Abstract
Air pollution is air contaminated by anthropogenic or naturally occurring substances in high concentrations for a prolonged time, resulting in adverse effects on human comfort and health as well as on ecosystems. Major air pollutants include particulate matters (PMs), ground-level ozone (O3), sulfur dioxide (SO2), nitrogen dioxides (NO2), and volatile organic compounds (VOCs). During the last three decades, air has become increasingly polluted in countries like China and India due to rapid economic growth accompanied by increased energy consumption. Various policies, regulations, and technologies have been brought together for remediation of air pollution, but the air still remains polluted. In this review, we direct attention to bioremediation of air pollutants by exploiting the potentials of plant leaves and leaf-associated microbes. The aerial surfaces of plants, particularly leaves, are estimated to sum up to 4 × 108 km2 on the earth and are also home for up to 1026 bacterial cells. Plant leaves are able to adsorb or absorb air pollutants, and habituated microbes on leaf surface and in leaves (endophytes) are reported to be able to biodegrade or transform pollutants into less or nontoxic molecules, but their potentials for air remediation has been largely unexplored. With advances in omics technologies, molecular mechanisms underlying plant leaves and leaf associated microbes in reduction of air pollutants will be deeply examined, which will provide theoretical bases for developing leaf-based remediation technologies or phylloremediation for mitigating pollutants in the air.
Keywords: air pollution, nitrogen dioxides, ozone, particulate matter, phylloremediation, phyllosphere, sulfur dioxide, volatile organic compounds
Introduction
Air pollution is referred to as the presence of harmful or poisonous substances in the earth's atmosphere, which cause adverse effects on human health and on the ecosystem. Major air pollutants include particulate matters (PMs), nitrogen oxides (NO2), sulfur dioxide (SO2), ground-level ozone (O3), and volatile organic compounds (VOCs) (Archibald et al., 2017). Various effects of some common air pollutants on human comfort and health are presented in Table 1, ranging from respiratory illness, cardiovascular disease to bladder and lung cancer (Kampa and Castanas, 2008).
Table 1.
Major air pollutants and their effects on human comfort and health.
| Pollutants | Adverse effects on human health | References |
|---|---|---|
| PMsa | Heart and lung cancer, risk of bladder cancer, nervous systems and respiratory illness | Anderson et al., 2012; Kumar et al., 2013; WHO, 2013; Kelly and Fussell, 2015 |
| O3 | Breathing problems, asthma, reduction of lung function, and lung diseases | Amann, 2008 |
| NO2 | Asthmatic bronchitis, reduced lung function growth, respiratory infections, and airway obstruction | Lambert et al., 1993; Lambert, 1996; Eberlein-Konig et al., 1998 |
| SO2 | Eye irritation, infections of the respiratory tract, coughing, mucus secretion, asthma and chronic bronchitis, and cardiac disease | Qin et al., 1993 |
| CO | Neuropsychological impairment, headache, fatigue, dizziness, and nausea, fetal damage, myocardial ischemia | Dahms et al., 1993 |
| PAHsb | Lung cancer | Fugas and Sega, 1995 |
| Volatile organic compounds | Asthma, nocturnal breathlessness, sensitization reactions, respiratory tract, mucous membrane irritation, central nervous system symptoms, headache, drowsiness, fatigue, confusion, lethargy, and dizziness | Molhave, 1991; Wallace, 1991; Wieslander et al., 1996 |
| Formaldehyde | Sneezing, coughing, minor eye irritation, irritant of the skin, respiratory tract, and nasopharyngeal cancer | Wieslander et al., 1996; Morgan, 1997; Eberlein-Konig et al., 1998 |
| Radon | Lung cancer, acute myeloid and acute lymphoblastic leukemia | Steindorf et al., 1995 |
| Tobacco smoke | Irritation of eye, nose, and throat, asthma, lung cancer, bronchitis, pneumonia, bronchiolitis, acute childhood lower respiratory tract illnesses, and tuberculosis | Lin et al., 2007 |
| Asbestos | Skin irritation, lung cancer, mesothelioma, and asbestosis | McDonald, 1991 |
Particulate matters.
Polycyclic aromatic hydrocarbons.
The world has experienced unprecedented urban growth during the last three decades. Urban population is expected to increase at 2.3% per year in developing countries from 2000 to 2030 (Brockherhoff, 2000; United Nations, 2000, 2004; UNFPA, 2004). Urbanization is often associated with rapid economic growth. For example, China's urbanization grew from 17.92% in 1978 to 52.57% in 2012, and China's gross domestic products (GDPs) increased from 454.6 billion Chinese Yuan in 1980 to 51,894.2 billion Yuan in 2012 (Zhao and Wang, 2015). The increased economic growth has been accompanied with elevated energy consumption. China's energy consumption, primarily fossil fuels like coal, increased from 602.75 million tons in 1980 to 3,617.32 million tons in 2012 (Zhao and Wang, 2015). The increased combustion of fossil fuels with relatively low combustion efficiency along with weak emission control measures have resulted in drastic increases in air pollutants, such as PMs, SO2, NO2, O3, and VOCs. Per unit of GDPs in 2006, China emitted 6–33 times more pollutants than the United States (US). As a result, air quality has become a major focus of environmental policy in China. India experiences similar situations as China. Urbanization coupled with rapid economic development in India increased energy consumption and also air pollution in some megacities (Gurjar et al., 2016). For example, PM10 in Delhi was almost 10 times of the maximum PM10 limit at 198 μg m−3 in 2011 (Rizwan et al., 2013). Concentrations of major pollutants in the air of some selected cities are present in Table 2.
Table 2.
Concentrations of some major air pollutants in the air of selected cities.
| City (country) | Year | PM2.5(μg m−3) | PM10(μg m−3) | O3(μg m−3) | NO2(μg m−3) | SO2(μg m−3) | CO (mg m−3) | References |
|---|---|---|---|---|---|---|---|---|
| Ho Chi Minh City (Vietnam) | 2007 | − | 74.00 | 40.00 | 18.90 | 30.30 | – | Phung et al., 2016 |
| Patras (Greece) | 2011 | − | 42.10 | 84.20 | 47.10 | – | 0.90 | Karagiannidis et al., 2014 |
| Quetta (Pakistan) | 2009 | 160.28 | 370.52 | – | 97.05 | 50.00 | 3.80 | Ilyas et al., 2009 |
| Beijing (China) | 2015 | 78.50 | 104.82 | – | 50.49 | 16.86 | 1.25 | Chen et al., 2016 |
| Shanghai (China) | 2015 | 55.54 | 75.64 | – | 45.23 | 18.40 | 0.86 | Chen et al., 2016 |
| Shenzhen (China) | 2015 | 32.83 | 55.37 | – | 32.94 | 8.08 | 1.01 | Chen et al., 2016 |
| Guangzhou (China) | 2015 | 44.38 | 65.91 | – | 45.46 | 14.67 | 0.99 | Chen et al., 2016 |
| Rome (Italy) | 2015 | 19.50 | 27.60 | 42.30 | 48.70 | 1.00 | 0.60 | Battista et al., 2016 |
| Lucknow City (India) | 2012 | 91.10 | 217.35 | – | 74.10 | 12.30 | 0.20 | Lawrence and Fatima, 2014 |
The World Health Organization (WHO) air quality guidelines stated that the mean limits for annual exposure to PM2.5 (particle diameters at 2.5 μm or less) and PM10 (particle diameter at 10 μm or less) are 10 μg m−3 and 25 μg m−3, respectively; and the limits for 24-h exposure are 25 μg m−3 and 50 μg m−3, respectively. The limit for 8-h exposure to O3 is 100 μg m−3. Annual mean for NO2 is 40 μg m−3 or 200 μg m−3 for 1 h, and 24-h exposure to SO2 is 20 μg m−3 or 500 μg m−3 for 10 min (WHO, 2006). The results presented in Table 2 suggest that residents in some of the listed cities were exposed to air contamination far beyond the limits set by WHO. PMs have become the most pressing environmental problems in China and India. For example, during the first quarter of 2013, China experienced extremely severe and persistent haze pollution that directly affected about 1.3 million km2 and about 800 million people (Huang et al., 2014). Of which daily average concentrations of PM2.5 measured at 74 major cities exceeded the Chinese pollution standard of 75 μg m−3, which is approximately twice that of the US EPA (United States Environmental Protection Agency) standard of 35 μg m−3, for 69% of days in January, with a record-breaking daily concentration of 772 μg m−3 (Huang et al., 2014).
Recent studies from the International Agency for Research on Cancer showed that there were 223,000 deaths in 2010 due to air pollution-resultant lung cancer worldwide, and air pollution has become the most widespread environmental carcinogen (International Agency for Research on Cancer, 2013). The WHO reported that around 7 million people died of air pollution exposure directly or indirectly in 2012. This data was more than double previous estimates and confirmed that air pollution has become a substantial burden to human health and is the world's largest single environmental health risk (WHO, 2014). Additionally, air pollution also harms animals, plants, and ecological resources including water and soils (Vallero, 2014; Duan et al., 2017).
Measures for reducing air pollution
To reduce air pollution, the first step is to eliminate or reduce anthropogenic-caused emissions. The second step is to remediate existing pollutants. Different strategies, policies, and models for air pollution abatement have been proposed or implemented (Macpherson et al., 2017). For example, the Chinese government has imposed restrictions on major pollution sources including vehicles, power plants, transport, and industry sectors (Liu et al., 2016) and promulgated the “Atmospheric Pollution Prevention and Control Action Plan” in September 2013, which was intended to reduce PM2.5 by 25% by 2017 relative to 2012 levels (Huang et al., 2014). Science-based technologies have been developed for control of air pollutants, such as diesel particulate filters (Tsai et al., 2011) and activated carbon filtering as adsorbent for xylene and NO2 (Guo et al., 2001). Catalytic oxidization and chemisorption methods have been used for indoor formaldehyde removal (Pei and Zhang, 2011; Wang et al., 2013). Photocatalysis as one of the most promising technologies has been used for eliminating VOCs (Huang et al., 2016).
Air pollutants can also be mitigated through biological means, commonly referred to as biological remediation or bioremediation. It is the use of organisms to assimilate, degrade or transform hazardous substances into less toxic or non toxic ones (Mueller et al., 1996). Plants have been used for remediation of pollutants from air, soils, and water, which has been termed as phytoremediation (Cunningham et al., 1995; Salt et al., 1995; Huang et al., 1997). Microbes such as bacteria and fungi are also capable of biodegrading or biotransforming pollutants into non toxic and less toxic substances, which is known as microbial biodegradation (Ward et al., 1980; Ma et al., 2016). Microbes as heterotrophs occur nearly everywhere, including plant roots and shoots. Both roots and shoots have been reported to be able to remediate air pollutants (Weyens et al., 2015; Gawronski et al., 2017), but little credit has been given to microbe activity.
Plant shoots or the above-ground organs of plants colonized by a variety of bacteria, yeasts, and fungi are known as phyllosphere (Last, 1955). However, most scientific work on phyllosphere microbiology has been focused on leaves (Lindow and Brandl, 2003). This review is intended to explore the potential of plant leaves and leaf-associated microbes in bioremediation of air pollutants, or simply known as phylloremediation. Phylloremediation was first coined by Sandhu et al. (2007), who demonstrated that surface-sterilized leaves took up phenol, and leaves with habiated microbes or a inoculated bacterium were able to biodegrade signficantly more phenol than leaves alone. Previous reports also documented that both plant leaves and leaf-associated microbes mitiagted air pollutants, such as azalea leaves and the leaf-associated Pseudomonas putida in reducing VOCs (De Kempeneer et al., 2004), leaves of yellow lupine plants along with endophytic Burkholderia cepacia for toluene reduction (Barac et al., 2004), and poplar leaves and the leaf-associated Methylobacterium sp. decreased xenobiotic compounds (Van Aken et al., 2004). Phyllo originated from Greek word of phullon, meaning leaf. Thus, phylloremediation should be defined as a natural process of bioremediation of air pollutants through leaves and leaf-associated microbes, not the microbes alone.
Plant leaves and phyllosphere
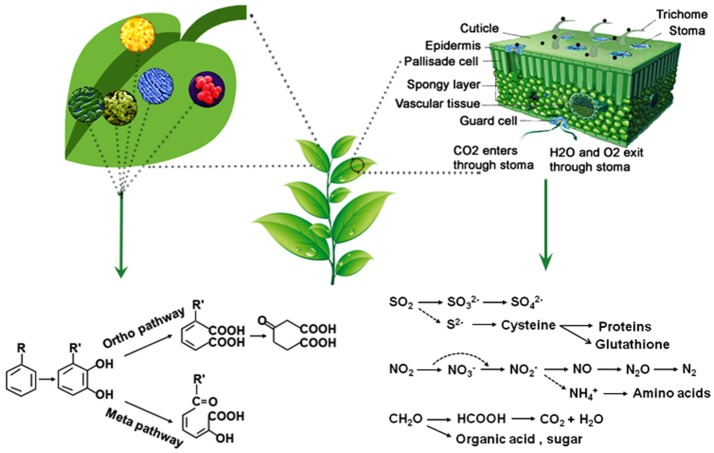
Leaves are the primary photosynthetic organs with distinctive upper surface (adaxial) and lower surface (abaxial) (Figure 1). The upper surface has a layer (<0.1–10 μm) of waxy cover called cuticle (Kirkwood, 1999). Wax contents and compositions frequently differ among plant species. The primary function of cuticle is to prevent evaporation of water from leaf surfaces, and it is also the first barrier for the penetration of xenobiotics. The leaf surface is filled with trichomes, which are epidermal outgrowths in various forms. Trichomes play roles in mechanical defense because of their physical properties and also in biochemical defense due to the secretion of secondary metabolites (Tian et al., 2017). Epidermis cells are directly underneath the cuticle layer in which stomata often occur. Xylem and phloem are situated within the veins of leaves as the plant vascular system, which are connected from root tips to leaf edges. There is a layer of compactly arranged cells around the vein called bundle sheath regulating substance circle around the xylem and the phloem. Xylem transports water and nutrients from roots to shoots, and phloem transports assimilated products from source and sink tissues. Under the epidermis, there are mesophyll cells in two layers: column-like palisade cells and loosely packed spongy cells. The air spaces among the spongy cells promote gas exchange, and photosynthesis takes place in chloroplasts packed in the mesophyll cells. The underside of leaves also has a layer of epidermal cells where most stomata are located. There are two guard cells surround the stomata, and stomatal pore opening and closure is regulated by changes in the turgor pressure of the guard cells. Stomata regulate the flow of gases in and out of leaves and also able to adsorb or absorb other chemicals.
Figure 1.
A schematic illustration of phyllosphere. The middle panel represent an aerial part of a plant. Right panel shows a magnified schematic cross section of a leaf where leaf surface and trichomes can retain particulate matter (PMs) and stomata adsorb or absorb PMs as well as how leaves can assimilate SO2, NO2, and CH2O (formaldehyde) to simple organic compounds, amino acids, or proteins. The left panel depict a magnified leaf surface with bacteria, which can biodegrade or transform volatile organic compounds to less toxic or nontoxic ones like benzene and its derivatives that can be degraded through Ortho pathway or Meta pathway.
Leaves also play pivotal roles in supporting phyllosphere microbes (Bringel and Couee, 2015). The phyllosphere is estimated to have area up to 4 × 108 km2 on the earth and is the home for up to 1026 bacterial cells (Kembel et al., 2014). Phyllosphere bacterial communities are generally dominated by Proteobacteria, such as Methylobacterium and Sphingomonas. Beijerinckia, Azotobacter, Klebsiella, and Cyanobacteria like Nostoc, Scytonema, and Stigonema also reside in the phyllosphere (Vacher et al., 2016). Population of γ-Proteobacteria such as Pseudomonas could be high as well (Delmotte et al., 2009; Fierer et al., 2011; Bodenhausen et al., 2013; Kembel et al., 2014). Dominant fungi in the phyllosphere include Ascomycota, of which the most common genera are Aureobasidium, Cladosporium, and Taphrina (Coince et al., 2013; Kembel and Mueller 2014). Basidiomycetous yeasts belonging to the genera Cryptoccoccus and Sporobolomyces are also abundant in phyllosphere (Cordier et al., 2012; Ottesen et al., 2013). The microbes can be epiphytic by living on the surface of plant organs and/or endophytic occurring within plant tissues without causing apparent disease.
Plant species significantly influence the composition of a phyllosphere community (Whipps et al., 2008). In a study of 56 different tree species, Redford et al. (2010) reported that different species harbor distinct microbial communities in phyllosphere. This principle was also confirmed for trees in temperate and tropical climates and for Mediterranean perennials (Lambais et al., 2006; Kim et al., 2012; Vokou et al., 2012; Kembel et al., 2014; Laforest-Lapointe et al., 2016). Using high-throughput sequencing technology, Kembel and Mueller (2014) studied fungal communities on leaves of 51 tree species in a lowland tropical rainforest in Panama and reported that fungal communities on leaves were dominated by the phyla Ascomycota, which accounted for 79% of all sequences, followed by Basidiomycota (11%) and Chytridiomycota (5%). More than half of the variation in fungal community composition could be explained by plant species differences. Leaf chemistry and morphology as well as plant growth status and mortality were closely related to fungal community structure (Kembel and Mueller, 2014). These results may suggest that different tree species host different fungal communities. Additionally, microbial compositions within plant species may differ due to geographic locations (Finkel et al., 2012; Qvit-Raz et al., 2012; Rastogi et al., 2012). The differences could be caused by climatic variation (Finkel et al., 2011) or due to the limited dispersal of the colonizing taxa (Finkel et al., 2012; Qvit-Raz et al., 2012). Furthermore, phyllosphere microbial community may differ between urban and non-urban locations (Jumpponen and Jones, 2010) and also differ by seasons (Redford and Fierer, 2009).
Roles of leaves and phyllosphere microbes in air remediation
The close association between plant species and specific microbial communities in the phyllosphere suggests their adaptation and coevolutionary relationships. Recent studies show that leaf bacterial diversity mediates plant diversity and ecosystem function relationships (Laforest-Lapointe et al., 2017). We hypothesize that a long-lasting exposure of leaves and leaf-associated microbes to air pollutants could result in plants or microbes individually or coordinately developing mechansims for adapting to the polluted substances. Such mechanisms may include leaf adsorption or absorption and pollutant assimilation as well as microbial biodegradation, transformation or metabolic assimilation of the substances. The coordination between leaves and micriobes could be synergistic or antagonistic. Table 3 presents plant-supported microbes that are able to biodegrade or biotransform air pollutants, primarily organic compounds. However, information regarding phyllospere microbes in remediation of PMs, SO2, NO2, and O3 is scarce, suggesting relatively limited research has been devoted to microbial roles. Thus, the current knowledge on phylloremediation of PM, SO2, NO2, and O3 is mostly come from plants.
Table 3.
Plant-supported microbes that are able to biodegrade or biotransform air pollutants.
| Plants | Microbes | Pollutant | References |
|---|---|---|---|
| Acorus calamus var. angustatus | Methylobacterium sp. | Methanol | Iguchi et al., 2012 |
| Amaranthus cruentus | Alcaligenes feacalis and Alcaligenes sp.11SO | Naphthalene and Phenanthrene | Ljs, 2016 |
| Annona muricana | Pseudomonas sp, Alcaligenes sp, and Microcccus roseus | Hydrocarbon | Ilori et al., 2006 |
| Arabidopsis thaliana | Hyphomicrobium sp, | Chloromethane | Nadalig et al., 2011 |
| Arabidopsis thaliana | Achromobacter xylosoxidans F3B | Phenolic pollutants | Ho et al., 2012 |
| Arachis hypogaea | Arthrobacter nitroguajacolicus, Achromobacter xylosoxidans, Basea thiooxidans, Microbacterium natoriense, Kocuria rosea, Dyadobacter fermentans, and Klebsiella pneumoniae | n-alkanes | Al-Awadhi et al., 2012 |
| Azalea indica | Pseudomonas putida TVA8 | Toluene | De Kempeneer et al., 2004 |
| Boerhavia diffusa | Rhodococcus corynebacteriodes and Exiguobacterium arabatum | n-alkanes | Al-Awadhi et al., 2012 |
| Bougainvillea buttiana | Enterobacter cloacae LSRC11, Staphylococcus sp. A1 and Pseudomonas aeruginos | Xylene | Sangthong et al., 2016 |
| Calystegia soldanella | Hyphomicrobium sp. | Methanol | Iguchi et al., 2012 |
| Chenopodium album | Burkholderia fungorum | n-alkanes | Al-Awadhi et al., 2012 |
| Chenopodium murale | Gordonia polyisoprenivorans | n-alkanes | Al-Awadhi et al., 2012 |
| Chenopodium murale | Flavobacterium sp., Halomonas sp., and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Chrysopogon zizanioides | Achromobacter xylosoxidans F3B | BTEX compounds (benzene, toluene, ethylbenzene, and xylene) | Ho et al., 2013 |
| Clitoria ternatea | Bacillus cereus | Formaldehyde | Khaksar et al., 2016b |
| Conocarpus lancifolius | Flavobacterium sp., Halomonas sp., and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Cucumis sativus | Arthrobacter ureafaciens, Arthrobacter aurescens, and Microbacterium natoriense | n-alkanes | Al-Awadhi et al., 2012 |
| Cynodon spp. | Rhodococcus sp. and Pseudomonas sp. | Volatile oil hydrocarbons crude oil, n-hexadecane, or phenanthrene | Sorkhoh et al., 2011 |
| Eichhornia crassipes | Methylophilus sp. | Methanol | Iguchi et al., 2012 |
| Ervatamia divaricata | Alcaligenes feacalis and Alcaligenes sp.11SO | Naphthalene and Phenanthrene | Ljs, 2016 |
| Fraxinus excelsior | Trichloroethylene, Toluene | Weyens et al., 2009a | |
| Fraxinus pennsyhanica | Acinetobacter sp., Alcaligenes sp., and Rhodococcus sp. | Phenol | Sandhu et al., 2009 |
| Gazania rigens | Methylobacterium populi, Gordonia lacunae, Dietzia maris, Microbacterium oleivorans, and Pseudomonas stutzeri | n-alkanes | Al-Awadhi et al., 2012 |
| Gazania rigens | Flavobacterium sp., Halomonas sp., and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Gossipium hirsutum ‘MCU12’ | Methylobacterium gossipiicola sp. nov. | Methanol and dichloromethane | Madhaiyan et al., 2012) |
| Hibiscus rosa-sinensis | Alcaligenes feacalis and Alcaligenes sp.11SO | Naphthalene and Phenanthrene | Ljs, 2016 |
| Hedera spp. | Hymenobacter sp. and Sphingomonadaceae sp. | PMs | Smets et al., 2016 |
| Ixora chinensis | Alcaligenes feacalis and Alcaligenes sp.11SO | Naphthalene and Phenanthrene | Ljs, 2016 |
| Ixora spp. | Acinetobacter sp. and Pseudomonas sp. Pseudoxanthomonas sp., and Mycobacterium sp. | Polycyclic aromatic hydrocarbons | Yutthammo et al., 2010 |
| Ixora spp. | Pseudomonas sp., Microbacterium sp., Rhizobium sp., and Deinococcus sp. | Phenanthrene | Waight et al., 2007 |
| Keteleeria davidiana | Methylobacterium sp. and Methylophilus sp. | Methanol | Iguchi et al., 2012 |
| Lolium multiflorum Lam. | Pseudomonas sp. Ph6-gfp | Phenanthrene | Sun et al., 2014 |
| Lolium multiflorum var. Taurus | Pseudomonas sp. ITSI10, ITRI15, ITRH76, and BTRH79 | Hydrocarbon | Yousaf et al., 2010 |
| Lolium perenne | Pseudomonas sp. | Petroleum hydrocarbons | Kukla et al., 2014 |
| Lotus corniculatus var. Leo | Pseudomonas sp. ITSI10, ITRI15, ITRH76, and BTRH79 | Hydrocarbon | Yousaf et al., 2010 |
| Magnifera indica | Pseudomonas sp., Alcaligenes sp., and Microcccus roseus | Hydrocarbon | Ilori et al., 2006 |
| Malus pumila | Arthrobacter sp. | 4-chlorophenol | Scheublin and Leveau, 2013 |
| Mattiola incana | Acinetobacter calcoacelicus, Pseudomonas putida, Planomicrobium glaciei,Arthrobacter agilis, Kucuria turfanensis, Pseudomonas geniculate, Chyseobacterium taeanense, Flavobacterium ahuensis, and Microbacterium oxydans | n-alkanes | Al-Awadhi et al., 2012 |
| Mesembryanthemum nodifloru | Nesterenkonia jeotgali, Nesterkonia lacusekhoensis, and Agrococcus terreus | n-alkanes | Al-Awadhi et al., 2012 |
| Oryza sativa | Methylobacterium sp. | Methanol | Knief et al., 2012 |
| Phaseolus vulgaris | Microbacterium sp. and Citrobacter freundii | Crude oil, phenanthrene and n-octadecane | Ali et al., 2012 |
| Phaseolus vulgaris | Arthrobacter chlorophenolicus | 4-chlorophenol, hydroquinone | Scheublin et al., 2014 |
| Phaseolus vulgaris | Pseudomonas sp. CF600 | Phenol | Sandhu et al., 2007 |
| Phragmites australis | Hyphomicrobium sp. | Methanol | Iguchi et al., 2012 |
| Picea abies | Trichloroacetic acid | Forczek et al., 2004 | |
| Pisum sativum | Pseudomonas putida VM1441(pNAH7) | Polyaromatic hydrocarbons (PAHs) | Germaine et al., 2009 |
| Pisum sativum | Microbacterium sp. and Rhodococcus sp. | Crude oil, phenanthrene and n-octadecane | Ali et al., 2012 |
| Pisum sativum | Agromyces fucosus, Agrococcus jenensis, Paenibacillus polymixa, Bacillus cereus, Bacillus megaterium, Brevibacillus brevis, Bacillus, Neolsonii, Bacillus subtilis, Arthrobacter ramosus, Microbacterium imperial, Bacillus endophyticus, and Cellulosimicrobium cellulans | n-alkanes | Al-Awadhi et al., 2012 |
| Poaceae spp. | Hyphomicrobium sp. | Methanol | Iguchi et al., 2012 |
| Populus deltoides | Pseudomonas putida W619-TCE | Trichloroethylene (TEC) | Weyens et al., 2009b |
| Populus deltoids × nigra | Methylobacterium sp. | 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine | Van Aken et al., 2004 |
| Populus trichocarpa deltoides | Burkholderia cepacia VM1468 | Toluene | Taghavi et al., 2005 |
| Pyrus calleryansa | Methylobacterium sp. and Methylophilus sp. | Methanol | Iguchi et al., 2012 |
| Quercus phillyraeoides | Methylobacterium sp. and Methylophilus sp. | Methanol | Iguchi et al., 2012 |
| Quercus robu | Trichloroethylene, toluene | Weyens et al., 2009a | |
| Salix discolor clone S-365 | Pseudomonas putida PD1 | Polycyclic aromatic hydrocarbons (PAHs) | Khan et al., 2014 |
| Salix purpurea clone 94006 | Pseudomonas putida PD1 | Polycyclic aromatic hydrocarbons (PAHs) | Khan et al., 2014 |
| Salsola baryosma | Halomonas marisflava and Salinococcus hispanicus | n-alkanes | Al-Awadhi et al., 2012 |
| Sonchus oleraceus | Flavobacterium sp., Halomonas sp., and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Tecoma stans | Afipia genosp and Microbacterium hydrocarbonoxydans and Bacillus subtilis | n-alkanes | Al-Awadhi et al., 2012 |
| Tecoma stans | Flavobacterium sp., Halomonas sp. and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Vicia faba | Flavobacterium sp., Halomonas sp. and Arthrobacter sp. | Volatile hydrocarbons | Ali et al., 2015 |
| Vicia faba | Rhodococcus sp. and Pseudomonas sp. | Volatile oil hydrocarbons crude oil, n-hexadecane, or phenanthrene | Sorkhoh et al., 2011 |
| Vigna unguiculata | Microbacterium arabinogalactanolyticum and Pseudomonas oryzihabtans | n-alkanes | Al-Awadhi et al., 2012 |
| Viola x wittrockiana | Methylobacterium sp. and Methylophilus sp. | Methanol | Iguchi et al., 2012 |
| W. religiosa | Acinetobacter sp., Pseudomonas sp., Pseudoxanthomonas sp. and Mycobacterium sp. | Polycyclic aromatic hydrocarbons | Yutthammo et al., 2010 |
| Zamioculcas zamiifolia | Bacillus cereus | Formaldehyde | Khaksar et al., 2016a |
| Zamioculcas zamiifolia | Pseudomonas aeruginosa and Bacillus cereus | Ethylbenzene | Toabaita et al., 2016 |
| Zea mays | Pseudomonas sp. CF600 | Phenol | Sandhu et al., 2007 |
Remediation of PMs
As mentioned above, PMs have become the most dangerous pollutants in some countries. Chemical species of PMs, derived from the available data over China included , , , organic carbon, and elemental carbon, which were in a range of 2.2–60.9, 0.1–35.6, 0.1–29.8, 1.5–102.3, 0.2–37.0 μg cm−3 in PM2.5, and 1.6–104.6, 0.5–46.6, 0.2–31.0, 1.7–98.7, and 0.3–26.8 μg cm−3 in PM10, respectively (Zhou et al., 2016). PM2.5 is the major component of PM10, accounting for 65%. PMs are also composed of microorganisms. In a study of PMs in Jeddah, Saudi Arabia (Alghamdi et al., 2014), the average concentrations of PM10 and PM2.5 were 159.9 and 60 μg cm−3, respectively and the concentrations of O3, SO2, and NO2 averaged 35.73, 38.1, and 52.5 μg cm−3, respectively. Microbial loads were higher in PM10 than PM2.5. Aspergillus fumigatus and Aspergillus niger were the common fungal species associated with PMs. Microbes were also found in PMs in Austria (Haas et al., 2013), including fungi from genera Aspergillus, Cladosporium, and Penicillium and aerobic mesophilic bacteria. Using metagenomic methods, Cao et al. (2014) identified 1,315 distinct bacterial and archaeal species from 14 PM samples collected from Beijing, China. The most abundant phyla were Actinobacteria, Proteobacteria, Chloroflexi, Firmicutes, Bacteroidetes, and Euryarchaeota. Among them, an unclassified bacterium in the nitrogen fixing, filamentous bacteria genus Frankia was the most abundant, and the most abundant classified bacterial species appeared to be Geodermatophilus obscures. The abundance of airborne bacteria was reported to be in a range from 104 to 106 cells m−3 depending on environmental conditions (Bowers et al., 2011), and materials of biological origin might account for up to 25% of the atmospheric aerosol (Jaenicke, 2005). Ammonia oxidizing archaea (AOA), ammonia oxidizing bacteria (AOB), and complete ammonia oxidizers (Comammox) were identified in PM2.5 collected from the Beijing-Tianjin-Heibei megalopolis, China (Gao et al., 2016). Of which Nitrosopumilus subcluster 5.2 was the most dominant AOA, Nitrosospira multiformis and Nitrosomonas aestuarii were the most dominant AOB, and the presence of Comammox was revealed by the occurrence of Candidatus Nitrospira inopinata. The mean cell numbers of AOA, AOB, and Ca. N. inopinata were 2.82 × 104, 4.65 × 103, and 1.15 × 103 cell m−3, respectively. The average maximum nitrification rate of PM2.5 was 0.14 μg (NH4+-N) [m3 air h]−1 (Gao et al., 2016). AOA might account for most of the ammonia oxidation, followed by Comammox, while AOB were responsible for a small part of ammonia oxidation. The assay of nitrification activity was performed in laboratory conditions (Gao et al., 2016). However, the nitrification potential of such bacteria in PMs after being deposited on leaf surfaces is unknown. We hypothesize that the nitrification process could be more active once such PM-containing bacteria settled on leaves. Further investigation on nitrification of PM-associated bacteria in the phyllosphere could provide insight into how the phyllosphere could potentially act as manufactories in the nitrification of ammonia.
The current literature regarding phylloremediation of PMs has been primarily focused on plant leaves. Plant canopy is a sink for PMs. This is due to the fact that leaves are in the air and they span more than 4 × 108 km2 on a global scale, which is about 78.4% of the total surface area of the earth; leaves thus physically act as a natural carrier for PMs. Leaves differ greatly in surface structure and metabolic secreted substances as well as microbial composition. The amount of surface waxes and compositions show different capacity to retain and embrace PMs. Sbø et al. (2012) studied leaves of 22 trees and 25 shrubs in accumulation of PMs in Norway and Poland and found that PM accumulation differed by 10 and 15 folds depending on plant species in the two locations and also positive correlations occurred among PM accumulation, leaf wax contents, and leaf hair density. Thirteen woody species were examined by Popek et al. (2013) during a 3-year period, and total amount of PMs captured by leaves ranged from 7.5 mg cm−2 by Catalpa bignonioides to 32 mg cm−2 by Syringa meyeri. Leaf wax contents were significantly correlated with the amount of PMs on leaves. Among the PMs captured, 60% was washable by water, and 40% could be washed by chloroform only, suggesting that the PMs were embraced in waxes. Using two photon excitation microscopy (TPEM), Terzaghi et al. (2013) investigated leaves of stone pine (Pinus pinea), cornel (Cornus mas), and maple (Acer pseudoplatanus) in capture and encapsulation of PMs. The authors found that particles ranging from 0.2 to 70.4 μm were visualized on leaves, of which PM2.6 was the dominant size across plant species. Particle less than 10.6 μm were encapsulated in the cuticle. Plant species differed in particle retention and encapsulation, which were attributed to leaf characteristics, cuticle chemical composition and structure.
Leaf physical characteristics such as leaf shape, hairs or trichomes, and stomata significantly affect PM accumulation. Needle leaves were reported to accumulate more PM2.5 than broad leaves (Terzaghi et al., 2013; Chen et al., 2017). The effectiveness was attributed to the higher capture efficiency and higher Stoke's numbers of needles compared to those of broad leaves (Beckett et al., 2000). Additionally, small individual leaf area and abundant wax layer also contribute to the effectiveness (Chen et al., 2017). Leaf trichomes have been shown to increase PM2.5 accumulation. The trichome density was positively correlated with amount of PM2.5 accumulated on leaves, and plant species with abundant hairs, such as Catalpa speciosa, Broussonetia papyrifera, and Ulmus pumila were able to retain more PM2.5 than those with fewer hairs (Chen et al., 2017). The adaxial surface of leaves accumulated more PMs than the abaxial leaf surface (Baldacchini et al., 2017), which is probably due to the fact that the abaxial surface in general has few trichomes and less rough surface. Stomata may play some roles in accumulation of PMs. The length of stomata ranges from 10 to 80 μm and densities varies from 5 to 1,000 mm−2 depending on plant species and environmental conditions (Hetherington and Woodward, 2003). Stomatal pore areas range from 46 to 125 μm2 (Peschel et al., 2003; Dow et al., 2014), thus stomata could retain or adsorb either PM2.5 or PM10. A study of PM deposition on leaves of five evergreen species in Beijing, China showed that PM diameter up to 2 μm was in the stomatal cavity (Song et al., 2015). Rai (2016) studied the effects of PMs on 12 common roadside plant species and found that stomatal sizes were reduced due to air dust deposition, but plant growth was not affected, suggesting the potential of plants in adsorbing air pollutants.
Growing evidence has suggested that plant leaves are able to capture PMs and act as biofilters. On average, the upper leaf surface of 11 plant species intercepted 1,531 particles per mm−2 (Wang et al., 2006). Needles of Pinus sylvestris accumulated 18,000 mineral particles per mm2 (Teper, 2009). Upper leaves of Hedera helix captured about 17,000 particles per mm2 (Ottele et al., 2010). Trees removed 1,261 tons of air pollutants in Beijing, of which 772 tons were PM10(Yang et al., 2005). In New Zealand, urban trees removed 1,320 tons of particular matter annually due to the existence of woodlands in Auckland (Cavanagh and Clemons, 2006). Nowak et al. (2014) showed trees within cities removed fine particles from the atmosphere and consequently improved air quality and human health. Tree effects on PM2.5 concentrations and human health are modeled for 10 U.S. cities. The total amount of PM2.5 removed by trees varied from 4.7 tons in Syracuse to 64.5 tons in Atlanta in the U.S annually. All the reported removal of PMs is attributed to plant leaves. It is unknown at this time if phyllosphere microbes could break down the PMs on leaves and if mineral elements released from the broken PMs could become plant nutrients. Considering the fact that the microbes can biodegrade a wide range of substances including petroleum, we hypothesize that some microbes should be able to break down PM. Future research in this regard will be conducted, and identified microbes could be used for PM reduction.
Remediation of SO2
Sulfur dioxide (SO2) was among the first air pollutants identified to harm human health and ecosystems. The combustion of fossil fuels has substantially increased SO2 in the air. China has contributed to about one-fourth of global SO2 emission since 1990 (Zhang et al., 2013). The emission of SO2 from Guangdong province totaled 1,177 Gg in 2007, of which 97% was emitted by power plants and industries (Lu et al., 2010). SO2 can be oxidized photochemically or catalytically to sulfur trioxide (SO3) and sulfate () in the air (Bufalini, 1971). With the presence of water, SO3 is converted rapidly to sulfuric acid (H2SO4), which is commonly known as acid rain. While in sulfur assimilation, is reduced to organic sulfhydryl groups (R-SH) by sulfate-reducing bacteria, fungi, and plants. Sulfur oxidizing bacteria such as Beggiatoa and Paracoccus are able to oxidize reduced sulfur compounds like H2S to inorganic sulfur, and thiosulfate to form sulfuric acid (Pokoma and Zabranska, 2015). Sulfate reducing bacteria like Archaeoglobus and Desulfotomaculum can convert sulfur compounds to hydrogen sulfide (H2S). Oxidation of H2S produces elemental sulfur (S°), which is completed by the photosynthetic green and purple sulfur bacteria and some chemolithothrophs. Further oxidation of elemental sulfur produces sulfate. Sulfate is assimilated through the sulfate activation pathway, which is consisted of three reactions: the synthesis of adenosine 5′-phosphorylation of (APS), the hydrolysis of GTp, and the 3′-phosphorylation of APS to produce 3′-phosphoadenosine 5′-phosphosulfate (PAPS) (Sun et al., 2005). In Mycobacterium tuberculosis, the entire sulfate activation pathway is organized into a single complex (Sun et al., 2005). Additionally, sulfate reducing bacteria have been shown to use hydrocarbons in pure cultures, which can be used for bioremediation of benzene, toluene, ethylbenzene, and xylene in contaminated soils (Muyzer and Stams, 2008). Such bacteria may also colonize leaf surfaces and could be used for remediation of air pollutants.
Plant leaves absorb SO2 via stomata. At apoplastic pH, it is hydrated and oxidized successively to sulfite and sulfate, both of which can inhibit photosynthesis and energy metabolism if they accumulate to a high concentration. Such inhibition can cause SO2 toxicity. Symptoms include interveinal chlorosis and necrosis in broad-leaved species, and chlorotic spots and brown tips in pine conifers (Rennenberg, 1984). Until the 1970s, SO2 was considered to be a key contributor of acid rain causing forest dieback (Bloem et al., 2015). Interestingly, when the Clean Air Acts came into action in the 1980s, the reduction in atmosphere SO2 resulted in sulfur (S) deficiency in crops, particularly Brassica species. The S deficiency was responsible for the increased incidence of disease caused by Pyrenopeziza brassicae (Bloem et al., 2015). The explanation is that plants could become injured in a SO2 concentration range from 131 to 1,310 μg m−3; plants, however, can rapidly assimilate SO2 and H2S into reduced sulfur pools such as cysteine and sulfates as illustrated in Figure 1. A recent transcriptome analysis of Arabidopsis responses to SO2 showed that plant adaptation to SO2 evokes a comprehensive reprogramming of metabolic pathways including NO and reactive oxygen species (ROS) signaling molecules, and also plant defense response pathways (Zhao and Yi, 2014). The importance of this study revealed that plant responses to SO2 stress is at the transcription level with initial activation of cross tolerance and followed by sulfur assimilation pathways. Cysteine metabolism in particular is associated with the network of plant stress responses, thus improving plant growth in soils where sulfur supply is limited (Bloem et al., 2015). It has been shown that an atmospheric level of 79 ng m−3 SO2 could contribute to 10–40% of leaf sulfur assimilation (De Kok et al., 2007; Zhao et al., 2008). Elevated SO2 concentrations around natural CO2 springs have been documented to enhance accumulation of sulfur metabolites and proteins in surrounding vegetation (Rennenberg, 1984). Therefore, plants can be selected for growing in SO2 polluted environments (Chung et al., 2010). In 2000, about 42.62 Mg of SO2 was removed from the atmosphere by urban trees in Guangzhou, China (Zhang et al., 2013). Additionally, S metabolism can be genetically engineered for improving plant resistance to SO2. Transgenic tobacco plants overexpressing cysteine synthase or serine acetyltransferase gene were highly tolerant to SO2 and sulfite (Noji et al., 2001).
Remediation of NOx
There are several oxides of nitrogen (N) in the atmosphere: nitrogen dioxide (NO2), nitric oxide (NO), nitrous oxide (N2O), nitrogen trioxide (N2O3), and nitrogen trioxide (N2O5). Among them, the USEPA regulates NO2 only because it is the most prevalent form of NOx generated anthropogenically (USEPA, 1999). NO2 also participates in the formation of ozone (O3) and NO. NOx emissions in China increased rapidly from 11.0 Mt in 1995 to 26.1 Mt in 2010. Power plants, industry, and transportation were major sources of NOx emissions, accounting for 28.4, 34.0, and 25.4% of the total NOx emissions in 2010, respectively (Zhou et al., 2013). The total NOx emissions in China are projected to increase 36% based on the 2010 value by 2030.
A group of bacteria like Azotobacter and Rhizobium and fungi such as mycorrhizas are capable of fixing atmospheric N. Cyanobacteria are able of using a variety of inorganic and organic sources of combined N, like nitrate, nitrite, ammonium, urea or some amino acids. These microbes are often associated with plant roots. Nitrifying bacteria including species from the genera Nitrosomonas, Nitrosococcus, Nitrobacter, and Nitrococcus oxidize ammonia to hydroxylamine, and nitrite oxidoreductase oxidizes nitrite to nitrate. Nitrifying bacteria thrive in soils, lakes, rivers, and streams with high inputs and outputs of sewage, wastewater and freshwater because of high ammonia content. Phyllosphere diazotrophic bacteria, like Beijerinckia, Azotobacter, and Klebsiella and also Cyanobacteria, such as Nostoc, Scytonema, and Stigonema can use atmospheric dinitrogen (N2) as a source of nitrogen (Whipps et al., 2008). N2 is fixed by the nitrogenase enzyme encoded by nif genes, and the gene nifH has been widely used for analysis of their community structure (Fürnkranz et al., 2008; Rico et al., 2014). The abundance of N2-fixing bacteria was also reported to improve drought tolerance, suggesting their adaptability to plants grown in different environmental conditions (Rico et al., 2014).
Plants absorb gaseous NO2 more rapidly than NO because NO2 reacts rapidly with water while NO is almost insoluble. The uptake of NO2 per unit leaf area was reported to be nearly three times that of NO when the two gases occurred in the same concentration (Law and Mansfield, 1982). As a result, NO2 has been considered to be more toxic than NO. Visible symptoms resulting from NO2 exposure are relatively large, irregular brown or black spots. However, phytotoxicity of NO2 is rare and much less than SO2 and O3. This is due to the fact that NOx are plant nutrients. When NO and NO2 are absorbed and dissolved in the extracellular solution of leaves, they form nitrate (NO3) and NO2 in equal amounts and proton (H+). NO3 is then utilized by plants in the same way as it is absorbed from roots and used as a nitrogen source for synthesizing amino acids and proteins (Figure 1). Foliar absorption of NO2 varies widely depending on plant species. Morikawa et al. (1998) studied 217 herbaceous and woody species in uptake of NO2 and found that plant species differed by 657 folds in NO2 uptake and assimilation. The most efficient woody plants included Eucalyptus viminalis, Populus nigra, Magnolia kobu, and Robinia pseudoacacia, and the most herbaceous plants include Erechtites hieracifolia, Crassocephalum crepidioides, and Nicotiana tabacum (Morikawa et al., 1998).
Nitrogen dioxide could be a plant signal molecule that improves plant growth. Morikawa et al. (2004) reported that about one-third of NO2-derived N absorbed by leaves was converted into a previously unknown Kjeldahl-unrecoverable organic nitrogen, which comprise a novel heterocyclic Δ2 1,2,3 thiadiazoline derivative and nitroso- and nitro-organic compounds (Miyawaki et al., 2004; Morikawa et al., 2005). These results indicate that NO2 is not only known as a pollutant or a supplemental source of N, but also acts as an airborne reactive nitrogen species signal (Morikawa et al., 2004, 2005). This is in agreement with the reports that endogenously produced NOx such as NO act as a vital plant signal (Wendehenne et al., 2001; Neill et al., 2003). To further analyze atmospheric NOx effects on plants, Morikawa et al. (2003) determined if plants could use NO2 as a fertilizer and concomitantly reduce NO2 concentrations. The authors found that application of 282 μg m−3 NO2, equivalent to the heavily polluted urban air, to plants for 10 weeks almost doubled the biomass, total leaf area, the contents of carbon (C), N, S, phosphorus (P), potassium (K), calcium (Ca), and magnesium (Mg) as well as free amino acid contents and crude proteins (Morikawa et al., 2003). The mass spectrometric analysis of the 15N/14N ratio showed that N derived from NO2 comprised less than 3% of total plant N, meaning that the contribution of NO2-N to total N was relatively low. These results imply that NO2 could be a multifunctional signal to stimulate plant growth, nutrient uptake, and metabolism (Takahashi et al., 2005).
Remediation of O3
Anthropogenic O3 is primarily generated from the reaction of atmospheric O2 with ground-state O (3P) radicals that result from the photolytic dissociation of ambient NO2. Thus, the presence of NO and NO2 in the lower atmosphere is closely linked with ground-level of O3. In China, O3 levels increased at a rate of 2.2 μg m−3 per year from 2001 to 2006. Average O3 concentrations in Beijing varied from 45 to 96.2 μg m−3 depending on locations (Wan et al., 2014). In Shanghai, 1-h average concentration of O3 was 54.2 μg m−3. O3 level increased during spring, reached the peak in late spring and early summer, and then decreased in autumn and finally dropped in winter. The highest monthly average O3 concentration (82.2 μg m−3) in June was 2.7 times greater than the lowest level (30.4 μg m−3) recorded in December (Zhao et al., 2015).
Ozone is considered an effective antimicrobial agent against some bacteria and fungi (Sharma and Hudson, 2008). There have been no reports on microbial-mediated O3 reduction. However, in a study of O3 effects on phyllosphere fungal populations, Fenn et al. (1989) found that a chronic exposure of mature Valencia orange trees (Citrus sinensis) to O3 or SO2 for 4 years decreased populations of phyllosphere fungi. In a same experiment conducted by the authors, a short-term fumigation of O3 to giant sequoia (Sequoiadendron giganteum) and California black oak (Quercus kelloggii) did not significantly affect the numbers of phyllospere fungi. Plant absorption of O3 is mainly through stomata, O3 is easily dissolved in water and reacts with apoplastic structures and plasma membranes to form reactive oxygen species (ROS), such as , H2O2, and OH radical. The O3 or ROS can disturb cell membrane integrity and attack sulfhydryl (SH) groups or ring amino acids of protein, thus causing phytotoxicity. Injury symptoms include white, yellow or brown flecks on the upper surface of leaves. The threshold concentrations that cause a 10% reduction in yield are 80μg m−3 for sensitive crops and 150 μg m−3 for the most resistant crops. Adaptation of plants to O3 stress has resulted in plants developing mechanisms against O3 toxicity. First, O3 can be removed from the air by chemical reactions with reactive compounds emitted by vegetation, particularly monoterpenes (Di Carlo et al., 2004). Second, semi-volatile organic compounds, such as different diterpenoids exuded by trichomes on leaves are an efficient O3 sink (Jud et al., 2016). Tobacco leaves can secret diterpenoid cis-abienol, which acts as a powerful chemical protection shield against stomatal O3 uptake by depleting O3 at the leaf surface. As a result, O3 flux through the open stomata is strongly reduced (Jud et al., 2016). As to O3 absorbed by leaves, an oxidative burst occurs as the initial reaction to O3, followed by activation of several signaling cascade and plant antioxidant systems including ascorbate-glutathione cycle and antioxidant enzymes to alleviate the oxidative burden resulting from O3 exposure (Vainonen and Kangasjarvi, 2015).
Remediation of VOCs
VOCs are organic chemicals that have a low boiling point and a high vapor pressure at room temperature causing large numbers of molecules to evaporate into the surrounding air. VOCs are numerous and ubiquitous including naturally occurring and anthropogenic chemical compounds. VOCs participate in atmospheric photochemical reactions contributing to O3 formation and also play a role in formation of secondary organic aerosols, which are found in PMs. The strong odor emitted by many plants consists of green leaf volatiles, a subset of VOCs called biogenic VOCs, which emit exclusively from plant leaves, the stomata in particular. Major species of biogenic VOCs include isoprene, terpenes, and alkanes.
Anthropogenic VOCs include large groups of organic chemicals, such as formaldehyde, polycyclic aromatic hydrocarbons (PAHs), and BTX (benzenes, toluene, and xylenes). The most significant sources of formaldehyde are engineered wood products made of adhesives that contain urea-formaldehyde (UF) resins. BTX come from painting and coating materials used for interior decoration and refurbishment. Motor-vehicle exhausts, tobacco smoke, and heating also contribute to the presence of VOCs. A great concern over VOCs has been indoor air quality. Indoor formaldehyde in recently renovated homes ranged from 0.14 to 0.61 mg m−3, and benzene, toluene, and xylenes were 124.0, 258.9, and 189.7 μg m−3, respectively (Hao et al., 2014). The formaldehyde concentration is 65–100% higher than indoor air quality standards of China. Formaldehyde and BTX as main indoor VOCs contribute to the so-called “sick building syndrome” (Brown et al., 1994; Wieslander et al., 1996; Wargocki et al., 2000; Berg et al., 2014). This review regarding VOCs is thus emphasized on indoor air quality.
As early as in the 1970s, NASA (U.S. National Aeronautics and Space Administration) conducted research on the use of foliage plants for remediation of air quality in space shuttles. Foliage plants are those with attractive foliage and/or flowers that are able to survive and grow indoors (Chen et al., 2005). Results showed that foliage plants removed nearly 87% of air pollutants from sealed chambers within 24 h (Wolverton et al., 1984, 1989; Cruz et al., 2014a). For example, each plant of peace lily (Spathiphyllum spp. ‘Mauna Loa’) removed 16 mg of formaldehyde, 27 mg of trichloroethylen, and 41 mg of benzene from sealed chambers after a 24-h exposure to the respective chemical. Generally, plants absorb gaseous pollutants via leaf stomata. Some of the VOCs are recognized as xenobiotics by plants, and they are detoxified through xenobiotic metabolism, involving oxidoreductase or hydrolases, bioconjugation with sugars, amino acids, organic acids, or peptides, and then removed from the cytoplasm for deposition in vacuoles (Edwards et al., 2011). In addition to plant leaves, rhizosphere microbes also contribute to reduction of VOCs under interior environments (Llewellyn and Dixon, 2011). Using a dynamic chamber technique, Xu et al. (2011) investigated formaldehyde removal by potted foliage plants and found that formaldehyde removal was attributed not only to the formaldehyde dehydrogenase activities of plant leaves but also to the absorption and metabolism by microorganisms in the rhizosphere. Such bacteria have been isolated from soils, water, and different tissues of plants in polluted environments. Many pure cultures of bacteria, including various strains of P. putida, have been evaluated for biodegradation of air pollutants. Some fungi strains are also able to use volatile aromatic hydrocarbons as sole source of carbon and catalyze degradation reactions (Prenafeta-Boldú et al., 2001; Kennes and Veiga, 2004; Jin et al., 2006). Here we mainly discuss phylloremediation of formaldehyde, benzene, toluene, and xylene as well as phenols and PAHS.
Formaldehyde
Formaldehyde is a colorless, flammable gas or liquid that has pungent and suffocating odor. It poses a significant danger to human health due to its high reactivity with proteins and DNA, thus formaldehyde is known to be a human carcinogen. Plants can directly absorb formaldehyde and transform it to organic acids, sugars or CO2 and H2O (Figure 1). Giese et al. (1994) exposed shoots of Chlorophytum comosum to 8.5 mg m−3 gaseous [14C]-formaldehyde over 24 h and found that about 88% of the recovered radioactivity was associated with plant metabolites as 14C, which had been incorporated into organic acids, amino acids, free sugars, lipids, and cell wall components. Formaldehyde responsive genes were identified from golden pothos (Epipremnum aureum) (Tada et al., 2010). Glutathione (GSH)-dependent formaldehyde dehydrogenase (FADH) and formate dehydrogenase (FDH) can detoxify formaldehyde to formate and further to carbon dioxide (Tada and Kidu, 2011). A wide range of foliage plants have been documented to be able to remove formaldehyde. Kim et al. (2010) exposed 86 species of foliage plants individually to 2 μl L−1 formaldehyde in sealed chambers and found that formaldehyde removed per cm2 leaf area in 5 h ranged from 0.1 to 6.64 mg m−3, depending on plant species. The most efficient species in removal of formaldehyde include Osmunda japonica, Selaginella tamariscina, Davallia mariesii, and Polypodium formosanum. Surprisingly, these efficient plants belong to pteridophytes, commonly known as ferns and fern allies. Why this group of plants is more efficient than the other foliage plants in formaldehyde removal deserves further investigation.
Formaldehyde can also be assimilated as a carbon source by bacteria (Vorholt, 2002). Such assimilation occurs in Methylobacterium extorquens through the reactions of the serine cycle (Smejkalova et al., 2010), in Bacillus methanolicus through the RuMP cycle (Kato et al., 2006), and in Pichia pastoris through the xylulose monophosphate cycle (Lüers et al., 1998). Some fungi also assimilate formaldehyde. Yu et al. (2015) isolated a fungal strain (Aspergillus sydowii HUA), which was able to grow in the presence of formaldehyde up to 2,400 mg l−1 and the specific activity of formaldehyde dehydrogenase and formate dehydrogenase were as high as 5.02 and 1.06 U mg−1, respectively, suggesting that this fungal isolate could have great potential for removing formaldehyde. Some of the bacteria and fungi used to colonize roots can also colonize leaves and could be used for phylloremediation of formaldehyde in the air (Khaksar et al., 2016a).
BTX
BTX refers to benzene, toluene, and three xylene isomers [ortho– (or o–), meta– (or m–), and para– (or p–)], which are major components of gasoline. Due to their low water solubility and acute toxicity and genotoxicity, BTX components have been classified as priority pollutants by the USEPA (Eriksson et al., 1998). Plants leaves can absorb BTX mainly through stomata, which are converted to phenol or pyrocatechol, and subsequently to muconic acid and fumaric acid (Ugrekhelidze et al., 1997). Foliage plants, such as Dracaena deremensis and Spathiphyllum spp. have been documented to remove BTX indoors (Wolverton et al., 1984, 1989; Wood et al., 2006; Mosaddegh et al., 2014). Liu et al. (2007) fumigated 73 plant species with 478.5 μg m−3 benzene gas and found that 23 of the 73 species showed inability to reduce fumigated benzene, the rest varied in benzene reduction, ranging from 0.1 to 80%. The most efficient plant species were Crassula portulacea, Hydrangea macrophylla, and Cymbidium ‘Golden Elf’. Foliage plants that are effective in removal of toluene include H. helix, Philodendron spp., Schefflera elegantisima, and Sansevieria spp. (Kim et al., 2011; Sriprapat et al., 2013; Cruz et al., 2014b). The wax of Sansevieria trifasciata and S. hyacinthoides is rich in hexadecanoic acid, which could pay an important role in absorption of toluene (Sriprapat et al., 2013). Sriprapat et al. (2014) also evaluated plant absorption of xylene. The tested 15 plant species were able to remove xylene with removal efficiency ranging from 59.1 to 88.2%, of which Zamioculcas zamiifolia was the most efficient species.
Bacteria including some strains of Rhodococcus rhodochrous (Deeb and Alvarez-Cohen, 1999), Alcaligenes xylosoxidans (Yeom and Yoo, 2002), and P. putida (Alagappan and Cowan, 2003) and also fungal cultures of Cladophialophora sp. (Prenafeta-Boldú et al., 2002) are able to degrade BTX (Figure 1). Many Pseudomonas species are leaf colonists and some are plant pathogens (Dulla et al., 2005). BTX are actual growth substrates for a number of organisms, such as P. putida (Inoue et al., 1991). In a study of bioremediation of airborne toluene, De Kempeneer et al. (2004) found that the time required for 95% reduction of the initial toluene concentration of 339 mg m−3 was 75 h by Azalea indica plants along. Such reduction by the plants inoculated with P. putida TVA8 under the identical conditions was only 27 h. Subsequent additions of toluene further increased the removal efficiency of plants inoculated with the bacterial strain, but the toluene-removal rate was comparably low in plants without inoculation. Hence, inoculation of the leaf surface with P. putida TVA8 was considered to be essential for rapid removal of toluene. These results clearly demonstrated the importance of both plant leaves and leaf-associated microbes in phylloremediation of indoor air pollutants. The genetics and biochemistry of strains F1 and mt-2 of P. putida have been intensively studied (Harayama and Rekik, 1990; Horn et al., 1991; Timmis et al., 1994; Aemprapa and Williams, 1998). Such information could be important for exploring these strains for effective removal of air pollutants.
Air borne phenols and polycyclic aromatic hydrocarbon (PAHs)
Air borne phenols are a class of chemical compounds containing a hydroxyl group bonded directly to an aromatic hydrocarbon group, whereas PAHs are hydrocarbon comprising only carbon and hydrogen with multiple aromatic rings. Phenol and PAHs are major air pollutants in urban areas, and some PAHs have been considered carcinogenic. It has been reported that Bacillus cereus can degrade phenol via meta-cleavage pathway (Banerjee and Ghoshal, 2010). Pseudomonas sp. CF600 can mineralize phenol on bean and maize leaves by dmp catabolic pathway (Sandhu et al., 2007). Sandhu et al. (2007) directly measured phenol degradation by natural phyllosphere communities. Leaves were collected from trees growing in an area that was known to have high concentrations of VOCs. Unsterilized and surface-sterilized leaves were then exposed to radiolabeled phenol in closed chambers for 24 h and the amount of phenol degradation was compared. The phenol degradation by the non-sterilized leaves was significantly greater than the degradation by the sterilized leaves, indicating that degradation of VOCs was enhanced by the presence of the phyllosphere communities. This work indicates that plant leaves can accumulate phenols, which may be subsequently available for bacteria in the phyllosphere for degradation.
Plant leaves can absorb atmospheric PAHs. A study on deciduous forest in Southern Ontario, Canada, confirmed that amounts of phenanthrene, anthracene, and pyrene were reduced within and above the forest canopy during bud break in early spring (Choi et al., 2008). Plant species differ in removal of PAHs, the differences could be attributed to specific morphological and chemical constitutions of plants as well as leaf-associated microbes. Phyllosphere bacteria on 10 ornamental plant species were studied based on their diversity and activity toward the removal of PAHs (Yutthammo et al., 2010). The phyllosphere hosted diverse bacterial species including Acinetobacter, Pseudomonas, Pseudoxanthomonas, Mycobacterium, and unculturable ones, of which PAH degrading bacteria accounted for about 1–10% of the total heterotrophic phyllosphere populations depending on plant species. The analysis of bacterial community structures using PCR and denaturing gradient gel electrophoresis showed that each plant species had distinct band patterns, suggesting that the bacterial communities are closely associated with leaf morphology and chemical characteristics of ornamental plant species. Furthermore, branches of fresh leaves of selected plant species were evaluated in sealed chambers for removal of a mixture of PAHs (acenaphthene, acenaphthylene, fluorene, and phenanthrene). Bacteria on unsterilized leaves of all tested plants showed an enhanced removal of phenanthrene. Bacteria on leaves of Wrightia religiosa in particular were able to reduce all the tested PAHs (Yutthammo et al., 2010). Therefore, phyllosphere bacteria on ornamental plants may play an important role in natural attenuation of airborne PAHs and plant species differ in supporting microbes in PAH removal.
Development of phylloremediation technologies
This review has documented that plant leaves and leaf-associated microbes individually can reduce air pollution and the combination of the two generally exhibits enhanced remediation of air pollutants. Since air pollution never before has become such an urgent problem in countries like China and India, now is the time to seriously consider all options for reducing the pollutants. Phylloremediation is a natural and environmentally friendly way of bioremediation of air contaminants. Our proposal for developing phylloremediation technologies is outlined in Figure 2, which includes (1) selection and evaluation of appropriate plant species and microorganisms that are tolerant to pollution and able to remove one or more air pollutants; (2) testing and analysis of the compatibility of plant leaf surfaces with isolated microbes for synergetic interactions in reduction of pollutants in laboratories, in simulated indoor environments, and in outdoor settings; (3) analysis of experimental data and development of phylloremediation technologies; and (4) implementation of the technologies for remediation of air in both indoor and outdoor environments.
Figure 2.
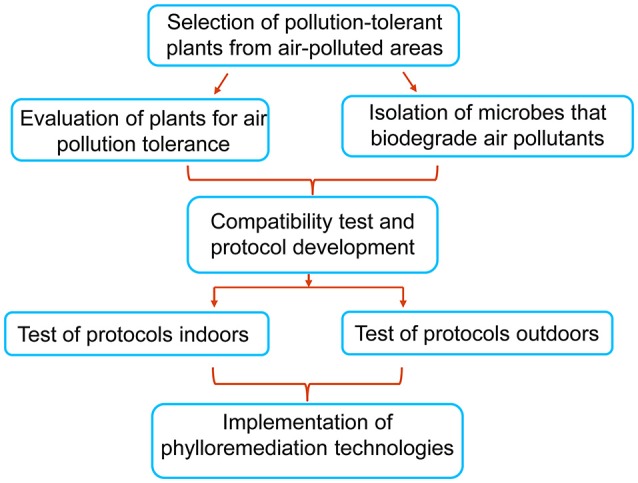
A general outline for developing phylloremediation technologies. Plants species and microbes should be selected from air polluted areas. Selected plants should be evaluated for their ability to adsorb or absorb air pollutants, and concurrently microbes are screened for biodegradation or biotransformation of pollutants. The selected plants and microbes are tested for synergistic effects on the reduction of particular air pollutants. Based on the test results, specific plant-microbe combinations that can remove one or more air pollutants are identified, and protocols are formulated for evaluating their effectiveness in removal pollutants indoors and outdoors. Effective protocols will be developed into phylloremediation technologies for use in reducing air pollutants.
Plant selection
Plants should be selected from four categories: (1) trees, (2) shrubs or small tress, and (3) ground cover plants for use in outdoor environments as well as (4) foliage plants for indoor environments. Trees are referred to as perennial plants with elongated stems or trunks, supporting branches and leaves. Shrubs (or small trees) are those small to medium-sized woody plants that grow under some degree of shaded conditions. Ground covers are any plants that can grow over an area of ground and they can grow below the shrub layer including turfgrass and other woody and herbaceous selections. Foliage plants are those which can grow and survive indoors for interior decoration.
Plant species not only differ greatly in adsorption, absorption, and assimilation of air pollutants but also vary significantly in pollution tolerance. Air pollution tolerance index has been used for evaluation of plants specie in response of pollutants (Singh et al., 1991). Information generated by the index is useful, but the index may require revision for better reflecting the ability of plants in tolerance of air pollutants. An initial large-scale evaluation of plants from the four categories should be conducted for identifying candidate species that are able to tolerate PMs, O2, SO2, NOx, and VOCs individually or collectively and can also substantially retain or assimilate these pollutants. Plants should also tolerate abiotic stresses, such as drought, heat, and cold, and biotic stresses like plant pathogens. Leaves of plants should be able to support one or more selected microbes. Trees should have a relatively fast growth rate. Needle-leaved plants should be particularly considered. As mentioned before, needles are rich in waxes for capturing PMs, and they are also used as as passive bio-samplers to determine polybrominated diphenyl ethers (Ratola et al., 2011). Broad-leaved plants should have more hairs or trichomes and more stomata with a large canopy. Leaf water and nutritional contents, leaf cuticular wax composition, hairs or trichomes, and surface physical characteristics should be suitable for microbial colonization. Shrubs and ground cover plants should have similar leaf physical and chemical properties but be able to tolerate slight shade. For foliage plants, they should substantially tolerate shade and can survive and grow under indoor low-light conditions.
Plant species possessing the aforementioned traits should be selected from particular regions where plants survive and thrive under heavily polluted environments. The rationale is that plants that are able to grow in the polluted environments may develop mechanisms for adaptation to the stressful conditions. Thus, some regions of China and India could be ideal locations for initial selection of plant species. Plants have been documented to tolerate multiple stresses, which include induced cross tolerances and the ability of particular variants to resist multiple distinct stresses. Reactive oxygen species are key molecular signals produced in response to multiple stresses, which are aimed at the maintenance of cellular equilibrium (Perez and Brown, 2014). Glutathione-S-transferase (GST) genes play an important role in the maintenance of ROS equilibrium. Salicylic acid, jasmonic acid, and ROS interplay in the transcriptional control of multiple stresses. Additionally, omics technologies should be used for identifying molecular mechanisms in regulation of plant responses to multiple stresses. Such information, particularly transcriptional factors, key regulatory genes or enzymes should be incorporated into the plant selection processes.
Genetic engineering is an option for improving plants to remediate air pollutants (Abhilash et al., 2009). Genes listed in Table 4 can be used for generating transgenic plants. Cysteine synthase is a key enzyme to utilize H2S and SO2 as a sulfur source to synthesize cysteine. Overexpression of cysteine synthase in rice was shown to enhance sulfur assimilation upon exposure to a high level of H2S (Yamaguchi et al., 2006). Nitrite reductase catalyzes the six-electron reduction of nitrite to ammonium. Transgenic Arabidopsis plants bearing chimeric spinach NiR gene enhanced nitrite reductase activity and NO2 assimilation (Takahashi and Morikawa, 2001). Cytochrome P450 2E1 has strong and specific capacity of decomposing organic pollutants in animal bodies. Transgenic tobacco plants overexpressing CYP2E1 gene showed increased ability to detoxify broad classes of pollutants such as chlorinated solvents and aromatic hydrocarbons (James et al., 2008). Unlike tobacco, poplar (Populus tremula × Populus alba) plants are a fast-growing tree species with large canopies. Poplar plants overexpressing a mammal CYP2E1 exhibited increased metabolism and enhanced removal of organic pollutants from hydroponic solution and the air (Doty et al., 2007). Some genes from microbes can also be used for engineering transgenic plants for phylloremediation. The ribulose monophosphate (RuMP) pathway is one of the formaldehyde-fixation pathways found in microorganisms (Orita et al., 2006). The key enzymes of this pathway are 3-hexulose-6-phosphate synthase (HPS), which fixes formaldehyde to D-ribulose 5-phosphate (Ru5P) to produce D-arabino-3-hexulose 6-phosphate (Hu6P) and 6-phospho-3-hexuloisomerase (PHI), and then converts Hu6P to fructose 6-phosphate (F6P) (Orita et al., 2006; Chen et al., 2010). Co-expression of HPS and PHI in tobacco plants resulted in 20% reduction of formaldehyde compared to the control plants (Chen et al., 2010). In another study, a chlorocatechol 1,2-dioxygenase gene (tfdC) derived from the bacteria Plesiomonas was introduced into Arabidopsis thaliana (Liao et al., 2006). Transgenic plants showed enhanced tolerances to catechol, an aromatic ring. Transgenic plants were also able to remove a large amount of catechol from their media and highly efficient in convertion of catechol to cis, cis-muconic acid, suggesting that degradative genes derived from microbes can be used to produce transgenic plants for bioremediation of aromatic pollutants in the environment (Liao et al., 2006).
Table 4.
Genes from different sources have been demonstrated to be able to remediate air pollutants in transgenic plants.
| Gene name | Source | Transgenic plants | Pollutant | References |
|---|---|---|---|---|
| 3-hexulose 6-phosphate and 6-phospho-3-hexuloisomerase | Mycobacterium gastri MB19 | Arabidopsis thaliana and Nicotiana tabacum | Formaldehyde | Chen et al., 2010 |
| Biphenyl-Chlorobiphenyl Dioxygenase | Burkholderia xenovorans | Nicotiana tabacum | 4-chlorobiphenyl | Mohammadi et al., 2007 |
| Chlorocatechol 1,2-dioxygenase | Plesiomonas spp. | Arabidopsis thaliana | Catechol | Liao et al., 2006 |
| CYP450 2E1 | Oryctolagus cuniculus | Populus tremula × populous alba | Trichloroethylene, benzene, and chloroform | Doty et al., 2007 |
| CYP450 2E1 | Homo sapiens | Nicotiana tabacum | Trichloroethylene, and ethylene dibromide | Doty et al., 2000 |
| CYP450 2E1 | Homo sapiens | Nicotiana tabacum | Trichloroethylene, benzene, toluene, vinyl chloride, chlorotoluene, and chloroform | James et al., 2008 |
| Cysteine synthase | Nicotiana tabacum | Nicotiana tabacum | SO2 | Noji et al., 2001 |
| Glutathione reductase | Escherichia coli | Populus sieboldii × P. grandidentata | SO2 | Endo et al., 1997 |
| Mn-peroxidase | Coriolus versicolor | Nicotiana tabacum | Phencyclidine | Iimura et al., 2002 |
| Nitrite reductase | Spinacia oleracea | Arabidopsis thaliana | NO2 | Takahashi and Morikawa, 2001 |
| O-acetylserine(thiol) lyase | Triticum aestivum | Nicotiana tabacum | Hydrogen sulfide | Youssefian et al., 1993 |
| O-acetylserine(thiol) lyase | Triticum aestivum | Nicotiana tabacum | SO2 | Youssefian et al., 2001 |
| Peroxidases | Lycopersicon esculentum | Nicotiana tabacum. | Phenol | Sosa Alderete et al., 2009 |
Selected plants should be evaluated in controlled environmental chambers to measure their capacity for tolerance and also assimilation of air pollutants. Seedlings could be exposed to particular pollutants or a mixture of pollutants in different concentrations and durations. Plant responses to the exposures could quickly evaluated based on stomatal conductance, net photosynthetic rate, the maximum quantum efficiency of photosystem II using the new LI-COR6800. Their morphological appearance, i.e., leaf greenness, leaf size, and plant height and canopy dimension compared to control treatments should be evaluated. The ability of plants to remove pollutants should be tested using GC-MS. For evaluation of plant responses to PM, in addition to the mentioned plant characteristics, leaf morphology, particularly leaf surface characters should be examined under microscopes and stomatal size and density recorded. If needed, isotopic labeling techniques could be used to track the fate of particular compounds. The evaluation results once analyzed and compared, plants that tolerate stresses and are able to adsorb or absorb or assimilate pollutants could be identified from each type of plants for subsequent compatiablity tests with selected microbes.
Microbe selection
Cultivable bacteria only account for a small fraction of the total diversity in the phyllosphere, which has greatly hampered the use of some valuable microbes. New approaches, such as the use of improved culture and advanced devices (i-Chip), co-culture with other bacteria, recreating the environment in the laboratory, and combining these approaches with microcultivation should be employed to convert more uncultivable bacteria into cultured isolates in the laboratory (Nichols et al., 2010; Stewart, 2012; Müller and Ruppel, 2014). Similar to plant selection, initial microbial selection could be carried out in areas where plants have been contaminated by air pollutants. In coordination with plant selection, microbes could be isolated from leaves of plants identified in plant selection. This is because the pollutants may exert selective pressures to phyllosphere microbial diversity. For example, bacterial communities hosted by Platanus × acerifolia leaves from different locations of Milan (Italy) were analyzed by high throughput sequencing. The results showed that biodiversity of bacterial communities decreased but hydrocarbon-degrading populations increased along the growing season, which suggest that air contaminants might play an important role in the selection of phyllospheric populations in urban areas (Gandolfi et al., 2017).
A particular attention should be given to endophytic microbes. There are about 300,000 plant species on the earth; each plant could host one or more endophytes (Petrini, 1991; Strobel and Daisy, 2003). Endophytes are resided inside plant tissues and generally have no harmful effects on plants. Endophytic bacteria that colonize leaves could be particularly desirable as they could not be washed away by precipitation. Recent advances in endophyte-assisted remediation have been reviewed (Khan and Doty, 2011; Stepniewska and Kuzniar, 2013; Ijaz et al., 2016; Syranidou et al., 2016). Endophytic B. cereus ZQN5 isolated from natural Zamioculcas zamiifolia leaves enhanced ethylbenzene removal rate on sterile Z. zamiifolia (Toabaita et al., 2016). Microbes could also be isolated from the rhizosphere of plants contaminated by air pollutants as more endophytism occurs in roots (Ijaz et al., 2016). Some of leaf endophytes could be initially established in roots and subsequently transported to shoots. Khaksar et al. (2016a) reported that some microbes isolated from roots can also colonize leaf surfaces. An endophytic strain of B. cereus ERBP from roots of Clitoria ternatea was able to colonize the leaf surface of Z. zamifolia. During a 20-d fumigation with formaldehyde, the inoculation of ERBP did not interfere with the natural shoot endophytic community of Z. zamiifolia. ERBP inoculated Z. zamiifolia exhibited a significantly higher formaldehyde removal efficiency when compared to the non-inoculated plants.
Microbes, once identified and cultured, could be engineered to improve phylloremediation capacity (Table 5). A pTOM toluene-degradation plasmid from B. cepacia G4 was introduced into Bacillus cepacia L.S.2.4, a natural endophyte from yellow lupine (Lupinus arboreus; Barac et al., 2004). After the engineered bacteria were inoculated into aseptic lupine seedlings, the recombinant endophytics degraded 50–70% more toluene and provided much more protection against the phytotoxic effects of toluene than that obtained from soil bacteria (Barac et al., 2004). Horizontal genes can transfer among plant-associated endophytic bacteria in plants. Poplar was inoculated with the yellow lupine endophyte B. cepacia VM1468, which contains the pTOM-Bu61 plasmid coding for constitutively expressed toluene degradation (Taghavi et al., 2005). Inoculated plant growth was enhanced in the presence of toluene, and the amount of toluene release via evapotranspiration was also reduced. Although no inoculated strains were detected in the endophytic community, there was horizontal gene transfer of pTOM-Bu61 to different members of the endogenous endophytic community (Taghavi et al., 2005). The TCE-degrading strain P. putida W619-TCE also can be engineered via horizontal gene transfer in poplar plants (Weyens et al., 2009b).
Table 5.
Genes from microbes have been demonstrated to be able to remediate pollutants in transgenic microbes.
| Gene name | Source | Transgenic organism | Pollutant | References |
|---|---|---|---|---|
| BphA1 | Burkholderia xenovorans LB400 | Pseudomonas pseudoalcaligenes | Aromatic hydrocarbons and pentachlorobenzene | Suenaga et al., 2010 |
| C23O | Pseudomonas aeruginosa zl1f4 | Bacillus subtilis | Phenol | Yang et al., 2012 |
| Camphor monooxygenase and a hybrid dioxygenase | Pseudomonas putida | Alcaligenes | Pentachloroaniline | Iwakiri et al., 2004 |
| Catechol 2, 3-dioxygenase | Pseudomonas aeruginosa SZH16 | Pseudomonas fluorescens P13 | Phenol | Yang et al., 2012 |
| Hemoglobin | Vitreoscilla sp. | Pseudomonas putida | Benzene, toluene and xylene | Kahraman and Geckil, 2005 |
| Hemoglobin | Vitreoscilla sp. | Xanthomonas maltophilia | Benzoic acid | Liu et al., 1996 |
| Phenol hydroxylase | Escherichia coli | Pseudomonas putida | Trichloroethylene | Fujita et al., 1995 |
| Phenol Hydroxylase | Ralstonia sp. | Ralstonia sp. | Trichloroethylene | Ishida and Nakamura, 2000 |
| Pro U operon | Escherichia coli | Microbial consortium | Hydrocarbon | Kapley et al., 1999 |
| Tod and xyl | Pseudomonas putida | Deinococcus radiodurans | Toluene | Brim et al., 2006 |
| Toluene dioxygenase | Deinococcus radiodurans | Deinococcus radiodurans | Toluene and Trichloroethylene | Lange et al., 1998 |
| Toluene o-monooxygenase | Burkholderia cepacia | Pseudomonas fluorescens | Trichloroethylene | Yee et al., 1998 |
| Xyl and lux gene cassette | Pseudomonas putida | Pseudomonas putida | Xylene | Kong et al., 2005 |
Efforts on microbe selection should also be placed on the identification of microbes that could remediate PM, SO2, NO2, and O3. As mentioned above, a group of microbes can assimilate SO2 and NO2, further research should explore those microbes for effective assimilation of the two pollutants. Thus far, it appears that no information is available regarding microbial remediation of PM and O3, which may not be the case in the nature. Extensive research should be conducted to determine if nature has offered microbes that can break down PMs and can also biodegrade or biotransform O3.
Selected microbes could be domesticated by growing them in different cultures varying in pH, carbon source, temperature, and O2 to identify appropriate culture media and conditions for maximizing their growth. Morphological characterization and internal transcribed spacer rDNA analysis should be conducted to determine their phylogenetic relationships with other microbes. Their ability to biodegrade particular or a group of air pollutants should be evaluated in the laboratory. Microbial characteristics including their utilization of organic compounds, decomposition rate of pollutants, adaptability, competition, and growth rate should be recorded and analyzed. Competitive strains that show promise in bioremediation should be identified. A series of bacterial and filamentous fungal genomes have been sequenced recently. More than hundreds of bacterial and fungal transcriptomic and proteomic datasets are available. With the advent of increasingly sophisticated bioinformatics and genetic manipulation tools, mechanisms underlying the biodegradation or transformation of pollutants by the isolated microbes could be elucidated. This information, in turn, will significantly improve our understanding of the microbes and provide us with molecular bases for manipulation of the microbes for enhancing phylloremediation.
Evaluation of the compatibility between plant leaves and microbes
Plants selected from the four categories should be inoculated with selected microbes to determine the compatibility of each selected microbe with each selected plant species. The test could begin first in laboratory settings using entire leaves in designated chambers or utilizing young seedlings in relative large growth chambers to evaluate if inoculated microbes could grow on leaf surfaces and if the specific inoculation affects plant growth. Compatible combinations would be exposed to pollutants at different concentrations and durations to determine the potential for pollutant reduction. A microbe that is compatible with one plant species may not be compatible with another. For example, B. cereus ERBP isolated from roots of C. ternatea was compatible with the leaf surface of Z. zamifolia but not with the leaf surface of Euphorbia milii. ERBP-colonized Z. zamifolia grew well and showed high efficiency in removal of formaldehyde, but ERBP-colonized E. milii were less effective in removal formaldehyde and the plants exhibited stress symptom (Khaksar et al., 2016a). Laboratory evaluation will generate a large number of plant-microbe combinations that are specifically effective in removal of a particular pollutant or a particular group of pollutants. Bacteria would be propagated using bioreactors and corresponding plants would be propagated through either cuttings or tissue culture. The plants would be transplanted into greenhouses or specific regions with air pollution for testing the effectiveness of the combinations in real-world situations.
Plants and microbe combinations that pass the real-world test will be investigated using the next-generation sequencing (NGS) technologies (metagenomics, metatranscriptomics, metaproteomics, and metabolomics) and the rapid evolution of SIP (Stable isotope probing) for identifying molecular mechanisms underlying microbial and plant interactions in facilitation of phylloremediation. The compatibility evaluation and molecular analysis would ultimately result in the development of protocols for culturing microbes and producing corresponding plants. Some protocols will be catered to trees, others used for shrubs or small trees. Some would be effective for improving groundcover plants, and some will be used for indoor foliage plants. Effectiveness of each protocol in remediation of particular or general pollutants would be determined using the model described by Nowak et al. (2006). If the test is to be conducted in a large scale, satellite image acquisition and analysis should be used. The analysis of the data will finally validate the protocols, i.e., particular plants can be inoculated with a specific group of microbes for use in remediation of a particular pollutant or a mixture of pollutants.
Implementation of phylloremediation technologies
The protocols will be implemented for phylloremediation. We propose three types of plantscape: (1) manufactory plantscape, (2) urban plantscape, and (3) interior plantscape. The plantscape for manufactories and cities should have three levels of greening: the sky with trees, the ground with groundcover plants, and shrubs in between. Additionally, climber plants can be used to build green walls and small trees and shrubs as well as groundcovers can be used to build green roofs. For interior plantscape, each room should have a minimum of one potted foliage plant. Foliage plants can also be used to install green walls in interior environments for enhance remediation of indoor air pollutants.
The implementation of phylloremediation technologies should also take landscape design concepts into consideration, resulting greenbelts, green parks, green walls that fulfill roles not only for air remediation but also for recreation. Depending on the occurrence of pollutants and the scale and degree of the overall pollution, relevant protocols to the particular situations would be implemented. The remediation efficiency could be monitored over time using specific models in connection with satellite imagine data to determine how much of individual pollutants have been removed.
Conclusion
Air pollution is real, and it is adversely affecting human comfort and health and jeopardizing the ecosystem. The causes are multidimensional including increased population, urbanization, and industrialization accompanied with increased energy consumption and economic growth along with weak regulation, deforestation, and climate change. A recent article published by Cai et al. (2017) suggested that circulation changes including the weakening of the East Asia winter monsoon induced by global greenhouse gas emission contribute to the increased frequency and persistence of the haze weather conditions in Beijing, China. This claim could be true. The fact is that air pollutants released anthropogenically has caused the global warming. Our attention nevertheless should focus on how to control the emissions and how to remediate the pollutants. Although rhizosphere (roots and root associated microbes) contributes greatly to remediation of air pollutants, in this review, we specifically discuss phylloremediation. The role of plant leaves and leaf-associated microbes in remediation of air pollutants has not been well explored. Using the Urban Forest Effects Model, Yang et al. (2005) studied the influence of the urban forest on air quality in Beijing, China and found that the 2.4 million trees in the central part of Beijing removed 1,261.4 tons of pollutants from the air in 2002, of which 720 tons were PM. Nowak et al. (2014) has shown that computer simulations with local environmental data reveal that trees and forests in the contiguous US removed 17.4 million tons (t) of air pollution in 2010, with human health effects valued at 6.8 billion US dollars. Such forest-aided remediation might have avoided more than 850 incidences of human mortality and 670,000 incidences of acute respiratory problems.
We believe that phylloremediation is an environmentally friendly, cost effective way of remediation of air pollutants. The key component of this technology lies in plants. It is plants that can adsorb or absorb pollutants and plants that support microbes in biodegradation or biotransformation of pollutants. To develop phylloremediation technologies, some basic questions should be addressed: (1) Anatomical, physiological, biochemical and molecular mechanisms underlying plant responses to each pollutant should be investigated. Previous research has documented plant responses to pollutants such as NOx, SO2, O3, and VOCs, but the research was largely intended to identify how plants were injured. We need to exploit why many plants are tolerant to the pollutants, what are the underling mechanisms, and how can we manipulate the mechanisms for increased tolerance and for use in phylloremediation. There is little information regarding plant responses to PM. Do plants simply adsorb PM? What are the fates of stomatal absorbed PM? (2) Phyllosphere microbes are still largely a mystery and many are not culturable. Methods for collection, identification, and cultivation should be developed. Some microbes isolated from the rhizosphere can also be used for leaf colonization. Mechanisms for biodegradation and transformation of pollutants have been mentioned in this review. However, we still do not know if there are microbes that can remediate PM and O3. An important question that should be immediately addressed is the roles of microbes within the PM. Do the microbes become active once settled on leaves? Do they have the ability to break down the PM? With the advances of omics, these questions will be answered, and new strains with high efficiency in breaking down pollutants are expected to be isolated and utilized. (3) A large scale and intensive test for the compatibility among identified plants and identified microbes should be carried out. Specific plant-microbe groups or combinations that can effectively reduce one or more pollutants should be identified, tested, and confirmed in real-world situations and corresponding protocols for using each combination should developed. (4) New methods for analyzing dynamic changes of air pollutants in the atmosphere should be developed and standardized for monitoring the effectiveness of the phyllosphere technologies. (5) Research and development of phyllosphere technologies is a multidisciplinary project requiring collaboration among researchers with different academic backgrounds at regional, national, and international levels. Nature has offered healthy alternatives for remediation of air pollution; we should collaborate with nature as a partner to restore nature's identity.
Author contributions
All authors contributed to the acquisition and interpretation of available literature and the conception of the work. JC, SL, and XW wrote the manuscript, and all authors reviewed and revised the manuscript and approved this final version. XW and SL contributed equally to this work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Fujian Science and Technology Key Projects (2013NZ0002-1B) for “Construction of High-level University program of Fujian Agriculture and Forestry University” and for “Construction of High-level Horticulture Science Discipline” (612014007) for supporting this study. The appreciation also extends to Mr. He Hong at College of Art and Landscape Architecture, Fujian Agriculture and Forestry University for assistance in preparation of Figure 1.
References
- Abhilash P. C., Jamil S., Singh N. (2009). Transgenic plants for enhanced biodegradation and phytoremediation of organic xenobiotics. Biotechnol. Adv. 27, 474–488. 10.1016/j.biotechadv.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Aemprapa S., Williams P. A. (1998). Implications of the xylQ gene of TOL plasmid pWW102 for the evolution of aromatic catabolic pathways. Microbiology 144, 1387–1396. 10.1099/00221287-144-5-1387 [DOI] [PubMed] [Google Scholar]
- Alagappan G., Cowan R. M. (2003). Substrate inhibition kinetics for toluene and benzene degrading pure cultures and a method for collection and analysis of respirometric data for strongly inhibited cultures. Biotechnol. Bioeng. 83, 798–809. 10.1002/bit.10729 [DOI] [PubMed] [Google Scholar]
- Al-Awadhi H., Al-Mailem D., Dashti N., Hakam L., Eliyas M., Radwan S. (2012). The abundant occurrence of hydrocarbon-utilizing bacteria in the phyllospheres of cultivated and wild plants in Kuwait. Int. Biodeter. Biodegr. 73, 73–79. 10.1016/j.ibiod.2012.05.016 [DOI] [Google Scholar]
- Alghamdi M. A., Shamy M., Redal M. A., Khoder M., Awad A. H., Elserougy S. (2014). Microorganisms associated particulate matter: a preliminary study. Sci. Total Environ. 479, 109–116. 10.1016/j.scitotenv.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Ali N., Al-Awadhi H., Dashti N., Khanafer M., El-Nemr I., Sorkhoh N., et al. (2015). Bioremediation of atmospheric hydrocarbons via bacteria naturally associated with leaves of higher plants. Int. J. Phytoremed. 17, 1160–1170. 10.1080/15226514.2015.1045125 [DOI] [PubMed] [Google Scholar]
- Ali N., Sorkhoh N., Salamah S., Eliyas M., Radwan S. (2012). The potential of epiphytic hydrocarbon-utilizing bacteria on legume leaves for attenuation of atmospheric hydrocarbon pollutants. J. Environ. Manage. 93, 113–120. 10.1016/j.jenvman.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Amann M. (2008). Health Risks of Ozone from Long-Range Transboundary Air Pollution. Copenhagen: WHO Regional Office Europe. [Google Scholar]
- Anderson J. O., Thundiyil J. G., Stolbach A. (2012). Clearing the air: a review of the effects of particulate matter air pollution on human health. J. Med. Toxicol. 8, 166–175. 10.1007/s13181-011-0203-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald A. T., Folberth G., Wade D. C., Scott D. (2017). A world avoided: impacts of changes in anthropogenic emissions on the burden and effects of air pollutants in Europe and North America. Farad. Discuss. [Epub ahead of print]. 10.1039/C7FD00004A [DOI] [PubMed] [Google Scholar]
- Baldacchini C., Castanheiro A., Maghakyan N., Sgrigna G., Verhelst J., Alonso R., et al. (2017). How does the amount and composition of PM deposited on Platanus acerfolia leaves change across different cities in Europe. Environ. Sci. Technol. 51, 1147–1156. 10.1021/acs.est.6b04052 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Ghoshal A. K. (2010). Phenol degradation by Bacillus cereus: pathway and kinetic modeling. Bioresour. Technol. 101, 5501–5507. 10.1016/j.biortech.2010.02.018 [DOI] [PubMed] [Google Scholar]
- Barac T., Taghavi S., Borremans B., Povoost A., Oeyen L., Colpaert J. V., et al. (2004). Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 22, 583–588. 10.1038/nbt960 [DOI] [PubMed] [Google Scholar]
- Battista G., Pagliaroli T., Mauri L., Basilicata C., De Lieto Vollaro R. (2016). Assessment of the air pollution level in the city of Rome (Italy). Sustainability 8:838 10.3390/su8090838 [DOI] [Google Scholar]
- Beckett K. P., Freer-Smith P., Taylor G. (2000). Particulate pollution capture by urban trees: effect of species and windspeed. Glob. Change Biol. 6, 995–1003. 10.1046/j.1365-2486.2000.00376.x [DOI] [Google Scholar]
- Berg G., Mahnert A., Moissl-Eichinger C. (2014). Beneficial effects of plant-associated microbes on indoor microbiomes and human health? Front. Microbiol. 5:15. 10.3389/fmicb.2014.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloem E., Haneklaus S., Schnug E. (2015). Milestones in plant sulfur research on sulfur-induced-resistance (SIR) in Europe. Front. Plant Sci. 5:779. 10.3389/fpls.2014.00779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N., Horton M. W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8:e56329. 10.1371/journal.pone.0056329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers R. M., McLetchie S., Knight R., Fierer N. (2011). Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5, 601–612. 10.1038/ismej.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim H., Osborne J. P., Kostandarithes H. M., Fredrickson J. K., Wackett L. P., Daly M. J. (2006). Deinococcus radiodurans engineered for complete toluene degradation facilitates Cr(VI) reduction. Microbiology 152, 2469–2477. 10.1099/mic.0.29009-0 [DOI] [PubMed] [Google Scholar]
- Bringel F., Couee I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6:486. 10.3389/fmicb.2015.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockherhoff M. P. (2000). An urbanizing world. Popul. Bull. 55, 3–44. [Google Scholar]
- Brown S., Sim M. R., Abramson M. J., Gray C. N. (1994). Concentrations of volatile organic compounds in indoor air–a review. Indoor Air 4, 123–134. 10.1111/j.1600-0668.1994.t01-2-00007.x [DOI] [Google Scholar]
- Bufalini M. (1971). Oxidization of sulfur dioxide in polluted atmospheres-a review. Environ. Sci. Technol. 5, 685–700. 10.1021/es60055a001 [DOI] [Google Scholar]
- Cai W., Li K., Liao H., Wang H., Wu L. (2017). Weather conditions coducive to Beigjing cerver haze more frequent sunder climate change. Nat. Climate Change 7, 257–262. 10.1038/nclimate3249 [DOI] [Google Scholar]
- Cao C., Jiang W., Wang B., Fang J., Lang J., Tian G., et al. (2014). Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe somg event. Environ. Sci. Technol. 48, 1499–1507. 10.1021/es4048472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J., Clemons J. (2006). Do urban forests enhance air quality. Aust. J. Environ. Manag. 13, 120–130. 10.1080/14486563.2006.10648678 [DOI] [Google Scholar]
- Chen J., McConnell D. B., Norman D. J., Henny R. J. (2005). The foliage plant industry, in Horticultural Reviews, ed Janick J. (Hoboken, NJ: John Wiley and Sons, Inc.), 45–110. [Google Scholar]
- Chen L., Liu C., Zhang L., Zou R., Zhang Z. (2017). Variation in tree species ability to capture and retain airborne fine particulate matter (PM2.5). Sci. Rep. 7:3206. 10.1038/s41598-017-03360-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. M., Yurimoto H., Li K. Z., Orita I., Akita M., Kato N., et al. (2010). Assimilation of formaldehyde in transgenic plants due to the introduction of the bacterial ribulose monophosphate pathway genes. Biosci. Biotechnol. Biochem. 74, 627–635. 10.1271/bbb.90847 [DOI] [PubMed] [Google Scholar]
- Chen Z., Barros C. P., Gil-Alana L. A. (2016). The persistence of air pollution in four mega-cities of China. Habitat Int. 56, 103–108. 10.1016/j.habitatint.2016.05.004 [DOI] [Google Scholar]
- Choi S. D., Staebler R. M., Li H., Su Y., Gevao B., Harner T., et al. (2008). Depletion of gaseous polycyclic aromatic hydrocarbons by a forest canopy. Atmos. Chem. Phys. 8, 4105–4113. 10.5194/acp-8-4105-2008 [DOI] [Google Scholar]
- Chung Y. C., Chung P. L., Liao S. W. (2010). Carbon fixation efficiency of plants influenced by sulfur dioxide. Environ. Monit. Assess. 173, 701–707. 10.1007/s10661-010-1416-5 [DOI] [PubMed] [Google Scholar]
- Coince A., Caël O., Bach C., Lengellé J., Cruaud C., Gavory F., et al. (2013). Below-ground fine-scale distribution and soil versus fine root detection of fungal and soil oomycete communities in a French beech forest. Fungal Ecol. 6, 223–235. 10.1016/j.funeco.2013.01.002 [DOI] [Google Scholar]
- Cordier T., Robin C., Capdevielle X., Fabreguettes O., Desprez-Loustau M. L., Vacher C. (2012). The composition of phyllosphere fungal assemblages of European beech (Fagus sylvatica) varies significantly along an elevation gradient. New Phytol. 196, 510–519. 10.1111/j.1469-8137.2012.04284.x [DOI] [PubMed] [Google Scholar]
- Cruz M. D., Christensen J. H., Thomsen J. D., Muller R. (2014a). Can ornamental potted plants remove volatile organic compounds from indoor air? A review. Environ. Sci. Pollut. Res. Int. 21, 13909–13928. 10.1007/s11356-014-3240-x [DOI] [PubMed] [Google Scholar]
- Cruz M. D., Muller R., Svensmark B., Pedersen J. S., Christensen J. H. (2014b). Assessment of volatile organic compound removal by indoor plants - a novel experimental setup. Environ. Sci. Pollut. Res. 21, 7838–7846. 10.1007/s11356-014-2695-0 [DOI] [PubMed] [Google Scholar]
- Cunningham S. D., Berti W. R., Huang J. W. (1995). Phytoremediation of contaminated soils. Trends Biotechnol. 13, 393–397. 10.1016/S0167-7799(00)88987-8 [DOI] [Google Scholar]
- Dahms T. E., Younis L. T., Wiens R. D., Zarnegar S., Byers S. L., Chaitman B. R. (1993). Effects of carbon monoxide exposure in patients with documented cardiac arrhythmias. J. Am. Coll. Cardiol. 21, 442–450. 10.1016/0735-1097(93)90687-V [DOI] [PubMed] [Google Scholar]
- Deeb R. A., Alvarez-Cohen L. (1999). Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotech. Bioeng. 62, 526–536. [DOI] [PubMed] [Google Scholar]
- De Kempeneer L., Sercu B., Vanbrabant W., Van Langenhove H., Verstraete W. (2004). Bioaugmentation of the phyllosphere for the removal of toluene from indoor air. Appl. Microbiol. Biotechnol. 64, 284–288. 10.1007/s00253-003-1415-3 [DOI] [PubMed] [Google Scholar]
- De Kok L. J., Durenkamp M., Yang L., Stulen I. (2007). Atmospheric sulfur, in Sulfur in Plants An Ecological Perspective, eds Hawkesford M. J., De Kok L. J. (Dordrecht: Springer; ), 91–106. [Google Scholar]
- Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 16428–16433. 10.1073/pnas.0905240106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo P., Brune W. H., Martinez M., Harder H., Lesher R., Ren X. (2004). Missing OH reactivity in a forest: evidence for unknown reactive biogenic VOCs. Science 304, 722–725. 10.1126/science.1094392 [DOI] [PubMed] [Google Scholar]
- Doty S. L., James C. A., Moore A. L., Vajzovic A., Singleton G. L., Ma C., et al. (2007). Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. Proc. Natl. Acad. Sci. U.S.A. 104, 16816–16821. 10.1073/pnas.0703276104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty S. L., Shang T. Q., Wilson A. M., Tangen J., Westergreen A. D., Newman L. A., et al. (2000). Enhanced metabolism of halogenated hydrocarbons in transgenic plants containing mammalian cytochrome P450 2E1. Proc. Natl. Acad. Sci. U.S.A. 97, 6287–6291. 10.1073/pnas.97.12.6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow G. J., Berry J. A., Bergmann D. C. (2014). The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsisi thaliana. New Phytol. 201, 1205–1217. 10.1111/nph.12586 [DOI] [PubMed] [Google Scholar]
- Duan K., Sun G., Zhang Y., Yahya K., Wang K. M., Madden J. M. (2017). Impact of air pollution induced climate change on water availability and ecosystem productivity in the conterminous United States. Clim. Chang. 140, 259–272. 10.1007/s10584-016-1850-7 [DOI] [Google Scholar]
- Dulla G., Marco M., Quinones B., Lindow S. (2005). A closer look at Psuedomonas syringe as leaf colonist- The pathogen P-syringae thrives on healthy plants by employing quorum sensing, virulence factors, and other traits. ASM News 71, 469–475. [Google Scholar]
- Eberlein-Konig B., Przybilla B., Kuhnl P., Pechak J., Gebefugi I., Kleinschmidt J., et al. (1998). Influence of airborne nitrogen dioxide or formaldehyde on parameters of skin function and cellular activation in patients with atopic eczema and control subjects. J. Allergy. Clin. Immunol. 101, 141–143. 10.1016/S0091-6749(98)70212-X [DOI] [PubMed] [Google Scholar]
- Edwards R., Dixon D. P., Cummins I., Brazier-Hicks M., Skipsey M. (2011). New perspectives on the metabolism and detoxification of synthetic compounds in plants, in Organic Xenobiotics and Plants, eds Schrode P., Collins C. D. (New York, NY: Springer; ), 125–148. [Google Scholar]
- Endo S., Matsunaga E., Yamada K., Evinuma H. (1997). Genetic engineering for air-pollutant resistance in hybrid aspen, in Micropropagation, Genetic Engineering, and Molecular Biology of Populus, eds Klopfenstein N. B., Chun Y. W., Kim M. S., Ahuja M. R. Gen Tech. Rep (Fort Collins, CO: USDA; ), 187–191. [Google Scholar]
- Eriksson M., Swartling A., Dalhammar G., Fäldt J., Borg-Karlson A.-K. (1998). Biological degradation of diesel fuel in water and soil monitored with solid-phase microextraction and GC-MS. Appl. Microbiol. Biotechnol. 50, 129–124. 10.1007/s002530051267 [DOI] [PubMed] [Google Scholar]
- Fenn M. E., Dunn P. H., Durall D. M. (1989). Effects of ozone and sulfur dioxide on phyllosphere fungi from three tree species. Appl. Environ. Microbiol. 55, 412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., McCain C. M., Meir P., Zimmermann M., Rapp J. M., Silman M. R., et al. (2011). Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92, 797–804. 10.1890/10-1170.1 [DOI] [PubMed] [Google Scholar]
- Finkel O. M., Burch A. Y., Elad T., Huse S. M., Lindow S. E., Post A. F., et al. (2012). Distance-decay relationships partially determine diversity patterns of phyllosphere bacteria on Tamrix trees across the Sonoran Desert. Appl. Environ. Microbiol. 78, 6187–6193. 10.1128/AEM.00888-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel O. M., Burch A. Y., Lindow S. E., Post A. F., Belkin S. (2011). Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Environ. Microbiol. 77, 7647–7655. 10.1128/AEM.05565-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forczek S. T., Uhlirova H., Gryndler M., Albrechtova J., Fuksova K., Vagner M., et al. (2004). Trichloroacetic acid in Norway spruce/soil-system. II. Distribution and degradation in the plant. Chemosphere 56, 327–333. 10.1016/j.chemosphere.2004.01.018 [DOI] [PubMed] [Google Scholar]
- Fugas M., Sega K. (1995). Assessment of human inhalation exposure to polycyclic aromatic hydrocarbons. J. Exp. Anal. Environ. Epidemiol. 6, 439–447. [PubMed] [Google Scholar]
- Fujita M., Ike M., Hioki J. I., Kataoka K., Takeo M. (1995). Trichloroethylene degradation by genetically engineered bacteria carrying cloned phenol catabolic genes. J. Ferment. Bioeng. 79, 100–106. 10.1016/0922-338X(95)94075-3 [DOI] [Google Scholar]
- Fürnkranz M., Wanek W., Richter A., Abell G., Rasche F., Sessitsch A. (2008). Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J. 2, 561–570. 10.1038/ismej.2008.14 [DOI] [PubMed] [Google Scholar]
- Gandolfi I., Canedoli C., Imperato V., Tagliaferri I., Gkorezis P., Vangronsveld J., et al. (2017). Diversity and hydrocarbon-degrading potential of epiphytic microbial communities on Platanus x acerifolia leaves in an urban area. Environ. Pollut. 220, 650–658. 10.1016/j.envpol.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Gao J., Fan X., Pan K., Li H., Sun L. (2016). Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Sci. Rep. 6:38785. 10.1038/srep38785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski S. W., Gawronska H., Lomnicki S., Saebo A., Vangronsveld J. (2017). Plants in air phytoremediation. Adv. Bot. Res. 83, 319–346. 10.1016/bs.abr.2016.12.008 [DOI] [Google Scholar]
- Germaine K. J., Keogh E., Ryan D., Dowling D. N. (2009). Bacterial endophyte-mediated naphthalene phytoprotection and phytoremediation. FEMS Microbiol. Lett. 296, 226–234. 10.1111/j.1574-6968.2009.01637.x [DOI] [PubMed] [Google Scholar]
- Giese M., Bauer-Doranth U., Langebartels C., Sandermann H., Jr. (1994). Detoxification of formaldehyde by the spider plant (Chlorophytum comosum L.) and by soybean (Glycine max L.) cell-suspension cultures. Plant Physiol. 104, 1301–1309. 10.1104/pp.104.4.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Xie Y., Hong I., Kim J. (2001). Catalytic oxidation of NO to NO 2 on activated carbon. Energy Convers. Manage. 42, 2005–2018. 10.1016/S0196-8904(01)00058-9 [DOI] [Google Scholar]
- Gurjar B. R., Ravindra K., Nagpure A. S. (2016). Air pollution trends over India megacities and their local-to-global implications. Atm. Environ. 142, 475–495. 10.1016/j.atmosenv.2016.06.030 [DOI] [Google Scholar]
- Haas D., Galler H., Luxner J., Zarfel G., Buzina W., Friedl H., et al. (2013). The concentrations of cultural microorganisms in relation to particulate matter in urban air. Atom. Environ. 65, 215–222. 10.1016/j.atmosenv.2012.10.031 [DOI] [Google Scholar]
- Hao J., Zhu T., Fan X. (2014). Indoor air pollution and its control in China, in Indoor Air Pollution, 2nd Edn., eds Pluschke P., Schleibinger H. (Berlin; Heidelberg: Springer-Verlag; ), 1–26. [Google Scholar]
- Harayama S., Rekik M. (1990). The meta cleavage operon of TOL degradative plasmid pWWO comprises 13 genes. Mol. Gen. Genet. 221, 113–120. 10.1007/BF00280375 [DOI] [PubMed] [Google Scholar]
- Hetherington A. M., Woodward F. I. (2003). The role of stomata in sensing and driving environmental change. Nature 424, 901–908. 10.1038/nature01843 [DOI] [PubMed] [Google Scholar]
- Ho Y. N., Hsieh J. L., Huang C. C. (2013). Construction of a plant-microbe phytoremediation system: combination of vetiver grass with a functional endophytic bacterium, Achromobacter xylosoxidans F3B, for aromatic pollutants removal. Bioresour. Technol. 145, 43–47. 10.1016/j.biortech.2013.02.051 [DOI] [PubMed] [Google Scholar]
- Ho Y. N., Mathew D. C., Hsiao S. C., Shih C. H., Chien M. F., Chiang H. M., et al. (2012). Selection and application of endophytic bacterium Achromobacter xylosoxidans strain F3B for improving phytoremediation of phenolic pollutants. J. Hazard. Mater. 219, 43–49. 10.1016/j.jhazmat.2012.03.035 [DOI] [PubMed] [Google Scholar]
- Horn J. M., Harayama S., Timmis K. N. (1991). DNA sequence determination of the TOL plasmid (pWW0) xylGFJ genes of Pseudomonas putida: implications for the evolution of aromatic catabolism. Mol. Microbiol. 5, 2459–2474. 10.1111/j.1365-2958.1991.tb02091.x [DOI] [PubMed] [Google Scholar]
- Huang J. W., Chen J., Berti W. R., Cunningham S. D. (1997). Phytoremediation of lead-contaminated soils: role of synthetic chelates in lead phytoextraction. Environ. Sci. Technol. 31, 800–805. 10.1021/es9604828 [DOI] [Google Scholar]
- Huang R.-J., Zhang Y., Bozzetti C., Ho K.-F., Cao J.-J., Han Y., et al. (2014). High secondary aerosol contribution to particulate pollution during haze events in China. Nature 514, 218–222. 10.1038/nature13774 [DOI] [PubMed] [Google Scholar]
- Huang Y., Ho S. S., Lu Y., Niu R., Xu L., Cao J., et al. (2016). Removal of indoor volatile organic compounds via photocatalytic oxidation: a short review and prospect. Molecules 21:56 10.3390/molecules21010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2013). Outdoor Air Pollution a Leading Environmental Cause of Cancer Deaths, No. 221. World Health Organization; Available online at: https://www.iarc.fr/en/media-centre/iarcnews/pdf/pr221_E.pdf [Google Scholar]
- Iguchi H., Sato I., Sakakibara M., Yurimoto H., Sakai Y. (2012). Distribution of methanotrophs in the phyllosphere. Biosci. Biotechnol. Biochem. 76, 1580–1583. 10.1271/bbb.120281 [DOI] [PubMed] [Google Scholar]
- Iimura Y., Ikeda S., Sonoki T., Hayakawa T., Kajita S., Kimbara K., et al. (2002). Expression of a gene for Mn-peroxidase from Coriolus versicolor in transgenic tobacco generates potential tools for phytoremediation. Appl. Environ. Microbiol. 59, 246–251. [DOI] [PubMed] [Google Scholar]
- Ijaz A., Imran A., Haq M. A., Khan Q. M., Afzal M. (2016). Phytoremediation: recent advances in plant-endophytic synergistic interactions. Plant Soil. 405, 179–195. 10.1007/s11104-015-2606-2 [DOI] [Google Scholar]
- Ilori M. O., Amund O. O., Ezeani C. J., Omoijiahina S., Adebusoye S. A. (2006). Occurrence and growth potentials of hydrocarbon degrading bacteria on the phylloplane of some tropical plants. Afr. J. Biotechnol. 5, 542–545. [Google Scholar]
- Ilyas S. Z., Khattak A. I., Nasir S. M., Qurashi T., Durrani R. (2009). Air pollution assessment in urban areas and its impact on human health in the city of Quetta, Pakistan. Clean Technolo. Environ. Policy. 12, 291–299. 10.1007/s10098-009-0209-4 [DOI] [Google Scholar]
- Inoue A., Yamamoto M., Horikoshi K. (1991). Pseudomonas putida which can grow in the presence of toluene. Appl. Environ. Microbiol. 57, 1560–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Nakamura K. (2000). Trichloroethylene degradation by Ralstonia sp. KN1-10A constitutively expressing phenol hydroxylase: transformation products, NADH limitation, and product toxicity. J. Biosci. Bioeng. 89, 438–445. 10.1016/S1389-1723(00)89093-3 [DOI] [PubMed] [Google Scholar]
- Iwakiri R., Yoshihira K., Ngadiman, Futagami T., Goto M., Furukawa K. (2004). Total degradation of pentachloroethane by an engineered Alcaligenes strain expressing a modified camphor monooxygenase and a hybrid dioxygenase. Biosci. Biotechnol. Biochem. 68, 1353–1356. 10.1271/bbb.68.1353 [DOI] [PubMed] [Google Scholar]
- Jaenicke R. (2005). Abundance of cellular material and proteins in the atmosphere. Science. 308, 73–73. 10.1126/science.1106335 [DOI] [PubMed] [Google Scholar]
- James C. A., Xin G., Doty S. L., Strand S. E. (2008). Degradation of low molecular weight volatile organic compounds by plants genetically modified with mammalian cytochrome P450 2E1. Environ. Sci. Technol. 42, 289–293. 10.1021/es071197z [DOI] [PubMed] [Google Scholar]
- Jin Y., Veiga M., Kennes C. (2006). Performance optimization of the fungal biodegradation of α-pinene in gas-phase biofilter. Process Biochem. 41, 1722–1728. 10.1016/j.procbio.2006.03.020 [DOI] [Google Scholar]
- Jud W., Fischer L., Canaval E., Wohlfahrt G., Tissier A., Hansel A. (2016). Plant surface reactions: an opportunistic ozone defence mechanism impacting atmospheric chemistry. Atmos. Chem. Phys. 16, 277–292. 10.5194/acp-16-277-2016 [DOI] [Google Scholar]
- Jumpponen A., Jones K. L. (2010). Seasonally dynamic fungal communities in the Quercus macrocarp phyllosphere differ between urban and nonurban environments. New Phytol. 186, 496–513. 10.1111/j.1469-8137.2010.03197.x [DOI] [PubMed] [Google Scholar]
- Kahraman H., Geckil H. (2005). Degradation of benzene, toluene and xylene by Pseudomonas aeruginosa Engineered with the Vitreoscilla Hemoglobin Gene. Eng. Life Sci. 5, 363–368. 10.1002/elsc.200520088 [DOI] [Google Scholar]
- Kampa M., Castanas E. (2008). Human health effects of air pollution. Environ. Pollut. 151, 362–367. 10.1016/j.envpol.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Kapley A., Purohit H. J., Chhatre S., Shanker R., Chakrabarti T., Khanna P. (1999). Osmotolerance and hydrocarbon degradation by a genetically engineered microbial consortium. Bioresour. Technol. 67, 241–245. 10.1016/S0960-8524(98)00121-7 [DOI] [Google Scholar]
- Karagiannidis A., Poupkou A., Giannaros T., Giannaros C., Melas D., Argiriou A. (2014). The air quality of a mediterranean urban environment area and its relation to major meteorological parameters. Water Air Soil Pollut. 226:2239 10.1007/s11270-014-2239-8 [DOI] [Google Scholar]
- Kato N., Yurimoto H., Thauer R. K. (2006). The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci. Biotechnol. Biochem. 70, 10–21. 10.1271/bbb.70.10 [DOI] [PubMed] [Google Scholar]
- Kelly F. J., Fussell J. C. (2015). Air pollution and public health: emerging hazards and improved understanding of risk. Environ. Geochem. Health. 37, 631–649. 10.1007/s10653-015-9720-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel S. W., Mueller R. C. (2014). Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92, 303–311. 10.1139/cjb-2013-0194 [DOI] [Google Scholar]
- Kembel S. W., O'connor T. K., Arnold H. K., Hubbell S. P., Wright S. J., Green J. L. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U.S.A. 111, 13715–13720. 10.1073/pnas.1216057111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennes C., Veiga M. C. (2004). Fungal biocatalysts in the biofiltration of VOC-polluted air. J. Biotechnol. 113, 305–319. 10.1016/j.jbiotec.2004.04.037 [DOI] [PubMed] [Google Scholar]
- Khaksar G., Treesubsuntorn C., Thiravetyan P. (2016a). Effect of endophytic Bacillus cereus ERBP inoculation into non-native host: potentials and challenges for airborne formaldehyde removal. Plant Physiol. Biochem. 107, 326–336. 10.1016/j.plaphy.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Khaksar G., Treesubsuntorn C., Thiravetyan P. (2016b). Endophytic Bacillus cereus ERBP-Clitoria ternatea interactions: potentials for the enhancement of gaseous formaldehyde removal. Environ. Exp. Bot. 126, 10–20. 10.1016/j.envexpbot.2016.02.009 [DOI] [Google Scholar]
- Khan Z., Doty S. (2011). Endophyte-assisted phytoremediation. Cur. Topics Plant Biol. 12, 97–105. [Google Scholar]
- Khan Z., Roman D., Kintz T., delas Alas M., Yap R., Doty S. (2014). Degradation, phytoprotection and phytoremediation of phenanthrene by endophyte Pseudomonas putida, PD1. Environ. Sci. Technol. 48, 12221–12228. 10.1021/es503880t [DOI] [PubMed] [Google Scholar]
- Kim K. J., Jeong M., Lee D. W., Song J. S., Kim H. D., Yoo E. H. (2010). Variation in formaldehyde removal efficiency among indoor plant species. HortScience. 45, 1485–1498. [Google Scholar]
- Kim K. J., Yoo E. H., Jeong M. I., Song J. S., Lee S. Y., Keys S. J. (2011). Changes in the phytoremediation potential of indoor plants with exposure to toluene. HortScience 46, 1646–1649. [Google Scholar]
- Kim M., Singh D., Lai-Hoe A., Go R., Abdul Rahim R., Ainuddin A. N., et al. (2012). Distinctive phyllosphere bacterial communities in tropical trees. Microb. Ecol. 63, 674–681. 10.1007/s00248-011-9953-1 [DOI] [PubMed] [Google Scholar]
- Kirkwood R. C. (1999). Recent developments in our understanding of the plant cuticle as a barrier to the foliar uptake of pesticides. Pestic. Sci. 55, 69–77. [DOI] [Google Scholar]
- Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. 10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong I. C., Park T. H., Ahn T. B., Burlage R. S. (2005). Bioluminescence activity and degradation kinetics of genetically engineered strain P-putida mt-2 RB 1401 in soil system with m-xylene. Soil Sediment Contam. 14, 135–142. 10.1080/15320380590911760 [DOI] [Google Scholar]
- Kukla M., Plociniczak T., Piotrowska-Seget Z. (2014). Diversity of endophytic bacteria in Lolium perenne and their potential to degrade petroleum hydrocarbons and promote plant growth. Chemosphere. 117, 40–46. 10.1016/j.chemosphere.2014.05.055 [DOI] [PubMed] [Google Scholar]
- Kumar S., Verma M. K., Srivastava A. K. (2013). Ultrafine particles in urban ambient air and their health perspectives. Rev. Environ. Health. 28, 117–128. 10.1515/reveh-2013-0008 [DOI] [PubMed] [Google Scholar]
- Laforest-Lapointe I., Messier C., Kembel S. W. (2016). Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome. 4:27. 10.1186/s40168-016-0174-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest-Lapointe I., Paquette A., Messier C., Kembel S. W. (2017). Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 546, 145–147. 10.1038/nature22399 [DOI] [PubMed] [Google Scholar]
- Lambais M., Crowley D., Cury J., Büll R., Rodrigues R. R. (2006). Bacterial diversity in tree canopies of the Atlantic forest. Science 312, 1917–1917. 10.1126/science.1124696 [DOI] [PubMed] [Google Scholar]
- Lambert W. (1996). Combustion pollution in indoor environments. Clin. Rev. Allergy. Imminol. 9, 83–104. [Google Scholar]
- Lambert W. E., Samet J. M., Hunt W. C., Skipper B. J., Schwab M., Spengler J. D. (1993). Nitrogen dioxide and respiratory illness in children. Part II: assessment of exposure to nitrogen dioxide. Res. Rep. Health Eff. Inst. 58, 33–50. [PubMed] [Google Scholar]
- Lange C. C., Wackett L. P., Minton K. W., Daly M. J. (1998). Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments. Nat. Biotechnol. 16, 929–933. 10.1038/nbt1098-929 [DOI] [PubMed] [Google Scholar]
- Last F. T. (1955). Seasonal incidence of Sporobolomyces on cereal leaves. Trans. Br. Mycol. Soc. 38, 221–239. 10.1016/S0007-1536(55)80069-1 [DOI] [Google Scholar]
- Law R. M., Mansfield T. A. (1982). Oxides of nitrogen and the greenhouse atmosphere, in Effects of Caseous Air Pollution in Agriculture and Horticuttiire, eds Unsworth M. H., Ormrod D. P. (London: Butterworth Scientific; ), 93–112. [Google Scholar]
- Lawrence A., Fatima N. (2014). Urban air pollution & its assessment in Lucknow City–the second largest city of North India. Sci. Total Environ. 488, 447–455. 10.1016/j.scitotenv.2013.10.106 [DOI] [PubMed] [Google Scholar]
- Liao Y., Zhou X., Yu J., Cao Y., Li X., Kuai B. (2006). The key role of chlorocatechol 1,2-dioxygenase in phytoremoval and degradation of catechol by transgenic Arabidopsis. Plant Physiol. 142, 620–628. 10.1104/pp.106.085936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. H., Ezzati M., Murray M. (2007). Tobacco smoke, indoor air pollution and tuberculosis: a systematic review and meta-analysis. PLoS Med. 4:e20. 10.1371/journal.pmed.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Brandl M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. 10.1128/AEM.69.4.1875-1883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Mauzerall D. L., Chen Q., Zhang Q., Song Y., Peng W., et al. (2016). Air polluant emissions from Chinese households: a major and underappreciated ambient pollutant source. Proc. Natl. Acad. Sci. U.S.A. 113, 7756–7761. 10.1073/pnas.1604537113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. C., Webster D. A., Wei M. B. (1996). Genetic engineering to contain the Vitreoscilla hemoglobin gene enhances degradation of benzoic acid by Xanthomonas maltophilia. Biotechnol. Bioeng. 49, 101–105. [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Mu Y. J., Zhu Y. G., Ding H., Arnes N. C. (2007). Which ornamental plant species effectively remove benzene from indoor air? Atm. Environ. 41, 650–654. 10.1016/j.atmosenv.2006.08.001 [DOI] [Google Scholar]
- Ljs U. (2016). Genetic Basis of Naphthalene and Phenanthrene Degradation by Phyllosphere Bacterial Strains Alcaligenes faecalis and Alcaligenes sp. 11SO. J. Bioremed. Biodeg. 7:333 10.4172/2155-6199.1000333 [DOI] [Google Scholar]
- Llewellyn D., Dixon M. (2011). Can plants really improve indoor air quality? in Comprehensive Biotechnology, 2nd Edn., ed Murray M. Y. (Burlington, ON: Academic Press; ), 331–338. [Google Scholar]
- Lüers G. H., Advani R., Wenzel T., Subramani S. (1998). The Pichia pastoris dihydroxyacetone kinase is a PTS1-containing, but cytosolic, protein that is essential for growth on methanol. Yeast 14, 759–771. [DOI] [PubMed] [Google Scholar]
- Lu Z., Streets D. G., Zhang Q., Wang S., Carmichael G. R., Cheng Y. F., et al. (2010). Sulfur dioxide emissions in China and sulfur trends in East Asia since 2000. Atmos. Chem. Phys. 10, 6311–6331. 10.5194/acp-10-6311-2010 [DOI] [Google Scholar]
- Ma Y., Oliveira R. S., Freitas H., Zhang C. (2016). Biochemical and molecular mechanisms of plant-microbe-metal interactions: revelance for phytoremediation. Front. Plant Sci. 7:918. 10.3389/fpls.2016.00918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson A. J., Simon H., Langdon R., Misenheimer D. (2017). A mixed integer programming model for national ambient air quality standards (NAAQS) attainment strategy analysis. Environ. Model. Soft. 91, 13–27. 10.1016/j.envsoft.2017.01.008 [DOI] [Google Scholar]
- Madhaiyan M., Poonguzhali S., Senthilkumar M., Lee J. S., Lee K. C. (2012). Methylobacterium gossipiicola sp. nov., a pink-pigmented, facultatively methylotrophic bacterium isolated from the cotton phyllosphere. Int. J. Syst. Evol. Microbiol. 62, 162–167. 10.1099/ijs.0.030148-0 [DOI] [PubMed] [Google Scholar]
- McDonald J. C. (1991). An epidemiological view of asbestos in buildings. Toxicol. Ind. Health. 7, 187–193. 10.1177/074823379100700521 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Suzuki H., Morikawa H. (2004). Attempted reduction of 1,2,3-thiadiazole-4-carboxylates with samarium/iodine in methanol. Unexpected ring enlargement to 1,2,5-trithiepan-4,6-dicarboxylates. Org. Biomol. Chem. 2, 2870–2873. 10.1039/b408195a [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Chalavi V., Novakova-Sura M., Laliberté J. F., Sylvestre M. (2007). Expression of bacterial biphenyl-chlorobiphenyl dioxygenase genes in tobacco plants. Biotechnol. Bioeng. 97, 496–505. 10.1002/bit.21188 [DOI] [PubMed] [Google Scholar]
- Molhave L. (1991). Indoor climate, air pollution, and human comfort. J. Expo. Anal. Environ. Epidemiol. 1, 63–81. [PubMed] [Google Scholar]
- Morgan K. T. (1997). A brief review of formaldehyde carcinogenesis in relation to rat nasal pathology and human health risk assessment. Toxicol. Pathol. 25, 291–307. 10.1177/019262339702500307 [DOI] [PubMed] [Google Scholar]
- Morikawa H., Higaki A., Nohno M., Takahashi M., Kamada M., Nakata M., et al. (1998). More than a 600-fold variation in nitrogen dioxide assimilation among 217 plant taxa. Plant Cell Environ. 21, 180–190. 10.1046/j.1365-3040.1998.00255.x [DOI] [Google Scholar]
- Morikawa H., Takahashi M., Kawamura Y. (2003). Metabolism and genetics of atmospheric nitrogen dioxide control using pollutant-philic plants, in Phytoremediation: Transformation and Control of Contaminants, eds McCutcheon S. C., Schnoor J. L. (Hoboken, NJ: John Wiley; ), 765–786. [Google Scholar]
- Morikawa H., Takahashi M., Sakamoto A., Matsubara T., Arimura G., Kawamura Y., et al. (2004). Formation of unidentified nitrogen in plants: an implication for a novel nitrogen metabolism. Planta 219, 14–22. 10.1007/s00425-003-1200-7 [DOI] [PubMed] [Google Scholar]
- Morikawa H., Takahashi M., Sakamoto A., Ueda-Hashimoto M., Matsubara T., Miyawaki K., et al. (2005). Novel metabolism of nitrogen in plants. Z. Naturforsch. 60C, 265–271 10.1515/znc-2005-3-411 [DOI] [PubMed] [Google Scholar]
- Mosaddegh M. H., Jafarian A., Ghasemi A., Mosaddegh A. (2014). Phytoremediation of benzens, toluene, ethylbenzene and xylene contaminated air by D. deremensis and O. microdasys plants. J. Environ. Health Sci. Eng. 12:39 10.1186/2052-336X-12-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J. G., Cerniglia C. E., Pritchard P. H. (1996). Bioremediation of environments contaminated by polycyclic aromatic hydrocarbons, in Bioremediation: Principles and Applications, eds Crawford E. L., Crawford D. L. (Cambridge: Cambridge University Press; ), 125–194. [Google Scholar]
- Müller T., Ruppel S. (2014). Progress in cultivation-indedependent phyllosphere microbiology. FEMS Microbiol. Ecol. 87, 2–17. 10.1111/1574-6941.12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., Stams A. J. M. (2008). The ecology and biotechnology of sulfate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. 10.1038/nrmicro1892 [DOI] [PubMed] [Google Scholar]
- Nadalig T., Farhan Ul Haque M., Roselli S., Schaller H., Bringel F., Vuilleumier S. (2011). Detection and isolation of chloromethane-degrading bacteria from the Arabidopsis thaliana phyllosphere, and characterization of chloromethane utilization genes. FEMS Microbiol. Ecol. 77, 438–448. 10.1111/j.1574-6941.2011.01125.x [DOI] [PubMed] [Google Scholar]
- Neill S. J., Desikan R., Hancock J. T. (2003). Nitric oxide signaling in plants. New Phytol. 159, 11–35. 10.1046/j.1469-8137.2003.00804.x [DOI] [PubMed] [Google Scholar]
- Nichols D., Cahoon N., Trakhtenberg E. M., Pham L., Mehta A., Belanger A., et al. (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 76, 2445–2450. 10.1128/AEM.01754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji M., Saito M., Nakamura M., Aono M., Saji H., Saito K. (2001). Cysteine synthase overexpression in tobacco confers tolerance to sulfur-containing environmental pollutants. Plant Physiol. 126, 973–980. 10.1104/pp.126.3.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D. J., Crane D. E., Stevens J. C. (2006). Air pollution removal by urban trees and shrubs in the United States. Urban For. Urban Gree. 4, 115–123. 10.1016/j.ufug.2006.01.007 [DOI] [Google Scholar]
- Nowak D. J., Hirabayashi S., Bodine A., Greenfield E. (2014). Tree and forest effects on air quality and human health in the United States. Environ. Pollut. 193, 119–129. 10.1016/j.envpol.2014.05.028 [DOI] [PubMed] [Google Scholar]
- Orita I., Sato T., Yurimoto H., Kato N., Atomi H., Imanaka T., et al. (2006). The rubulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the Archaeon Thermococcus kodakaraensis. J. Bacteriol. 188, 4698–4704. 10.1128/JB.00492-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottele M., can Bohemen H. D., Fraaij A. L. A. (2010). Quantifying the deposition of particulate matter on climber vegetation on living walls. Ecol. Eng. 36, 154–162. 10.1016/j.ecoleng.2009.02.007 [DOI] [Google Scholar]
- Ottesen A. R., Gonzalez P. A., White J. R., Pettengill J. B., Li C., et al. (2013). Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13:14 10.1186/1471-2180-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J. J., Zhang J. S. S. (2011). Critical review of catalytic oxidization and chemisorption methods for indoor formaldehyde removal. HVAC&R Res. 17, 476–503. 10.1080/10789669.2011.587587 [DOI] [Google Scholar]
- Perez I. B., Brown P. J. (2014). The role of ROS signaling in cross-tolerance: from model to crops. Front. Plant Sci. 5:754. 10.3389/fpls.2014.00754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel S., Beyer M., Knoche M. (2003). Surface characteristics of sweet cherry fruit: stomata-number, distribution, functionality and surface wetting. Sci. Hortic. 97, 265–278. 10.1016/S0304-4238(02)00207-8 [DOI] [Google Scholar]
- Petrini O. (1991). Fungal endophytes of tree leaves, in Microbial Ecology of the Leaves, eds Fokkema N. J., van den Heuvel I. (Cambridge: Cambridge University Press; ), 185–187. [Google Scholar]
- Phung D., Hien T. T., Linh H. N., Luong L. M., Morawska L., Chu C., et al. (2016). Air pollution and risk of respiratory and cardiovascular hospitalizations in the most populous city in Vietnam. Sci. Total Environ. 557, 322–330. 10.1016/j.scitotenv.2016.03.070 [DOI] [PubMed] [Google Scholar]
- Pokoma D., Zabranska J. (2015). Sulfur-oxidizing bacteria in environmental technology. Biotechnol. Adv. 33, 1246–1259. 10.1016/j.biotechadv.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Popek R., Gawronska H., Wrochna M., Gawronski S. W., Saebo A. (2013). Particulate matter on foliage of 13 woody species: deposition on surfaces and phytostabilisation in waxes–a 3-year study. Int. J. Phytoremed. 15, 245–256. 10.1080/15226514.2012.694498 [DOI] [PubMed] [Google Scholar]
- Prenafeta-Boldú F. X., Kuhn A., Luykx D. M. A. M., Anke H., van Groenestijn J. W., de Bont J. A. M. (2001). Isolation and characterisation of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol. Res. 105, 477–484. 10.1017/S0953756201003719 [DOI] [Google Scholar]
- Prenafeta-Boldú F. X., Vervoort J., Grotenhuis J. T. C., vanGroenestijn J. W. (2002). Substrate interactions during the biodegradation of benzene, toluene, ethylbenzene, and xylene (BTEX) hydrocarbons by the fungus Cladophialophora sp. strain T1. Appl. Environ. Microbiol. 68, 2660–2665. 10.1128/AEM.68.6.2660-2665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Zhang X., Jin H., Liu Y., Fan D., Cao Z., et al. (1993). Effects of indoor air pollution on respiratory illness of school children, in Proceedings of the Sixth International Conference on Indoor Air Quality and Climate, eds Jantunen M., Kalliokoski P., Kukkonen E., Saarela K., Seppänen A. (Helsinki: Indoor Air 93; ), 477–482. [Google Scholar]
- Qvit-Raz N., Finkel O. M., Al-Deeb T. M., Malkawi H. I., Hindiyeh M. Y., Jurkevitch E., et al. (2012). Biogeographical diversity of leaf-associated microbial communities from salt-secreting Tamarix trees of the Dead Sea region. Res. Microbiol. 163, 142–150. 10.1016/j.resmic.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Rai P. K. (2016). Impacts of particulate matter pollution on plants: implications for environmental biomonitoring. Ecotoxicol. Environ. Saf. 129, 120–136. 10.1016/j.ecoenv.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Rastogi G., Sbodio A., Tech J. J., Suslow T. V., Coaker G. L., Leveau J. H. (2012). Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 6, 1812–1822. 10.1038/ismej.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratola N., Alves A., Santos L., Lacorte S. (2011). Pine needles as passive bio-samplers to determine polybrominated diphenyl ether. Chemosphere 85, 207–252. 10.1016/j.chemosphere.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Redford A. J., Bowers R. M., Knight R., Linhart Y., Fierer N. (2010). The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 12, 2885–2893. 10.1111/j.1462-2920.2010.02258.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford A. J., Fierer N. (2009). Bacterial succession on the leaf surface: a novel system for studying succesional dynamics. Microb. Ecol. 58, 189–198. 10.1007/s00248-009-9495-y [DOI] [PubMed] [Google Scholar]
- Rennenberg H. (1984). The fate of excess sulfur in higher plants. Annu. Rev. Plant Physiol. 35, 121–153. 10.1146/annurev.pp.35.060184.001005 [DOI] [Google Scholar]
- Rico L., Ogaya R., Terradas J., Penuelas J. (2014). Community structures of N 2-fixing bacteria associated with the phyllosphere of a Holm oak forest and their response to drought. Plant Biol. 16, 586–593. 10.1111/plb.12082 [DOI] [PubMed] [Google Scholar]
- Rizwan S. A., Nongkynrih B., Gupta S. K. (2013). “Air pollution in Delhi”: its magnitude and effects on health. Indian J. Communtiy Med. 38, 4–8. 10.4103/0970-0218.106617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbø A., Popek R., Nawrot B., Hanslin H. M., Gawronska H., Gawronska S. W. (2012). Plant species differences in particulate matter accumulation on leaf surfaces. Sci. Total Environ. 427, 347–354. 10.1016/j.scitotenv.2012.03.084 [DOI] [PubMed] [Google Scholar]
- Salt D. E., Blaylock M., Kumer N. P. B. A., Dushenkov V., Ensley B. D., Chet I., et al. (1995). Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. Biotechnology 13, 468–474. 10.1038/nbt0595-468 [DOI] [PubMed] [Google Scholar]
- Sandhu A., Halverson L. J., Beattie G. A. (2007). Bacterial degradation of airborne phenol in the phyllosphere. Environ. Microbiol. 9, 383–392. 10.1111/j.1462-2920.2006.01149.x [DOI] [PubMed] [Google Scholar]
- Sandhu A., Halverson L. J., Beattie G. A. (2009). Identification and genetic characterization of phenol-degrading bacteria from leaf microbial communities. Microb. Ecol. 57, 276–285. 10.1007/s00248-008-9473-9 [DOI] [PubMed] [Google Scholar]
- Sangthong S., Suksabye P., Thiravetyan P. (2016). Air-borne xylene degradation by Bougainvillea buttiana and the role of epiphytic bacteria in the degradation. Ecotoxicol. Environ. Saf. 126, 273–280. 10.1016/j.ecoenv.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Scheublin T. R., Deusch S., Moreno-Forero S. K., Muller J. A., van der Meer J. R., Leveau J. H. (2014). Transcriptional profiling of Gram-positive Arthrobacter in the phyllosphere: induction of pollutant degradation genes by natural plant phenolic compounds. Environ. Microbiol. 16, 2212–2225. 10.1111/1462-2920.12375 [DOI] [PubMed] [Google Scholar]
- Scheublin T. R., Leveau J. H. (2013). Isolation of Arthrobacter species from the phyllosphere and demonstration of their epiphytic fitness. Microbiologyopen 2, 205–213. 10.1002/mbo3.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Hudson J. B. (2008). Ozone gas is an effective and practical antibacterial agent. Am. J. Infect Control. 36, 559–563. 10.1016/j.ajic.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Singh S. K., Rao D. N., Agrawal M., Pandey J., Narayan D. (1991). Air pollution tolerance index of plants. J. Environ. Manag. 32, 45–55. 10.1016/S0301-4797(05)80080-5 [DOI] [Google Scholar]
- Smejkalova H., Erb T. J., Fuchs G. (2010). Methanol assimilation in Methylobacterium extorquens AM1: demonstration of all enzymes and their regulation. PLoS ONE 5:e13001. 10.1371/journal.pone.0013001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets W., Wuyts K., Oerlemans E., Wuyts S., Denys S., Samson R., et al. (2016). Impact of urban land use on the bacterial phyllosphere of ivy (Hedera sp.). Atm. Envion. 147, 376–383. 10.1016/j.atmosenv.2016.10.017 [DOI] [Google Scholar]
- Song Y., Maher B. A., Li F., Wang X., Sun X., Zhang H. (2015). Particulate matter deposited on leaf of five evergreen species in Beijing, China: source identification and size distribution. Atm. Envion. 105, 53–60. 10.1016/j.atmosenv.2015.01.032 [DOI] [Google Scholar]
- Sorkhoh N. A., Al-Mailem D. M., Ali N., Al-Awadhi H., Salamah S., Eliyas M., et al. (2011). Bioremediation of volatile oil hydrocarbons by epiphytic bacteria associated with American grass (Cynodon sp.) and broad bean (Vicia faba) leaves. Int. Biodeterior. Biodegrad. 65, 797–802. 10.1016/j.ibiod.2011.01.013 [DOI] [Google Scholar]
- Sosa Alderete L. G., Talano M. A., Ibanez S. G., Purro S., Agostini E., Milrad S. R., et al. (2009). Establishment of transgenic tobacco hairy roots expressing basic peroxidases and its application for phenol removal. J. Biotechnol. 139, 273–279. 10.1016/j.jbiotec.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Sriprapat W., Boraphech P., Thiravetyan P. (2013). Factor affecting xylenecontaminated air removal by the ornamental plants Zamioculcas zamiifolia. Environ. Sci. Pollut. Res. 21, 2603–2610. 10.1007/s11356-013-2175-y [DOI] [PubMed] [Google Scholar]
- Sriprapat W., Suksabye P., Areephak S., Klantup P., Waraha A., Sawattan A., et al. (2014). Uptake of tolune and ethylbenzern by plants: removal of volatile indoor air contaminants. Ecotoxicol. Environ. Safety 102, 147–151. 10.1016/j.ecoenv.2014.01.032 [DOI] [PubMed] [Google Scholar]
- Steindorf K., Lubin J., Wichmann H. E., Becher H. (1995). Lung cancer deaths attributable to indoor radon exposure in West Germany. Int. J. Epidemiol. 24, 485–492. 10.1093/ije/24.3.485 [DOI] [PubMed] [Google Scholar]
- Stepniewska Z., Kuzniar A. (2013). Endophytic microorganisms-promising applications in bioremediation of greenhouse gases. Appl. Microbiol. Biotechnol. 97, 9589–9596. 10.1007/s00253-013-5235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. 10.1128/JB.00345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G., Daisy B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. R. 67, 491–502. 10.1128/MMBR.67.4.491-502.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga H., Nonaka K., Fujihara H., Goto M., Furukawa K. (2010). Hybrid pseudomonads engineered by two-step homologous recombination acquire novel degradation abilities toward aromatics and polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 88, 915–923. 10.1007/s00253-010-2840-8 [DOI] [PubMed] [Google Scholar]
- Sun K., Liu J., Gao Y., Jin L., Gu Y., Wang W. (2014). Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp. Ph6-gfp. Sci. Rep. 4:5462. 10.1038/srep05462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Andreassi A. K., Liu S., Pinto R., Triccas J. A., Leyh T. S. (2005). The trifunctional sulfate-activating complex (SAC) of Mycobacterium tuberculosis. J. Biol. Chem. 280, 7861–7866. 10.1074/jbc.M409613200 [DOI] [PubMed] [Google Scholar]
- Syranidou E., Christofilopoulos S., Gkavrou G., Thijs S., Weyens N., Vangronsveld J., et al. (2016). Exploitation of endophytic bacteria to enhance the phytoremediationpPotential of the wetland helophyte Juncus acutus. Front. Microbiol. 7:1016. 10.3389/fmicb.2016.01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Kidu Y. (2011). Glutathione-dependent formaldehyde dehydrogenase from golden pothos (Epipremnum aureum) and the production of formaldehyde detoxifying plants. Plant Biotechnol. 28, 373–378. 10.5511/plantbiotechnology.11.0620a [DOI] [Google Scholar]
- Tada Y., Matsuzaki T., Tanaka Y. (2010). Isolation and characterization of formaldehyde-responsive genes from golden pothos (Epipremnum aureum). Plant Biotechnol. 27, 325–331. 10.5511/plantbiotechnology.27.325 [DOI] [Google Scholar]
- Taghavi S., Barac T., Greenberg B., Borremans B., Vangronsveld J., van der Lelie D. (2005). Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl. Environ. Microbiol. 71, 8500–8505. 10.1128/AEM.71.12.8500-8505.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Morikawa H. (2001). Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol. 126, 731–741. 10.1104/pp.126.2.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Nakagawa M., Sakamoto A., Ohsumi C., Matsubara T., Morikawa H. (2005). Atmospheric nitrogen dioxide gas is a plant vitalization signal to increase plant size and the contents of cell constituents. New Phytol. 168, 129–154. 10.1111/j.1469-8137.2005.01493.x [DOI] [PubMed] [Google Scholar]
- Teper E. (2009). Dust-particle migration around flotation tailings ponds: pine needles as passive samplers. Environ. Monit. Assess. 154, 383–391. 10.1007/s10661-008-0405-4 [DOI] [PubMed] [Google Scholar]
- Terzaghi E., Wild E., Zacchello G., Cerabolini E. L., Jones K. V., Di Guardo A. (2013). Forest filter effect: role of leaves in capturing/releasing air particulate matter and its associated PAHS. Atm. Environ. 74, 378–384. 10.1016/j.atmosenv.2013.04.013 [DOI] [Google Scholar]
- Tian N., Liu F., Wang P., Zhang X., Li X., Wu G. (2017). The molecular basis of glandular trichome development and secondary metabolism in plants. Plant Gene. 12, 1–12. 10.1016/j.plgene.2017.05.010 [DOI] [Google Scholar]
- Timmis K. N., Steffan R. J., Unterman R. (1994). Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol. 48, 525–557. 10.1146/annurev.mi.48.100194.002521 [DOI] [PubMed] [Google Scholar]
- Toabaita M., Vangnai A. S., Thiravetyan P. (2016). Removal of ethylbenzene from contaminated air by Zamioculcas Zamiifolia and microorganisms associated on Z. Zamiifolia leaves. Water Air Soil Pollut. 227, 1–11. 10.1007/s11270-016-2817-z [DOI] [Google Scholar]
- Tsai Y. I., Yang H. H., Wang L. C., Huan J. L., Young L. H., Cheng M. T., et al. (2011). The influences of diesel particulate filter installation on air pollutant emissions for used vehicles. Aerosol Air Qual. Res. 11, 578–583. 10.4209/aaqr.2011.05.0066 [DOI] [Google Scholar]
- Ugrekhelidze D., Korte F., Kveitdze G. (1997). Uptake and transformation of benzene and toluene by plant leaves. Ecotoxicol. Environ. Saf. 37, 24–29. 10.1006/eesa.1996.1512 [DOI] [PubMed] [Google Scholar]
- UNFPA (2004). “State of World Population 2004: The Cairo Consensus at Ten: Population, Reproductive Health and the Global Effort to End Poverty: Chapter 4 Migration and Urbanization. New York, NY: United Nations Population Fund. [Google Scholar]
- United Nations (2000). World Urbanization Prospects (the 1999 Revision). New York, NY: Population Division, Department of Economic and Social Affairs, United Nations. [Google Scholar]
- United Nations (2004). World Urbanization Prospects (the 2003 Revision). New York, NY: Population Division, Department of Economic and Social Affairs, United Nations. [Google Scholar]
- USEPA (1999). Nitrogen Oxides (NOx), Why and How They are Controlled. Washington, DC: United States Environmental Protection Agency, Publication No. EPA-456/F-99-006R. [Google Scholar]
- Vacher C., Hampe A., Porte A. J., Sauer U., Compant S., Morris C. E. (2016). The phyllosphere: microbial jungle at the plant-climate interface. Ann. Rev. Ecol. Evol. Systemat. 47, 1–24. 10.1146/annurev-ecolsys-121415-032238 [DOI] [Google Scholar]
- Vainonen J. P., Kangasjarvi J. (2015). Plant signaling in acute ozone exposure. Plant Cell Environ. 38, 240–252. 10.1111/pce.12273 [DOI] [PubMed] [Google Scholar]
- Vallero D. A. (2014). Fundamentals of Air Pollution, 5nd Edn. San Diego, CA: Elsevier. [Google Scholar]
- Van Aken B., Yoon J. M., Schnoor J. L. (2004). Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoides × nigra DN34). Appl. Environ. Microbiol. 70, 508–517. 10.1128/AEM.70.1.508-517.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokou D., Vareli K., Zarali E., Karamanoli K., Constantinidou H. I., Monokrousos N., et al. (2012). Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria. Microb. Ecol. 64, 714–724. 10.1007/s00248-012-0053-7 [DOI] [PubMed] [Google Scholar]
- Vorholt J. A. (2002). Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178, 239–249. 10.1007/s00203-002-0450-2 [DOI] [PubMed] [Google Scholar]
- Waight K., Pinyakong O., Luepromchai E. (2007). Degradation of phenanthrene on plant leaves by phyllosphere bacteria. J. Gen. Appl. Microbiol. 53, 265–272. 10.2323/jgam.53.265 [DOI] [PubMed] [Google Scholar]
- Wallace L. A. (1991). Comparison of risks from outdoor and indoor exposure to toxic chemicals. Environ. Health Perspect. 95, 7–13. 10.1289/ehp.91957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W., Manning W. J., Wang X., Zhang H., Sun X., Zhang Q. (2014). Ozone and ozone injury on plants in and around Beijing, China. Environ. Pollut. 191, 215–222. 10.1016/j.envpol.2014.02.035 [DOI] [PubMed] [Google Scholar]
- Wang L., Liu L., Gao S., Hasi E., Wang Z. (2006). Physicochemical characteristics of ambient particles seeling upon leaf surfaces of urban plants in Beijing. J. Environ. Sci. 18, 921–926. 10.1016/S1001-0742(06)60015-6 [DOI] [PubMed] [Google Scholar]
- Wang Z., Pei J., Zhang J. (2013). Catalytic oxidization of indoor formaldehyde at room temperature – Effect of operation conditions. Build. Environ. 65, 49–57. 10.1016/j.buildenv.2013.03.007 [DOI] [Google Scholar]
- Ward D. M., Atlas R. M., Boehm P. D., Calder J. A. (1980). Microbial biodegradation and chemical evolution of oil from the Amoco spill. Ambio 9, 277–283. [Google Scholar]
- Wargocki P., Wyon D. P., Sundell J., Clausen G., Fanger P. (2000). The effects of outdoor air supply rate in an office on perceived air quality, sick building syndrome (SBS) symptoms and productivity. Indoor air. 10, 222–236. 10.1034/j.1600-0668.2000.010004222.x [DOI] [PubMed] [Google Scholar]
- Wendehenne D., Pugin A., Klessig D. F., Durner J. (2001). Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6, 177–183. 10.1016/S1360-1385(01)01893-3 [DOI] [PubMed] [Google Scholar]
- Weyens N., Taghavi S., Barac T., van der Lelie D., Boulet J., Artois T., et al. (2009a). Bacteria associated with oak and ash on a TCE-contaminated site: characterization of isolates with potential to avoid evapotranspiration of TCE. Environ. Sci. Pollut. Res. Int. 16, 830–843. 10.1007/s11356-009-0154-0 [DOI] [PubMed] [Google Scholar]
- Weyens N., Thijs S., Popek R., Witters N., Przybysz A., Espenshade J., et al. (2015). The role of plant-microbe interactions and their exploitation for phytoremediation of air pollutants. Int. J. Mol. Sci. 16, 25576–25604. 10.3390/ijms161025576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyens N., Van Der Lelie D., Artois T., Smeets K., Taghavi S., Newman L., et al. (2009b). Bioaugmentation with engineered endophytic bacteria improves contaminant fate in phytoremediation. Environ. Sci. Tech. 43, 9413–9418. 10.1021/es901997z [DOI] [PubMed] [Google Scholar]
- Whipps J. M., Hand P., Pink D., Bending G. D. (2008). Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 105, 1744–1755. 10.1111/j.1365-2672.2008.03906.x [DOI] [PubMed] [Google Scholar]
- WHO (2006). WHO Air Quality Guidelines for Particulate Matter, Ozone, Nitrogen Dioxide and Sulfure Dioxide. Geneva: WHO Press, World Health Organization. [PubMed] [Google Scholar]
- WHO (2013). Health Effects of Particulate Matter. Available online at: http://www.euro.who.int/__data/assets/pdf_file/0006/189051/Health-effects-of-particulate-matter-final-Eng.pdf
- WHO (2014). 7 million Premature Deaths Annually Linked to Air Pollution. Available online at: http://www.who.int/phe/eNews_63.pdf?ua=1
- Wieslander G., Norbäck D., Björnsson E., Janson C., Boman G. (1996). Asthma and the indoor environment: the significance of emission of formaldehyde and volatile organic compounds from newly painted indoor surfaces. Int. Arch. Occup. Environ. Health. 69, 115–124. 10.1007/s004200050125 [DOI] [PubMed] [Google Scholar]
- Wolverton B. C., Johnson A., Bounds K. (1989). Interior Landscape Plants for Indoor Air Pollution Aabatement. Final Report. National Aeronautics and Space Administration, John C. Stennis Space Center, MO.
- Wolverton B. C., McDonald R. C., Watkins E. A. (1984). Foliage plants for removing indoor air pollutants from energy efficient homes. Econ. Bot. 38, 224–229. 10.1007/BF02858837 [DOI] [Google Scholar]
- Wood R. A., Burchett M. D., Alquezar R., Orwell R. L., Tarran J., Torpy F. (2006). The potted-plant microcosm substantially reduces indoor air VOC pollution: I. Office field-study. Water Air Soil Pollut. 175, 163–180. 10.1007/s11270-006-9124-z [DOI] [Google Scholar]
- Xu Z., Wang L., Hou H. (2011). Formaldehyde removal by potted plant-soil systems. J. Hazard. Mater. 192, 314–318. 10.1016/j.jhazmat.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Nakamura T., Tatsumi Y., Nakamura K., Sano H. (2006). Efficient assimilation of sulfide by transgenic rice plants over-expressing a rice cysteine synthase. Plant Biotechnol. 23, 117–122. 10.5511/plantbiotechnology.23.117 [DOI] [Google Scholar]
- Yang J., McBride J., Zhou J., Sun Z. (2005). The urban forest in Beijing and its role in air pollution reduction. Urban For. Urban Green. 3, 65–78. 10.1016/j.ufug.2004.09.001 [DOI] [Google Scholar]
- Yang Q. L., Liu Y. F., Yan-Ru J. I., Dong Y., Gao Y. (2012). Construction of phenol degradation genetically engineered bacteria Bacillus subtilis dqly-2. Biotechnology 22, 65–68. [Google Scholar]
- Yee D. C., Maynard J. A., Wood T. K. (1998). Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl. Environ. Microbiol. 64, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom S. H., Yoo Y. J. (2002). Analysis of microbial adaptation at enzyme level for enhancing biodegradation rate of BTX. Korean J. Chem. Eng. 19, 780–782. 10.1007/BF02706967 [DOI] [Google Scholar]
- Yousaf S., Ripka K., Reichenauer T. G., Andria V., Afzal M., Sessitsch A. (2010). Hydrocarbon degradation and plant colonization by selected bacterial strains isolated from Italian ryegrass and birdsfoot trefoil. J. Appl. Microbiol. 109, 1389–1401. 10.1111/j.1365-2672.2010.04768.x [DOI] [PubMed] [Google Scholar]
- Youssefian S., Nakamura M., Orudgev E., Kondo N. (2001). Increased cysteine biosynthesis capacity of transgenic tobacco overexpressing an O-acetylserine(thiol) lyase modifies plant responses to oxidative stress. Plant Physiol. 126, 1001–1011. 10.1104/pp.126.3.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S., Nakamura M., Sano H. (1993). Tobacco plants transformed with the O-acetylserine (thiol) lyase gene of wheat are resistant to toxic levels of hydrogen sulphide gas. Plant J. 4, 759–769. 10.1046/j.1365-313X.1993.04050759.x [DOI] [PubMed] [Google Scholar]
- Yu D. S., Song G., Song L. L., Wang W., Guo C. H. (2015). Formaldehyde degradation by a newly isolate fungus Aspergillus sp. Hua. Intl. J. Environ. Sci. Technol. 12, 247–254. 10.1007/s13762-013-0411-0 [DOI] [Google Scholar]
- Yutthammo C., Thongthammachat N., Pinphanichakarn P., Luepromchai E. (2010). Diversity and activity of PAH-degrading bacteria in the phyllosphere of ornamental plants. Microb. Ecol. 59, 357–368. 10.1007/s00248-009-9631-8 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhou P., Zhang W., Zhang W., Wang Y. (2013). Selection of landscape tree species of tolerant to sulfur dioxide pollution in subtropical China. Open J. Forestry 3, 104–108. 10.4236/ojf.2013.34017 [DOI] [Google Scholar]
- Zhao F. J., Tausz M., De Kok L. J. (2008). Role of sulfur for plant production in agricultural and natural ecosystems, in Sulfur Metabolism in Phototrophic Organisms, eds Hell R., Dahl C., Knaff D. B., Leustek T. (Dordrecht: Springer; ), 417–435. [Google Scholar]
- Zhao J., Yi H. (2014). Genome-wide transcriptome analysis of Arabidopsis reponse to sulfur dioxide fumigation. Mol. Genet. Genomics. 289, 989–999. 10.1007/s00438-014-0870-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang S. (2015). The relationship between urbanization, economic growth and energy consumption in China: an econometric perspective analysis. Sustainability 7, 5609–5627. 10.3390/su7055609 [DOI] [Google Scholar]
- Zhao H., Wang S., Wang W., Liu R., Zhou B. (2015). Investigation of ground-level ozone and high-pollution episodes in a megacity of eastern China. PLoS ONE 10:e0131878. 10.1371/journal.pone.0131878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. B., Zou P., Sun P. S., Bi X. Y., Wang J., Chen J. Q., et al. (2013). Effects of the external carbon sources on the microbes in the simultaneous biological removal of SO2 and NO (x) process. Environ. Earth Sci. 70, 2381–2386. 10.1007/s12665-013-2452-6 [DOI] [Google Scholar]
- Zhou X., Cao Z., Ma Y., Wang L., Wu R., Wang W. (2016). Concentrations, correlations and chemical species of PM2.5/PM10 based on published data in China: potential implications for the revised particulate standard. Chemosphere 144, 518–526. 10.1016/j.chemosphere.2015.09.003 [DOI] [PubMed] [Google Scholar]