ is dramatically reduced in Hs3st1−/− left ventricles.
(A) Heart cryosections (~5 μm) from Hs3st1+/+ (WT) and Hs3st1−/− (KO) mice were co-stained with AT-647 (green), a fluorescent AT conjugate that selectively detects HSAT+ [51], and anti-podocalyxin to detect endothelial cells (red). The yellow color indicates co-expression of podocalyxin and HSAT+. Fields are from the left ventricle and show capillaries (the bar indicates 10 μm). The insets show coronary arterioles and the bar indicates 50 μm. Representative images from duplicate experiments are shown. (B) HSAT+ content of HS isolated from the left ventricles was determined as in vitro anti-Xa activity, where HS containing AT-binding sites catalyzes AT neutralization of factor Xa. Activity was calibrated to USP units of a heparin standard. Data are the mean ± SEM of 3 animals, P < 0.001 by Student’s t-test.
2.1.4 HSAT+-deficient mice show enhanced sensitivity to TNF
Sepsis involves high circulating levels of TNF (Fig. 1) that contribute to myocardial depression [52, 53]. To determine if TNF contributes to cardiogenic shock in our model, Hs3st1−/− and Hs3st1+/+ mice were pretreated with the TNF-trap etanercept, a fusion protein containing a dimerized ligand-binding domain of the human p75 TNF receptor [54]. For both genotypes, pretreatment with etanercept dramatically prolonged survival time (Fig. 4A). Most importantly, etanercept pretreatment nullified genotype-specific lethality (P = 0.24). This result may be due to either differential production of TNF or differential response to TNF. The first scenario was ruled out, as the time course of plasma TNF in response to LPS was equivalent between genotypes (Fig. 4B). In contrast, Hs3st1−/− mice, compared with Hs3st1+/+ littermates, were more susceptible to death upon TNF infusion (Fig. 4C). Moreover, the Hs3st1−/− genotype hazard ratio for TNF of 3.30 (1.24–8.79) was comparable to that observed for LPS (4.42, 95% CI 2.21–9.05). Hemodynamic analysis confirmed that TNF, like LPS, induced lethal cardiogenic shock (not shown). Combined, these data indicate that Hs3st1−/− mice, compared with Hs3st1+/+, are more susceptible to LPS-lethality because they are more sensitive to TNF.
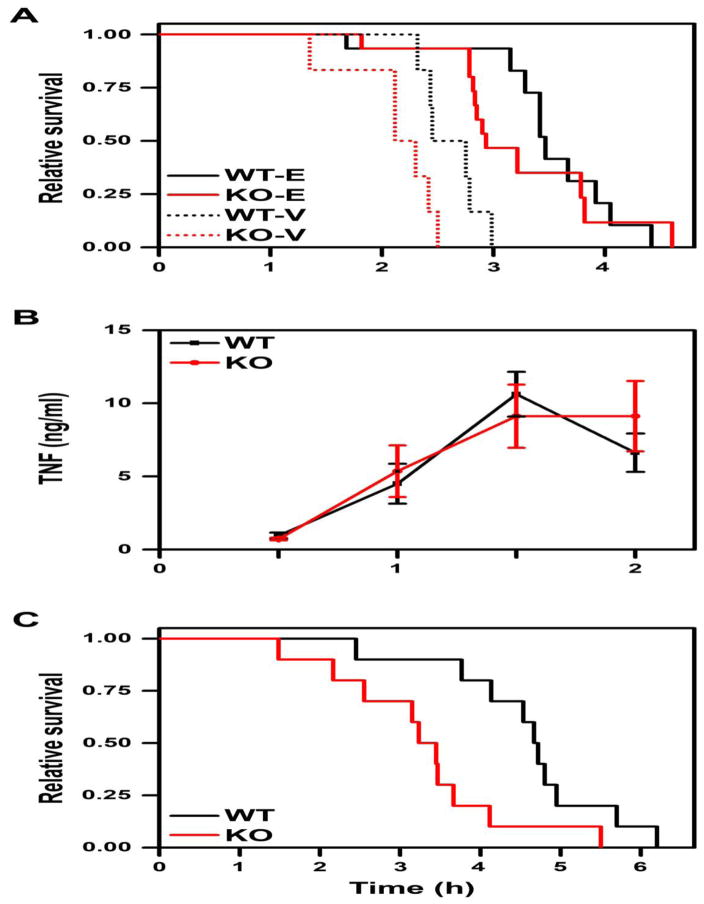
Fig. 4. Hs3st1−/− mice show enhanced TNF-lethality.
(A) Genotype effect for LPS-lethality is abolished by etanercept treatment. Shown is Kaplan-Meier survival analysis for mice intravenously administered vehicle (WT-V and KO-V, 6 mice per genotype) or 10 mg/kg etanercept (WT-E and KO-E, 15 mice per genotype) followed 30 min later by 50 mg/kg LPS (T = 0). Data were evaluated by log-rank with Holm-Sidak pairwise analysis. Within each genotype, the etanercept effect was significant (P < 0.0001 each). The genotype effect was significant within vehicle (P < 0.05) but not for etanercept treatment (P = 0.24). (B) Hs3st1−/− mice exhibit normal TNF production in response to LPS. Hs3st1+/+ and Hs3st1−/− littermates were injected with 50 mg/kg LPS and plasma TNF levels were determined over time. Data are the mean ± SEM of 5 animals per genotype. (C) Kaplan-Meier survival analysis for mice administered 120 ng/g of recombinant mouse TNF over the initial 3 h period; n = 10 mice per genotype. P = 0.012 for a genotype effect by log rank analysis.
2.2 AT effects are reversed in Hs3st1−/− mice
2.2.1 AT enhances LPS-lethality in HSAT+-deficient mice
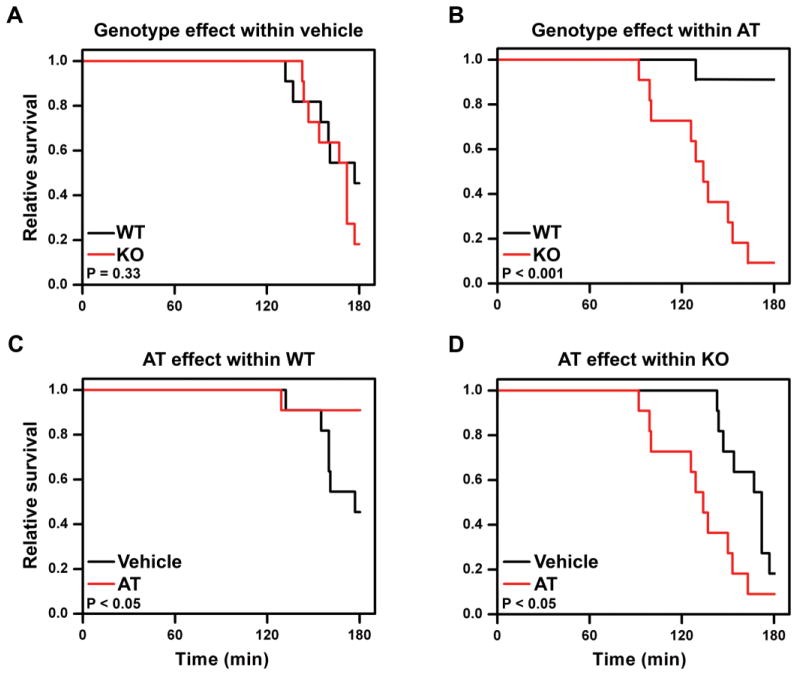
AT treatment reduces morbidity and/or mortality in animal and non-human primate models of sepsis [9, 30, 38–40, 55]. Thus, the pro-inflammatory phenotype of Hs3st1−/− mice suggests that HSAT+ mediates an anti-inflammatory activity of endogenous AT. To determine if this mechanism is operable, we examined the ability of exogenous AT to ameliorate LPS-lethality in Hs3st1+/+ and Hs3st1−/− mice. We examined the efficacy of AT treatment using physiologically comparable doses of LPS (50 mg/kg for Hs3st1+/+; 25 mg/kg for Hs3st1−/− mice) that induced an equivalent rate of death in both genotypes (Fig. 5A).
Fig. 5. AT rescue of LPS-lethality is reversed in Hs3st1−/− mice.
Kaplan-Meier survival analyses for mice (n = 11 per group) administered a physiologically comparable dose of LPS (50 mg/kg for Hs3st1+/+; 25 mg/kg for Hs3st1−/−) at T = 0. At T = −30 min, mice were injected with 250 U/kg AT or AT-vehicle. Between group significance was evaluated by log rank analysis. (A) In vehicle-treated mice, LPS-lethality was comparable between genotypes. (B) For AT treatment, Hs3st1−/− mice, compared with Hs3st1+/+, were more sensitive to LPS. (C) In Hs3st1+/+ mice, AT treatment reduced LPS-lethality. (D) In Hs3st1−/− mice, AT treatment enhanced LPS-lethality.
With AT pretreatment (Fig. 5B), Hs3st1−/− mice exhibited a dramatically reduced survival against LPS with a hazard ratio of 19.8 (2.5–158). The large difference in mortality rates reflects opposite responses in the two genotypes; AT pretreatment improved Hs3st1+/+ survival against LPS (Fig. 5C), but reduced Hs3st1−/− survival against LPS (Fig. 5D). AT versus vehicle pretreatment induced similar hemodynamic alterations in Hs3st1−/− mice at T = Pmax50% (Supplementary Table 1). Thus, for Hs3st1−/− mice, AT pretreatment does not alter the mechanism of death; instead, it enhances susceptibility to LPS. In the absence of HSAT+, AT exhibits a pro-inflammatory tone that enhances LPS-lethality. This concept is consistent with findings that AT is a modulator of inflammation and the net effect (enhancement or inhibition) is highly context dependent [36, 37, 56, 57]. Together, these results suggest that HSAT+ mediates the anti-inflammatory activity of AT.
2.2.2 AT enhances leukocyte-endothelial interactions in HSAT+-deficient mice
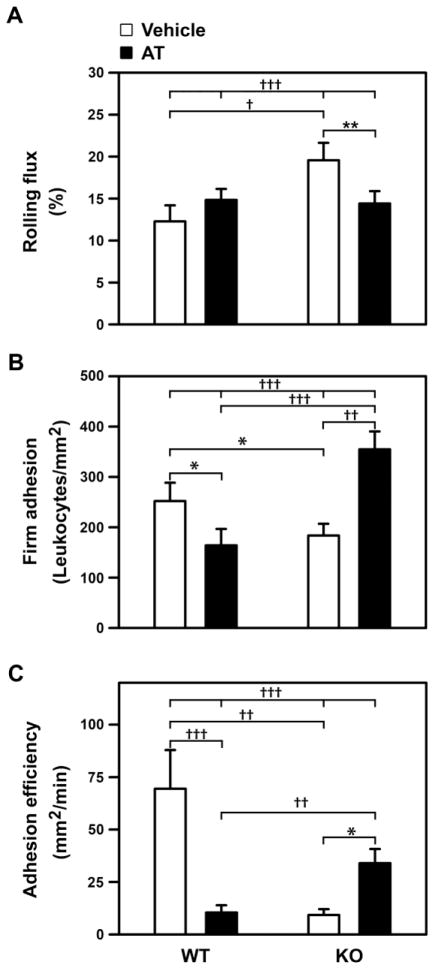
AT’s anti-inflammatory effects include inhibition of leukocyte-endothelial cell interactions [10, 58–61]. To determine if the paradoxic AT effects, in HSAT+-deficient versus wild-type mice, are due to differential modulation of inflammation, we examined the influence of AT or vehicle treatment on LPS-induced leukocyte-endothelial interactions within cremaster muscle venules. We found strong AT-treatment by genotype interactions for all of the examined parameters (P < 0.0001 for each, Fig. 6). In Hs3st1+/+ mice, AT treatment reduced firm adhesion (Fig. 6B) and adhesion efficiency (Fig. 6C) by 35% and 85%, respectively, both of which are anti-inflammatory effects. In contrast, AT treatment produced opposite effects in Hs3st1−/− mice – enhancing LPS induced leukocyte firm adhesion and adhesion efficiency by 1.9- and 3.7-fold, respectively. Overall, AT-treated Hs3st1−/− mice, compared with Hs3st1+/+, exhibited comparable LPS-induced rolling fluxes (Fig. 6A), but a 2.2-fold enhanced firm adhesion, and a 3.3-fold elevated adhesion efficiency. Combined, these results indicate that AT’s net effects are anti-inflammatory in Hs3st1+/+ mice but pro-inflammatory in Hs3st1−/− mice, which parallels AT’s effects on LPS-lethality (Fig. 5).
Fig. 6. LPS-treated Hs3st1−/− mice show diametrically opposite AT effects on leukocyte-endothelial interactions.
Hs3st1+/+ and Hs3st1−/− littermates were pretreated with vehicle or AT (T = −2 h), administered an intraperitoneal injection of LPS (T = 0), and 2 h later leukocyte-endothelial interactions in cremaster muscle venules were measured. (A) Rolling flux: leukocytes rolling on endothelium standardized to total WBC flux through the vessel. (B) Firm adhesion: leukocytes stationary on endothelium standardized to vessel surface area. (C) Adhesion efficiency: stationary leukocytes standardized to rolling leukocytes. Results are expressed as the mean ± SEM from 31, 45, 38, and 68 vessels per group for WT-vehicle, WT-AT, KO-vehicle, and KO-AT, respectively. Rank order normalized data were analyzed by ANOVA with Holm-Sidak pairwise analysis: P < 0.05 (*); P < 0.01 (**); P < 0.002 (†); P < 0.001 (††); P < 0.0001 (†††). Genotype by treatment interactions are indicated by the four-way comparisons (at top of graphs).
2.2.3 HSAT+ selectively mediates the vasodilatory activity of AT
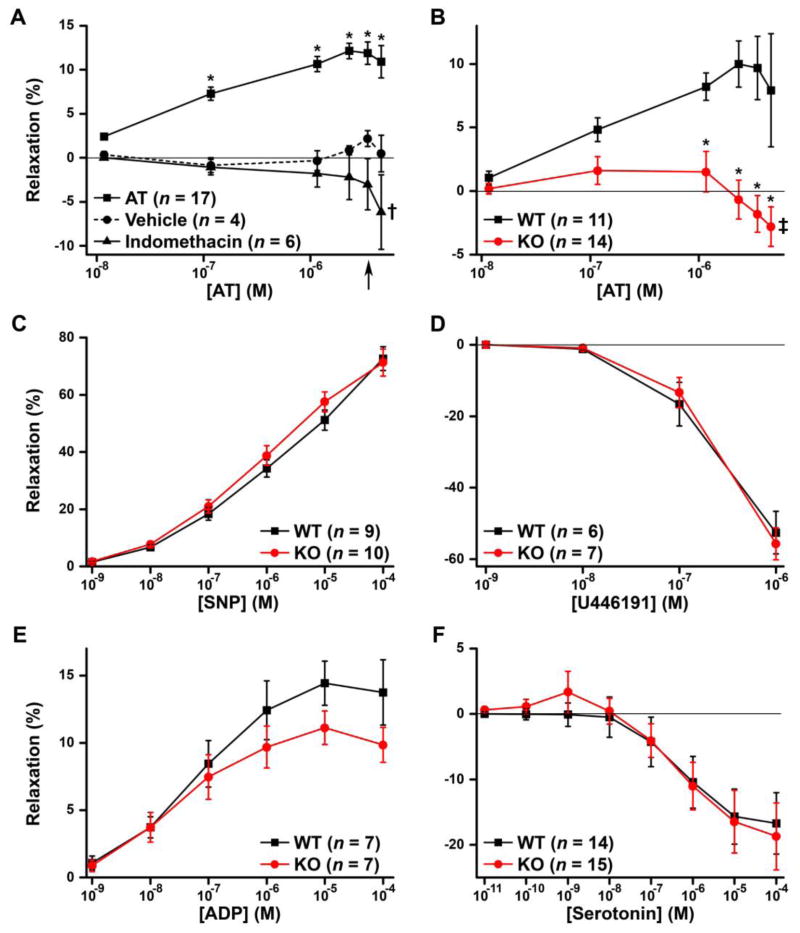
AT’s beneficial effects in sepsis involve the induction of prostacyclin (Fig. 1), which is also a potent vasodilator [41, 45, 62]. If HSAT+ is required for prostacyclin induction, then Hs3st1−/− mice should exhibit altered vasomotor responses to AT. This possibility was tested by first characterizing AT vasomotor activity using coronary arterioles from C57BL/6 mice. Isolated vessels were incubated with increasing concentrations of AT, equivalent volumes of vehicle, or pre-incubated for 30 min with indomethacin, to block prostaglandin/prostacyclin production, followed by AT exposure (Fig. 7A). Vasomotor responses were dramatically different between treatments (P < 10−6 for AT-concentration by treatment interaction). AT concentrations above 12 nM produced significant vasodilation, compared with vehicle and indomethacin treatments. This activity was maximal near the normal plasma concentration for AT (~3.4 μM) [63] (Fig. 7A, arrow). Conversely, in the presence of indomethacin, AT elicited the opposite effect, vasoconstriction, which achieved significance at 4.6 μM AT. Combined, the results indicate that AT exhibits two distinct vasomotor activities. At low concentrations AT is a vasodilator, but when this activity is blocked with indomethacin, high concentrations of AT stimulate vasoconstriction.
Fig. 7. HSAT+ selectively mediates AT vasodilatory activity.
Ex vivo vasomotor responses of precontracted coronary microvessels isolated from C57BL/6 mice (A) or Hs3st1+/+ and Hs3st1−/− mice on a C57BL/6 X 129S4/SvJae background (B–F). Vessels were exposed to increasing concentrations of AT (A, B), sodium nitroprusside (SNP) (C), a thromboxane A2 analog, U44619 (D), ADP (E), or serotonin (F). Results were analyzed by repeated measures ANOVA (with Holm-Sidak pairwise tests) and are presented as the mean ± SEM; n indicates number of vessels analyzed. Only one coronary vessel was used per mouse. (A) Responses to AT, vehicle for AT, or AT plus 10 μM indomethacin, as indicated. The arrow indicates the human plasma concentration of AT; * indicates P < 0.005 compared with vehicle or indomethacin; † indicates P < 0.05 compared with vehicle. (B) AT concentration effects were significant (P < 0.001 for each genotype); * indicates P < 0.005 compared with Hs3st1+/+; ‡ indicates P < 0.01 compared with maximal Hs3st1−/− relaxation (at 0.12 and 1.2 μM AT).
The vasodilatory activity occurred over the affinity range of AT for HSAT+ (Kd≈25 nM), consistent with involvement of HSAT+. Indeed, Hs3st1−/− and Hs3st1+/+ vessels responded oppositely to AT (P < 10−8 for genotype by AT-concentration interaction); AT stimulated vasodilation in Hs3st1+/+ and vasoconstriction in Hs3st1−/− microvessels (Fig. 7B). To rule out the possibility that Hs3st1−/− arterioles have a globally altered vasomotor response, we treated vessels with endothelium-independent agents (Fig. 7C and D) and endothelium-dependent agents (Fig. 7E and F). Hs3st1−/− and Hs3st1+/+ vessels exhibited comparable responses to each vasomotor agent, showing that the genotype effect is selective to AT. Combined, our results reveal that Hs3st1 genotypes differentially affect the vasomotor activities of AT.
2.3 An HS3ST1 SNP associates with cardiovascular disease
2.3.1 HSAT+ is expressed at arterial branch points
Potentially, HSAT+ mediation of AT’s anti-inflammatory activity protects against atherosclerosis, which involves inflammation within blood vessel walls. This protective role requires that HSAT+ be expressed in large arteries, the prime site where atherosclerotic plaques form. Thus, we examined HSAT+ expression in mouse aorta whole mounts. Integrity of the endothelium was confirmed by staining with podocalyxin, which is localized to the endothelial lumenal surface. Staining with AT-biotin•streptavidin-Qdot 647 complexes revealed that endothelial cells throughout the aorta express HSAT+; however, expression was profoundly elevated at aortic branch points (Supplementary Fig. 2). Such sites in the human arterial tree are preferentially susceptible to the development of atherosclerosis [64], consistent with the hypothesis that HSAT+ is atheroprotective.
To test for a causal role, we attempted to generate HSAT+ deficiency in atherosclerosis prone mice (ApoE−/− or Ldr−/−;ApoBec1−/−) on a pure C57BL/6 background. Unfortunately, Hs3st1−/− mice were not viable in these crosses. This is consistent with our prior finding that the Hs3st1−/− genotype does not survive on inbred genetic backgrounds [33]. We also bred the ApoE−/− genotype onto the genetic background in which Hs3st1−/− mice are viable (F1 hybrids of C57BL/6 x 129S4/SVJae), but this background completely prevented the formation of atherosclerosis. Given these limitations in the mouse experiments, we examined if a single nucleotide polymorphism (SNP) in the human HS3ST1 gene associates with atherosclerosis.
2.3.2 rs16881446G/G associates with atherosclerotic cardiovascular disease
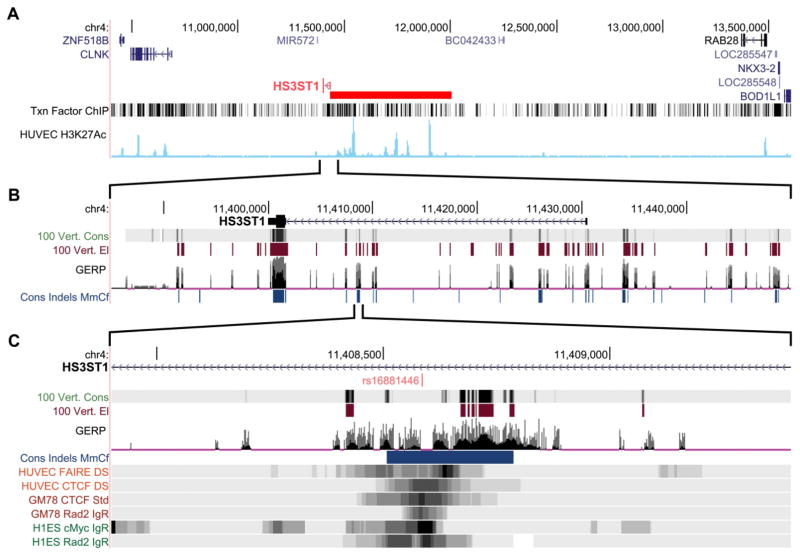
Bioinformatic analyses (section 4.2.2) of transcriptional regulatory regions identified rs16881446 as a putatively functional SNP within the sole HS3ST1 intron (Fig. 8). Relative to the sense orientation of HS3ST1, the rs16881446 allelic nucleotides are A and G. To determine if this SNP associates with severity of CAD and cardiovascular events, we genotyped >2000 coronary angiography patients. Within this population, the rs16881446G allele exhibited a frequency of 0.26, which is comparable to the minor allele frequency (MAF = 0.27) in the HapMap CEU population (Utah residents with ancestry from northern and western Europe). The rs16881446 genotypes were in Hardy-Weinberg equilibrium (P = 0.66).
Fig. 8. HS3ST1 intronic SNP rs16881446 localizes to a functional regulatory region.
The chromosome 4 region of the HS3ST1 gene (which has an antiparallel orientation) was examined using the UCSC Genome Browser on the hg19 assembly. (A) The putative transcriptional control region (red bar) is revealed by ChIP-seq analysis for 161 transcription factors in 91 cell types (Txn FactorChIP) and for H3K27Ac in HUVECs. (B) The transcribed region of HS3ST1 extends from ~11.43 Mb to 11.4 Mb. The top indicates HS3ST1 hnRNA with all coding sequences occurring in a single exon at ~11.4 Mb (thick box). Regions and regulatory elements that exhibit sequence conservation among 100 vertebrates are indicated by 100 Vert Cons and 100 Vert El, respectively. Putative functional mammalian DNA elements (evolutionarily constrained elements) were determined by genomic evolutionary rate profiling (GERP; comparing the hg19 build to 35 other mammalian genomes) and by insertion-deletion based conservation (Cons Indels MmCf; comparing human, mouse and dog genomes), which provides a sequence independent evaluation of conservation; dark blue indicates extremely conserved regions. (C) The rs16881446 SNP occurs in the center of a region with high conservation and overlaps markers of active chromatin. Formaldehyde-assisted isolation of regulatory elements (FAIRE) on HUVECs reveals open chromatin regions. ChIP-seq analysis of HUVECS, lymphoblastoid GM12878 (GM78), and embryonic stem cells (h1ES) reveals chromatin regions containing the transcription factors CTCF, RAD2, and c-Myc.
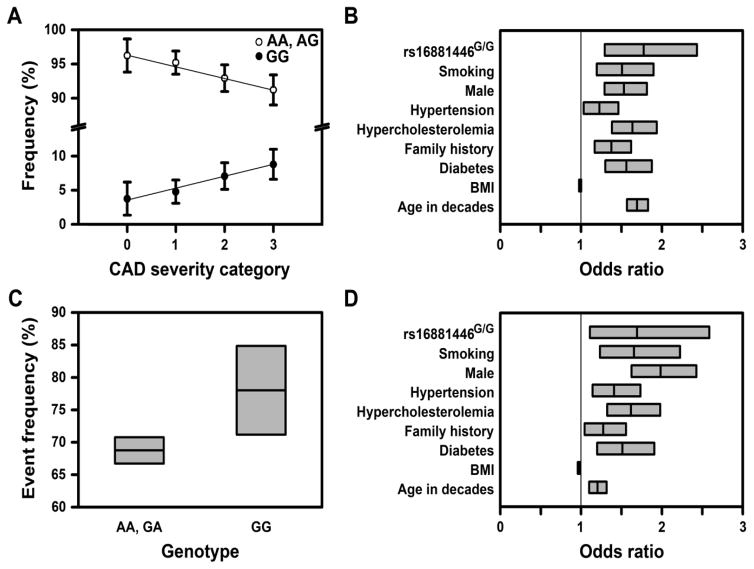
The frequency of the rs16881446G/G genotype was higher in the more severe CAD categories (Fig. 9A); the across group OR was 1.71 (1.25–2.33), P = 0.00075. The average CAD severity score was higher in the GG genotype group than the non-GG group (P = 0.0007). Relative to non-carriers, the genotype specific ORs (across CAD severity) for heterozygotic and homozygotic carriers were 1.09 (0.93–1.28) versus 1.77 (1.29–2.43). Thus, the minor allele of rs16881446 associated with CAD in a recessive fashion. The magnitude of this association was not altered by adjustment for major cardiovascular risk factors (Fig. 9B); the across group adjusted OR was 1.77 (1.29–2.43), P = 0.0004. Indeed, rs16881446G/G exhibited the greatest effect size of all covariates.
Fig. 9. The rs16881446G/G genotype independently associates with severity of CAD and atherosclerotic cardiovascular events.
(A) The frequency of the rs16881446G/G genotype increases with CAD severity. Shown are rs16881446 genotype frequencies ± 95% CI within each CAD severity group, P = 0.0008 by ordinal logistic regression. (B) The rs16881446G/G genotype independently associates with CAD severity. Shown are the adjusted, across ordered group ORs for the rs16881446G/G genotype and CAD clinical risk factors, as determined by ordinal logistic regression. (C) The frequency of atherosclerotic cardiovascular events is higher in the rs16881446G/G genotype group. Shown are event frequencies ± 95% CI, P = 0.021 by chi-square test. (D) The rs16881446G/G genotype independently associates with the combined endpoint of Any Cardiovascular Event, analysis by logistic regression. Data in (B) and (D) are OR ± 95% CI. The broad CI for rs16881446G/G reflects the low population frequency. The above analyses were also conducted by including variables for sex and sex-by-rs16881446G/G interaction. In all cases, the interaction term was insignificant (P > 0.05); thus, rs16881446G/G associations are independent of sex (see also Supplementary Fig. 6).
Thus, rs16881446G/G exhibited a strong and independent association to CAD severity, which matches the behavior of major cardiovascular risk factors (Table 2). The rs16881446G/G genotype also associated with the combined endpoint of Any Cardiovascular Event (Fig. 9C). The rs16881446G/G group exhibited a higher frequency of cardiovascular events, OR of 1.61 (1.07–2.42) and P = 0.021. Again, the magnitude of this association was not affected by adjustment for classical cardiovascular risk factors (Fig. 9D). The adjusted OR for rs16881446G/G was 1.69 (1.11–2.59), P = 0.015, which was second only to male sex in effect size. Thus, rs16881446G/G exhibited a strong and independent association to atherosclerotic cardiovascular events.
Table 2.
Characteristics of cardiovascular patients across CAD severity categories
| Parameter | CAD Severity Category | P-valuea | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| n | 239 | 605 | 664 | 636 | |
| Males n | 128 | 418 | 461 | 448 | |
| Females n | 111 | 187 | 203 | 188 | |
| Male SYNTAX range | 0 | 0–6 | 7–17 | 17.5–118 | |
| Female SYNTAX range | 0 | 0–2 | 3–12 | 13–90 | |
| Male | 53.6% | 69.1% | 69.5% | 70.5% | <0.001 |
| Female | 46.4% | 30.9% | 30.5% | 29.5% | <0.001 |
| Age | 57.8 ± 0.8 | 60.8 ± 0.5 | 63.7 ± 0.4 | 68.5 ± 0.4 | <0.001 |
| Hypercholesterolemia | 49.0% | 60.7% | 64.3% | 75.0% | <0.001 |
| Diabetes | 18.0% | 23.0% | 28.8% | 34.3% | <0.001 |
| Family history | 33.9% | 42.3% | 44.9% | 42.0% | 0.030 |
| Presently smoking | 12.6% | 18.0% | 14.9% | 12.0% | 0.018 |
| BMI | 30.9 ± 0.5 | 30.2 ± 0.3 | 29.6 ± 0.2 | 29.3 ± 0.2 | 0.003 |
by nonparametric test for trend across ordered groups
2.3.3 The rs16881446G allele associates with reduced HS3ST1 expression
Within its linkage group (Supplementary Fig. 3), rs16881446 is the most likely SNP to be functional (Appendix B). To test if rs16881446 genotype associates with HS3ST1 expression, we screened several human primary endothelial cells and identified heterozygotic cells, rs16881446A/G, from human lung (HLMEC). (For pulmonary endothelial cells, HS also protects against TNF-stimulated leukocyte adhesion and lethality [19].) We then tested for allele specific expression in an equivalent cellular setting.
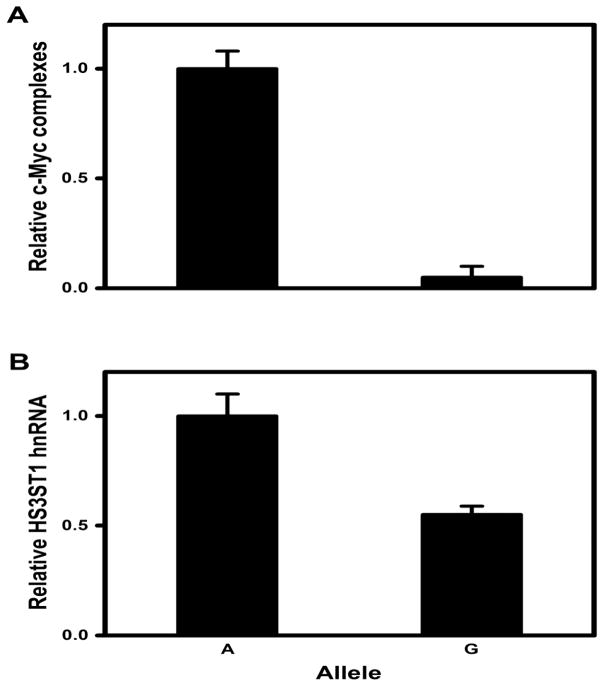
Our bioinformatic analysis indicated that the rs16881446 region directs the formation of c-Myc containing transcription complexes (Fig. 8C). To determine if the rs16881446G allele influences regional formation of c-Myc containing chromatin, we performed ChIP analysis on HLMECs. DNA for rs16881446A was recovered but rs16881446G DNA was undetectable (Fig. 10A). Thus, the rs16881446 region can direct the incorporation of c-Myc into chromatin complexes only when the major allele is present. Since rs16881446 is intronic, we also measured accumulation of HS3ST1 nuclear transcripts. The rs16881446A nuclear transcripts were 2-fold more abundant than that of rs16881446G (Fig. 10B). Together, these data indicate that the minor allele of rs16881446 associates with reduced HS3ST1 expression.
Fig. 10. The rs16881446G allele associates with reduced HS3ST1 expression.
(A) Crosslinked chromatin extracted from HLMEC was immunoprecipitated with anti-Myc antibodies. DNA was recovered and relative levels of rs16881446 alleles were quantified by real time PCR with allelic discrimination Taq-ManR probes. The rs16881446G levels in ChIP DNA were comparable to background controls. (B) Nuclear RNA was isolated from HLMEC; HS3ST1 hnRNA levels were determined by reverse transcription followed by real time PCR with allelic discrimination Taq-ManR probes; P < 0.01 by Student’s t-test. All data are expressed as mean ± SEM from three independent experiments.
3 Discussion
The AT-binding pentasaccharide within HSAT+ has been studied for more than 30 years [48], yet the biologic role of HSAT+ has remained enigmatic. It was long predicted that endothelial HSAT+ would convey an antithrombotic tone to the blood vessel wall by catalyzing AT’s anticoagulant activity. However, our prior analyses of Hs3st1−/− mice revealed that hemostasis is unaltered, despite their very low levels of HSAT+ [33]. Consequently, we sought to determine if HSAT+ might be involved in AT’s inflammomodulatory activities. Our studies show that Hs3st1−/− mice, compared with Hs3st1+/+, are more susceptible to LPS-lethality due to increased sensitivity to TNF. In Hs3st1+/+ mice, AT treatment reduced LPS-lethality, diminished leukocyte firm adhesion to endothelial cells, and dilated isolated coronary arterioles. Surprisingly, in Hs3st1−/− mice AT induced diametrically opposite effects. Combined, these results indicate that HSAT+ selectively mediates AT’s anti-inflammatory activity. To the best of our knowledge, we are the first to report the biologic role of HSAT+ and Hs3st1 genotype dependent actions.
An AT•HSAT+ anti-inflammatory pathway may also be relevant to chronic vascular inflammation. Our finding that rs16881446 associates with reduced HS3ST1 expression, increased CAD severity, and increased atherosclerotic cardiovascular events raises the possibility that endothelial HSAT+ may be atheroprotective. Thus, HSAT+ may serve as a novel target for the development of therapeutics to combat acute and chronic inflammatory disorders.
Our findings are supported by prior studies of AT’s inflammomodulatory properties. First, numerous approaches have shown that AT’s anti-inflammatory actions require HSPGs and have implicated syndecan-4, a cell surface HSPG with inherent signaling properties [37, 41, 43, 65]. Interestingly, syndecan-4 deficient mice also show enhanced death in response to LPS [66]. We have previously shown that syndecan-4 is a major endothelial HSPG that can bear HSAT+ [30, 67]. Herein, we have identified the specific species of HS (HSAT+) that is essential for AT’s anti-inflammatory effects. Second, in vitro studies have found that the engagement of AT with endothelial HSPGs stimulates signaling pathways that block TNF activation of NF-κB (Fig. 1) [10]. Loss of these pathways should produce increased sensitivity to TNF–a key feature of Hs3st1−/− mice. Third, in vitro and in vivo approaches have demonstrated that AT’s anti-inflammatory effects involve endothelial cell synthesis of prostacyclin (a potent vasodilator), and AT’s beneficial effects are prevented by treatment with indomethacin, an inhibitor of prostaglandin synthesis [9, 35, 62]. Using isolated coronary vessels, we showed that AT is an HSAT+-dependent vasodilator and this activity is blocked by indomethacin. Fourth, in vitro studies show that AT can exert context-dependent pro-inflammatory effects, which include macrophage activation, leukocyte chemotaxis, and smooth muscle cell elaboration of pro-inflammatory molecules [36, 37, 56, 57]. Our in vivo study demonstrates that HSAT+ selectively mediates AT’s anti-inflammatory effects; when HSAT+ levels are reduced (Hs3st1−/− mice), AT’s latent pro-inflammatory activities are revealed, as evidenced by increased LPS-lethality and endothelial-leukocyte interactions. It is important to note that AT exerts anti-inflammatory effects on various leukocyte populations [10, 41]. Thus, we presently cannot determine if the in vivo, HSAT+-dependent effects of AT involve endothelial cells, leukocytes, or both cell-types.
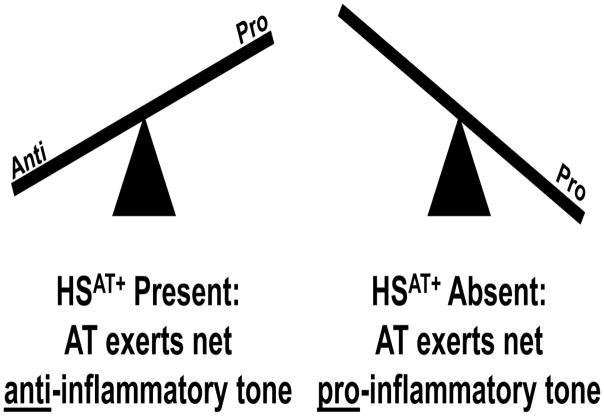
Based on the above findings we propose that HSAT+ serves to regulate the inflammomodulatory tone of AT (Fig. 11). At the extremes (high vs low HSAT+) AT provides either a net anti- or pro-inflammatory tone, respectively. This mechanism might occur at the level of a single cell-type such as endothelial cells, which highly express HSAT+ [28, 30]. Alternatively, differential effects might occur across multiple cell-types, including various types of leukocytes [10, 37, 68]. We hypothesize that intermediate levels of HSAT+ provide fine-tuning that defines the precise inflammatory tone.
Fig. 11. HSAT+ is essential for AT’s anti-inflammatory activity.
AT is a bifunctional regulator of inflammation that exhibits HSAT+-dependent anti-inflammatory and HSAT-independent proinflammatory activities. AT’s net in vivo action is determined by the balance between these two opposing activities.
The involvement of HSAT+ in AT’s anti-inflammatory activity is quite surprising given that the pentasaccharide sites in HSAT+ (Kd≈25 nM) should be saturated at normal plasma levels of AT (~3.5 μM). How then could HSAT+ be involved in a process that is induced by supraphysiologic plasma levels of AT (~7 μM) [48]? On one hand, this scenario suggests that HSAT+ may not function as a “receptor”, as it would not “sense” changes in AT plasma levels. Potentially, HSAT+ serves in a permissive fashion. Such a mechanism occurs in FGF signaling, where HS participates in docking the ligand into its receptor and thereby creates a functional ternary signaling complex [13, 69]. On the other hand, perhaps only a small subpopulation of AT molecules exhibits anti-inflammatory activity. AT is not homogeneous; it can bear four or three N-linked glycans (αAT or βAT, respectively). Compared with αAT, βAT is 10- to 20-fold less abundant in plasma, has a greater affinity for HS, and may have far greater anti-inflammatory activity [10, 70, 71]. Potentially, administration of supraphysiologic levels of exogenous AT increases HSAT+ occupancy by βAT. Much more extensive analyses will be required to assess these and other potential mechanisms.
Resolution of the above issues must also take into account our finding that Hs3st1−/− mice, without exogenous AT treatment, exhibited a pro-inflammatory phenotype. In particular, Hs3st1−/− mice, compared with Hs3st1+/+, were more susceptible to LPS- and TNF-induced death. These results indicate that an AT•HSAT+ anti-inflammatory pathway may be constitutive – operable under physiologic levels of AT. This possibility offers potential insights into the pathophysiology of genetic AT deficiency. Mutations in the heparin binding site (AT-HBS), which reduce AT binding to HSAT+, are far more likely to cause arterial thrombosis than mutations that reduce AT levels/activity [72]. Based on this finding, it has been proposed that HSAT+ functions as an anticoagulant preferentially in arteries; however, such a role could not be demonstrated in Hs3st1−/− mice [33]. In contrast, loss of a constitutive AT•HSAT+ anti-inflammatory pathway in endothelial cells could lead to a pro-inflammatory endothelial phenotype featuring upregulation of pro-coagulant factors (e.g. tissue factor) and down regulation of anticoagulant proteins (e.g. thrombomodulin) [73]. Thus, arterial thrombosis in AT-HBS patients may, at least in part, be secondary to a pro-inflammatory endothelial cell phenotype. Potentially, management of such patients might include treatment with statins, which exert anti-inflammatory effects on endothelial cells [74].
A constitutive AT•HSAT+ anti-inflammatory pathway in arteries may protect against chronic diseases involving vascular inflammation, such as atherosclerosis. Indeed, our analysis of rs16881446 suggests that HSAT+ may be atheroprotective. Intriguingly, the minor allele (G) is also the ancestral allele. The UCSS Genome Browser shows that rs16881446G is the reference allele for 9 of 11 non-human primate species and for the two Neandertal samples in which this region has been sequenced. It is also the reference allele for most mammalian species (Supplementary Fig. 4, Supplementary Table 2). Thus, the rs16881446A allele appears to have arisen early in the human lineage. These data raise the possibility that the emergent rs16881446A conveyed a functional benefit and became the major allele through selection; however, a founder effect cannot be excluded.
Our study design enhanced the likelihood of detecting an association. On one hand, we first used a bioinformatic approach to identify a potentially functional SNP rather than analyzing random SNPs. On the other hand, our primary outcome, CAD severity, provides a semi-quantitative index of plaque pathology (burden/stage). This outcome provides greater sensitivity than binary clinical events (such as myocardial infarction) that are dependent on plaque pathology plus multiple additional factors (such as environmental and physiologic precipitators of plaque rupture). One limitation of our study is that we have examined a clinical population enriched in cardiovascular risk factors, which selects for individuals with specific environmental exposures, life-styles, and genetic variants. Moreover, although our data are consistent with HSAT+ being atheroprotective, they do not reveal if the role of HSAT+ is anti-inflammatory, anticoagulant, or both. However, our study demonstrates a clear association with CAD that warrants further investigation to resolve these mechanistic issues.
Since our study involved individuals at high disease risk, further investigations will be required to extend our findings to the general population. Moreover, prospective studies will be required to determine if HS3ST1 SNPs are risk factors for atherosclerotic cardiovascular disease. Such studies should consider the extremely large putative regulatory region of the HS3ST1 gene (~600 kb), which likely contains numerous additional functional SNPs. The net impact of multiple regulatory SNPs on HS3ST1 expression may generate far greater effects than any individual SNP. Thus, the elucidation of HS3ST1 regulatory regions that harbor functional SNPs would be a critical first step towards evaluating if HS3ST genotype is a risk factor for atherosclerosis.
In summary, our analyses of an acute sepsis model in mice reveal that HSAT+ is an anti-inflammatory molecule that inhibits TNF-responsiveness. HSAT+ is essential for AT’s anti-inflammatory effects, but not AT’s pro-inflammatory effects. It follows that the level of HSAT+ can tightly regulate AT’s inflammomodulatory tone. An AT•HSAT+ anti-inflammatory pathway appears to be constitutive; Hs3st1−/− mice are more susceptible to LPS- and TNF-lethality. Such a constitutive pathway might protect against atherosclerosis; rs16881446G/G associates with reduced HS3ST1 expression and increased CAD/atherosclerotic cardiovascular events. Thus, our study identifies HS3ST1/HSAT+ as targets for the development of novel diagnostics/therapeutics to combat acute and chronic inflammatory disorders.
4 Methods
4.1 Mouse Studies
4.1.1 Mice
All animal experiments were performed following review and approval by Institutional Animal Care and Use Committees of all participating institutes. Hs3st1−/− mice, deficient in HSAT+, were previously generated by gene targeting [33]. The Hs3st1−/− genotype is lethal on inbred strains, so heterozygous mice (Hs3st1+/−) were maintained by backcrossing >15 generations against C57BL/6 mice or >10 generations against 129S4/SvJae mice. Viable Hs3st1−/− mice and wild-type littermates (Hs3st1+/+) were generated as F1 hybrids by crossing the isocongenic heterozygotic strains (male 129S4/SvJae-Hs3st1+/− X female C57BL/6-Hs3st1+/−). This approach ensured that the resulting mice were viable and had a uniform genetic background. Animals were housed under diurnal lighting (12 h light, 12 h dark) and allowed free access to food and water. Hs3st1+/− exhibit minimal phenotype [33], so only Hs3st1−/− mice were analyzed. Each experiment used mice between 15 and 22 weeks old (mean age 17 weeks), with ages being balanced between experimental groups.
4.1.2 LPS-induced sepsis model
Mice were anesthetized with isoflurane (2.5%) in room air and placed on a heated surgical bed (CWI Inc., IL, USA) that was feedback regulated (37 ± 0.2°C) by a rectal thermometer (Thermistor probe, TH-2Kmr) connected to a TC-1000 temperature controller. The right jugular vein was catheterized with PE-10 tubing to enable i.v. delivery of all treatments. A 1.2F pressure-volume catheter with 4.5 mm electrode spacing (Scisence, ON, Canada) was inserted via the right carotid artery into the left ventricle. Mice were allowed to breath spontaneously during the experiment. Catheter volume calibration was validated by echocardiography with a Vevo 770 ultrasound (VisualSonics).
After surgical instrumentation, the isoflurane concentration was reduced to 1.25% and steady-state hemodynamics were allowed to establish over a period of at least 20 min. Human clinical grade AT (Cutter Biologic) was reconstituted on the day of the experiment. At T = −30 min, mice were injected (jugular vein) with 250 U/kg AT/an equivalent volume of AT-vehicle, or 10 mg/kg etanercept (Immunex Corporation, Thousand Oaks, CA)/an equivalent volume of etanercept-vehicle. At T = 0, an automatic syringe pump (Syringe Pump 11plus, Harvard Apparatus, Holliston, MA) was used to deliver 10 mg/ml LPS in sterile saline (25 or 50 mg/kg), or an equivalent volume of vehicle, over 20 min. Alternatively, 30 ng/g/h of 20 μg/ml TNF, (recombinant mouse, Life Technologies, Grand Island, NY, USA) in PBS with 1 mg/ml BSA, was injected over 3 h. Also at T = 0, the body temperature set point was increased to 38.5 ± 0.2°C, so as to mimic the mild fever in sepsis. Real time conductance and pressure data were recorded throughout the experiment with a 308T 24-bit data acquisition system (iWorx Systems, Inc., Dover, NH). Hemodynamic parameters were extracted with Labscribe software (iWorx Systems Inc., Dover, NH).
4.1.3 Fluorescent detection of HSAT+
The preparation of AT conjugated to Alexa Flour 647 (AT-647) has been previously described [51]. This procedure was also used to conjugate biotin to AT (AT-biotin). Euthanized mice were sequentially perfused through the left ventricle with ice-cold solutions (20 ml) of PBS, 4% paraformaldehyde, and PBS. Hearts were isolated and embedded in OCT; whereas, isolated aortas were processed as below. Cryosections (~5 μm) were co-stained with AT-647 and goat anti-mouse-podocalyxin antibody followed by Alexa Fluor 568-labeled donkey anti-goat antibody, as previously described [51]. Endothelial cells are usually identified with CD31, but this epitope is destroyed by fixation; consequently, we used podocalyxin, which exhibits comparable specificity to detect endothelial cells in our target tissues.
For en face staining, aortas were opened longitudinally, pinned down, and biotin blocked (Molecular Probes, E21390). Samples were incubated at room temperature in blocking solution (Perkin Elmer, NEL746B) supplemented as indicated below and washed with 1% BSA in PBS (w/v) (5 min per wash). Tissues were blocked for 30 min, washed three times, incubated for 30 min with 200 nM AT-biotin and 5 μg/ml goat anti-mouse podocalyxin (R&D Systems, AF1556), washed three times, incubated for 1 h with 20 nM streptavidin-conjugated Qdot 647 (Molecular Probes #1016-1) and 4 μg/ml donkey anti-goat Alexa Fluor 568 (Molecular Probes), and washed three times. Images were obtained using a Leica fluorescence dissecting-microscope.
4.1.4 HSAT+ specific activity of left ventricle HS
Total HS was isolated from left ventricle tissue and quantified by dye binding, as previously described [33]. HSAT+ specific activity was assessed as the in vitro activity of HS to enhance AT neutralization of factor Xa with monitoring of S2765 cleavage by factor Xa, as previously described. Activity was calibrated against a standard curve of porcine heparin (179 USP U/mg) (H-3393; Sigma Aldrich).
4.1.5 Measurement of plasma TNF
Blood (~1 ml) was collected by cardiac puncture with a syringe containing 100 μl of 3.2% sodium citrate. Plasma was prepared by two sequential centrifugations (1000 x g for 15 min), then frozen on liquid nitrogen and stored at −80°C. TNF was measured by ELISA for mouse TNF (#KMC3012; Invitrogen, Camarillo, CA).
4.1.6 Leukocyte-endothelial interactions
Mice were anesthetized with intraperitoneal ketamine (125 mg/kg, Ayerst, Guelph, ON, Canada), xylazine (12.5 mg/kg, Bayer, Toronto, ON), and atropine (0.25 mg/kg, Ormond Veterinary Supply, Ancaster, ON) and the cremaster muscle was exteriorized. To maintain anesthesia, Nembutal (Abbott Laboratories, Toronto, ON) was administered through a cannula placed in the jugular vein. AT or its vehicle was given i.v. (250 U/kg in less than 150 μl) 2 h before administration of LPS. To ensure that all mice would survive throughout the experiment, a sublethal dose of LPS (10 mg/kg) was employed. After 2 h, leukocyte-endothelial interactions in venules of the cremaster muscle were measured by intravital videomicroscopy.
4.1.7 Intravital videomicroscopy
The intravital technique, adapted from Ley et al. [75], is based upon the previously described analysis of leukocyte rolling in the mouse cremaster muscle [76, 77]. Cremaster venules were visualized using an Olympus BX51WI microscope with a 40X 0.8 NA water immersion objective. Centerline erythrocyte velocity (Vrbc) was measured in real-time with a photo-diode velocimeter running a computerized digital cross-correlation program (Microvessel Velocity OD-RT, CircuSoft Instrumentation, Hockessin DE). Images were acquired using a Sony Model SSC-S20 CCD camera connected to a monitor and a VHS recorder (Sony, SVA-9500MD). Blood was obtained by carotid cannulation. Systemic leukocyte counts were obtained using Zap-OGLOBIN and a hemocytometer. The volumetric blood flow, total leukocyte flux, wall shear rate, and critical velocity for each vessel were determined, as previously described [77].
Data were analyzed blinded to group and treatment. One minute video recordings of each vessel were analyzed offline for 1) the number of rolling leukocytes passing through a plane perpendicular to the blood flow, 2) the number of firmly adherent leukocytes, defined as being stationary for over 30 seconds, and 3) the length (Lv) and diameter (Dv) of the venule segment examined. In cases of a visible high velocity cell, the velocity of the cell was estimated to determine whether such a cell fit the criteria for rolling, having a velocity less than the critical velocity for that vessel [78]. Vessels were only included if they fell within the following ranges: diameter of 20–50 μm, centerline velocity of 0.5–3.3 mm/s, and shear rate of 200–800 s−1. These parameters were balanced between groups and did not confound outcomes. To compensate for variations in the systemic leukocyte count, leukocyte rolling in the cremaster muscle is presented as the leukocyte rolling flux, which is the number of rolling leukocytes in the vessel as a percentage of the total leukocyte flux. To compensate for the length of vessel analyzed, firm adhesion is presented as leukocytes per mm2, calculated by dividing the number of firmly adherent leukocytes by the area of vessel wall analyzed (π x Dv x Lv). To relate the number of adherent cells to the pool of available rolling leukocyte in the same vessel, we also determined the adhesion efficiency; the number of firmly adherent leukocytes per mm2 were normalize to the number of rolling cells per min [79].
4.1.8 Coronary arteriole vasomotor activity
From each mouse heart, a single segment of the left anterior descending coronary artery (70–180 μm internal diameter) lacking side branches was isolated. The vessel segment was placed in an organ bath, cannulated with micropipettes on both ends and secured with 10-0 nylon suture, inflated at 40 mmHg, and monitored with a video camera calibrated to reveal vessel diameter, as previously described [80]. All experimental agents were added to 37°C extravascular buffer, which was oxygenated and recirculated. After equilibration, microvessels were preconstricted to 30–50% of the baseline diameter with U46619 (a thromboxane A2 analog). Vessels were then exposed to increasing concentrations of analytes. Between treatments, vessels were washed for 15–30 min with three exchanges of Krebs buffered solution. Responses were expressed as percent relaxation from the U-46619 constricted microvessel diameter.
4.2 Human Studies
4.2.1 Bioinformatic Analysis of HS3ST1 Gene Region
Structural and functional features of the human HS3ST1 gene region were identified with UCSC Gene Browser (http://genome.ucsc.edu/) [81] programs (Supplementary Table 3), relative to hg19 coordinates. Linkage disequilibrium of SNPS and regional recombination rates were determined with SNAP v 2.2 (https://www.broadinstitute.org/mpg/snap/doc.php) [82], using 1000 Genomes Pilot 1 phased genotypes for 179 individuals of the HapMap CEU panel.
4.2.2 Bioinformatic identification of a putatively functional HS3ST1 SNP – rs16881446
To maximize the possibility of detecting a genetic association, we sought to identify a putatively functional SNP in HS3ST1. An initial survey of the protein-coding region with the NCBI Variation Viewer revealed 36 non-synonymous SNPs. However, the most common, Lys295Arg, (MAF of ~3%) is a very conservative substitution and the remainder were too rare for analysis (MAF < 0.2%). Given the high conservation of the protein coding region, we focused on identifying a candidate regulatory SNP. Analysis with the UCSC Genome Browser revealed that the HS3ST1 gene occurs in a ~2.6 Mb gene desert (chr4:10.75–13.35 Mb, hg19 coordinates) (Fig. 8A). ChIP-seq analysis for a marker of enhancer elements (H3K27Ac) and for 161 transcription factors suggests a large putative transcriptional regulatory region (~600 kb, from 11.4–12.0 Mb). This region contains ~46,000 SNPs; thus, it is likely that one or more SNPs may affect the expression of HS3ST1.
To simplify the search for a putatively functional SNP and to enable detection of recessive associations, we only examined SNPs with a MAF >0.2. In addition, we narrowed our focus to a 60 kb span that encompasses the HS3ST1 primary transcript (Fig. 8B). We identified SNPs within regions of high phylogenetic conservation, which implies functionality. The best location that conformed to all of these criteria was within the gene’s sole intron, ~7 kb upstream of the protein-coding region. The resulting ~400 bp span contained rs16881446 (Fig. 8C). The rs16881446 minor allele (G) is present in all 26 human populations of the 1000 Genomes Project (Phase 3) with an overall frequency of 0.26 (individual population MAFs range from 0.14–0.42). In HUVECs, sequences from this region are recovered by FAIRE, which indicates an open chromatin configuration. In HUVECs and additional cell lines that express HS3ST1, ChIP-seq analysis reveals that chromatin of this region contains the transcription regulator c-Myc [83] and the factors CTCF and Rad2 (Fig. 8C), which participate in both chromatin organization and transcriptional control [84, 85]. Combined, these data strongly suggest that rs16881446 occurs in a region that regulates HS3ST1 expression.
4.2.3 Study population
The study was approved by the Institutional Review Board of Dartmouth-Hitchcock Medical Center (DHMC)/Dartmouth College and all participants provided written informed consent. Consecutive cardiology patients (>95% Caucasian) undergoing diagnostic or therapeutic coronary angiography at DHMC were enrolled in batches from June 2003 to May 2013. From each subject, ten milliliters of whole blood (EDTA-preserved) was obtained for extraction of genomic DNA (Sorenson Genomics). Patients in shock or unable to consent were excluded. All subjects underwent a complete medical history and physical examination. The study population was a subset from the Dartmouth Dynamic Registry (DDR), a large, prospective, clinical registry of consecutive patients undergoing diagnostic and/or interventional cardiovascular catheterization procedures at DHMC [86]. The registry collects detailed data on demographics and medical history including cardiovascular risk factors, coronary anatomy and function, and procedural indications and processes. Most subjects had multiple coronary angiograms; we obtained 3596 blood draws from 2,200 unique individuals. To obtain a uniform data set, we extracted all registry records per participant and selected the earliest complete data set for each patient (Supplementary Fig. 5). From this group, we successfully genotyped 2,144 individuals for rs16881446 and rs1047389, which were used as the study population. For association to CAD severity, the sample size (2,144) and genotype frequency (6.58%) enable detection of an across group OR of 1.54 at a power greater than 0.83 (α = 0.05).
4.2.4 Determination of CAD severity
CAD severity was categorically defined based on a modification of the SYNTAX score, which describes complex coronary disease and is predictive of coronary outcomes [87, 88]. The DDR documents >90% of the parameters required to calculate a SYNTAX score. Specifically, angiographic findings are recorded by dividing the coronary arterial tree into 27 segments (Coronary Artery Surgery Study, CASS, criteria) [89] with scoring of each segment for primary criteria of the ACC/AHA lesion classification system [90], which includes extent and length of stenosis as well as morphologic features such as calcification, thombosis, eccentricity, and tortuosity. The presence of bridging collaterals, restenosis, bypass grafts, and dissection are also recorded. These parameters were used to approximate the SYNTAX score. This approach was validated by selecting 27 angiogram cines for direct SYNTAX scoring by an experienced invasive cardiologist, blinded to case and CAD severity score. The database derived SYNTAX score closely correlated with direct scoring from angiogram cines (r = 0.85). CAD severity was then categorized in four levels progressively ranging from angiographically undetectable CAD to severe disease. The SYNTAX system does not score lesions producing <50% occlusion, which do not warrant clinical intervention. Consequently, a SYNTAX score of 0 encompasses a mix of detectable and undetectable pathologies. To distinguish between these two categories, all cases with a SYNTAX score of 0 and undetectable coronary occlusion were assigned to CAD severity level 0. The remaining subjects were grouped by lower, middle, and upper tertiles (per sex) of the SYNTAX score and were assigned to CAD severity levels 1–3, respectively. The primary cardiovascular risk factors of male sex, age, hypercholesterolemia, hypertension, and diabetes progressively increased with rising CAD severity (Table 2). This graded association between risk factors and extent of CAD indicates that our categorization method provides a valid metric for CAD severity.
4.2.5 Definition of Any Cardiovascular Event
Patients were assigned to this event group if they had a past or present history of at least one of the following: heart attack, percutaneous coronary intervention (angioplasty, atherectomy, thrombolysis, or stent insertion), coronary artery bypass graft, carotid endarterectomy, transient ischemic attack, stroke, or peripheral occlusive vascular disease. The risk factors of male sex, age, hypercholesterolemia, hypertension, diabetes, and family history were all more prevalent in patients with at least one atherosclerotic cardiovascular event (Table 3), which confirms the validity of this metric.
Table 3.
Patient characteristics by Any Cardiovascular Event group.
| Parameter | No event | Any CV event | P-valuea |
|---|---|---|---|
| n | 657 | 1487 | |
| Male | 58.8% | 72.0% | <0.001 |
| Female | 41.2% | 28.8% | <0.002 |
| Age | 62.0 ± 0.5 | 64.4 ± 0.3 | <0.001 |
| Hypercholesterolemia | 55.4% | 68.9% | <0.001 |
| Diabetes | 22.0% | 30.0% | <0.001 |
| Family history | 38.6% | 43.2% | 0.048 |
| Presently smoking | 12.6% | 15.5% | 0.078 |
| BMI | 30.5 ± 0.3 | 29.5 ± 0.2 | 0.002 |
by Student’s t-tes
4.2.6 Genotyping
Genotyping was performed by TaqManR allelic variant discrimination using an Applied Biosystems 7500 Fast Real-Time PCR System. The rs16881446 SNP was amplified with 5’-dCCTGCCAAGGATGAAAATCTG and 5’-dCATGTCTTCACTTTCATCTTTGCA and then detected with 5’-6-FAM-dTGAGGTGCTCTTCCGTA-MGB (minor allele) and 5’-VIC-dTGAGGTGCTCTTCCGTA-MGB (major allele). The rs1047389 SNP was amplified with 5’-dGCCTCTACCACGTGCACATG and 5’-dGGTCGCCGTCCACAATG and then detected with 5’-6-FAM-dTGCGCTTCTTCCCGCT-MGB (minor allele) and 5’-VIC-dCGCTTTTTCCCGC-MGB (major allele).
4.2.7 ChIP analysis
HLMEC cells were obtained from Lonza (cc-2527, Lot 6F3754) and maintained in EGM-2MV (cc-3202). ChIP assays were performed with the ChIP Assay kit (Upstate Biotechnology), according to the manufacturer's instructions. Nucleoprotein complexes from 40 x 106 HLMECs were cross-linked in vivo by incubating monolayers with 1% formaldehyde for 10 min. Cells were harvested, pelleted, and resuspended in 350 μl SDS lysis buffer. Chromatin DNA was fragmented by sonication, and oligonucleosomal-protein complexes were immunoprecipitated using a polyclonal anti-c-Myc antibody (N262, Santa Cruz Biotech) and a 1:1 mixture of protein A and protein G Novex Dynabeads. Precipitated oligonucleosomal-protein complexes were washed, protein was removed by Proteinase K digestion followed by phenol/chloroform extraction. Recovered DNA fragments were analyzed by TaqManR allelic variant discrimination PCR to measure relative levels of rs16881446 alleles, similar to section 4.2.8 quantitation. ChIP background was determined by immunoprecipitation reactions that contained all components except primary antibody.
4.2.8 Nuclear RNA analysis
Cells were harvested by trypsinization and pellets were resuspended in 200 μl of lysis buffer (0.14 M NaCl, 1.5 mM MgCl2, 0.5% Triton X-100, 10 mM Tris•Cl at pH 7.0, and 10 mM vanadyl-ribonucleoside complexes) on ice with repeated vortexing over 5 min. Nuclei were pelleted at 500 x g, then resuspended in 200 μl of lysis buffer. Nuclei were disrupted by the addition of 200 μl of 2% sodium dodecyl sulfate, 0.3 M NaCl, 25 mM EDTA, 0.1 M 10 mM Tris•Cl at pH 7.0, and 0.4 mg/ml proteinase K, then the mixture was incubated at 65°C for 1 h. The solution was extracted against 1 ml of phenol:chloroform:isoamyl alcohol (25:24:1, v/v), then 320 μl of isopropanol was added, and nucleic acids were harvested by centrifugation. DNA was removed by treatment with RNase free DNase, the samples were phenol extracted, and DNA was recovered by ethanol precipitation. First strand synthesis was performed as previously described [91]. Levels of allele specific transcripts were measured by TaqManR allelic variant discrimination, as described in section 4.2.6. To determine true allelic ratios, samples were calibrated against standards (heterozygotic DNA as well as various mixtures of rs16881446A/A DNA and rs16881446G/G DNA).
4.2.9 Statistics
Parametric data are expressed as the mean ± SEM and were evaluated by the Student’s t-test, logistic regression, or repeated measures ANOVA; survival data were evaluated by log-rank analysis or by Cox proportional hazard test; categorical data were evaluated by Fisher exact probability or chi-square test; ordinal data were evaluated by nonparametric test for trend across ordered groups or ordered logistic regression test, as appropriate. As required, continuous data were transformed by rank order normalization. The Holm-Sidak method was used for post-hoc pairwise analysis. Effect sizes are expressed as hazard ratio (95% CI) or OR (95% CI). All P-values and 95% CIs are two-sided. Differences with P < 0.05 were considered significant. Statistics were performed with Stata/IC v12.1 or Sigma Plot 11.0.
Supplementary Material
Hemodynamic parameters were examined at the mid-point of cardiovascular collapse, T = Pmax50%, where the maximum systolic pressure fell to 50% of the starting value, at T = 0.
Enface view of the endothelial surface of a mouse aorta at the location of an intercostal vessel side branch. HSAT+ was stained with AT-biotin•streptavidin-Qdot 647 complexes (green); whereas, podocalyxin was indirectly immunostained using a secondary antibody labeled with Alexa Fluor 568 (red). The bar indicates 20 μm.
Shown are all CEU SNPs within the indicated region and LD score (R2) to rs16881446 (green). Also shown is the recombination rate across this region (blue). Analysis and image by SNP Annotation and Proxy Search 2.2.
Comparison of the 11 nucleotide region centered on rs16881446 from 57 mammalian species. Upper left, color coded clades (per UCSC Genome Browser) are arranged from most recent (top) to oldest emergence (bottom). For each indicated group, the consensus sequence of the rs16881446 region is shown. The dendrogram was based on pairwise comparison of the motif nucleotide identities and largely conforms to evolutionary groupings. The human region (motif 1) is most similar to that of chimp and gorilla (motif 2), then to motif 3 (gibbon, rhesus monkey, macaque, baboon, and green monkey). Most species including the earliest emerging (clade Mammal) contain motifs with the rs16881446G allele. The results suggest that the rs16881446A allele arose in three separate lineages: humans (motif 1), a small subgroup of Laurasiatheria (motif 7), and in the rodent branch of Euarchontoglires (motif 9). The examined species are listed in Supplementary Table 2.
The process for selecting the study population ensured that each individual was represented only once and clinical data were extracted from the earliest complete record in the DDR.
Data from Fig. 9 are plotted stratified for sex and all genotypes. Moreover, to justify our recessive model, we compared the effect of the AG genotype to the AA genotype, for all Fig. 9 analyses. The AG genotype OR for: CAD severity adjusted for CV risk factors was 1.00 (0.82–1.28), for CV events was 1.04 (0.86–1.26), and for CV events adjusted for CV risk factors was 1.00 (0.82–1.20).
Highlights.
Hs3st1−/− mice had increased LPS-lethality due to enhanced TNF-sensitivity.
HSAT+ is essential for AT’s anti- but not pro- inflammatory effects.
The rs16881446G allele associated with reduced HS3ST1 expression.
rs16881446G/G associated with cardiovascular disease severity and events.
An HSAT+AT pathway may protect against acute and chronic inflammatory disorders.
Acknowledgments
We are grateful to members of the Heart and Vascular Research Center for their insightful comments. We thank Anne R. Atalig for excellent technical assistance. This work was supported by National Institutes of Health grants RO1HL079104, RO1AG023590, and S10RR023436 to NWS; RO1LM010098 to SMW; R21CA175592, S10D010330, and R21CA172983 to RVS; RO1HL46716 to FWS; a Bayer/Talecris Canadian Blood Services Hema-Quebec Partnership Award to PLG; and the Netherlands Organization for Scientific Research grant 825.10.008 to NCS. Confocal microscopy was carried out at Geisel School of Medicine at Dartmouth in the Microscopy Resource, which is supported by the Norris Cotton Cancer Center Core Grant (P30CA023108) from the National Cancer Institute. We are indebted to Dr. Linda Werling for proofreading.
Abbreviations
- AT
Antithrombin
- AT-HBS
AT with a mutation in the heparin binding site
- CAD
coronary artery disease
- CEU
Utah residents with ancestry from northern and western Europe
- DDR
Dartmouth Dynamic Registry
- DHMC
Dartmouth-Hitchcock Medical Center
- FAIRE
formaldehyde-assisted isolation of regulatory elements
- HS
heparan sulfate
- HLMEC
human lung microvascular endothelial cells
- HSPG
HS proteoglycan
- HSAT+
HS with pentasaccharide
- HSAT−
HS without pentasaccharide
- HS3ST1 or Hs3st1
human or murine gene for HS 3-O-sulfotransferase-1, respectively
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
Footnotes
5 Interest Disclosure
All of the authors have no conflicting financial interests.
6 Author contributions
NCS and TK designed, conducted and analyzed experiments and participated in manuscript preparation. PKS, SS, JAI, and DJE performed experiments. RVS, GJT, FWS, PLG and MDC designed experiments and analytics as well as participated in manuscript preparation. JTD, AVK, and JFR developed analytics and analyzed coronary angiograms. SMW evaluated genetic analyses and participated in manuscript preparation. NWS conceived and coordinated the study, designed and conducted experiments, analyzed data, and participated in manuscript preparation. All authors have reviewed the results and approved the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- 2.Packard RR, Lichtman AH, Libby P. Innate and adaptive immunity in atherosclerosis. Semin Immunopathol. 2009;31:5–22. doi: 10.1007/s00281-009-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balk RA. Optimum treatment of severe sepsis and septic shock: evidence in support of the recommendations. Dis Mon. 2004;50:168–213. doi: 10.1016/j.disamonth.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu D, Olson J, Cole JN, van Wijk XM, Brinkmann V, Zychlinsky A, Nizet V, Esko JD, Chang YC. Heparan Sulfate Modulates Neutrophil and Endothelial Function in Antibacterial Innate Immunity. Infect Immun. 2015;83:3648–3656. doi: 10.1128/IAI.00545-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marki A, Esko JD, Pries AR, Ley K. Role of the endothelial surface layer in neutrophil recruitment. J Leukoc Biol. 2015;98:503–515. doi: 10.1189/jlb.3MR0115-011R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosadini CV, Kagan JC. Early innate immune responses to bacterial LPS. Curr Opin Immunol. 2016;44:14–19. doi: 10.1016/j.coi.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri B, Malur A, Bonfield TL, Abraham S, Schilz RJ, Farver CF, Kavuru MS, Arroliga AC, Thomassen MJ. The prostacyclin analogue treprostinil blocks NFkappaB nuclear translocation in human alveolar macrophages. J Biol Chem. 2002;277:33344–33348. doi: 10.1074/jbc.M203567200. [DOI] [PubMed] [Google Scholar]
- 9.Isobe H, Okajima K, Uchiba M, Harada N, Okabe H. Antithrombin prevents endotoxin-induced hypotension by inhibiting the induction of nitric oxide synthase in rats. Blood. 2002;99:1638–1645. doi: 10.1182/blood.v99.5.1638. [DOI] [PubMed] [Google Scholar]
- 10.Oelschlager C, Romisch J, Staubitz A, Stauss H, Leithauser B, Tillmanns H, Holschermann H. Antithrombin III inhibits nuclear factor kappaB activation in human monocytes and vascular endothelial cells. Blood. 2002;99:4015–4020. doi: 10.1182/blood.v99.11.4015. [DOI] [PubMed] [Google Scholar]
- 11.Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity. 2013;38:1092–1104. doi: 10.1016/j.immuni.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, Kimata K, Spear PG, Shworak NW, Nakato H. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J Cell Biol. 2004;166:1069–1079. doi: 10.1083/jcb.200403077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg RD, Shworak NW, Liu J, Schwartz JJ, Zhang L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J Clin Invest. 1997;99:2062–2070. doi: 10.1172/JCI119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iozzo RV. Heparan sulfate proteoglycans: intricate molecules with intriguing functions. J Clin Invest. 2001;108:165–167. doi: 10.1172/JCI13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart MD, Sanderson RD. Heparan sulfate in the nucleus and its control of cellular functions. Matrix Biol. 2014;35:56–59. doi: 10.1016/j.matbio.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christianson HC, Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014;35:51–55. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Wang B, Li JP. Implications of heparan sulfate and heparanase in neuroinflammation. Matrix Biol. 2014;35:174–181. doi: 10.1016/j.matbio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt EP, Yang Y, Janssen WJ, Gandjeva A, Perez MJ, Barthel L, Zemans RL, Bowman JC, Koyanagi DE, Yunt ZX, Smith LP, Cheng SS, Overdier KH, Thompson KR, Geraci MW, Douglas IS, Pearse DB, Tuder RM. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat Med. 2012;18:1217–1223. doi: 10.1038/nm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanderson RD, Elkin M, Rapraeger AC, Ilan N, Vlodavsky I. Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J. 2016;284:42–55. doi: 10.1111/febs.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peysselon F, Ricard-Blum S. Heparin-protein interactions: from affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. 2014;35:73–81. doi: 10.1016/j.matbio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Marcum JA, Fritze L, Galli SJ, Karp G, Rosenberg RD. Microvascular heparin-like species with anticoagulant activity. Am J Physiol. 1983;245:H725–733. doi: 10.1152/ajpheart.1983.245.5.H725. [DOI] [PubMed] [Google Scholar]
- 24.Thacker BE, Xu D, Lawrence R, Esko JD. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014;35:60–72. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Agostini AI, Watkins SC, Slayter HS, Youssoufian H, Rosenberg RD. Localization of anticoagulantly active heparan sulfate proteoglycans in vascular endothelium: antithrombin binding on cultured endothelial cells and perfused rat aorta. J Cell Biol. 1990;111:1293–1304. doi: 10.1083/jcb.111.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordts PL, Esko JD. Heparan sulfate proteoglycans fine-tune macrophage inflammation via IFN-beta. Cytokine. 2015;72:118–119. doi: 10.1016/j.cyto.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Gordts PL, Foley EM, Lawrence R, Sinha R, Lameda-Diaz C, Deng L, Nock R, Glass CK, Erbilgin A, Lusis AJ, Witztum JL, Esko JD. Reducing macrophage proteoglycan sulfation increases atherosclerosis and obesity through enhanced type I interferon signaling. Cell Metab. 2014;20:813–826. doi: 10.1016/j.cmet.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shworak NW, Liu J, Fritze LMS, Schwartz JJ, Zhang L, Logeart D, Rosenberg RD. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J Biol Chem. 1997;272:28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 29.Shworak NW, Fritze LM, Liu J, Butler LD, Rosenberg RD. Cell-free synthesis of anticoagulant heparan sulfate reveals a limiting activity which modifies a nonlimiting precursor pool. J Biol Chem. 1996;271:27063–27071. [PubMed] [Google Scholar]
- 30.Shworak NW, Shirakawa M, Colliec-Jouault S, Liu J, Mulligan RC, Birinyi LK, Rosenberg RD. Pathway-specific regulation of the synthesis of anticoagulantly active heparan sulfate. J Biol Chem. 1994;269:24941–24952. [PubMed] [Google Scholar]
- 31.Lindahl U, Bäckström G, Thunberg L, Leder IG. Evidence for a 3-O-sulfated D-glucosamine residue in the antithrombin-binding sequence of heparin. Proc Natl Acad Sci U S A. 1980;77:6551–6555. doi: 10.1073/pnas.77.11.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 33.HajMohammadi S, Enjyoji K, Princivalle M, Christi P, Lech M, Beeler D, Rayburn H, Schwartz JJ, Barzegar S, Ide Agostini A, Post MJ, Rosenberg RD, Shworak NW. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J Clin Invest. 2003;111:989–999. doi: 10.1172/JCI15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiedermann Ch J, Romisch J. The anti-inflammatory actions of antithrombin--a review. Acta Med Austriaca. 2002;29:89–92. doi: 10.1046/j.1563-2571.2002.02012.x. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi T, Umeda F, Inoguchi T, Nawata H. Antithrombin III stimulates prostacyclin production by cultured aortic endothelial cells. Biochem Biophys Res Commun. 1989;163:1404–1411. doi: 10.1016/0006-291x(89)91135-2. [DOI] [PubMed] [Google Scholar]
- 36.Kwak JY, Park SY, Han MK, Lee HS, Sohn MH, Kim UH, McGregor JR, Samlowski WE, Yim CY. Receptor-mediated activation of murine peritoneal macrophages by antithrombin III acts as a costimulatory signal for nitric oxide synthesis. Cell Immunol. 1998;188:33–40. doi: 10.1006/cimm.1998.1337. [DOI] [PubMed] [Google Scholar]
- 37.Kaneider NC, Reinisch CM, Dunzendorfer S, Romisch J, Wiederman CJ. Syndecan-4 mediates antithrombin-induced chemotaxis of human peripheral blood lymphocytes and monocytes. J Cell Sci. 2002;115:227–236. doi: 10.1242/jcs.115.1.227. [DOI] [PubMed] [Google Scholar]
- 38.Uchiba M, Okajima K. Antithrombin III (AT III) prevents LPS-induced pulmonary vascular injury: novel biological activity of AT III. Semin Thromb Hemost. 1997;23:583–590. doi: 10.1055/s-2007-996140. [DOI] [PubMed] [Google Scholar]
- 39.Minnema MC, Chang AC, Jansen PM, Lubbers YT, Pratt BM, Whittaker BG, Taylor FB, Hack CE, Friedman B. Recombinant human antithrombin III improves survival and attenuates inflammatory responses in baboons lethally challenged with Escherichia coli. Blood. 2000;95:1117–1123. [PubMed] [Google Scholar]
- 40.Dickneite G, Kroez M. Treatment of porcine sepsis with high-dose antithrombin III reduces tissue edema and effusion but does not increase risk for bleeding. Blood Coagul Fibrinolysis. 2001;12:459–467. doi: 10.1097/00001721-200109000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Mizutani A, Okajima K, Uchiba M, Isobe H, Harada N, Mizutani S, Noguchi T. Antithrombin reduces ischemia/reperfusion-induced renal injury in rats by inhibiting leukocyte activation through promotion of prostacyclin production. Blood. 2003;101:3029–3036. doi: 10.1182/blood-2002-08-2406. [DOI] [PubMed] [Google Scholar]
- 42.Horie S, Ishii H, Kazama M. Heparin-like glycosaminoglycan is a receptor for antithrombin III-dependent but not for thrombin-dependent prostacyclin production in human endothelial cells. Thromb Res. 1990;59:895–904. doi: 10.1016/0049-3848(90)90113-q. [DOI] [PubMed] [Google Scholar]
- 43.Hirose K, Okajima K, Uchiba M, Nakano KY, Utoh J, Kitamura N. Antithrombin reduces the ischemia/reperfusion-induced spinal cord injury in rats by attenuating inflammatory responses. Thromb Haemostasis. 2004;91:162–170. doi: 10.1160/TH03-06-0385. [DOI] [PubMed] [Google Scholar]
- 44.Harada N, Okajima K, Uchiba M, Kushimoto S, Isobe H. Antithrombin reduces ischemia/reperfusion-induced liver injury in rats by activation of cyclooxygenase-1. Thromb Haemostasis. 2004;92:550–558. doi: 10.1160/TH03-07-0460. [DOI] [PubMed] [Google Scholar]
- 45.Harada N, Okajima K, Kushimoto S, Isobe H, Tanaka K. Antithrombin reduces ischemia/reperfusion injury of rat liver by increasing the hepatic level of prostacyclin. Blood. 1999;93:157–164. [PubMed] [Google Scholar]
- 46.Streusand VJ, Bjork I, Gettins PG, Petitou M, Olson ST. Mechanism of acceleration of antithrombin-proteinase reactions by low affinity heparin. Role of the antithrombin binding pentasaccharide in heparin rate enhancement. J Biol Chem. 1995;270:9043–9051. doi: 10.1074/jbc.270.16.9043. [DOI] [PubMed] [Google Scholar]
- 47.Beeler D, Rosenberg R, Jordan R. Fractionation of low molecular weight heparin species and their interaction with antithrombin. J Biol Chem. 1979;254:2902–2913. [PubMed] [Google Scholar]
- 48.Shworak NW, Kobayashi T, de Agostini A, Smits NC. Anticoagulant heparan sulfate to not clot--or not? Prog Mol Biol Transl Sci. 2010;93:153–178. doi: 10.1016/S1877-1173(10)93008-1. [DOI] [PubMed] [Google Scholar]
- 49.Saia RS, Anselmo-Franci JA, Carnio EC. Hypothermia during endotoxemic shock in female mice lacking inducible nitric oxide synthase. Shock. 2008;29:119–126. doi: 10.1097/shk.0b013e31805cdc96. [DOI] [PubMed] [Google Scholar]
- 50.Mabley JG, Horvath EM, Murthy KG, Zsengeller Z, Vaslin A, Benko R, Kollai M, Szabo C. Gender differences in the endotoxin-induced inflammatory and vascular responses: potential role of poly(ADP-ribose) polymerase activation. J Pharmacol Exp Ther. 2005;315:812–820. doi: 10.1124/jpet.105.090480. [DOI] [PubMed] [Google Scholar]
- 51.Girardin EP, Hajmohammadi S, Birmele B, Helisch A, Shworak NW, de Agostini AI. Synthesis of anticoagulantly active heparan sulfate proteoglycans by glomerular epithelial cells involves multiple 3-O-sulfotransferase isoforms and a limiting precursor pool. J Biol Chem. 2005;280:38059–38070. doi: 10.1074/jbc.M507997200. [DOI] [PubMed] [Google Scholar]
- 52.Krishnagopalan S, Kumar A, Parrillo JE, Kumar A. Myocardial dysfunction in the patient with sepsis. Curr Opin Crit Care. 2002;8:376–388. doi: 10.1097/00075198-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 53.Tang J, Hu JJ, Lu CH, Liang JN, Xiao JF, Liu YT, Lin CS, Qin ZS. Propofol inhibits lipopolysaccharide-induced tumor necrosis factor-alpha expression and myocardial depression through decreasing the generation of superoxide anion in cardiomyocytes. Oxid Med Cell Longev. 2014;2014:157376. doi: 10.1155/2014/157376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li P, Schwarz EM, O'Keefe RJ, Ma L, Looney RJ, Ritchlin CT, Boyce BF, Xing L. Systemic tumor necrosis factor alpha mediates an increase in peripheral CD11bhigh osteoclast precursors in tumor necrosis factor alpha-transgenic mice. Arthritis Rheum. 2004;50:265–276. doi: 10.1002/art.11419. [DOI] [PubMed] [Google Scholar]
- 55.Okajima K. Antithrombin prevents endotoxin-induced pulmonary vascular injury by inhibiting leukocyte activation. Blood Coagul Fibrinolysis. 1998;9(Suppl 2):S25–37. [PubMed] [Google Scholar]
- 56.Feistritzer C, Mosheimer BA, Tancevski I, Kaneider NC, Sturn DH, Patsch JR, Wiedermann CJ. Src tyrosine kinase-dependent migratory effects of antithrombin in leukocytes. Exp Cell Res. 2005;305:214–220. doi: 10.1016/j.yexcr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Totzke G, Schobersberger W, Schloesser M, Czechowski M, Hoffmann G. Effects of antithrombin III on tumor necrosis factor-alpha and interleukin-1beta synthesis in vascular smooth muscle cells. J Interferon Cytokine Res. 2001;21:1063–1069. doi: 10.1089/107999001317205196. [DOI] [PubMed] [Google Scholar]
- 58.Uchiba M, Okajima K, Kaun C, Wojta J, Binder BR. Inhibition of the endothelial cell activation by antithrombin in vitro. Thromb Haemostasis. 2004;92:1420–1427. doi: 10.1160/TH04-03-0139. [DOI] [PubMed] [Google Scholar]
- 59.Yamashiro K, Kiryu J, Tsujikawa A, Honjo M, Nonaka A, Miyamoto K, Honda Y, Tanihara H, Ogura Y. Inhibitory effects of antithrombin III against leukocyte rolling and infiltration during endotoxin-induced uveitis in rats. Invest Ophthalmol Vis Sci. 2001;42:1553–1560. [PubMed] [Google Scholar]
- 60.Nishijima K, Kiryu J, Tsujikawa A, Honjo M, Nonaka A, Yamashiro K, Kamizuru H, Ieki Y, Tanihara H, Honda Y, Ogura Y. Inhibitory effects of antithrombin III on interactions between blood cells and endothelial cells during retinal ischemia-reperfusion injury. Invest Ophthalmol Vis Sci. 2003;44:332–341. doi: 10.1167/iovs.02-0493. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmann JN, Vollmar B, Romisch J, Inthorn D, Schildberg FW, Menger MD. Antithrombin effects on endotoxin-induced microcirculatory disorders are mediated mainly by its interaction with microvascular endothelium. Crit Care Med. 2002;30:218–225. doi: 10.1097/00003246-200201000-00031. [DOI] [PubMed] [Google Scholar]
- 62.Uchiba M, Okajima K, Murakami K, Okabe H, Takatsuki K. Effects of antithrombin III (AT III) and Trp49-modified AT III on plasma level of 6-keto-PGF1 alpha in rats. Thromb Res. 1995;80:201–208. doi: 10.1016/0049-3848(95)00168-q. [DOI] [PubMed] [Google Scholar]
- 63.Hockin MF, Jones KC, Everse SJ, Mann KG. A model for the stoichiometric regulation of blood coagulation. J Biol Chem. 2002;277:18322–18333. doi: 10.1074/jbc.M201173200. [DOI] [PubMed] [Google Scholar]
- 64.Asakura T, Karino T. Flow patterns and spatial distribution of atherosclerotic lesions in human coronary arteries. Circ Res. 1990;66:1045–1066. doi: 10.1161/01.res.66.4.1045. [DOI] [PubMed] [Google Scholar]
- 65.Kaneider NC, Egger P, Dunzendorfer S, Wiedermann CJ. Syndecan-4 as antithrombin receptor of human neutrophils. Biochem Biophys Res Commun. 2001;287:42–46. doi: 10.1006/bbrc.2001.5534. [DOI] [PubMed] [Google Scholar]
- 66.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Iwase M, Yoshikai Y, Yanada M, Yamamoto K, Matsushita T, Nishimura M, Kusugami K, Saito H, Muramatsu T. Syndecan-4 deficiency leads to high mortality of lipopolysaccharide-injected mice. J Biol Chem. 2001;276:47483–47488. doi: 10.1074/jbc.M106268200. [DOI] [PubMed] [Google Scholar]
- 67.Kojima T, Shworak NW, Rosenberg RD. Molecular cloning and expression of two distinct cDNA encoding heparan sulfate proteoglycan core proteins from a rat endothelial cell line. J Biol Chem. 1992;267:4870–4877. [PubMed] [Google Scholar]
- 68.Vlodavsky I, Blich M, Li JP, Sanderson RD, Ilan N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013;32:241–251. doi: 10.1016/j.matbio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanderson RD, Yang Y, Kelly T, MacLeod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies. J Cell Biochem. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- 70.Romisch J, Donges R, Stauss H, Inthorn D, Muhlbayer D, Jochum M, Hoffmann J. Quantification of antithrombin isoform proportions in plasma samples of healthy subjects, sepsis patients, and in antithrombin concentrates. Pathophysiol Haemost Thromb. 2002;32:143–150. doi: 10.1159/000065218. [DOI] [PubMed] [Google Scholar]
- 71.Brennan SO, George PM, Jordan RE. Physiological variant of antithrombin-III lacks carbohydrate sidechain at Asn 135. FEBS Lett. 1987;219:431–436. doi: 10.1016/0014-5793(87)80266-1. [DOI] [PubMed] [Google Scholar]
- 72.Luxembourg B, Pavlova A, Geisen C, Spannagl M, Bergmann F, Krause M, Alesci S, Seifried E, Lindhoff-Last E. Impact of the type of SERPINC1 mutation and subtype of antithrombin deficiency on the thrombotic phenotype in hereditary antithrombin deficiency. Thromb Haemostasis. 2014;111:249–257. doi: 10.1160/TH13-05-0402. [DOI] [PubMed] [Google Scholar]
- 73.Levi M, van der Poll T. Inflammation and coagulation. Crit Care Med. 2010;38:S26–34. doi: 10.1097/CCM.0b013e3181c98d21. [DOI] [PubMed] [Google Scholar]
- 74.Stach K, Nguyen XD, Lang S, Elmas E, Weiss C, Borggrefe M, Fischer J, Kalsch T. Simvastatin and atorvastatin attenuate VCAM-1 and uPAR expression on human endothelial cells and platelet surface expression of CD40 ligand. Cardiology J. 2012;19:20–28. doi: 10.5603/cj.2012.0005. [DOI] [PubMed] [Google Scholar]
- 75.Ley K. Gene-targeted mice in leukocyte adhesion research. Microcirculation. 1995;2:141–150. doi: 10.3109/10739689509146762. [DOI] [PubMed] [Google Scholar]
- 76.Ng DS, Chu T, Esposito B, Hui P, Connelly PW, Gross PL. Paraoxonase-1 deficiency in mice predisposes to vascular inflammation, oxidative stress, and thrombogenicity in the absence of hyperlipidemia. Cardiovasc Path. 2008;17:226–232. doi: 10.1016/j.carpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Hirata T, Croce K, Merrill-Skoloff G, Tchernychev B, Williams E, Flaumenhaft R, Furie BC, Furie B. Targeted gene disruption demonstrates that P-selectin glycoprotein ligand 1 (PSGL-1) is required for P-selectin-mediated but not E-selectin-mediated neutrophil rolling and migration. J Exp Med. 1999;190:1769–1782. doi: 10.1084/jem.190.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ley K, Cerrito M, Arfors KE. Sulfated polysaccharides inhibit leukocyte rolling in rabbit mesentery venules. Am J Physiol. 1991;260:H1667–1673. doi: 10.1152/ajpheart.1991.260.5.H1667. [DOI] [PubMed] [Google Scholar]
- 79.Jung U, Norman KE, Scharffetter-Kochanek K, Beaudet AL, Ley K. Transit time of leukocytes rolling through venules controls cytokine-induced inflammatory cell recruitment in vivo. J Clin Invest. 1998;102:1526–1533. doi: 10.1172/JCI119893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sellke FW, Li J, Stamler A, Lopez JJ, Thomas KA, Simons M. Angiogenesis induced by acidic fibroblast growth factor as an alternative method of revascularization for chronic myocardial ischemia. Surgery. 1996;120:182–188. doi: 10.1016/s0039-6060(96)80286-8. [DOI] [PubMed] [Google Scholar]
- 81.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 84.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brown JR, Malenka DJ, DeVries JT, Robb JF, Jayne JE, Friedman BJ, Hettleman BD, Niles NW, Kaplan AV, Schoolwerth AC, Thompson CA. Transient and persistent renal dysfunction are predictors of survival after percutaneous coronary intervention: insights from the Dartmouth Dynamic Registry. Catheter Cardiovasc Interv. 2008;72:347–354. doi: 10.1002/ccd.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sianos G, Morel MA, Kappetein AP, Morice MC, Colombo A, Dawkins K, van den Brand M, Van Dyck N, Russell ME, Mohr FW, Serruys PW. The SYNTAX Score: an angiographic tool grading the complexity of coronary artery disease. EuroIntervention. 2005;1:219–227. [PubMed] [Google Scholar]
- 88.Serruys PW, Onuma Y, Garg S, Sarno G, van den Brand M, Kappetein AP, Van Dyck N, Mack M, Holmes D, Feldman T, Morice MC, Colombo A, Bass E, Leadley K, Dawkins KD, van Es GA, Morel MA, Mohr FW. Assessment of the SYNTAX score in the Syntax study. EuroIntervention. 2009;5:50–56. doi: 10.4244/eijv5i1a9. [DOI] [PubMed] [Google Scholar]
- 89.T. Principal Investigators of CASS and their associates; Killip, Fisher LD, Mock MB, editors. National Heart, Lung, and Blood Institute Coronary Artery Surgery Study (CASS) Circulation. 1981;63(suppl I):I–1. [Google Scholar]
- 90.Ryan TJ, Faxon DP, Gunnar RM, Kennedy JW, King SB, 3rd, Loop FD, Peterson KL, Reeves TJ, Williams DO, Winters WL, Jr, et al. Guidelines for percutaneous transluminal coronary angioplasty. A report of the American College of Cardiology/American Heart Association Task Force on Assessment of Diagnostic and Therapeutic Cardiovascular Procedures (Subcommittee on Percutaneous Transluminal Coronary Angioplasty) Circulation. 1988;78:486–502. doi: 10.1161/01.cir.78.2.486. [DOI] [PubMed] [Google Scholar]
- 91.Shworak NW, O'Connor T, Wong NC, Gedamu L. Distinct TATA motifs regulate differential expression of human metallothionein I genes MT-IF and MT-IG. J Biol Chem. 1993;268:24460–24466. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hemodynamic parameters were examined at the mid-point of cardiovascular collapse, T = Pmax50%, where the maximum systolic pressure fell to 50% of the starting value, at T = 0.
Enface view of the endothelial surface of a mouse aorta at the location of an intercostal vessel side branch. HSAT+ was stained with AT-biotin•streptavidin-Qdot 647 complexes (green); whereas, podocalyxin was indirectly immunostained using a secondary antibody labeled with Alexa Fluor 568 (red). The bar indicates 20 μm.
Shown are all CEU SNPs within the indicated region and LD score (R2) to rs16881446 (green). Also shown is the recombination rate across this region (blue). Analysis and image by SNP Annotation and Proxy Search 2.2.
Comparison of the 11 nucleotide region centered on rs16881446 from 57 mammalian species. Upper left, color coded clades (per UCSC Genome Browser) are arranged from most recent (top) to oldest emergence (bottom). For each indicated group, the consensus sequence of the rs16881446 region is shown. The dendrogram was based on pairwise comparison of the motif nucleotide identities and largely conforms to evolutionary groupings. The human region (motif 1) is most similar to that of chimp and gorilla (motif 2), then to motif 3 (gibbon, rhesus monkey, macaque, baboon, and green monkey). Most species including the earliest emerging (clade Mammal) contain motifs with the rs16881446G allele. The results suggest that the rs16881446A allele arose in three separate lineages: humans (motif 1), a small subgroup of Laurasiatheria (motif 7), and in the rodent branch of Euarchontoglires (motif 9). The examined species are listed in Supplementary Table 2.
The process for selecting the study population ensured that each individual was represented only once and clinical data were extracted from the earliest complete record in the DDR.
Data from Fig. 9 are plotted stratified for sex and all genotypes. Moreover, to justify our recessive model, we compared the effect of the AG genotype to the AA genotype, for all Fig. 9 analyses. The AG genotype OR for: CAD severity adjusted for CV risk factors was 1.00 (0.82–1.28), for CV events was 1.04 (0.86–1.26), and for CV events adjusted for CV risk factors was 1.00 (0.82–1.20).