Abstract
Idiosyncratic drug-induced liver injury (IDILI) is a rare but potentially severe adverse drug reaction that should be considered in patients who develop laboratory criteria for liver injury secondary to the administration of a potentially hepatotoxic drug. Although currently used liver parameters are sensitive in detecting DILI, they are neither specific nor able to predict the patient's subsequent clinical course. Genetic risk assessment is useful mainly due to its high negative predictive value, with several human leucocyte antigen alleles being associated with DILI. New emerging biomarkers which could be useful in assessing DILI include total keratin18 (K18) and caspase-cleaved keratin18 (ccK18), macrophage colony-stimulating factor receptor 1, high mobility group box 1 and microRNA-122. From the numerous in vitro test systems that are available, monocyte-derived hepatocytes generated from patients with DILI show promise in identifying the DILI-causing agent from among a panel of coprescribed drugs. Several computer-based algorithms are available that rely on cumulative scores of known risk factors such as the administered dose or potential liabilities such as mitochondrial toxicity, inhibition of the bile salt export pump or the formation of reactive metabolites. A novel DILI cluster score is being developed which predicts DILI from multiple complimentary cluster and classification models using absorption–distribution–metabolism–elimination-related as well as physicochemical properties, diverse substructural descriptors and known structural liabilities. The provision of more advanced scientific and regulatory guidance for liver safety assessment will depend on validating the new diagnostic markers in the ongoing DILI registries, biobanks and public–private partnerships.
Keywords: HEPATOBILIARY DISEASE, DRUG INDUCED HEPATOTOXICITY, BILE ACID, PHARMACOGENETICS, ADVERSE DRUG REACTIONS
Importance of DILI diagnosis
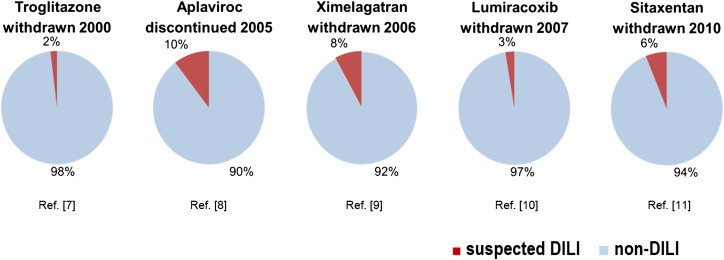
Drug-induced liver injury (DILI) accounts for <1% of cases of acute liver injury seen by gastroenterologists, but is the most common cause for acute liver failure in the USA and Europe.1–3 According to surveys in France and Iceland, DILI occurs with an annual incidence of about 14–19 per 100 000 inhabitants.4 5 DILI is also a leading cause of attrition of compounds in drug development and one of the two most frequent causes for drug withdrawals, restrictions and project terminations (figure 1).6–11 Of 76 drugs withdrawn from the market between 1969 and 2002, 12 were attributable to liver damage.12 Whereas liver signals that escape detection during drug approval result in postmarketing restrictions (eg, pazopanib, temozolomide and flupirtine in 2013), the risk of false-positive DILI adjudication may lead to unnecessary attrition, thereby contributing to the considerable economic issues associated with DILI.
Figure 1.
Impact of idiosyncratic drug-induced liver injury (IDILI) on drug attrition. Pie charts showing the occurrence of liver test abnormalities in clinical trials with drugs withdrawn or stopped due to DILI. Blue: percentage of study participants with normal liver tests and Red: percentage of patients with possibly drug-related liver enzyme elevations.
Patients who consume acetaminophen (APAP) at a single dose exceeding 7.5 g experience acute liver toxicity, notably if plasma concentrations exceed 200 or 100 μg/L 4 or 8 hours after ingestion, respectively. APAP intake at the licensed dose of 4 g/day over a period of 2 weeks results in elevations of alanine aminotransferase (ALT) above 3× the upper limit of normal (ULN) in one-third of patients.13 This form of dose-dependent APAP-induced toxicity is termed intrinsic DILI: it is predictable, reproducible in preclinical models and much insight has been gained into the underlying mechanisms.14 15
In contrast to intrinsic DILI, the onset of idiosyncratic DILI (IDILI), which is very rare but nonetheless responsible for about 10–15% of acute liver failures in the USA,16 is almost impossible to predict. IDILI is characterised by a variable latency to onset (weeks to months) and a lack of clear dose dependency.17 Drug–protein adducts, formed by drugs or their metabolites that interact with host proteins, are presented as neoantigens by major histocompatibility complex class II, thereby triggering an immunoallergic reaction. Individuals with underlying hepatic injury such as viral hepatitis or inflammatory conditions may be more susceptible to immunoallergic injury.18 Following the initial insult, additional mechanisms such as inhibition of transporters, mitochondrial injury, endoplasmic reticulum and oxidative stress and proinflammatory cytokines can further amplify the injury mechanisms that lead to acute DILI.19 Identification of host factors that render an individual susceptible is the focus of ongoing pharmacogenetic studies.20 This review article focuses on IDILI and the subsequent use of the term DILI essentially implies IDILI.
A major problem in drug development is the frequency of adverse hepatic reactions induced by the newer molecular targeted agents (MTAs) in oncology. Hepatotoxicity occurs in one-third of patients treated with a protein kinase inhibitor, with fatal outcome reported for pazopanib, sunitinib and regorafenib.21 Ten per cent of patients treated with immune checkpoint inhibitors, notably ipilimumab, develop liver injury with high rates of recurrent liver injury upon rechallenge.21 The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) gefitinib is associated with a 18.5% frequency of hepatotoxicity and casualties have occurred for all EGFR TKIs.22 The oncology population that is treated with MTAs is more likely to have multiple comorbidities and comedications and is therefore at risk for hepatotoxicity. The Food and Drug Administration (FDA) issues detailed recommendations in drug labels as to liver test monitoring intervals and stopping rules; however, intensive postmarketing surveillance of MTA-associated liver injury is required. The standard recommendations contained in the current FDA Guidance to Industry23 are not applicable to the oncology population and new methods of liver safety assessment are required.24 For the management of IDILI, a key question is whether a patient in whom DILI has been diagnosed will progress to severe liver injury with potentially fatal outcome or recover spontaneously after cessation of the causative agent. A diagnostic algorithm that allows identification of risk factors and prediction of the subsequent clinical course would greatly facilitate the management of acute DILI. The current lack of predictive safety testing before administration of a potentially hepatotoxic compound reinforces the need for rapid identification of a high-risk DILI situation that requires intensified surveillance. Novel biomarkers such as those evaluated in the European Union Innovative Medicines Initiative (IMI) Safer and Faster Evidence-based Translation (SAFE-T) Consortium have gained regulatory support for systematic implementation by sponsors in future trials.25–27
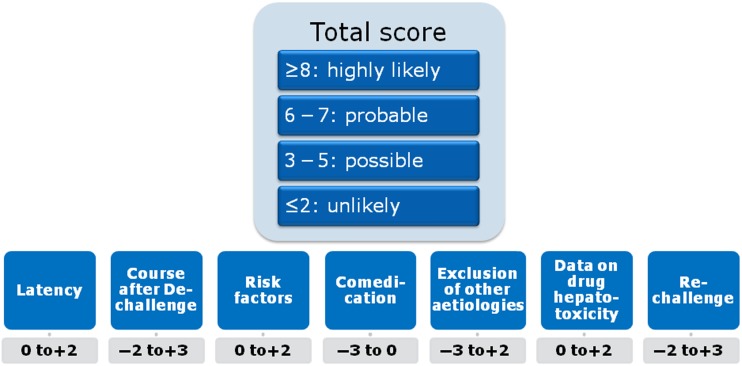
Current diagnosis of IDILI depends on expert opinion that is based on patient data and the typical ‘signatures’ associated with certain drugs.28 Causality scores such as the Roussel-Uclaf Causality Assessment Method (RUCAM, figure 2) are intended to confirm or exclude the suspicion of DILI.29 Limitations of such scoring algorithms are poor inter-rater reliability and arbitrary scoring, for example, for alcohol use.30 This can be mitigated by a consensus process such as the one employed by the US Drug-Induced Liver Injury Network (DILIN), even though consensus opinion carries the risk of overruling a more insightful minority opinion.31 32 Liver injury caused by herbal and dietary supplements presents unique challenges to hepatotoxicity assessment and its incidence is increasing.33 34 Due to the lack of a reliable diagnostic in vitro test, there is no objective method beyond expert opinion that assesses causality of a given drug in individual cases.
Figure 2.
Roussel-Uclaf Causality Assessment Method (RUCAM) diagnostic score.
Standard of diagnosis: role of currently performed liver tests in assessing DILI
DILI most often presents as an acute viral hepatitis-like syndrome, without symptoms that specifically point to the drug aetiology unless rash or other cutaneous manifestations35 reinforce the suspicion of drug toxicity. The clinical spectrum of DILI can mimic almost every other liver disorder. Accompanying blood eosinophilia is uncommon in large series of patients with DILI,36 37 but is clearly suggestive of drug allergy. Histopathological findings in DILI can resemble many other liver disorders, thereby limiting the value of liver biopsy in DILI diagnosis. However, biopsy can be of use to establish an alternative diagnosis when the underlying liver disease worsens (ie, alcoholic hepatitis, autoimmune hepatitis (AIH))38 (table 1).
Table 1.
Examples of host and environmental variables influencing the diagnostic workup in patients assessed for suspected DILI
| Factor | Alternative diagnosis | Diagnostic appraisal |
|---|---|---|
| Age | ||
| <40 years | Wilson's disease | Ceruloplasmin, copper in 24-hour urine, ABCB7 genetic testing |
| >60 years (DILI is most often cholestatic regardless of the drug) | Benign and malignant biliary obstruction | MRI and/or ERCP If inconclusive and damage persists, consider liver biopsy |
| Type of injury | ||
| Cholestatic/mixed | Benign and malignant biliary obstruction | MRI and/or ERCP If inconclusive and damage persists, consider liver biopsy |
| Comorbidities | ||
| 1. Cardiovascular disease (right/congestive heart failure, coronary artery disease) | Ischaemic hepatitis | Towering AST/ALT Search for prior hypotensive episodes Echocardiogram |
| 2. Hyperthyroidism (untreated) | Thyrotoxic hepatitis | T3, T4, TSH |
| 3. Type 1 diabetes mellitus (poorly controlled) | Glycogenic hepatopathy | Consider liver biopsy |
| 4. Pre-existing liver disease (AIH, ALD, NASH, HBV, HCV) | Flare-up of underlying liver disease | Consider liver biopsy |
| Subject behaviour and local burden of infectious diseases | ||
| 1. Sexual transmission | Syphilis | Serology for acute infection |
| 2. Tropical and developing areas (±underlying HIV infection) | Malaria, dengue, tuberculosis, typhoid fever, leptospirosis and others | Specific serology |
| 3. Hepatitis E (exposure to farm animals, consumption of undercooked pork) | Differential diagnosis in acute hepatitis suspected to be DILI39 | Specific serology (anti-HEV IgM and IgG, HEV PCR) |
ALD, alcoholic liver disease; AIH, autoimmune hepatitis; ALT, alanine aminotransferase; AST, aspartate aminotransferase; DILI, drug-induced liver injury; ERCP, endoscopic retrograde cholangio-pancreatography; HEV, hepatitis E virus; NASH, nonalcoholic steatohepatitis; TSH, thyroid-stimulating hormone.
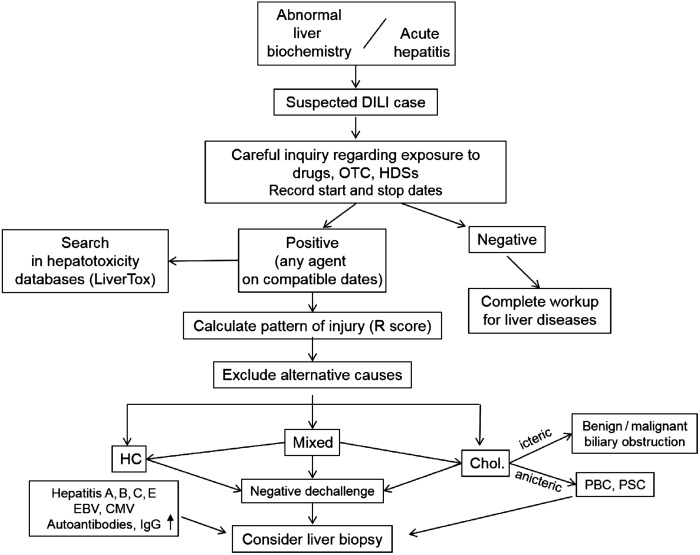
A diagnostic workflow for assessing cases of suspected DILI is shown in figure 3. Serum aminotransferases, that is, ALT and aspartate aminotransferase (AST), alkaline phosphatase (ALP) and total bilirubin (TB) levels, although not specific for DILI, remain the hallmark for detecting and classifying liver damage.40 Minor increases in aminotransferases that can result from adaptive and reversible liver responses to the drug (eg, statins) or from pre-existing liver disease (eg, fatty liver) should not be classified as DILI. An international expert group41 proposed the following thresholds for a diagnosis of DILI: (a) ALT value ≥5× ULN, (b) ALP value ≥2× ULN or (c) ALT value ≥3× ULN and TB ≥2× ULN. The latter constellation defines ‘Hy's Law’, which anticipates a 10% risk of mortality/liver transplantation,42 as confirmed in large databases of DILI cases.36 43 The FDA guidance for DILI extends the interpretation of Hy's Law by stating that ‘there should not be a prominent cholestatic component’ in the hepatocellular nature of the liver injury,23 suggesting that a cholestatic component as defined by elevated ALP levels is associated with less risk of progression. However, a recent analysis from the Spanish DILI Registry showed that raised ALP >2 ULN did not decrease the risk of acute liver failure in cases fulfilling Hy's Law.44 A marked increase of AST and an AST/ALT ratio >1.5 at DILI recognition also predict a worse prognosis.43 44
Figure 3.
Flow diagram of diagnostic workup of drug-induced liver injury (DILI). The phenotypes of liver injury are categorised according to the R value, defined as the ratio ALT/ULN:ALP/ULN. An R value of ≥5 indicates hepatocellular injury, ≤2 cholestatic injury and 2–5 mixed-type injury. ALP, alkaline phosphatase; ALT, alanine aminotransferase; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HC, hepatocellular; HDSs, herbal and dietary supplements; OTC, over-the-counter drugs; PBC, primary biliary cirrhosis; PSC, primary sclerosing cholangitis; ULN, upper limit of normal.
The presence of autoimmune features such as antinuclear antibodies (ANAs), smooth muscle antibodies (SMAs) and elevated IgG levels as well as histological features of AIH may cause diagnostic confusion in DILI.45 Screening for autoantibodies and serum IgG in hepatocellular injury is mandatory, although the typical laboratory and pathological features of AIH may also be drug induced. Moreover, recurrent DILI induced by a different drug tends to present with an AIH phenotype.46 DILI with autoimmune features should be clearly distinguished from idiopathic AIH and typically resolves after stopping the causative drug. If treated with corticosteroids, a lack of recurrence following corticosteroid cessation supports a diagnosis of drug-induced AIH rather than idiopathic AIH.47 As yet there are no diagnostic tests to differentiate idiopathic from drug-related AIH, although histological findings can help in the differential diagnosis.48 49
Rechallenge with the suspected drug, although considered the gold standard for diagnostic confirmation,50 carries ethical and practical issues. First, it confers a risk that is only justified when an alternative drug is not available. Second, the definition of a ‘positive rechallenge’ is controversial regarding the required threshold, if any, of liver enzyme elevation and symptoms, partly due to the lack of data on ‘negative rechallenge’.51 In the RUCAM score, the re-exposure test is positive if ALT is ≥2× baseline upon re-exposure, provided that ALT was below 5× ULN before re-exposure, and negative if one or both criteria are not fulfilled.29 Follow-up in patients with DILI must include routine liver biochemistry until complete normalisation. Rapid normalisation of aminotransferases supports the diagnosis, whereas slow or incomplete resolution suggests alternative causes. In such instances, a liver biopsy can be helpful (figure 3). Persistently elevated TB and ALP 30–60 days after the initial DILI diagnosis can predict a chronic outcome.52
In vitro and in silico tools for the assessment and prediction of DILI
The risk of IDILI has hung like the sword of Damocles over the drug approval process since decades, leading to fatal liver failures and subsequent market withdrawals and creating nervousness on the part of sponsors and regulators alike. These signals may occur in only very few individuals, making it impossible to identify the risk in premarketing registration trials. Thus, the quest for predictive tools that would allow an a priori identification of both host factors as well as compound features conferring a DILI risk has led to the development of numerous cell-based systems, animal models and in silico algorithms. Whereas none of these has lived up to the crystal ball promises that frequently accompany marketing initiatives, the spectrum of predictive tools available today may allow implementation of a panel of select methodologies which, in combination, yield new insight into IDILI.
Cell-based assays include primary human hepatocytes, immortalised hepatocytes, hepatoma cell lines and induced pluripotent stem cell-derived hepatocytes.53–57 These systems have been reviewed in detail and the development of coculture systems with non-parenchymal cells as well as 3D organoids has allowed the in vitro analysis of several mechanisms of DILI, including mitochondrial toxicity, oxidative and endoplasmic reticulum stress and inhibition of transporters such as the bile salt export pump (BSEP).58 59 Animal models used to study mechanisms of DILI include the heterozygous superoxide dismutase (Sod2)±mouse, panels of inbred mouse strains and chimeric mice with humanised livers.60–62 These models have proven useful to elucidate mechanisms such as troglitazone-induced mitochondrial toxicity.63 The prediction of IDILI in a susceptible individual has not been made possible by any of these in vitro systems.
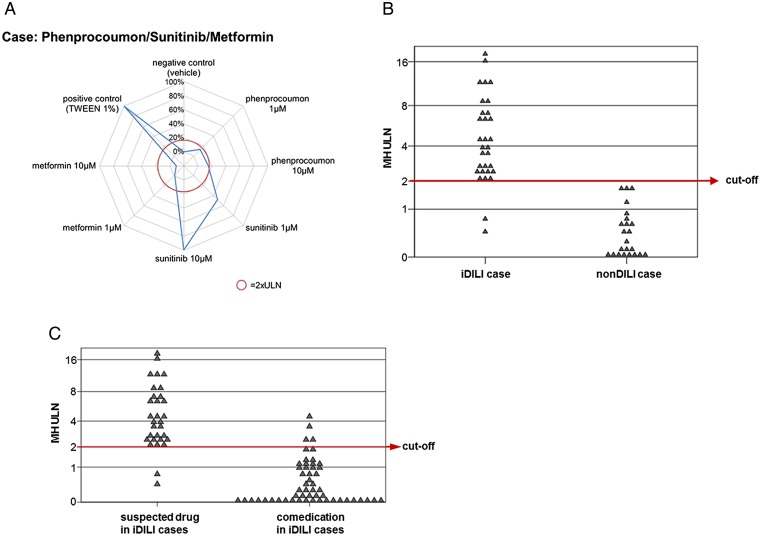
A frequent problem in assessing causality in patients with polypharmacy who have experienced DILI is which of the drugs, especially if some of them are known to be potential hepatotoxins, was causative in the given patient. A test system called MetaHeps allows the identification of the DILI-causing drug out of a panel of comedications. This test uses monocyte-derived hepatocyte-like cells (MH cells) from the affected patient (figure 4). MH cells possess donor-specific hepatocyte characteristics when compared with primary human hepatocytes derived from the same donor.64 MH cells from patients with IDILI are more susceptible to toxicity induced by the causative drug than MH cells from patients with non-drug liver injury or healthy donors.65 66 MH cells show high sensitivity and specificity for the diagnosis and exclusion of IDILI and outperform the RUCAM score67 in identifying the causative drug in cases of polypharmacy. MH cells may furthermore be useful in assessing the role of drug–drug interactions in the onset of IDILI. The identification of true positives among patients with multiple comedications could help to develop more specific biomarkers that identify patients at risk of progressing to more severe DILI.
Figure 4.
(A) Example for a monocyte-derived hepatocyte-like (MH) cell test result from a patient with acute liver injury during treatment with sunitinib (for renal cell carcinoma), phenprocoumon (for atrial fibrillation) and metformin (for diabetes type II). MH cell toxicity is shown in a spiderweb graph. Sunitinib exerts marked toxicity in MH cells of this patient, whereas phenprocoumon and metformin do not show any effects. The red circle represents the individual cut-off for test positivity. (B) MH cell test results in 31 patients with idiosyncratic drug-induced liver injury (IDILI) and 23 patients with acute liver injury of other origin (non-DILI) using the drugs most likely to have caused liver injury in these cases. The MH cell test correctly identifies 29 of the 31 IDILI cases and shows no false-positive results. (C) MH cell test results using all drugs involved in the IDILI cases. Only four of the 84 comedications show positive results, suggesting that the MH cell test could be useful to identify the causative drug in complex IDILI cases. TWEEN, polyethylene glycol sorbitan monolaurate; ULN, upper limit of normal.
In addition to studying the biological effects that drugs have in ex vivo test systems, predictive algorithms are being developed that correlate structural and chemical properties of the parent drug as well as of its metabolites with the clinical risk of DILI. The FDA's Liver Toxicity Knowledge Database (LTKB) contains a benchmark data set of drugs whose potential to cause DILI is categorised into most-DILI-concern drugs (boxed warning or withdrawn from the market due to hepatotoxicity), less-DILI-concern drugs (DILI risk mentioned in the label) and no-DILI-concern drugs (no DILI indication in the label).68 This DILI classification has been refined by incorporating the causality assessment from clinical studies together with drug labelling information to improve its accuracy.69 FDA investigators reported the Rule-of-2 which identified lipophilicity, that is, an octanol–water partitioning coefficient (logP) of ≥3, as well as a daily dose of ≥100 mg as risk factors for DILI.70 By analysing data on 254 orally administered drugs in the LTKB benchmark data set, the FDA group found that drugs that are substrates of cytochrome P450 (CYP) enzymes have a higher likelihood of causing DILI irrespective of the administered dose, whereas mere inhibitors of P450 enzymes were only associated with a risk of DILI at high daily doses.71 By factoring in the formation of reactive metabolites, the predictive value of logP and daily dose could be improved, as shown in an analysis of 159 clinical cases collected from the National Institutes of Health's LiverTox Database.72
Other groups have applied inhibition of BSEP and mitochondrial toxicity as parameters to the most-concern, less-concern and no-concern-DILI drugs in the LTKB and found that dual potency as mitochondrial and BSEP inhibitors was highly associated with more severe human DILI as well as with an exposure–safety correlation represented by the maximum plasma concentration Cmax.73 The role of BSEP inhibition as a mechanism of DILI first became evident from transport studies using isolated membrane vesicles from Sf9 insect cells that overexpress Bsep.74 This technique was employed by various industry groups to systematically correlate the risk of DILI of selected compounds with their inhibitory potential towards BSEP as a function of their exposure.75 76 The Critical Path Institute's Predictive Safety Testing Consortium (C-Path PSTC) hosted a webinar in 2016 focused on BSEP inhibition and perturbation of bile acid homeostasis as mechanisms of DILI and a broad industry-wide consensus was reached on the importance of testing lead compounds in BSEP inhibition assays so as to identify potential DILI liabilities at an early stage.77
Based on the knowledge about mechanisms which cause DILI, in silico algorithms are being developed that allow modelling of various parameters to extrapolate the risk of DILI in vivo. The DILIsym model, for example, predicts that the BSEP inhibitor bosentan, but not the BSEP inhibitor telmisartan, causes mild hepatocellular ATP decline and serum ALT elevation in a simulated population.78 The catechol-O-methyltransferase inhibitors tolcapone and entacapone both cause mitochondrial impairment and inhibit BSEP, but liver injury has only been associated with the use of tolcapone. DILIsym identified patient-specific risk factors for tolcapone-induced liver injury and in a simulated population (SimPops) increases in ALT were predicted in 2.2% of the population.79 The Virtual Liver Network is a German research initiative that bridges investigations from the subcellular level to patient and healthy volunteer studies in an integrated workflow to generate validated computer models of human liver physiology.80 These in silico approaches rely on cumulative scores of known risk factors such as the administered dose or on potential liabilities such as mitochondrial toxicity, BSEP inhibition or the formation of reactive metabolites which can be measured in vitro. The major challenge when constructing predictive DILI models is to account for the broad range of chemotypes which have been associated with clinically relevant liver findings as well as the various mechanistic (pathway) considerations which translate into different clinical phenotypes of liver injury. A ‘DILI cluster score’ is being developed at Novartis that correlates a comprehensive set of several hundred compound properties against validated clinical scores as obtained from an extended version of the LTKB Database. Predictions are obtained from multiple complimentary cluster and classification models using calculated and measured compound properties related to absorption–distribution–metabolism–elimination and physicochemical properties, diverse substructural descriptors and known structural liabilities. This also allows for successful prediction of compounds which may not be classified based on typical risk factor profiles or are administered at fairly low doses (eg, methotrexate). The current algorithm is limited to orally administered drugs given over a prolonged period or in a chronic dosing regimen.
Novel biomarkers
There have been recent efforts mainly by public–private partnerships such as the IMI SAFE-T Consortium together with C-Path PSTC and DILIN to develop and qualify new liver safety biomarkers that outperform current standard markers in terms of sensitivity, specificity and predictivity. From the new markers investigated by IMI SAFE-T and PSTC (table 2), a subset has recently received regulatory support from both European Medicines Agency (EMA) and FDA for more systematic use in an exploratory development setting,26 27 which will ultimately enable full qualification of the most promising markers. Once qualified in well-controlled trials, regulatory guidance will then also have to account for the new markers and incorporate them into existing guidelines.
Table 2.
Selected biomarkers of DILI investigated by the IMI SAFE-T and the C-Path PSTC Consortia
| Marker | Origin of biomarker | Summary |
|---|---|---|
| MicroRNA (miR)-122 | Liver specific | miR-122 is an early and specific marker of hepatocellular injury and a sensitive marker of DILI.81–86 |
| High mobility group box 1 (HMGB1) | Detectable in numerous tissues | In acetaminophen (APAP)-induced liver injury, hyperacetylated HMGB1 is significantly elevated in patients who die or require a liver transplant, whereas in spontaneous survivors it is not significantly different from healthy volunteers.87 |
| Cytokeratin 18 full length | Epithelial cells | The full-length protein is released from necrotic cells. It is significantly elevated in APAP overdose patients who die/require a liver transplant compared with spontaneous survivors.83–85 87 |
| Cytokeratin 18 caspase-cleaved fragment (caspase-cleaved keratin 18 (ccK18)) | Epithelial cells | The caspase-cleaved fragment is released from apoptotic cells and helps define the type of cytotoxicity. ccK18 fragments in blood predict severity of disease in NASH and in hepatitis C.83–85 88 |
| Cadherin 5 | Endothelial cells | Cadherin 5 is a calcium-dependent cell adhesion protein (also called vascular endothelial cadherin) that is specific to endothelial cells. Initial results indicate a potential use as a susceptibility marker for DILI.89 |
| Liver fatty acid-binding protein (L-FABP) | Primarily liver; lower levels in the kidneys and small intestine | L-FABP is a sensitive marker for hepatocellular injury following liver transplantation and in hepatitis C.90 91 In heparin-induced DILI, L-FABP levels correlate well with serum ALT levels.89 |
| Glutamate dehydrogenase (GLDH) | Mitochondrial matrix; primarily in the centrilobular region of the liver; lower levels in the kidney and brain | A sensitive biomarker of liver toxicity with hepatocellular damage in preclinical species; shown to be elevated in humans with hepatic ischaemia or hepatitis; shown to correlate with ALT in patients with a broad range of clinically demonstrated liver injuries, including APAP-induced liver injury and to detect mild hepatocyte necrosis in patients treated with heparin. Marker for mitochondrial injury or cellular injury in multiple clinical DILI and acute liver failure studies.84 92 93 |
| Glutathione S-transferase (GSTα) | Centrilobular region of the liver; multiple tissues | Hepatotoxicity biomarker shown in rats to have enhanced specificity and sensitivity compared with ALT; humans with APAP overdose show elevated GSTα levels earlier than ALT; GSTα may offer a better assessment of rapid changes in liver damage due to the shorter half-life of plasma GSTα compared with ALT or AST.94 95 |
| Osteopontin (OPN) | Multiple tissue and cell types, including liver | Elevated serum levels of OPN are detectable in patients with severe liver damage. Increased levels of serum OPN are associated with a poor prognosis. OPN is associated with inflammatory cell activation and with liver regeneration due to activation of hepatic stem cells.96 Hepatocytes are a major source of OPN and HMGB1 signalling to hepatic stellate cells, thereby promoting collagen-1 production. OPN is upstream of HMGB1 and both play a major role in the pathogenesis of liver fibrosis.97 |
| Macrophage colony-stimulating factor receptor 1(MCSFR1) | Cytokine receptor on macrophages/monocytes | Data from the ximelagatran biomarker discovery study suggest that MCSFR1 is shed from macrophages during DILI. MCSFR1 serum/plasma levels may have value as a prognostic marker for liver disease associated with inflammation.98 |
| Sorbitol dehydrogenase (SDH) | Multiple tissue and cell types, including liver | Sensitive enzymatic serum marker of liver toxicity in preclinical species. Shown to be elevated in humans with various liver diseases and to detect mild hepatocyte necrosis in patients treated with heparin. The biomarker serves two purposes:
|
| Bile acids | Synthesised by the liver |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; C-Path PSTC, Critical Path Institute's Predictive Safety Testing Consortium; DILI, drug-induced liver injury; IMI, Innovative Medicines Initiative; SAFE-T, Safer and Faster Evidence-based Translation.
Several new biomarkers have been studied in the context of APAP-induced DILI.99 MicroRNA-122 (miR-122) is a hepatocyte-specific miRNA that is elevated in the plasma of patients within hours of an APAP overdose. Together with high mobility group box-1 (HMGB1) and keratin-18, it has been shown to predict the subsequent onset of liver injury at an early time point before ALT is elevated.83 Previous studies in mice already showed that miR-122 and miR-192 are enriched in liver tissue and exhibit dose-dependent and exposure-dependent changes in plasma that parallel serum aminotransferase levels and the histopathology of liver degeneration.81
HMGB1 is a chromatin-binding protein released by necrotic cells. HMGB1 subsequently targets Toll-like receptors and the receptor for advanced glycation end products (RAGE), thus acting as a damage-associated molecular pattern molecule.100 A hyperacetylated form is released from immune cells and acts as a marker of immune activation. Given the role of immune activation in IDILI, HMGB1 has been studied in the context of APAP-induced (intrinsic) DILI and as a marker of IDILI in the IMI SAFE-T Consortium DILI cohort.
Another marker of immune activation is macrophage colony-stimulating factor receptor 1 (MCSFR1). In 10 cases of IDILI caused by the centrally acting non-opioid analgesic flupirtine, the use of which has been restricted by the EMA because of hepatotoxicity, MCSFR1 levels were considerably higher than in 19 cases of APAP-induced DILI, despite ALT levels being markedly higher in APAP-induced DILI. Furthermore, both MCSFR1 and the biomarker osteopontin (table 2) were higher in 31 patients with DILI that fulfilled Hy's Law criteria compared with 70 patients with DILI who did not fulfil Hy's Law criteria (SAFE-T Consortium, unpublished data).
Serum bile acids have traditionally been considered to have little use in the workup of liver disease with the possible exception of intrahepatic cholestasis of pregnancy, given the multitude of analytes and the complexity of bile acid metabolism. Glycodeoxycholic acid (GDCA) has been shown to have prognostic value in predicting the outcome of acute liver failure induced by APAP, with GDCA levels being considerably higher in non-surviving patients with acute liver failure (ALF).101 With the availability of new analytical multiplexing methods based on liquid chromatography-tandem mass spectrometry (LC-MS/MS) and gas chromatography-mass spectrometry (GC/MS), circulating bile acid (BA) profiles are now being evaluated as biomarkers for hepatotoxicity.102 A targeted LC-MS/MS approach identified cholic acid, glycocholic acid and taurocholic acid (TCA) as potential biomarkers of liver injury in rodent models.103 In the IMI SAFE-T Consortium DILI cohort, several bile acids were markedly elevated in flupirtine-induced DILI, including glycochenodeoxycholic acid, taurochenodeoxycholic acid and TCA. This was not simply the result of cholestasis since ALP was normal. This suggests that selected bile acids could be markers of IDILI.
Serum autoantibodies and pyrrole–protein adducts
Certain drugs causing IDILI are associated with the formation of serum autoantibodies. Examples include anti-CYP 2C9 in tielinic acid-induced hepatitis, anti-epoxide hydrolase in germander-induced liver injury, anti-CYP1A2 in dihydralazine hepatitis, anti-CYP3A in anticonvulsant hepatitis and anti-CYP2E1 in halothane hepatitis.104 105 Autoimmune reactions involving CYP2E1 are a feature of DILI induced by halogenated hydrocarbons and isoniazid (INH), but are also detectable in about one-third of patients with alcoholic liver disease and chronic hepatitis C.106 From 19 patients enrolled in the Acute Liver Failure Study Group for INH-induced acute liver failure, eight tested positive for anti-INH antibodies.107 These were not detectable in patients with only mild INH-induced liver injury, suggesting that mild cases of INH DILI resolve with immune tolerance.
In pyrrolizidine-induced liver injury, blood pyrrole–protein adducts (PPAs) predict the onset of sinusoidal obstruction syndrome with a positive predictive value of 96% and a negative predictive value of 100%.108 The blood PPA concentration is related to the severity and clinical outcome of pyrrolizidine alkaloid-associated hepatic sinusoidal obstruction syndrome.
Genetic testing in the assessment of DILI
In keeping with their reputation as ‘hypothesis generating’ research methodology, genome-wide association studies (GWASs) have unearthed a number of novel associations, in particular between human leucocyte antigen (HLA) class I and II alleles and DILI.
In contrast to GWASs focused on other complex traits, those investigating DILI have identified risk alleles with substantially higher risk ratios for susceptibility to DILI. As demonstrated in table 3 (modified from ref. 120), there is substantial overlap among the risk alleles associated with clinically varied phenotypes of toxicities due to structurally dissimilar compounds. For example, DRB1*0701 is a risk allele for flucloxacillin, ximelagatran and lapatinib-related DILI, while DRB1*1501 is associated with DILI secondary to amoxicillin-clavulanate and lumiracoxib. Conversely, DRB1*1501 is associated with reduced risk of flucloxacillin DILI and DRB1*0701 is protective of amoxicillin-clavulanate DILI.121 Such associations extend beyond DILI into a variety of other adverse reactions, including cutaneous hypersensitivity and drug-induced pancreatitis. For example, carriage of HLA-B*5701 allele increases by 80-fold the risk of flucloxacillin-induced DILI and the same allele is also strongly associated with hypersensitivity due to abacavir.111 Another example of common genetic factors underlying different organ toxicities is the link between DRB1*0701 and pancreatitis induced by thiopurine immunosuppressants as well as DILI due to a number of drugs listed above.122 Recently, GWASs led by the international DILI Consortium demonstrated HLA-A*33:01 as a risk factor for a cholestatic or mixed pattern of DILI when these are considered as a single phenotype irrespective of the causative drugs.118
Table 3.
Genetic susceptibility for DILI identified in GWASs
| Drug studied | Cohort (ethnicity) | Association described | SNP (gene)109 | OR |
|---|---|---|---|---|
| Ximelagatran110 thrombin inhibitor |
74 cases, 130 T controls (European) | HLA-DRB1*07 | HLA-DRB1 | 4.4 |
| Flucloxacillin111 β-lactam antibiotic |
51 cases, 282 P controls (European) | HLA-B*5701 | rs2395029 HCP5 |
45.0 |
| ST6GAL1 | rs10937275 ST6GAL1 | 4.1 | ||
| OR5H2 | rs1497546 OR5H8P—OR5K4 | 6.6 | ||
| ALG10B | rs6582630 ALG10B—CPNE8 | 2.8 | ||
| MCTP2 | rs4984390 MCTP2 |
3.3 | ||
| C9orf82 (CAAP1) | rs10812428 FAM71BP1—CAAP1 | 2.9 | ||
| Lumiracoxib112 Cyclo-oxygenase-2 inhibitor |
41 cases, 176 T controls; Replic: 24 cases (European†) | HLA-DRB1 | rs3129900 C6orf10 | 7.5 |
| Lapatinib113 kinase inhibitor |
37 cases, 1071 T controls, (European†) |
HLA-DRB1*0701 Perfect linkage disequilibrium with DQA1*0201 |
NR | NR |
| Lapatinib114 kinase inhibitor |
34 cases, 810 T controls, (European†) | HLA-DRB1*0701 | NR | NR |
| Amoxicillin-clavulanate115 antibiotic |
201 cases, 532 P controls (European) | HLA-DQB1*0602 | rs9274407 HLA-DQB1 |
3.1 |
| HLA-A*0201 | rs2523822 TRNAI25 | 2.3 | ||
| Multiple (Diclofenac116 non-steroidal anti-inflammatory drug) | 783 cases (30 diclofenac) 3001 P controls (European) | PPARG‡ | rs17036170 PPARG |
11.3 |
| Multiple (Flupirtine117 non-opioid analgesic) | 614 cases (6 flupirtine) 10 588 P controls (European) | HLA-DRB1*16:01-DQB1*05:02 | HLA-DRB1 | 18.7 |
| Multiple118 | 862 cases (21 terbinafine; 7 fenofibrate; 5 ticlopidine cases) 10 588 P controls (European) | HLA-A*33:01 | rs114577328 | 40.5; 58.7; 163.1 |
| Minocycline antibiotic119 | 25 cases 10 588 P controls (European) | HLA-B*35:02 | HLA-B*35:02 | 29.6 |
†Predominantly.
‡Associated with diclofenac DILI only.
DILI, drug-induced liver injury; GWASs, genome-wide association studies; NR, not reported; P, population control group; Replic, replication cohort; SNP, single nucleotide polymorphism; T, treated control group.
Interaction between drugs and HLA molecules
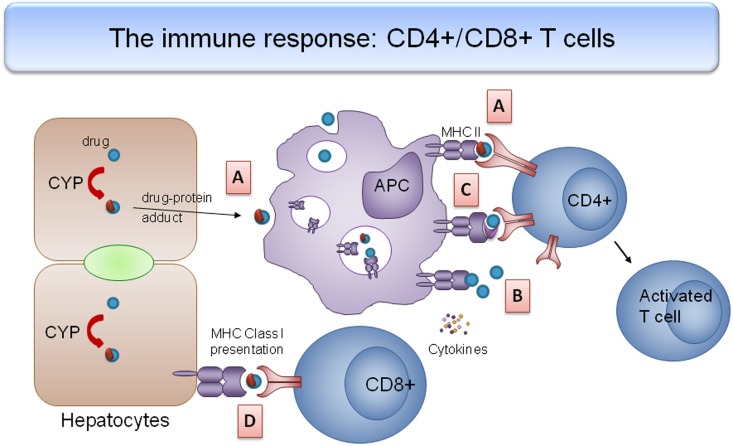
These investigations highlight the role of the adaptive immune response in DILI pathogenesis.123 The ‘hapten hypothesis’ states that an individual's susceptibility to DILI is determined by the covalent binding of a drug or its metabolites to a cellular or circulating protein and the interaction of the resultant complex with the peptide-binding groove of a specific HLA molecule (figure 5). Alternatively, the ‘pharmacological interaction (pi) concept’ proposes that a drug or drug metabolite can directly bind to the HLA molecule (as in the case of ximelagatran and HLA-DRB1*0701)110 or T cell receptor to trigger T cell activation, leading to immune-mediated liver injury. Recently, it has been proposed that drugs may make van der Waals contacts with the antigen-binding cleft (as in the case of minocycline and HLA-B*3502119) and alter the shape and chemistry of the antigen-binding cleft, thus altering the repertoire of endogenous peptides that subsequently bind to it, which in turn leads to an adaptive immune response.
Figure 5.
Interaction between drugs and human leucocyte antigen (HLA) molecule leading to an adaptive immune response. (A) Hapten hypothesis: drug–protein adducts (blue circles with red semicircle) released from dying hepatocytes are phagocytosed by antigen-presenting cells (APCs) and presented with major histocompatibility complex (MHC) II molecules. These hapten–carrier complexes bind to the peptide-binding groove on T cell receptors, leading to CD4+ cell activation and an effector T cell response. (B) Pharmacological interaction concept: drugs or metabolites can bind to HLA molecules directly and activate T cells. (C) Altered repertoire model: drug changes the shape and chemistry of the antigen-binding cleft, altering the repertoire of endogenous peptides that subsequently bind; the ‘altered self’ activates drug-specific T cells. (D) CD8+ cells recognise drug–protein adducts on the plasma membrane of hepatocytes when presented with MHC I molecules, leading to immunological destruction of hepatocytes.
Clinical applications
The majority of HLA alleles associated with DILI have a very high negative predictive value of >0.95. Therefore, genotyping can be used to rule out adverse hepatic reactions due to particular drugs (listed in table 3) so that alternative diagnoses are considered.20 The HLA-B*35:02 genotype is a useful diagnostic test in the setting of suspected minocycline DILI, especially in distinguishing it from idiopathic AIH as both conditions share similar serological markers such as ANAs and SMAs.124 A high negative predictive value of a genetic test can also be used to identify the correct agent underlying DILI when the patient has been exposed to two concomitant medications.
Overall, the strength of association between HLA genotypes and DILI has raised controversy (eg, in relation to lumiracoxib) regarding the use of genetic testing in risk stratification.125 The incidence of DILI is less than 1 in 10 000 for most drugs used in clinical practice126 and thus too low for preprescription genotyping to be cost-effective at present. It is foreseeable, however, that personal genetic information such as the HLA profile may become routinely accessible to assist precision medicine and to minimise adverse drug reactions.
Expert summary
DILI has raised less awareness in routine patient care than it has in the regulatory and industry setting, where DILI is a leading cause of drug attrition and a major safety issue. Acute liver failure induced by a drug in clinical practice requires immediate supportive management of the patient and referral to a liver transplantation unit if the clinical situation deteriorates. Even with a test system in place that could accurately predict a patient's risk to develop liver failure, the likelihood that this would be routinely employed is low given the rarity of the event. This is in marked contrast to the situation in drug development, where pharmaceutical industry and regulators alike are frequently confronted with liver safety issues requiring expert assessment to quantify the risk and to implement an appropriate action scheme. Several examples of drug failures during development over the last 20 years underscore the need to develop new diagnostic tools and predictive systems which help to manage the challenge imposed by DILI. Genetic testing has identified HLA alleles that increase the risk of idiosyncratic reactions and in this sense has strengthened the pathophysiological concept. In a next step, diagnostic tools are required that assess this immunological risk. There is agreement that preclinical species are not useful for assessing the risk of idiosyncratic reactions, although certain intrinsic mechanisms of toxicity may be reproducible. In vitro tools which may predict a risk of DILI prior to first-in-human studies rely on human-derived cell assays to assess mitochondrial toxicity, inhibition of transporters, induction of oxidative stress and other endpoints. These test systems are used as supportive evidence but rarely trigger a decision with respect to the further development of a drug. Novel computer-based algorithms that integrate these in vitro readouts with structural properties are still at an early stage of development, but may offer potential as learning systems that correlate well-characterised compounds with clinical outcome.
This leaves the DILI community with the task of validating new biomarkers and in vitro tools for causality assessment which classify the type of injury and the risk associated with the observed biomarker pattern. How should a transient but rapidly reversible elevation of ALT to >20-fold ULN be interpreted? Biomarkers that inform us whether this rise in ALT was accompanied by immune activation would help us to classify the incident as an idiosyncratic reaction. Are all cases of DILI that fulfil the Hy's Law criteria in the same risk category or can new biomarkers help to define subcategories? Systematic measurement of new predictive biomarkers should be performed in patients in whom liver injury can be attributed to a specific causative drug by use of the RUCAM score. The choice of biomarkers is a major challenge that is being taken up by numerous initiatives such as the IMI Consortia, DILIN, Pro-Euro DILI and dedicated DILI groups within the International Consortium for Innovation and Quality in Pharmaceutical Development (IQ DILI) and the Council for International Organizations of Medical Sciences (CIOMS). Constructive dialogue and close collaboration of these networks with regulatory and academic DILI experts is but one example of the steps required to advance scientific and regulatory guidance for liver safety assessment and management.
Acknowledgments
Authors thank all partners of the DILI work package in the IMI SAFE-T Consortium that evaluated new biomarkers in DILI.
Footnotes
Contributors: GAK-U planned and wrote parts of the manuscript and contributed table 2; RJA wrote parts of the manuscript and contributed table 1 and figure 3; MM and PE wrote parts of the manuscript; AB contributed figures 1, 2 and 4; ALG wrote parts of the manuscript and GPA wrote parts of the manuscript and contributed table 3 and figure 5.
Funding: This work was supported by Swiss National Science Foundation grant no. 320030_144193 (to GAK-U).
Competing interests: AB and ALG have equity in the company MetaHeps GmbH. PE and MM are employees and GAK-U is a contractor of Novartis Pharma.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Lee WM. Drug-induced acute liver failure. Clin Liver Dis 2013;17:575–86, viii 10.1016/j.cld.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larrey D, Pageaux GP. Drug-induced acute liver failure. Eur J Gastroenterol Hepatol 2005;17:141–3. 10.1097/00042737-200502000-00002 [DOI] [PubMed] [Google Scholar]
- 3.Fontana RJ, Hayashi PH, Gu J, et al. Idiosyncratic drug-induced liver injury is associated with substantial morbidity and mortality within 6 months from onset. Gastroenterology 2014;147:96–108.e4. 10.1053/j.gastro.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sgro C, Clinard F, Ouazir K, et al. Incidence of drug-induced hepatic injuries: a French population-based study. Hepatology 2002;36:451–5. 10.1053/jhep.2002.34857 [DOI] [PubMed] [Google Scholar]
- 5.Björnsson ES, Bergmann OM, Björnsson HK, et al. Incidence, presentation, and outcomes in patients with drug-induced liver injury in the general population of Iceland. Gastroenterology 2013;144:1419–25, 25.e1–3; quiz e19–20 10.1053/j.gastro.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 6.Stevens JL, Baker TK. The future of drug safety testing: expanding the view and narrowing the focus. Drug Discov Today 2009;14:162–7. 10.1016/j.drudis.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 7.Lewis JH. ‘Hy's law,’ the ‘Rezulin Rule,’ and other predictors of severe drug-induced hepatotoxicity: putting risk-benefit into perspective. Pharmacoepidemiol Drug Saf 2006;15:221–9. 10.1002/pds.1209 [DOI] [PubMed] [Google Scholar]
- 8.Nichols WG, Steel HM, Bonny T, et al. Hepatotoxicity observed in clinical trials of aplaviroc (GW873140). Antimicrob Agents Chemother 2008;52:858–65. 10.1128/AAC.00821-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee WM, Larrey D, Olsson R, et al. Hepatic findings in long-term clinical trials of ximelagatran. Drug Saf 2005;28:351–70. 10.2165/00002018-200528040-00006 [DOI] [PubMed] [Google Scholar]
- 10.Schnitzer TJ, Burmester GR, Mysler E, et al. Comparison of lumiracoxib with naproxen and ibuprofen in the Therapeutic Arthritis Research and Gastrointestinal Event Trial (TARGET), reduction in ulcer complications: randomised controlled trial. Lancet 2004;364:665–74. 10.1016/S0140-6736(04)16893-1 [DOI] [PubMed] [Google Scholar]
- 11.Benza RL, Barst RJ, Galie N, et al. Sitaxsentan for the treatment of pulmonary arterial hypertension: a 1-year, prospective, open-label observation of outcome and survival. Chest 2008;134:775–82. 10.1378/chest.07-0767 [DOI] [PubMed] [Google Scholar]
- 12.Wysowski DK, Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969–2002: the importance of reporting suspected reactions. Arch Intern Med 2005;165:1363–9. 10.1001/archinte.165.12.1363 [DOI] [PubMed] [Google Scholar]
- 13.Watkins PB, Kaplowitz N, Slattery JT, et al. Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: a randomized controlled trial. JAMA 2006;296:87–93. 10.1001/jama.296.1.87 [DOI] [PubMed] [Google Scholar]
- 14.McGill MR, Jaeschke H. Metabolism and disposition of acetaminophen: recent advances in relation to hepatotoxicity and diagnosis. Pharm Res 2013;30:2174–87. 10.1007/s11095-013-1007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGill MR, Sharpe MR, Williams CD, et al. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest 2012;122:1574–83. 10.1172/JCI59755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuben A, Koch DG, Lee WM, Acute Liver Failure Study Group. Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology 2010;52:2065–76. 10.1002/hep.23937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan L, Kaplowitz N. Mechanisms of drug-induced liver injury. Clin Liver Dis 2013;17:507–18, vii 10.1016/j.cld.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov 2005;4:489–99. 10.1038/nrd1750 [DOI] [PubMed] [Google Scholar]
- 19.Russmann S, Jetter A, Kullak-Ublick GA. Pharmacogenetics of drug-induced liver injury. Hepatology 2010;52:748–61. 10.1002/hep.23720 [DOI] [PubMed] [Google Scholar]
- 20.Aithal GP. Pharmacogenetic testing in idiosyncratic drug-induced liver injury: current role in clinical practice. Liver Int 2015;35:1801–8. 10.1111/liv.12836 [DOI] [PubMed] [Google Scholar]
- 21.Lee KW, Chan SL. Hepatotoxicity of targeted therapy for cancer. Expert Opin Drug Metab Toxicol 2016;12:789–802. 10.1080/17425255.2016.1190831 [DOI] [PubMed] [Google Scholar]
- 22.Takeda M, Okamoto I, Nakagawa K. Pooled safety analysis of EGFR-TKI treatment for EGFR mutation-positive non-small cell lung cancer. Lung Cancer 2015;88:74–9. 10.1016/j.lungcan.2015.01.026 [DOI] [PubMed] [Google Scholar]
- 23. https://www.fda.gov/downloads/Drugs/.../Guidances/UCM174090.pdf.
- 24.Kullak-Ublick GA, Merz M, Griffel L, et al. Liver safety assessment in special populations (hepatitis B, C, and oncology trials). Drug Saf 2014;37(Suppl 1):S57–62. 10.1007/s40264-014-0186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiler S, Merz M, Kullak-Ublick GA. Drug-induced liver injury: the dawn of biomarkers? F1000Prime Rep 2015;7:34 10.12703/P7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. http://www.fda.gov/Drugs/DevelopmentApprovalProcess/ucm434382.htm.
- 27. http://www.ema.europa.eu/docs/en_GB/document_library/Other/2016/09/WC500213479.pdf.
- 28.Chalasani NP, Hayashi PH, Bonkovsky HL, et al. ACG Clinical Guideline: the diagnosis and management of idiosyncratic drug-induced liver injury. Am J Gastroenterol 2014;109:950–66; quiz 67 10.1038/ajg.2014.131 [DOI] [PubMed] [Google Scholar]
- 29.Danan G, Teschke R. RUCAM in drug and herb induced liver injury: the update. Int J Mol Sci 2015;17:E14 10.3390/ijms17010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Cortés M, Stephens C, Lucena MI, et al. Spanish Group for the Study of Drug-Induced Liver Disease (Grupo de Estudio para las Hepatopatías Asociadas a Medicamentos GEHAM). Causality assessment methods in drug induced liver injury: strengths and weaknesses. J Hepatol 2011;55:683–91. 10.1016/j.jhep.2011.02.007 [DOI] [PubMed] [Google Scholar]
- 31.Hayashi PH. Drug-induced liver injury network causality assessment: criteria and experience in the United States. Int J Mol Sci 2016;17:201 10.3390/ijms17020201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockey DC, Seeff LB, Rochon J, et al. Causality assessment in drug-induced liver injury using a structured expert opinion process: comparison to the Roussel-Uclaf causality assessment method. Hepatology 2010;51:2117–26. 10.1002/hep.23577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teschke R, Schwarzenboeck A, Eickhoff A, et al. Clinical and causality assessment in herbal hepatotoxicity. Expert Opin Drug Saf 2013;12:339–66. 10.1517/14740338.2013.774371 [DOI] [PubMed] [Google Scholar]
- 34.Navarro VJ, Barnhart H, Bonkovsky HL, et al. Liver injury from herbals and dietary supplements in the U.S. Drug-Induced Liver Injury Network. Hepatology 2014;60:1399–408. 10.1002/hep.27317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devarbhavi H, Raj S, Aradya VH, et al. Drug-induced liver injury associated with Stevens-Johnson syndrome/toxic epidermal necrolysis: patient characteristics, causes, and outcome in 36 cases. Hepatology 2016;63:993–9. 10.1002/hep.28270 [DOI] [PubMed] [Google Scholar]
- 36.Andrade RJ, Lucena MI, Fernández MC, et al. Drug-induced liver injury: an analysis of 461 incidences submitted to the Spanish registry over a 10-year period. Gastroenterology 2005;129:512–21. 10.1016/j.gastro.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 37.Chalasani N, Bonkovsky HL, Fontana R, et al. Features and Outcomes of 899 Patients With Drug-Induced Liver Injury: The DILIN Prospective Study. Gastroenterology 2015;148:1340–52.e7. 10.1053/j.gastro.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrade RJ, Robles M, Fernández-Castañer A, et al. Assessment of drug-induced hepatotoxicity in clinical practice: a challenge for gastroenterologists. World J Gastroenterol 2007;13:329–40. 10.3748/wjg.v13.i3.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davern TJ, Chalasani N, Fontana RJ, et al. Acute hepatitis E infection accounts for some cases of suspected drug-induced liver injury. Gastroenterology 2011;141:1665–72.e1–9. 10.1053/j.gastro.2011.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega-Alonso A, Stephens C, Lucena MI, et al. Case characterization, clinical features and risk factors in drug-induced liver injury. Int J Mol Sci 2016;17:E714 10.3390/ijms17050714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aithal GP, Watkins PB, Andrade RJ, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther 2011;89:806–15. 10.1038/clpt.2011.58 [DOI] [PubMed] [Google Scholar]
- 42.Zimmerman HJ. The spectrum of hepatotoxicity. Perspect Biol Med 1968;12:135–61. 10.1353/pbm.1968.0004 [DOI] [PubMed] [Google Scholar]
- 43.Björnsson E, Olsson R. Outcome and prognostic markers in severe drug-induced liver disease. Hepatology 2005;42:481–9. 10.1002/hep.20800 [DOI] [PubMed] [Google Scholar]
- 44.Robles-Diaz M, Lucena MI, Kaplowitz N, et al. Use of Hy's law and a new composite algorithm to predict acute liver failure in patients with drug-induced liver injury. Gastroenterology 2014;147:109–18.e5. 10.1053/j.gastro.2014.03.050 [DOI] [PubMed] [Google Scholar]
- 45.de Boer YS, Kosinski AS, Urban TJ, et al. Features of autoimmune hepatitis in patients with drug-induced liver injury. Clin Gastroenterol Hepatol 2017;15:103–112.e2. 10.1016/j.cgh.2016.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucena MI, Kaplowitz N, Hallal H, et al. Recurrent drug-induced liver injury (DILI) with different drugs in the Spanish Registry: the dilemma of the relationship to autoimmune hepatitis. J Hepatol 2011;55:820–7. 10.1016/j.jhep.2010.12.041 [DOI] [PubMed] [Google Scholar]
- 47.deLemos AS, Foureau DM, Jacobs C, et al. Drug-induced liver injury with autoimmune features. Semin Liver Dis 2014;34:194–204. 10.1055/s-0034-1375959 [DOI] [PubMed] [Google Scholar]
- 48.Suzuki A, Brunt EM, Kleiner DE, et al. The use of liver biopsy evaluation in discrimination of idiopathic autoimmune hepatitis versus drug-induced liver injury. Hepatology 2011;54:931–9. 10.1002/hep.24481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foureau DM, Walling TL, Maddukuri V, et al. Comparative analysis of portal hepatic infiltrating leucocytes in acute drug-induced liver injury, idiopathic autoimmune and viral hepatitis. Clin Exp Immunol 2015;180:40–51. 10.1111/cei.12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrade RJ, Robles M, Lucena MI. Rechallenge in drug-induced liver injury: the attractive hazard. Expert Opin Drug Saf 2009;8:709–14. 10.1517/14740330903397378 [DOI] [PubMed] [Google Scholar]
- 51.Senior JR. Can rechallenge be done safely after mild or moderate drug-induced liver injury? Hepatology 2016;63:691–3. 10.1002/hep.28353 [DOI] [PubMed] [Google Scholar]
- 52.Medina-Caliz I, Robles-Diaz M, Garcia-Muñoz B, et al. Definition and risk factors for chronicity following acute idiosyncratic drug-induced liver injury. J Hepatol 2016;65:532–42. 10.1016/j.jhep.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LeCluyse EL, Alexandre E, Hamilton GA, et al. Isolation and culture of primary human hepatocytes. Methods Mol Biol 2005;290:207–29. [DOI] [PubMed] [Google Scholar]
- 54.Gómez-Lechón MJ, Lahoz A, Gombau L, et al. In vitro evaluation of potential hepatotoxicity induced by drugs. Curr Pharm Des 2010;16:1963–77. 10.2174/138161210791208910 [DOI] [PubMed] [Google Scholar]
- 55.Anson BD, Kolaja KL, Kamp TJ. Opportunities for use of human iPS cells in predictive toxicology. Clin Pharmacol Ther 2011;89:754–8. 10.1038/clpt.2011.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Einhorn S, Venkatarangan L, et al. Morphological and functional characterization and assessment of iPSC-derived hepatocytes for in vitro toxicity testing. Toxicol Sci 2015;147:39–54. 10.1093/toxsci/kfv117 [DOI] [PubMed] [Google Scholar]
- 57.Atienzar FA, Blomme EA, Chen M, et al. Key challenges and opportunities associated with the use of in vitro models to detect human DILI: integrated risk assessment and mitigation plans. Biomed Res Int 2016;2016: 9737920 10.1155/2016/9737920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godoy P, Hewitt NJ, Albrecht U, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013;87:1315–530. 10.1007/s00204-013-1078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schadt S, Simon S, Kustermann S, et al. Minimizing DILI risk in drug discovery—a screening tool for drug candidates. Toxicol In Vitro 2015;30:429–37. 10.1016/j.tiv.2015.09.019 [DOI] [PubMed] [Google Scholar]
- 60.Tateno C, Miya F, Wake K, et al. Morphological and microarray analyses of human hepatocytes from xenogeneic host livers. Lab Invest 2013;93:54–71. 10.1038/labinvest.2012.158 [DOI] [PubMed] [Google Scholar]
- 61.Xu D, Wu M, Nishimura S, et al. Chimeric TK-NOG mice: a predictive model for cholestatic human liver toxicity. J Pharmacol Exp Ther 2015;352:274–80. 10.1124/jpet.114.220798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boelsterli UA, Hsiao CJ. The heterozygous Sod2(+/-) mouse: modeling the mitochondrial role in drug toxicity. Drug Discov Today 2008;13:982–8. 10.1016/j.drudis.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 63.Lee YH, Chung MC, Lin Q, et al. Troglitazone-induced hepatic mitochondrial proteome expression dynamics in heterozygous Sod2(+/-) mice: two-stage oxidative injury. Toxicol Appl Pharmacol 2008;231:43–51. 10.1016/j.taap.2008.03.025 [DOI] [PubMed] [Google Scholar]
- 64.Benesic A, Rahm NL, Ernst S, et al. Human monocyte-derived cells with individual hepatocyte characteristics: a novel tool for personalized in vitro studies. Lab Invest 2012;92:926–36. 10.1038/labinvest.2012.64 [DOI] [PubMed] [Google Scholar]
- 65.Benesic A, Gerbes AL. Drug-induced liver injury and individual cell models. Dig Dis 2015;33:486–91. 10.1159/000374094 [DOI] [PubMed] [Google Scholar]
- 66.Benesic A, Leitl A, Gerbes AL. Monocyte-derived hepatocyte-like cells for causality assessment of idiosyncratic drug-induced liver injury. Gut 2016;65:1555–63. 10.1136/gutjnl-2015-309528 [DOI] [PubMed] [Google Scholar]
- 67.Danan G, Benichou C. Causality assessment of adverse reactions to drugs—I. A novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol 1993;46:1323–30. [DOI] [PubMed] [Google Scholar]
- 68.Chen M, Vijay V, Shi Q, et al. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov Today 2011;16:697–703. 10.1016/j.drudis.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 69.Chen M, Suzuki A, Thakkar S, et al. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov Today 2016;21:648–53. 10.1016/j.drudis.2016.02.015 [DOI] [PubMed] [Google Scholar]
- 70.Chen M, Borlak J, Tong W. High lipophilicity and high daily dose of oral medications are associated with significant risk for drug-induced liver injury. Hepatology 2013;58:388–96. 10.1002/hep.26208 [DOI] [PubMed] [Google Scholar]
- 71.Yu K, Geng X, Chen M, et al. High daily dose and being a substrate of cytochrome P450 enzymes are two important predictors of drug-induced liver injury. Drug Metab Dispos 2014;42:744–50. 10.1124/dmd.113.056267 [DOI] [PubMed] [Google Scholar]
- 72.Chen M, Borlak J, Tong W. A model to predict severity of drug-induced liver injury in humans. Hepatology 2016;64:931–40. 10.1002/hep.28678 [DOI] [PubMed] [Google Scholar]
- 73.Aleo MD, Luo Y, Swiss R, et al. Human drug-induced liver injury severity is highly associated with dual inhibition of liver mitochondrial function and bile salt export pump. Hepatology 2014;60:1015–22. 10.1002/hep.27206 [DOI] [PubMed] [Google Scholar]
- 74.Stieger B, Fattinger K, Madon J, et al. Drug- and estrogen-induced cholestasis through inhibition of the hepatocellular bile salt export pump (Bsep) of rat liver. Gastroenterology 2000;118:422–30. 10.1016/S0016-5085(00)70224-1 [DOI] [PubMed] [Google Scholar]
- 75.Morgan RE, Trauner M, van Staden CJ, et al. Interference with bile salt export pump function is a susceptibility factor for human liver injury in drug development. Toxicol Sci 2010;118:485–500. 10.1093/toxsci/kfq269 [DOI] [PubMed] [Google Scholar]
- 76.Dawson S, Stahl S, Paul N, et al. In vitro inhibition of the bile salt export pump correlates with risk of cholestatic drug-induced liver injury in humans. Drug Metab Dispos 2012;40:130–8. 10.1124/dmd.111.040758 [DOI] [PubMed] [Google Scholar]
- 77. https://c-path.org/current-trends-in-bsep-inhibition-and-perturbation-to-bile-acid-homeostasis-as-mechanisms-of-drug-induced-liver-injury/
- 78.Woodhead JL, Yang K, Siler SQ, et al. Exploring BSEP inhibition-mediated toxicity with a mechanistic model of drug-induced liver injury. Front Pharmacol 2014;5:240 10.3389/fphar.2014.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Longo DM, Yang Y, Watkins PB, et al. Elucidating differences in the hepatotoxic potential of tolcapone and entacapone with DILIsym(®), a mechanistic model of drug-induced liver injury. CPT Pharmacometrics Syst Pharmacol 2016;5:31–9. 10.1002/psp4.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuepfer L, Kerb R, Henney AM. Clinical translation in the virtual liver network. CPT Pharmacometrics Syst Pharmacol 2014;3:e127 10.1038/psp.2014.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009;106:4402–7. 10.1073/pnas.0813371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Starkey Lewis PJ, Dear J, Platt V, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011;54:1767–76. 10.1002/hep.24538 [DOI] [PubMed] [Google Scholar]
- 83.Antoine DJ, Dear JW, Lewis PS, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology 2013;58:777–87. 10.1002/hep.26294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thulin P, Nordahl G, Gry M, et al. Keratin-18 and microRNA-122 complement alanine aminotransferase as novel safety biomarkers for drug-induced liver injury in two human cohorts. Liver Int 2014;34:367–78. 10.1111/liv.12322 [DOI] [PubMed] [Google Scholar]
- 85.Singhal R, Harrill AH, Menguy-Vacheron F, et al. Benign elevations in serum aminotransferases and biomarkers of hepatotoxicity in healthy volunteers treated with cholestyramine. BMC Pharmacol Toxicol 2014;15:42 10.1186/2050-6511-15-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Starkey Lewis PJ, Merz M, Couttet P, et al. Serum microRNA biomarkers for drug-induced liver injury. Clin Pharmacol Ther 2012;92:291–3. 10.1038/clpt.2012.101 [DOI] [PubMed] [Google Scholar]
- 87.Antoine DJ, Jenkins RE, Dear JW, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol 2012;56:1070–9. 10.1016/j.jhep.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Joka D, Wahl K, Moeller S, et al. Prospective biopsy-controlled evaluation of cell death biomarkers for prediction of liver fibrosis and nonalcoholic steatohepatitis. Hepatology 2012;55:455–64. 10.1002/hep.24734 [DOI] [PubMed] [Google Scholar]
- 89.Mikus M, Drobin K, Gry M, et al. Elevated levels of circulating CDH5 and FABP1 in association with human drug-induced liver injury. Liver Int 2017;37:132–40. 10.1111/liv.13174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pelsers MM, Morovat A, Alexander GJ, et al. Liver fatty acid-binding protein as a sensitive serum marker of acute hepatocellular damage in liver transplant recipients. Clin Chem 2002;48:2055–7. [PubMed] [Google Scholar]
- 91.Akbal E, Köklü S, Koçak E, et al. Liver fatty acid-binding protein is a diagnostic marker to detect liver injury due to chronic hepatitis C infection. Arch Med Res 2013;44:34–8. 10.1016/j.arcmed.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 92.Harrill AH, Roach J, Fier I, et al. The effects of heparins on the liver: application of mechanistic serum biomarkers in a randomized study in healthy volunteers. Clin Pharmacol Ther 2012;92:214–20. 10.1038/clpt.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schomaker S, Warner R, Bock J, et al. Assessment of emerging biomarkers of liver injury in human subjects. Toxicol Sci 2013;132:276–83. 10.1093/toxsci/kft009 [DOI] [PubMed] [Google Scholar]
- 94.Beckett GJ, Foster GR, Hussey AJ, et al. Plasma glutathione S-transferase and F protein are more sensitive than alanine aminotransferase as markers of paracetamol (acetaminophen)-induced liver damage. Clin Chem 1989;35:2186–9. [PubMed] [Google Scholar]
- 95.Bailey WJ, Holder D, Patel H, et al. A performance evaluation of three drug-induced liver injury biomarkers in the rat: alpha-glutathione S-transferase, arginase 1, and 4-hydroxyphenyl-pyruvate dioxygenase. Toxicol Sci 2012;130:229–44. 10.1093/toxsci/kfs243 [DOI] [PubMed] [Google Scholar]
- 96.Ramaiah SK, Rittling S. Pathophysiological role of osteopontin in hepatic inflammation, toxicity, and cancer. Toxicol Sci 2008;103:4–13. 10.1093/toxsci/kfm246 [DOI] [PubMed] [Google Scholar]
- 97.Arriazu E, Ge X, Leung TM, et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut Published online first: 27 January 2016 10.1136/gutjnl-2015-310752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersson U, Lindberg J, Wang S, et al. A systems biology approach to understanding elevated serum alanine transaminase levels in a clinical trial with ximelagatran. Biomarkers 2009;14:572–86. 10.3109/13547500903261354 [DOI] [PubMed] [Google Scholar]
- 99.McGill MR, Jaeschke H. Mechanistic biomarkers in acetaminophen-induced hepatotoxicity and acute liver failure: from preclinical models to patients. Expert Opin Drug Metab Toxicol 2014;10:1005–17. 10.1517/17425255.2014.920823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clarke JI, Dear JW, Antoine DJ. Recent advances in biomarkers and therapeutic interventions for hepatic drug safety—false dawn or new horizon? Expert Opin Drug Saf 2016;15:625–34. 10.1517/14740338.2016.1160057 [DOI] [PubMed] [Google Scholar]
- 101.Woolbright BL, McGill MR, Staggs VS, et al. Glycodeoxycholic acid levels as prognostic biomarker in acetaminophen-induced acute liver failure patients. Toxicol Sci 2014;142:436–44. 10.1093/toxsci/kfu195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schadt HS, Wolf A, Pognan F, et al. Bile acids in drug induced liver injury: key players and surrogate markers. Clin Res Hepatol Gastroenterol 2016;40:257–66. 10.1016/j.clinre.2015.12.017 [DOI] [PubMed] [Google Scholar]
- 103.Luo L, Schomaker S, Houle C, et al. Evaluation of serum bile acid profiles as biomarkers of liver injury in rodents. Toxicol Sci 2014;137:12–25. 10.1093/toxsci/kft221 [DOI] [PubMed] [Google Scholar]
- 104.De Berardinis V, Moulis C, Maurice M, et al. Human microsomal epoxide hydrolase is the target of germander-induced autoantibodies on the surface of human hepatocytes. Mol Pharmacol 2000;58:542–51. [DOI] [PubMed] [Google Scholar]
- 105.Obermayer-Straub P, Strassburg CP, Manns MP. Target proteins in human autoimmunity: cytochromes P450 and UDP- glucuronosyltransferases. Can J Gastroenterol 2000;14:429–39. 10.1155/2000/910107 [DOI] [PubMed] [Google Scholar]
- 106.Sutti S, Rigamonti C, Vidali M, et al. CYP2E1 autoantibodies in liver diseases. Redox Biol 2014;3:72–8. 10.1016/j.redox.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Metushi IG, Sanders C, Acute Liver Study Group, Lee WM, et al. Detection of anti-isoniazid and anti-cytochrome P450 antibodies in patients with isoniazid-induced liver failure. Hepatology 2014;59:1084–93. 10.1002/hep.26564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao H, Ruan JQ, Chen J, et al. Blood pyrrole-protein adducts as a diagnostic and prognostic index in pyrrolizidine alkaloid-hepatic sinusoidal obstruction syndrome. Drug Des Devel Ther 2015;9:4861–8. 10.2147/DDDT.S87858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hindorff LA, Morales J, Junkins HA, et al. A Catalog of Published Genome-Wide Association Studies. http://www.genome.gov/gwastudies
- 110.Kindmark A, Jawaid A, Harbron CG, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J 2008;8: 186–95. 10.1038/sj.tpj.6500458 [DOI] [PubMed] [Google Scholar]
- 111.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet 2009;41: 816–19. 10.1038/ng.379 [DOI] [PubMed] [Google Scholar]
- 112.Singer JB, Lewitzky S, Leroy E, et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet 2010;42:711–14. 10.1038/ng.632 [DOI] [PubMed] [Google Scholar]
- 113.Spraggs CF, Budde LR, Briley LP, et al. HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol 2011;29:667–73. 10.1200/JCO.2010.31.3197 [DOI] [PubMed] [Google Scholar]
- 114.Parham LR, Briley LP, Li L, et al. Comprehensive genome-wide evaluation of lapatinib-induced liver injury yields a single genetic signal centered on known risk allele HLA-DRB1*07:01. Pharmacogenomics J 2016;16:180–5. 10.1038/tpj.2015.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lucena MI, Molokhia M, Shen Y, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology 2011;141:338–47. 10.1053/j.gastro.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Urban TJ, Shen Y, Stolz A, et al. Limited contribution of common genetic variants to risk for liver injury due to a variety of drugs. Pharmacogenet Genomics 2012;22:784–95. 10.1097/FPC.0b013e3283589a76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nicoletti P, Werk AN, Sawle A, et al. HLA-DRB1*16: 01-DQB1*05: 02 is a novel genetic risk factor for flupirtine-induced liver injury. Pharmacogenet Genomics 2016;26:218–24. 10.1097/FPC.0000000000000209 [DOI] [PubMed] [Google Scholar]
- 118.Nicoletti P, Aithal GP, Bjornsson ES, et al. Association of liver injury from specific drugs, or groups of drugs, with polymorphisms in HLA and other genes in a genome-wide association study. Gastroenterology 2016. doi:10.1053/j.gastro.2016.12.016 [Epub ahead of print 30 Dec 2016]. 10.1053/j.gastro.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Urban TJ, Nicoletti P, Chalasani N, et al. Minocycline hepatotoxicity: clinical characterization and identification of HLA-B 35:02 as a risk factor. Hepatology 2015;62(Suppl 1):1149A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aithal GP, Grove JI. Genome-wide association studies in drug-induced liver injury: step change in understanding the pathogenesis. Semin Liver Dis 2015;35:421–31. 10.1055/s-0035-1567829 [DOI] [PubMed] [Google Scholar]
- 121.Donaldson PT, Daly AK, Henderson J, et al. Human leucocyte antigen class II genotype in susceptibility and resistance to co-amoxiclav-induced liver injury. J Hepatol 2010;53:1049–53. 10.1016/j.jhep.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 122.Heap GA, Weedon MN, Bewshea CM, et al. HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat Genet 2014;46:1131–4. 10.1038/ng.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grove JI, Aithal GP. Human leukocyte antigen genetic risk factors of drug-induced liver toxicology. Expert Opin Drug Metab Toxicol 2015;11:395–409. 10.1517/17425255.2015.992414 [DOI] [PubMed] [Google Scholar]
- 124.Björnsson E, Aithal G. Immune-mediated drug-induced liver injury. In: Gershwin ME, Vierling JM, Manns MP, eds. Liver immunology. Springer International Publishing, 2014:401–12. [Google Scholar]
- 125.Aithal GP, Daly AK. Preempting and preventing drug-induced liver injury. Nat Genet 2010;42:650–1. 10.1038/ng0810-650 [DOI] [PubMed] [Google Scholar]
- 126.Tujios S, Fontana RJ. Mechanisms of drug-induced liver injury: from bedside to bench. Nat Rev Gastroenterol Hepatol 2011;8:202–11. 10.1038/nrgastro.2011.22 [DOI] [PubMed] [Google Scholar]