Abstract
Controlling plant disease has been a struggle for mankind since the advent of agriculture. Studies of plant immune mechanisms have led to strategies of engineering resistant crops through ectopic transcription of plants’ own defence genes, such as the master immune regulatory gene NPR11. However, enhanced resistance obtained through such strategies is often associated with significant penalties to fitness2, making the resulting products undesirable for agricultural applications. To remedy this problem, we sought more stringent mechanisms of expressing defence proteins. Based on our latest finding that translation of key immune regulators, such as TBF13, is rapidly and transiently induced upon pathogen challenge (accompanying manuscript), we developed “TBF1-cassette” consisting of not only the immune-inducible promoter but also two pathogen-responsive upstream open reading frames (uORFsTBF1) of the TBF1 gene. We demonstrate that inclusion of the uORFsTBF1-mediated translational control over the production of snc1 (an autoactivated immune receptor) in Arabidopsis (At) and AtNPR1 in rice enables us to engineer broad-spectrum disease resistance without compromising plant fitness in the laboratory or in the field. This broadly applicable new strategy may lead to reduced use of pesticides and lightening of selective pressure for resistant pathogens.
To meet the demand for food production caused by the explosion in world population while limiting the use of pesticides, which are potential pollutants, new strategies must be developed to control crop diseases. As an alternative to the traditional chemical and breeding methods, studies of plant immune mechanisms have made it possible to engineer resistance through ectopic expression of plants’ own resistance-conferring genes4. The first line of active defence in plants involves recognition of microbial/damage-associated molecular patterns (M/DAMPs) by host pattern-recognizing receptors (PRRs) in pattern-triggered immunity (PTI)5. Ectopic expression of PRRs for MAMPs6, 7 and the DAMP signal eATP8, as well as in vivo release of the DAMP molecules, oligogalacturonides9, have all been shown to enhance resistance in transgenic plants. Besides PRR-mediated basal resistance, plant genomes encode hundreds of intracellular nucleotide-binding and leucine-rich repeat (NB-LRR) immune receptors (also known as “R proteins”) to detect the presence of pathogen effectors delivered inside plant cells10. Individual or stacked R genes have been transformed into plants to confer effector-triggered immunity (ETI)11, 12. In addition to PRR and R genes, NPR1 is another favourite gene used in engineering plant resistance4. Unlike immune receptors that are activated by specific MAMPs and pathogen effectors, NPR1 is a positive regulator of broad-spectrum resistance induced by a general plant immune signal, salicylic acid1. Overexpression of the Arabidopsis NPR1 (AtNPR1) could enhance resistance against a variety of pathogens in diverse plant families such as rice13–15.
A major challenge in engineering disease resistance, however, is to overcome the associated fitness costs2. In the absence of specialized immune cells, immune induction in plants involves switching from growth-related activities to defence3, 16. Plants normally avoid autoimmunity by tightly controlling transcription, mRNA nuclear export and degradation of defence proteins17. However, only transcriptional control has been used prevalently so far in engineering disease resistance2. Based on our global translatome analysis (accompanying manuscript), we discovered translation to be a fundamental layer of regulation during immune induction which can be explored to allow more stringent pathogen-inducible expression of defence proteins.
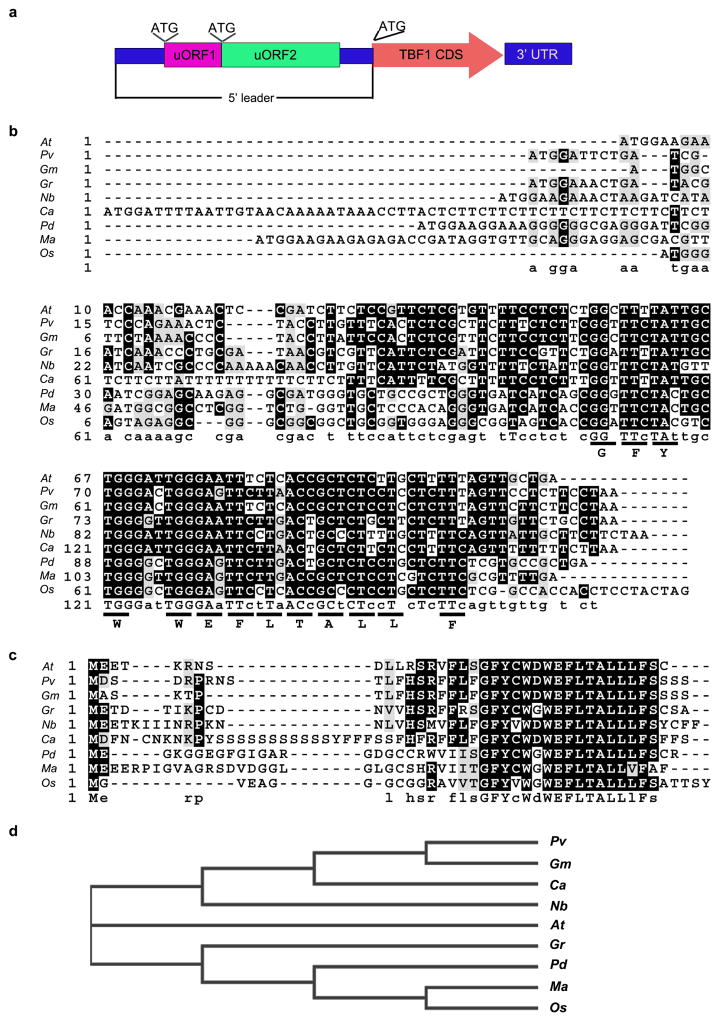
To test our hypothesis that tighter control of defence protein translation can minimize the fitness penalties associated with enhanced disease resistance, we used the TBF1 promoter (TBF1p) and the 5′ leader sequence (before the start codon for TBF1), which we designated as “TBF1-cassette”. TBF1 is an important transcription factor for the growth-to-defence switch upon immune induction. Translation of TBF1 is normally suppressed by two uORFs within the 5′ leader sequence3. BLAST analysis showed that uORF2TBF1, the major mRNA feature conferring the translational suppression (accompanying manuscript and ref3), is conserved across plant species (> 50% identity) (Extended Data Fig. 1), suggesting an evolutionarily conserved control mechanism and a potential use of TBF1-cassette to regulate defence protein production in plant species other than Arabidopsis.
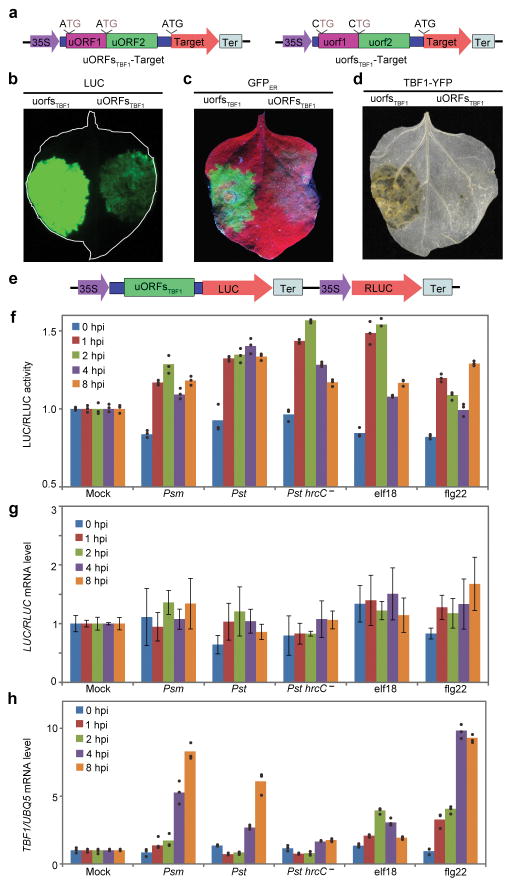
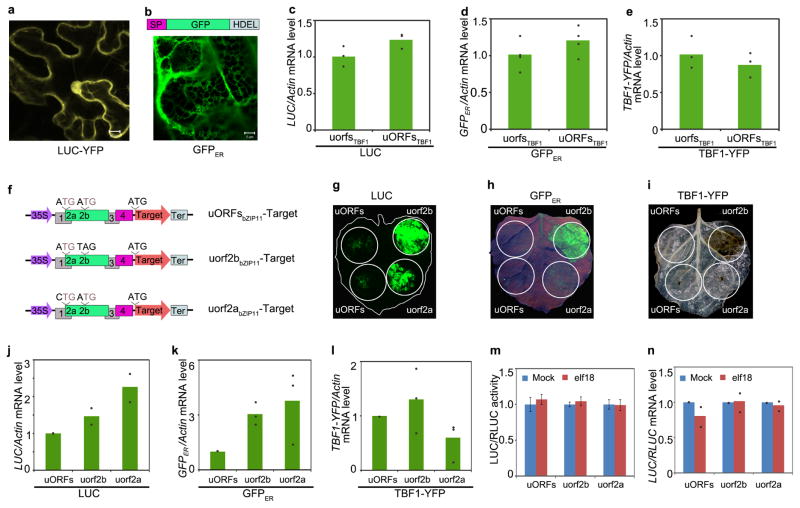
To explore the application of uORFsTBF1, we first demonstrated its capacity to control both cytosol- and ER-synthesized proteins (“Target”) using the firefly luciferase (LUC; Extended Data Fig. 2a) and GFPER (Extended Data Fig. 2b), respectively, as proxies through transient expression in Nicotiana benthamiana (N. benthamiana) (Fig. 1a–c, Extended Data Fig. 2c, d). This uORFsTBF1-mediated translational suppression was tight enough to prevent cell death induced by overexpression of TBF1 (TBF1-YFP) observed in 35S:uorfsTBF1-TBF1-YFP (Fig. 1d, Extended Data Fig. 2e). A similar repression activity was observed for uORF2bbZIP11 of the sucrose-responsive bZIP11 gene18 (Extended Data Fig. 2f–l). However, unlike uORFsTBF1, the uORF2bZIP11-mediated repression could not be alleviated by the MAMP signal elf18 (Extended Data Fig. 2m, n). These results support the potential utility of uORFsTBF1 in providing stringent control of cytosol- and ER-synthesized defence proteins specifically for engineering disease resistance.
Figure 1. uORFsTBF1-mediated translational and TBF1 promoter-mediated transcriptional regulation.
a, Schematics of WT uORFsTBF1 or mutant uorfsTBF1. b–d, LUC activity (b), GFPER fluorescence (c) and cell death induced by TBF1-YFP (d), representative of 6 images. e, Dual-luciferase system. f, Translational changes of the reporter to different treatments. Mean of the LUC/RLUC activity ratios normalized to Mock (n = 3). g, LUC/RLUC mRNA levels in (f). Mean ± s.d. of LUC/RLUC mRNA normalized to Mock (n = 6). h, Endogenous TBF1 mRNA levels (n = 3). UBQ5, internal control. Solid circles, individual biological replicates. See Extended Data Fig. 2.
To monitor the effect of uORFsTBF1 on translational efficiency (TE), a dual-luciferase system was constructed to calculate the ratio of LUC activity to the control renilla luciferase (RLUC) activity (Fig. 1e). The resulting transgenic plants were tested for responsiveness to bacterial pathogens Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326), Ps pv. tomato (Pst) DC3000, and the corresponding mutant of the type III secretion system Pst DC3000 hrcC−, as well as to MAMP signals, elf18 and flg22. The equally rapid induction in the reporter TE by all treatments suggests that it is likely a part of PTI, which does not involve bacterial type III effectors (Fig. 1f). The transient increases in translation were not correlated with significant changes in mRNA levels (Fig. 1g). In parallel, the endogenous TBF1 mRNA level was elevated at later time points than the translational increases observed using the reporter (Fig. 1h), suggesting that in response to pathogen challenge, translational induction may precede transcriptional reprogramming in plants.
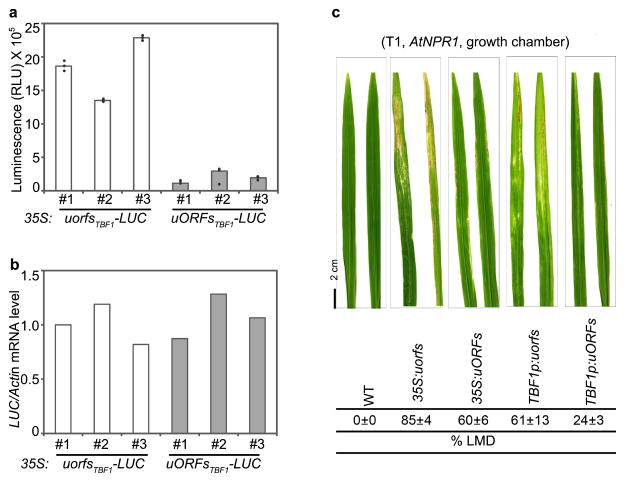
To engineer resistant plants using TBF1-cassette we picked two candidates from Arabidopsis, snc1-119 and NPR113. The Arabidopsis snc1-1 (for simplicity, snc1 from here on) is an autoactivated point mutant of the NB-LRR immune receptor SNC1. Even though the snc1 mutant plants have constitutively elevated resistance to various pathogens, their growth is significantly retarded19. Such a growth defect is also prevalent in transgenic plants ectopically expressing the WT SNC1 by either the 35S promoter or its native promoter20, 21, limiting the utility of SNC1, and perhaps other R genes, in engineering resistant plants. To overcome the fitness penalty associated with the snc1 mutant, we put it under the control of uORFsTBF1 driven by either the 35S promoter or TBF1p to create 35S:uORFsTBF1-snc1 and TBF1p:uORFsTBF1-snc1, respectively. As controls, we also generated 35S:uorfsTBF1-snc1 and TBF1p:uorfsTBF1-snc1, in which the start codons of the uORFs were mutated. The first generation of transgenic Arabidopsis (T1) with these four constructs displayed three distinct developmental phenotypes: Type I plants were small in rosette diameter, dwarf and with chlorosis; Type II plants were healthier but still dwarf; and Type III plants were indistinguishable from WT (Extended Data Fig. 3). We found that regulating either transcription or translation of snc1 significantly improved plant growth as judged by the increased percentage of Type III plants. The highest percentage of Type III plants were found in TBF1p:uORFsTBF1-snc1 transformants, in which snc1 was regulated by TBF1-cassette at both transcriptional and translational levels. The absence of Type I plants in these transformants clearly demonstrated the stringency of TBF1-cassette (Extended Data Fig. 3).
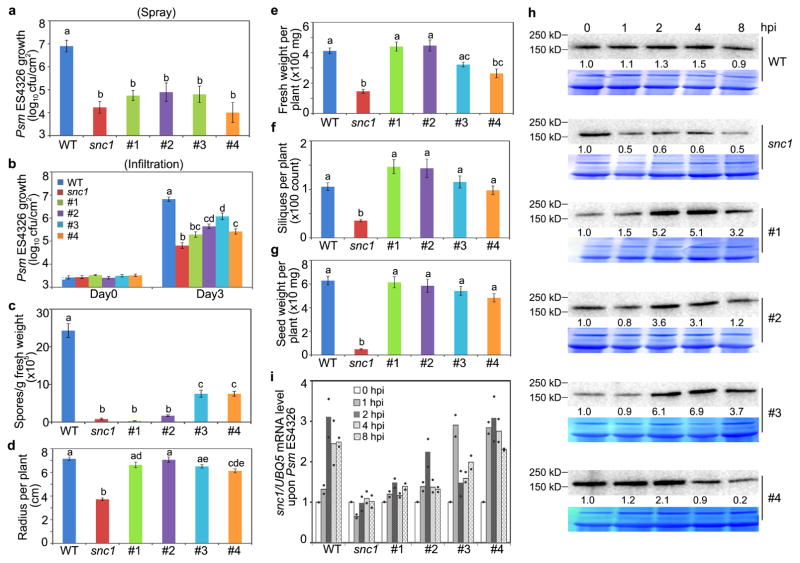
We propagated the transformants to obtain homozygotes for the transgene. For the TBF1p:uorfsTBF1-snc1 and 35S:uORFsTBF1-snc1 lines, homozygosity caused most of the Type III plants in T1 to show the Type II phenotype in T2. But for TBF1p:uORFsTBF1-snc1 transformants, they maintained their normal growth phenotype as homozygotes. We then picked four independent TBF1p:uORFsTBF1-snc1 lines for further disease resistance and fitness tests (Fig. 2a, b). We first showed that these transgenic lines indeed had elevated resistance to Psm ES4326 by either spray inoculation or infiltration (Fig. 2c, d, Extended Data Fig. 4a, b). They also displayed enhanced resistance to Hyaloperonospora arabidopsidis Noco2 (Hpa Noco2), an oomycete pathogen which causes downy mildew in Arabidopsis (Fig. 2e, f, Extended Data Fig. 4c). However, in contrast to snc1, these transgenic lines showed almost the same fitness as WT, including total seed weight per plant (Fig. 2g–i, Extended Data Fig. 4d–g). Upon Psm ES4326 challenge, we detected significant increases in the snc1 protein within 2 hpi in all four TBF1p:uORFsTBF1-snc1 transgenic lines, but not in WT or snc1 (Extended Data Fig. 4h, i). These data provide a proof of concept that adding pathogen-inducible translational control is an effective way to enhance plant resistance without fitness costs.
Figure 2. Effects of controlling transcription and translation of snc1 in Arabidopsis.
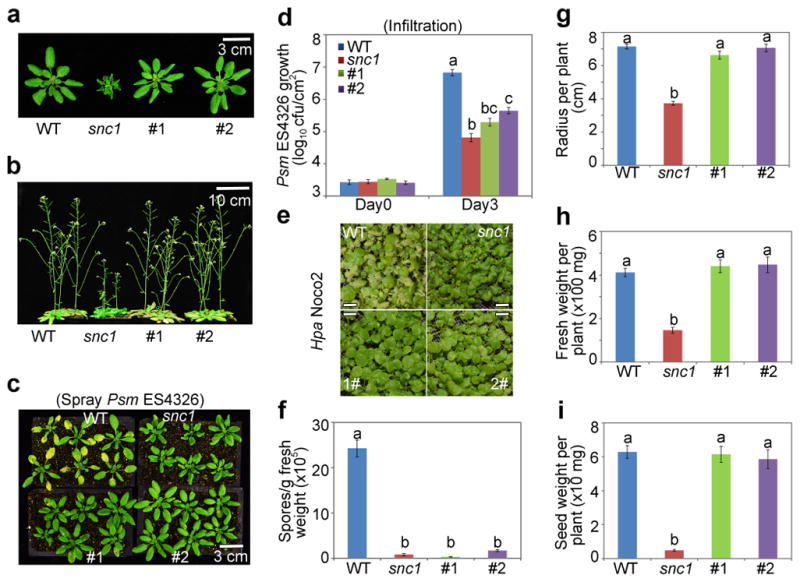
a, b, Effects on vegetative and reproductive growth. snc1, autoactivated mutant. #1 and #2, independent lines carrying TBF1p:uORFsTBF1-snc1. Representative of 5 images. c, d, Psm ES4326 growth after inoculation by spray (c) or infiltration (d). e, f, Photos (representative of 6 images; scale bar, 0.5 cm) and quantification of Hpa Noco2. g–i, Rosette radius, fresh weight and total seed weight. Mean ± s.e.m.. Letters above indicate significant differences (P < 0.05). See Source Data for sample size (n) and Extended Data Fig. 4 for two additional lines.
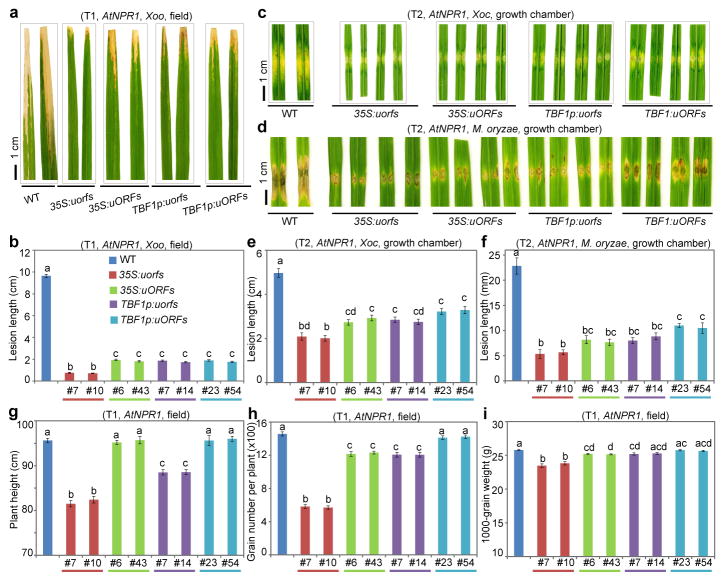
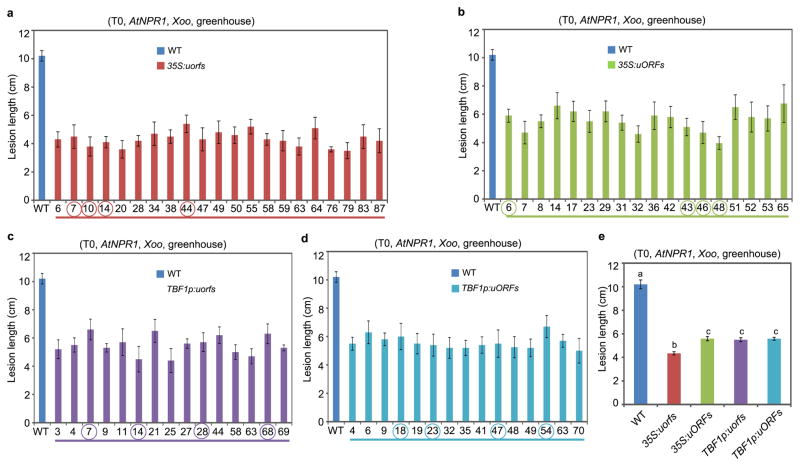
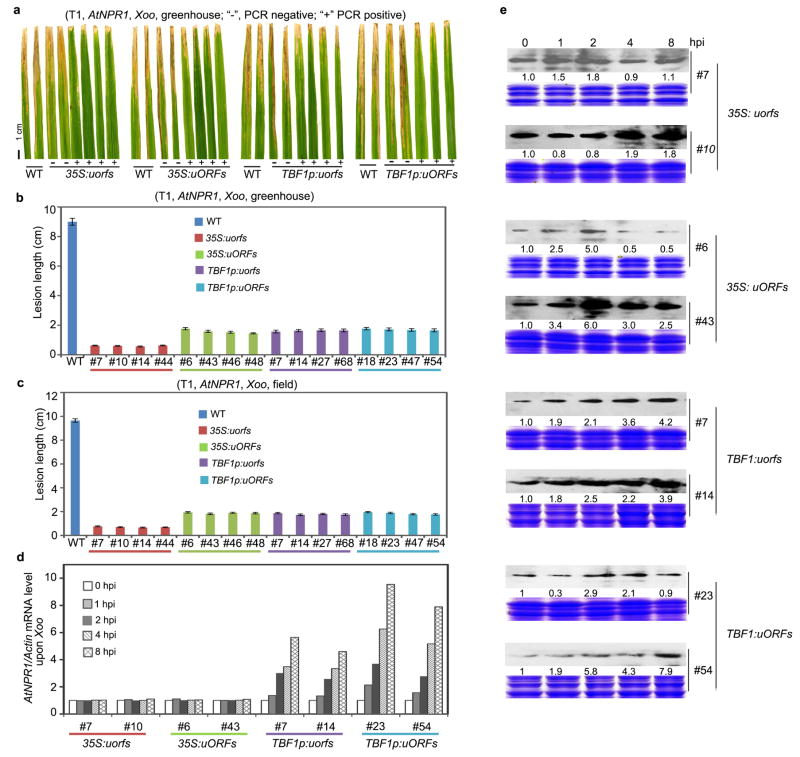
We next applied TBF1-cassette to engineering resistance in rice, which is one of the most important staple crops in the world. Using 35S:uORFsTBF1-LUC and 35S:uorfsTBF1-LUC (Fig. 1b), we first showed that the Arabidopsis uORFsTBF1 could suppress translation without significantly influencing mRNA levels in the rice (Oryza sativa) cultivar ZH11 (Extended Data Fig. 5a, b). We then chose Arabidopsis NPR1 (AtNPR1)1, which has been shown to confer broad-spectrum disease resistance in a variety of plants, as the transgene. However, it is known that overexpressing AtNPR1 in rice by the maize ubiquitin promoter caused growth retardation, seed size reduction and development of the so-called lesion mimic disease (LMD) phenotype under certain environmental conditions14, 22. To remedy the fitness problem, we expressed the AtNPR1-EGFP fusion gene under the following four regulatory systems: 35S:uorfsTBF1-AtNPR1-EGFP, 35S:uORFsTBF1-AtNPR1-EGFP, TBF1p:uorfsTBF1-AtNPR1-EGFP and TBF1p:uORFsTBF1-AtNPR1-EGFP. These four constructs were assigned different codes for blind testing of resistance and fitness phenotypes. Under growth chamber conditions, either the TBF1p-mediated transcriptional or the uORFsTBF1-mediated translational control largely decreased the ratio and the severity of rice plants with LMD (Extended Data Fig. 5c). However, the best results were obtained using TBF1-cassette with both transcriptional and translational control. Next, we tested plant resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), the causal agent for rice blight, in the first (T0 in rice research) and the second (T1) generations of transformants under the greenhouse conditions where LMD was not observed even for 35S:uorfsTBF1-AtNPR1. Unsurprisingly, the 35S:uorfsTBF1-AtNPR1 plants displayed the highest level of resistance to Xoo, due to the constitutive transcription and translation of AtNPR1 (Extended Data Figs. 6, 7a, b). However, similar levels of resistance were also observed in plants with either transcriptional or translational control or with both. Excitingly, these resistance results were faithfully reproduced in the field (Fig. 3a, b, Extended Data Fig. 7c). In response to Xoo challenge, transgenic lines with functional uORFsTBF1 displayed transient AtNPR1 protein increases which peaked around 2 hpi, even in the absence of significant changes in mRNA levels (e.g., 35S:uORFsTBF1-AtNPR1 in Extended Data Fig. 7d, e).
Figure 3. Effects of controlling transcription and translation of AtNPR1 in rice.
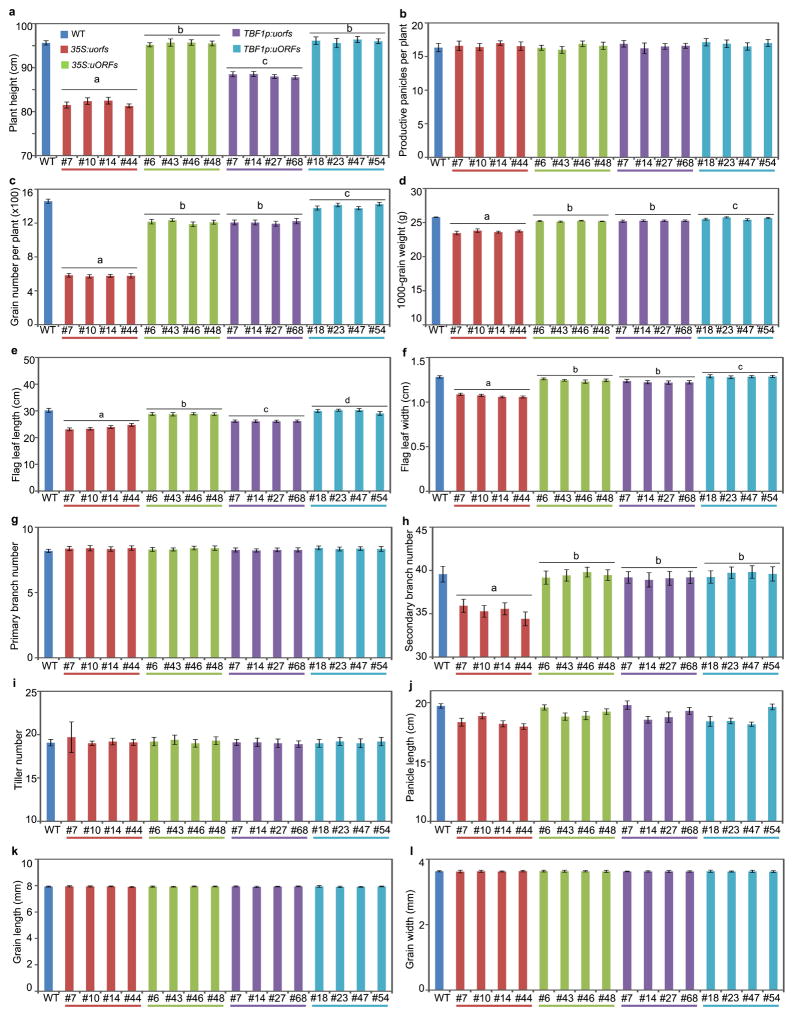
a, b, Symptoms and quantification after Xoo inoculation in field-grown T1 plants. c–f, Symptoms and quantification after Xoc (c, e, water-soaking) and M. oryzae (d, f) in T2 plants. g–i, Fitness under field conditions, including plant height (g), the number of grains per plant (h) and 1000-grain weight (i). Mean ± s.e.m.. Different letters above indicate significant differences (P < 0.05). See Source Data for sample size (n) and Extended Data Figs. 7, 8 for data from two additional lines and for more fitness parameters.
To determine the spectrum of AtNPR1-mediated resistance, we inoculated the third generation of transgenic rice plants (T2) with Xanthomonas oryzae pv. oryzicola (Xoc) and Magnaporthe oryzae (M. oryzae), the causal pathogens for rice bacterial leaf streak and fungal blast, respectively. We observed similar patterns of enhanced resistance against Xoc and M. oryzae in growth chambers designated for these controlled pathogens (Fig. 3c–f) as for Xoo, confirming the broad spectrum of AtNPR1-mediated resistance. The lack of significant variation among the different transgenic lines suggests that they all had saturating levels of AtNPR1 in conferring resistance.
We then performed detailed fitness tests on these transgenic plants in the field and found that constitutive transcription and translation of AtNPR1 in 35S:uorfsTBF1-AtNPR1 plants clearly had fitness penalties (Fig. 3g–i, Extended Data Fig. 8). Addition of transcriptional or/and translational control of AtNPR1 significantly reduced costs to agronomically important traits, with combination of both transcriptional and translational control performed the best in eliminating cost on yield based on the number of grains per plant and 1000-grain weight (Fig. 3h, i).
Using TBF1-cassette, we established a new strategy of enhancing broad-spectrum disease resistance with minimal adverse effects on plant growth and development. The ubiquitous presence of uORFs in mRNAs of organisms ranging from yeast (13% of all mRNA)23 to humans (49% of all mRNA)24 suggests the potentially broad utility of these mRNA features for the precise control of transgene expression.
Methods
Plasmid construction
The 35S promoter with duplicated enhancers was amplified from pRNAi-LIC25 and flanked with PstI and XbaI sites using primers P1/P2. The NOS terminator was amplified from pRNAi-LIC and flanked with KpnI and EcoRI sites using primers P3/P4. Gateway cassette with LIC adapter sequences was amplified and flanked with KpnI and AflII sites using primers P5/P6/P7 (the PCR fragment by P5/P6 was used as template for P5/P7) from pDEST375 (GenBank: KC614689.1). The NOS terminator, the 35S promoter, and the Gateway cassette were sequentially ligated into pCAMBIA1300 (GenBank: AF234296.1) via KpnI/EcoRI, PstI/XbaI and KpnI/AflII, respectively. The resultant plasmid was used as an intermediate plasmid. The 5′ leader sequences of TBF1 (upstream of the ATG start codon of TBF1) with WT uORFs and mutant uorfs were amplified with P8/P9 and P8/P10 from the previously published plasmids3 carrying uORF1-uORF2-GUS and uorf1-uorf2-GUS, respectively, and cloned into the intermediate plasmid via XbaI/KpnI. The resultant plasmids were designated as pGX179 (35S:uORFsTBF1-Gateway-NOS) and pGX180 (35S:uorfsTBF1-Gateway-NOS). TBF1p was amplified from the Arabidopsis genomic DNA and flanked with HindIII/AscI using primers P11/P1, and the TBF1 5′ leader sequence was amplified from pGX180 and flanked with AscI/KpnI using primers P8/P13. The TBF1 promoter (P11/P12) and the TBF1 5′ leader sequence (P8/P13) were digested with AscI, ligated, and used as template for PCR and introduction of HindIII/KpnI using primer P11/P8. The 35S promoter in pGX179 was replaced by the TBF1 promoter to produce pGX1 (TBF1p:uORFsTBF1-Gateway-NOS). The TBF1 promoter was amplified from the Arabidopsis genomic DNA and flanked with HindIII/SpeI using primers P14/P15 and ligated into pGX179, which was cut with HindIII/XbaI, to generate pGX181 (TBF1p:uorfsTBF1-Gateway-NOS). LUC, GFPER and snc1 were amplified from pGWB23526, GFP-HDEL27 and the snc1 mutant genomic DNA, respectively. TBF1-YFP and NPR1-EGFP were fused together through PCR, cloned via ligation independent cloning25. EFR was amplified from U21686 (TAIR), fused with EGFP and controlled by the 35S promoter. The 5′ leader sequence of bZIP11 (containing uORFsbZIP11) was amplified from the Arabidopsis genomic DNA with G904/G905. The start codons (ATG) for uORF2a and uORF2b in the 5′ leader sequence were mutated to CTG and TAG, respectively, to generate uorf2abZIP11 and uorf2bbZIP11 by PCR using primers containing point mutations. Primer and plasmid information can be found in Supplementary Table 1.
Arabidopsis growth, transformation, and pathogen infection
The Arabidopsis Col-0 accession was used for all experiments. Plants were grown on soil (Metro Mix 360) at 22 °C with 55% relative humidity (RH) and under 12/12-h light/dark cycles for bacterial growth assay and measurements of plant radius and fresh weight or 16/8-h light/dark cycles for seed weight and silique number measurements. Floral dip method25 was used to generate transgenic plants. The BGL2:GUS reporter line19 was used for snc1-related transformation. For infection, bacteria were first grown on the King’s Broth medium plate at 28 °C for 2 d before resuspended in 10 mM MgCl2 solution for infiltration. The antibiotic selection for Psm ES4326 was 100 μg/ml streptomycin, for Pst DC3000 25 μg/ml rifampicin, and for Pst DC3000 hrcC− 25 μg/ml rifampicin and 30 μg/ml chloramphenicol. For spray inoculation, Psm ES4326 was transferred to liquid King’s Broth with 100 μg/ml streptomycin, grown for another 8 to 12 h to OD600nm = 0.6 to 1.0 and sprayed at OD600nm = 0.4 in 10 mM MgCl2 with 0.02 % Silwet L-77. Infected leaf samples were collected on day 0 (4 biological replicates with 3 leaf discs each) and day 3 (8 replicates with 3 leaf discs each). For Hpa Noco2 infection, 12-day-old plants grown under 12/12-h light/dark cycles with 95% RH were sprayed with 4×104 spores/ml and incubated for 7 d. Spores were collected by suspending infected plants in 1 ml water and counted in a hemocytometer under a microscopy.
Transient expression in N. benthamiana
N. benthamiana plants were grown at 22°C under 12/12-h light/dark cycles before used for Agrobacterium-mediated transient expression. Agrobacterium GV3101 transformed with each construct was grown in LB with kanamycin (50 μg/ml), gentamycin (50 μg/ml) and rifampicin (25 μg/ml) at 28°C overnight. Cells were resuspended in the infiltration buffer [10 mM 2-(N-morpholino) ethanesulfonic acid (MES), 10 mM MgCl2, 200 μM acetosyringone] at OD600nm = 0.1 and incubated at room temperature for 4 h before infiltration. Activity of cytosol-synthesized firefly luciferase was detected after spraying 1 mM luciferin and displayed by chemiluminescence with pseudo colour after transient expression in N. benthamiana for 2 d. Fluorescence of ER-synthesized GFPER was detected under UV after transient expression in N. benthamiana for 2 d. Cell death induced by overexpression of TBF1-YFP fusion was examined by clearing with ethanol after transient expression in N. benthamiana for 3 d. For elf18 induction in N. benthamiana, the Agrobacterium harbouring the elf18 receptor-expressing construct (pGX664) was coinfiltrated with the Agrobacterium carrying the test construct at 1:1 ratio. 20 h later, the same leaves were infiltrated with 10 mM MgCl2 (Mock) solution or 10 μM elf18 before leaf disc collection 2 h later.
Dual-luciferase assay
The MgCl2 solution (10 mM), Psm ES4326 (OD600nm = 0.02), Pst DC3000 (OD600nm = 0.02), Pst DC3000 hrcC− (OD600nm = 0.02), elf18 (10 μM) or flg22 (10 μM) was infiltrated. Leaf discs were collected at the indicated time points. LUC and RLUC activities were measured as CPS (counts per second) using the Victor3 plate reader (PerkinElmer) according to the kit from Promega (E1910).
Real-time polymerase chain reaction (PCR)
~100 mg leaf tissue was collected for total RNA extraction with TRIzol (Ambion). DNase I (Ambion) treatment was performed before reverse transcription with SuperScript® III Reverse Transcriptase (Invitrogen) using oligo (dT). Real-time PCR was done using FastStart Universal SYBR Green Master (Roche). Primers used are listed in Supplementary Table 1.
Rice growth, transformation, and pathogen infection
For LMD phenotype observation, rice was grown in greenhouse for 6 weeks and moved to a growth chamber for 3 weeks (12/12-h light/dark cycles, 28°C and 90% RH). For fitness test, rice was grown during the normal rice growing season (From Nov. 2015 to May 2016) under field conditions in Lingshui, Hainan (18° N latitude). Agrobacterium-mediated transformation into the Oryza sativa cultivar ZH11 was used to obtain transgenic rice plants26. For Xoo infection in the greenhouse (performed in year 2016), rice was grown for 3 weeks from Feb. 2 and inoculated on Feb. 23 with data collection on Mar. 8. For Xoo infection in the field (performed in year 2016), rice was grown on May 10 in the Experimental Stations of Huazhong Agricultural University, Wuhan, China (31° N latitude) and inoculated on July 20 with data collection on Aug. 4. Xoo strains PXO347 and PXO99 were grown on nutrient agar medium (0.1% yeast extract, 0.3% beef extract, 0.5% polypeptone, and 1% sucrose) at 28 °C for 2 d before resuspension in sterile water and dilution to OD600nm = 0.5 for inoculation. 5 to 10 leaves of each plant were inoculated by the leaf-clipping method at the booting (panicle development) stage27, 28. Disease was scored by measuring the lesion length at 14 d post inoculation (dpi). PCR was performed using primer rice-F and rice-R (Supplementary Table 1) for identification of AtNPR1 transgenic plants. Both PCR positive and negative T1 plants were scored. For Xoc infection in the growth chamber (performed in year 2016), rice was grown on Oct. 20 and inoculated on Nov. 15 with data collection on Nov. 29. Xoc strain RH3 was grown on nutrient agar medium (0.1% yeast extract, 0.3% beef extract, 0.5% polypeptone, and 1% sucrose) at 28 °C for 2 d before resuspension in sterile water and dilution to OD600nm = 0.5 for inoculation. 5 to 10 leaves of each plant were inoculated by the penetration method using a needleless syringe at the tillering stage27. Disease was scored by measuring the lesion length at 14 dpi. For M. oryzae infection in the growth chamber (performed in year 2016), rice was grown on Oct. 15 and inoculated on Nov. 16 with data collection on Nov. 23. M. oryzae isolate M229 was cultured on oatmeal tomato agar (OTA) medium (40 g oat, 150 ml tomato juice, 20 g agar for 1 L culture medium) at 28 °C. 10 μl of the conidia suspension (5.0×105 spores/ml) containing 0.05% Tween-20 was dropped to the press-injured spots on 5 to 10 fully expanded rice leaves and then wrapped with cellophane tape. Plants were maintained in darkness at 90% RH for one day and were grown under 12/12-h light/dark cycles with 90% RH. Disease was scored by measuring the lesion length at 7 dpi. For Xoc and M. oryzae, 3 independent transgenic lines for each construct were tested, with data from 2 lines shown in Fig. 3 and from the third line in Source Data of Fig. 3. For Xoo infection and fitness, 4 independent transgenic lines for each construct were tested, with data from 2 lines shown in Fig. 3 and from all four lines in Extended Data Figs. 7, 8 and in Source Data.
Immunoblot
Arabidopsis tissue (100 mg) infected by Psm ES4326 (OD600nm = 0.02) was collected and lysed in 200 μl lysis buffer [50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, protease inhibitor cocktail (Roche, 1 tablet for 10 mL)] before centrifugation at 12,000 rpm for the supernatant. The same protocol was used to extract proteins from rice infected by Xoo (PXO99, at OD600nm = 0.5) using a slightly different lysis buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, 2 mM EDTA, 0.1 % Triton X-100, protease inhibitor cocktail (Roche, 1 tablet for 10 mL)]. Antibody information and the experimental conditions can be found in Supplementary Table 1.
Statistical analyses
Normal distribution was tested using the Shapiro-Wilk test. Two-sided one-way ANOVA together with Tukey test was used for multiple comparisons. Sample size can be found in Source Data. Unless specifically stated, sample size n means biological replicates. Experiments have been done three times with similar results for all the Arabidopsis experiments. GraphPad Prism 6 was used for all the statistical analyses.
Data availability
The authors declare that the main data supporting the findings of this study are available within the article and its Source Data files. Extra data are available from the corresponding author upon request.
Extended Data
Extended Data Figure 1. Conservation of uORF2TBF1 nucleotide and peptide sequences in plant species.
a, Schematic of TBF1 mRNA structure. The 5′ leader sequence contains two uORFs, uORF1 and uORF2. CDS, coding sequence. b–d, Alignment of uORF2 nucleotide sequences (b) and alignment (c) and phylogeny (d) of uORF2 peptide sequences in different plant species. The corresponding triplets encoding the conserved amino acids among these species are underlined. Identical residues (black background), similar residues (grey background) and missing residues (dashes) were identified using Clustlw2. At (Arabidopsis thaliana; AT4G36988), Pv (Phaseolus vulgaris; XP_007155927), Gm (Glycine max; XP_006600987), Gr (Gossypium raimondii; CO115325), Nb (Nicotiana benthamiana; CK286574), Ca (Cicer arietinum; XP_004509145), Pd (Phoenix dactylifera; XP_008797266), Ma (Musa acuminata subsp. Malaccensis; XP_009410098), Os (Oryza sativa; Os09g28354).
Extended Data Figure 2. Characterization of uORFsTBF1 and uORFsbZIP11 in translational control, related to Fig. 1.
a, Subcellular localization of the LUC-YFP fusion (a) and GFPER (b). SP, signal peptide from Arabidopsis basic chitinase; HDEL, ER retention signal. Representative of 8 images. Scale bar, 10 μm. c–e, mRNA levels of LUC in (Fig. 1b; n = 3), GFPER in (Fig. 1c; n = 4), and TBF1-YFP in (Fig. 1d; n = 3) 2 dpi before cell death was observed in plants expressing TBF1. f, Schematics of the 5′ leader sequences used in studying the translational activities of WT uORFsbZIP11, mutant uorf2abZIP11 (ATG to CTG) or uorf2bbZIP11 (ATG to TAG). g–i, uORFsbZIP11-mediated translational control of cytosol-synthesized LUC (g; chemiluminescence with pseudo colour); ER-synthesized GFPER (h; fluorescence under UV); and cell death induced by overexpression of TBF1-YFP fusion (i; cleared using ethanol) after transient expression in N. benthamiana for 2 d (g, h) and 3 d (i), respectively. Representative of 4 images. j–l, mRNA levels of LUC in (g; n = 2 experiments with 3 technical replicates), GFPER in (h; n = 3 experiments with 3 technical replicates), and TBF1-YFP in (i; n = 3 experiments with 3 technical replicates). m, TE changes in LUC controlled by the 5′ leader sequence containing WT uORFsbZIP11, mutant uorf2abZIP11 or uorf2bbZIP11 in response to elf18 in N. benthamiana. Mean of the LUC/RLUC activity ratios (n = 12). n, LUC/RLUC mRNA changes in (m). Mean of LUC/RLUC mRNA normalized to Mock from 2 experiments with 3 technical replicates. Bar with solid circles, mean with individual biological replicates.
Extended Data Figure 3. Three developmental phenotypes observed in primary Arabidopsis transformants expressing snc1.
The three developmental phenotypes observed in T1 (i.e., the first generation) Arabidopsis transgenic lines carrying 35S:uorfsTBF1-snc1, 35S:uORFsTBF1-snc1, TBF1p:uorfsTBF1-snc1 and TBF1p:uORFsTBF1-snc1 (above). Representative of 5 images. Fisher’s exact test was used for the pairwise statistical analysis (below). Different letters in “Total” indicate significant differences between Type III versus Type I+Type II (P < 0.01).
Extended Data Figure 4. Effects of controlling transcription and translation of snc1 on defence and fitness in Arabidopsis, related to Fig. 2.
a, b, Psm ES4326 growth in WT, snc1, transgenic lines #1–4 after inoculation by spray (a) or infiltration (b). Mean ± s.e.m.. c, Hpa Noco2 growth as measured by spore counts 7 dpi. Mean ± s.e.m.. d–g, Analyses of plant radius (d), fresh weight (e), silique number (f) and total seed weight (g). Mean ± s.e.m.. h, i, Relative levels of Psm ES4326-induced snc1 protein (h; numbers below immunoblots; see Supplementary Figure 1 for gel source data) and mRNA (i; mean from 2 experiments with 3 technical replicates). Solid circles, individual biological replicates. #1–4, four independent transgenic lines carrying TBF1p:uORFsTBF1-snc1 with #1 and #2 shown in Fig. 2. hpi, hours after Psm ES4326 infection; CBB, Coomassie Brilliant Blue. See Source Data for sample size (n). Different letters above bar graphs indicate significant differences (P < 0.05).
Extended Data Figure 5. Functionality of uORFsTBF1 in rice.
a, b, LUC activity (a) and mRNA levels (b) in three independent primary transgenic rice lines (called “T0” in rice research) carrying 35S:uorfsTBF1-LUC and 35S:uORFsTBF1-LUC. Mean of LUC activities (RLU, relative light unit) of 3 biological replicates. Solid circles, individual biological replicates; and mean of LUC mRNA levels of 3 technical replicates after normalization to the 35S:uorfsTBF1-LUC line #1. c, Representative lesion mimic disease (LMD) phenotypes (above) and percentage of AtNPR1-transgenic rice plants showing LMD in the second generation (T1) grown in the growth chamber (below).
Extended Data Figure 6. Effects of controlling transcription and translation of AtNPR1 on defence in T0 rice, related to Fig. 3.
a–d, Lesion length measurements after infection by Xoo strain PXO347 in primary transformants (T0) for 35S:uorfsTBF1-AtNPR1 (a), 35S:uORFsTBF1-AtNPR1 (b), TBF1p:uorfsTBF1-AtNPR1 (c) and TBF1p:uORFsTBF1-AtNPR1 (d). Lines further analysed in T1 and T2 are circled. e, Average leaf lesion lengths. WT, recipient Oryza sativa cultivar ZH11. Mean ± s.e.m.. Different letters above indicate significant differences (P < 0.05). See Source Data for sample size (n).
Extended Data Figure 7. Effects of controlling transcription and translation of AtNPR1 on defence in T1 rice, related to Fig. 3.
a, b, Representative symptoms observed in T1 AtNPR1-transgenic rice plants grown in the greenhouse (a) after Xoo inoculation and corresponding leaf lesion length measurements (b). PCR was performed to detect the presence (+) or the absence (-) of the transgene gene. c, Quantification of leaf lesion length of 4 lines for Xoo inoculation in field-grown T1 AtNPR1-transgenic rice plants. Mean ± s.e.m.. See Source Data for sample size (n). Different letters above indicate significant differences (P < 0.05). d, e, Relative levels of AtNPR1 mRNA (d) and protein (e; numbers below immunoblots; see Supplementary Figure 1 for gel source data) in response to Xoo infection. Mean of AtNPR1 mRNA levels of 3 technical replicates after normalization to 0 hpi (d). Solid circles, individual biological replicates.
Extended Data Figure 8. Effects of controlling transcription and translation of AtNPR1 on fitness in T1 rice under field conditions, related to Fig. 3.
Mean ± s.e.m.. See Source Data for sample size (n). Different letters above indicate significant differences among constructs (P < 0.05).
Supplementary Material
Acknowledgments
This study was supported by grants from NIH 5R01 GM069594-11 and Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (through Grant GBMF3032) to X. Dong, National Natural Science Foundation of China (31371926) to M. Yuan and the National Key Research and Development Program of China (2016YFD0100903) to S. Wang. We thank J. Motley, J. Marqués, P. Zwack and S. Zebell for comments on this manuscript.
Footnotes
Author Contributions
G.X. and X.D. designed the research. G.X. performed the Arabidopsis-related experiments with help from E.Z. for fitness tests. L.L. isolated snc1 genomic DNA. S.K. maintained Hpa Noco2 strain in the lab and helped with inoculation; M.Y., C.A. and G.X. carried out and S.W. supervised the rice-related experiments. G.X. and X.D. wrote the manuscript with input from all authors.
The authors declare competing financial interests: A patent based on this study has been filed by Duke University with G.X. and X.D. as inventors.
Supplementary Information is available in the online version of the paper.
References
- 1.Fu ZQ, Dong XN. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 2.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol. 2005;23:283–290. doi: 10.1016/j.tibtech.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Pajerowska-Mukhtar KM, et al. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol. 2012;22:103–112. doi: 10.1016/j.cub.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol. 2005;23:275–282. doi: 10.1016/j.tibtech.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 6.Schwessinger B, et al. Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. Plos Pathog. 2015;11 doi: 10.1371/journal.ppat.1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacombe S, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010;28:365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- 8.Bouwmeester K, et al. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol J. 2014;12:10–16. doi: 10.1111/pbi.12111. [DOI] [PubMed] [Google Scholar]
- 9.Benedetti M, et al. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc Natl Acad Sci USA. 2015;112:5533–5538. doi: 10.1073/pnas.1504154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 11.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH, Qi D, Ashfield T, Helm M, Innes RW. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science. 2016;351:684–687. doi: 10.1126/science.aad3436. [DOI] [PubMed] [Google Scholar]
- 13.Chern MS, et al. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 2001;27:101–113. doi: 10.1046/j.1365-313x.2001.01070.x. [DOI] [PubMed] [Google Scholar]
- 14.Quilis J, Penas G, Messeguer J, Brugidou C, Segundo BS. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact. 2008;21:1215–1231. doi: 10.1094/MPMI-21-9-1215. [DOI] [PubMed] [Google Scholar]
- 15.Molla KA, et al. Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci. 2016;250:105–114. doi: 10.1016/j.plantsci.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson KCM, Dong OX, Huang Y, Li X. A rolling stone gathers no moss, but resistant plants must gather their moses. Cold Spring Harb Symp Quant Biol. 2012;77:259–268. doi: 10.1101/sqb.2013.77.014738. [DOI] [PubMed] [Google Scholar]
- 18.Rahmani F, et al. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Clarke JD, Zhang YL, Dong XN. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 20.Li YQ, Yang SH, Yang HJ, Hua J. The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol Plant Microbe Interact. 2007;20:1449–1456. doi: 10.1094/MPMI-20-11-1449. [DOI] [PubMed] [Google Scholar]
- 21.Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald HA, Chern MS, Navarre R, Ronald PC. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 23.Lawless C, et al. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu GY, et al. One-step, zero-background ligation-independent cloning intron-containing hairpin RNA constructs for RNAi in plants. New Phytol. 2010;187:240–250. doi: 10.1111/j.1469-8137.2010.03253.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 27.Xu G, et al. Plant ERD2-like proteins function as endoplasmic reticulum luminal protein receptors and participate in programmed cell death during innate immunity. Plant J. 2012;72:57–69. doi: 10.1111/j.1365-313X.2012.05053.x. [DOI] [PubMed] [Google Scholar]
- 28.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- 30.Yuan M, et al. A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. Elife. 2016;5 doi: 10.7554/eLife.19605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan YX, et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J. 2007;5:313–324. doi: 10.1111/j.1467-7652.2007.00243.x. [DOI] [PubMed] [Google Scholar]
- 32.Kang H, et al. Dissection of the genetic architecture of rice resistance to the blast fungus Magnaporthe oryzae. Mol Plant Pathol. 2016;17:959–972. doi: 10.1111/mpp.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Source Data files. Extra data are available from the corresponding author upon request.