Abstract
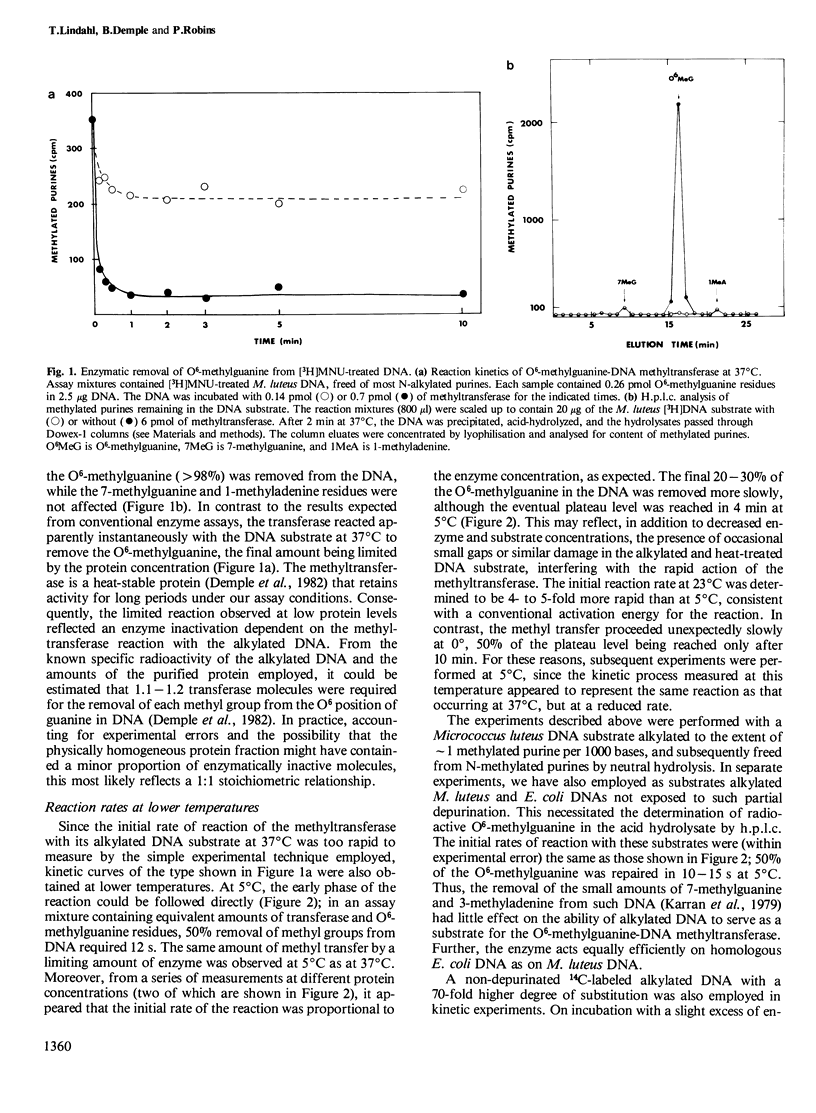
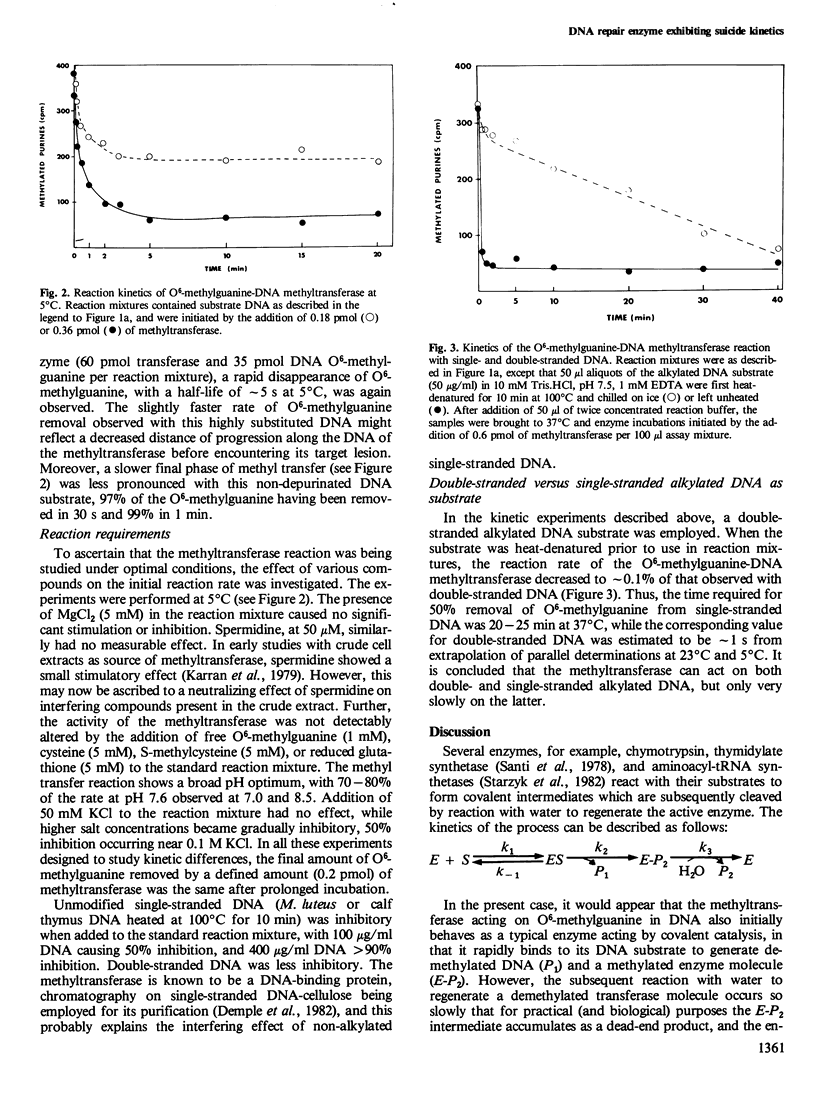
The O6-methylguanine-DNA methyltransferase of Escherichia coli acts rapidly and stoichiometrically to convert a mutagenic O6-methylguanine residue in DNA to unsubstituted guanine. Even at low protein concentrations and in the absence of any cofactors, the transfer of a methyl group to one of the protein's own cysteine residues occurs in less than 2 s at 37 degrees C. The entire kinetic process can be followed experimentally at 5 degrees C. Formation of S-methylcysteine in the protein is accompanied by loss of activity and accounts for the exceptional suicide kinetics of this enzyme as well as for the sharp saturation of O6-methylguanine repair observed in vivo. The enzyme can remove greater than 98% of the methyl groups from O6-methylguanine present in alkylated DNA, but leaves N-alkylated purines untouched. Single-stranded DNA containing O6-methylguanine is a poor substrate, with the methyl transfer occurring at approximately 0.1% of the rate for duplex DNA. This latter observation may explain the high frequency of mutations induced by alkylating agents at DNA replication forks.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrows L. R., Magee P. N. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3(3):349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- Cairns J. Efficiency of the adaptive response of Escherichia coli to alkylating agents. Nature. 1980 Jul 10;286(5769):176–178. doi: 10.1038/286176a0. [DOI] [PubMed] [Google Scholar]
- Cairns J., Robins P., Sedgwick B., Talmud P. The inducible repair of alkylated DNA. Prog Nucleic Acid Res Mol Biol. 1981;26:237–244. doi: 10.1016/s0079-6603(08)60408-0. [DOI] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Hanawalt P. C., Guerola N. Mutagenesis of the replication point by nitrosoguanidine: map and pattern of replication of the Escherichia coli chromosome. J Mol Biol. 1968 May 14;33(3):705–719. doi: 10.1016/0022-2836(68)90315-x. [DOI] [PubMed] [Google Scholar]
- Flockhart D. A., Corbin J. D. Regulatory mechanisms in the control of protein kinases. CRC Crit Rev Biochem. 1982 Feb;12(2):133–186. doi: 10.3109/10409238209108705. [DOI] [PubMed] [Google Scholar]
- Foote R. S., Mitra S., Pal B. C. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem Biophys Res Commun. 1980 Nov 28;97(2):654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- Hince T. A., Neale S. A comparison of the mutagenic action of the methyl and ethyl derivatives of nitrosamides and nitrosamidines on Escherichia coli. Mutat Res. 1974 Sep;24(3):383–387. doi: 10.1016/0027-5107(74)90183-3. [DOI] [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Marinus M. G. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982 Apr 29;296(5860):868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- Medcalf A. S., Lawley P. D. Time course of O6-methylguanine removal from DNA of N-methyl-N-nitrosourea-treated human fibroblasts. Nature. 1981 Feb 26;289(5800):796–798. doi: 10.1038/289796a0. [DOI] [PubMed] [Google Scholar]
- Mehta J. R., Ludlum D. B., Renard A., Verly W. G. Repair of O6-ethylguanine in DNA by a chromatin fraction from rat liver: transfer of the ethyl group to an acceptor protein. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6766–6770. doi: 10.1073/pnas.78.11.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeda K., Horiuchi T., Sekiguchi M. The uvrD gene of E. coli encodes a DNA-dependent ATPase. Nature. 1982 Jul 1;298(5869):98–100. doi: 10.1038/298098a0. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Pegg A. E., Perry W. Stimulation of transfer of methyl groups from O6-methylguanine in DNA to protein by rat liver extracts in response to hepatotoxins. Carcinogenesis. 1981;2(11):1195–1200. doi: 10.1093/carcin/2.11.1195. [DOI] [PubMed] [Google Scholar]
- Raggenbass M., Caro L. Intermediates of chromosomal DNA replication in Escherichia coli. J Mol Biol. 1982 Aug 5;159(2):273–301. doi: 10.1016/0022-2836(82)90496-x. [DOI] [PubMed] [Google Scholar]
- Rando R. R. Mechanism-based irreversible enzyme inhibitors. Methods Enzymol. 1977;46:28–41. doi: 10.1016/s0076-6879(77)46007-5. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Robins P., Cairns J. Quantitation of the adaptive response to alkylating agents. Nature. 1979 Jul 5;280(5717):74–76. doi: 10.1038/280074a0. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. Genetic mapping of ada and adc mutations affecting the adaptive response of Escherichia coli to alkylating agents. J Bacteriol. 1982 May;150(2):984–988. doi: 10.1128/jb.150.2.984-988.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B., Lindahl T. A common mechanism for repair of O6-methylguanine and O6-ethylguanine in DNA. J Mol Biol. 1982 Jan 5;154(1):169–175. doi: 10.1016/0022-2836(82)90424-7. [DOI] [PubMed] [Google Scholar]
- Sedgwick B., Robins P. Isolation of mutants of Escherichia coli with increased resistance to alkylating agents: mutants deficient in thiols and mutants constitutive for the adaptive response. Mol Gen Genet. 1980;180(1):85–90. doi: 10.1007/BF00267355. [DOI] [PubMed] [Google Scholar]
- Singer B., Kuśmierek J. T. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Sklar R., Brady K., Strauss B. Limited capacity for the removal of O6-methylguanine and its regeneration in a human lymphoma line. Carcinogenesis. 1981;2(12):1293–1298. doi: 10.1093/carcin/2.12.1293. [DOI] [PubMed] [Google Scholar]
- Sklar R. Enchancement of nitrosoguanidine mutagenesis by chloramphenicol in Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):460–462. doi: 10.1128/jb.136.1.460-462.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar R., Strauss B. Role of the uvrE gene product and of inducible O6-methylguanine removal in the induction of mutations by N-methyl-N'-nitro-N-nitrosoguanidine in Escherichia coli. J Mol Biol. 1980 Nov 15;143(4):343–362. doi: 10.1016/0022-2836(80)90217-x. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]
- Swann P. F., Mace R. Changes in O6-methylguanine disappearance from rat liver DNA during chronic dimethylnitrosamine administration. A possible similarity between the system removing O6-methylguanine from DNA in rat liver and in Escherichia coli adapted to N-methyl-N'-nitro-N-nitrosoguanidine. Chem Biol Interact. 1980 Aug;31(2):239–245. doi: 10.1016/0009-2797(80)90012-5. [DOI] [PubMed] [Google Scholar]
- Teo I. A., Karran P. Excision of O6-methylguanine from DNA by human fibroblasts determined by a sensitive competition method. Carcinogenesis. 1982;3(8):923–928. doi: 10.1093/carcin/3.8.923. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Hutchison C. A., 3rd, Baker M. S. DNA precursors in chemical mutagenesis: a novel application of DNA sequencing. Nature. 1982 Aug 26;298(5877):863–865. doi: 10.1038/298863a0. [DOI] [PubMed] [Google Scholar]
- Waldstein E. A., Cao E. H., Bender M. A., Setlow R. B. Abilities of extracts of human lymphocytes to remove O6-methylguanine from DNA. Mutat Res. 1982 Aug;95(2-3):405–416. doi: 10.1016/0027-5107(82)90274-3. [DOI] [PubMed] [Google Scholar]
- Waldstein E. A., Cao E. H., Miller M. E., Cronkite E. P., Setlow R. B. Extracts of chronic lymphocytic leukemia lymphocytes have a high level of DNA repair activity fo O6-methylguanine. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4786–4790. doi: 10.1073/pnas.79.15.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]



