Abstract
Identification of novel therapeutics in pelvic high-grade serous carcinoma (HGSC) has been hampered by a paucity of actionable point mutations in target genes. The aim of the present study was to investigate the extent of amplification of the therapeutically targetable NSD3-CHD8-BRD4 pathway in pelvic HGSC, and to determine whether amplification is associated with worse prognosis. The Cancer Genome Atlas (TCGA) ovarian and endometrial cancer cohorts were retrospectively analyzed via online data-mining tools to test the association of NSD3, CHD8 and BRD4 genomic alterations with survival of pelvic HGSC patients. It was demonstrated that amplification of the NSD3-CHD8-BRD4 pathway in the ovarian HGSC cohort (observed in 18% of the cases, 88/489) was significantly associated with worse overall and progression-free survival compared with non-amplified cases. In addition, amplification of NSD3, CHD8 and BRD4 also occurred in 9% (21/232) of overall endometrial cancer TCGA cases, which was associated with worse overall survival. In the endometrial cancer TCGA cohort, NSD3, CHD8 and BRD4 amplification occurred specifically in the serous carcinoma (25%, 13/53) and ‘serous-like’ copy number high endometrial carcinoma (33%, 20/60) subgroups, compared with the polymerase e (0%, 0/17), microsatellite instability high (0%, 0/65) or low copy number (1%, 1/90) subgroups. These findings support the hypothesis that amplification of the NSD3-BRD4-CDH8 axis is frequent in pelvic HGSC of both ovarian and endometrial origin, and that this pathway is potentially targetable in a subset of HGSC patients.
Keywords: ovarian, endometrial, serous carcinoma, BRD4, NSD3, CHD8
Introduction
Pelvic high-grade serous carcinoma (HGSC) of either tubo-ovarian or endometrial origin is an aggressive and deadly gynecological malignancy with few available targeted therapies, due in part to the absence of high-frequency oncogenic point mutations in drug target genes (1–4). Recently, The Cancer Genome Atlas (TCGA) projects have advanced our understanding of the genomic landscape of ovarian and endometrial HGSC and confirmed a prevalence of somatic tumor protein p53 mutations and extensive copy number alterations, such as numerous DNA amplifications and deletions (1,5).
Gene amplification in neoplastic cells may be a mechanism for overexpression of cancer-promoting driver genes with potential ‘druggable’ properties (6). In ovarian HGSC, it has been previously demonstrated that BRD4, a BET bromo-domain-containing protein that is associated with acetylated chromatin and transcriptional activation, is amplified in a small subset (12%) of cases (7–10), and BRD4 amplification in ovarian HGSC may be associated with worse overall and progression-free survival (9). Recently, BRD4 has become an attractive target for cancer therapy, as specific small-molecule BRD4/BET inhibitors, such as JQ1 and OTX015/MK-8628, are currently in clinical trials for cancer (ClinicalTrials.gov identifier: NCT01713582) (11–13). In ovarian cancer cell lines and patient-derived xenograft model systems of HGSC, suppression of BRD4 using the small-molecule BET inhibitors JQ1 and/or I-BET151 exerted robust antitumor effects (7,10), thereby providing a rationale for further investigating genomic alterations of the BRD4 pathway and the clinical benefits of BRD4-specific inhibitors in pelvic HGSC.
In several tumor types, BRD4 has been shown to selectively regulate transcription of key oncogenic drivers, such as CMYC (14–17), by specifically targeting cell type-specific enhancer sequences (i.e., super enhancers) (18). In addition, the histone methyltransferase NSD3 short isoform is an adaptor that couples BRD4 to the chromatin remodeling factor CHD8 to activate transcription (19). In leukemia cells, genetic targeting of NSD3 or CHD8 mimics the effects of BRD4 inhibition. Furthermore, BRD4, NSD3 and CHD8 co-localize across the genome, and BRD4-NSD3-CHD8 protein complexes are evicted from chromatin super-enhancer regions upon chemical inhibition by BRD4 small-molecule inhibitors (19). The goal of this study was to investigate the extent of amplification of the NSD3-CHD8-BRD4 axis in pelvic HGSC of both tubo-ovarian and endometrial origin, and to determine whether amplification of this pathway is associated with worse prognosis and survival in pelvic HGSC patients.
Materials and methods
Study type and design
An observational retrospective secondary data analysis of clinically annotated multi-platform ovarian and endometrial cancer-omics datasets from the TCGA project was performed. Publicly available online tools were used to data mine the ovarian and endometrial carcinoma TCGA datasets and to investigate the survival of pelvic HGSC patients in association with genomic and transcriptomic alterations of NSD3, BRD4 and CHD8.
TCGA cohorts, gene amplification and survival analysis
Clinical data from the ovarian carcinoma TCGA study (1), or the endometrial carcinoma TCGA study (5), were analyzed with respect to NSD3, BRD4 and CHD8 amplification via the cBio Cancer Genomics Portal (http://cbioportal.org; Memorial Sloan Kettering Cancer Center, New York, NY, USA) (20,21). In the present study, copy number gain was defined as gain of 1 gene copy number, while amplification was defined as gain of ≥2 gene copy numbers.
From the ovarian carcinoma TCGA study, the following cohorts of patients were queried: i) All tumors (n=557), ii) all complete tumors (n=316), iii) cases with available copy number alteration data (n=489), iv) cases with complete response to primary therapy (n=276) and v) cases that were platinum-sensitive (n=197). The cohorts were queried specifically for NSD3 (also referred to as WHSC1L1), BRD4 and CHD8 gene amplification in the cBio portal by using the advanced Onco Query Language (WHSC1L1: AMP; BRD4: AMP; CHD8: AMP). Kaplan-Meier overall and disease-free survival analyses were obtained via the cBio portal, with significance estimated with the log-rank test (20,21).
A similar survival analysis via the cBio portal and via the advanced Onco Query Language was performed with the endometrial carcinoma TCGA study, in which the following cohorts were analyzed: i) Complete endometrioid-type tumors (n=232), ii) copy number high tumors (n=60), iii) copy number low tumors (n=90), iv) microsatellite instability (MSI; hypermutated) tumors (n=65), v) polymerase e (POLE; ultramutated) tumors (n=17) and vi) tumors with specific serous-type histology (n=53).
mRNA expression analysis
TCGA cohorts were queried specifically for NSD3 (WHSC1L1) and CHD8 mRNA overexpression in the cBio portal by using the advanced Onco Query Language. To identify significant NSD3 and CHD8 mRNA overexpression with Z scores of either +2.0 or +3.0, the following advanced Onco Query Language was used respectively (WHSC1L1: EXP>2; CHD8: EXP>2) or (WHSC1L1: EXP>3; CHD8: EXP>3). Kaplan-Meier overall and disease-free survival analysis were obtained via the cBio portal, with significance estimated using the log-rank test.
Results
Amplification of NSD3, CHD8, BRD4 and other BRD4-associated genes in ovarian HGSC
Clinical data from the ovarian carcinoma TCGA cohort, specifically 489 cases with copy number alteration data, was integrated with NSD3, CHD8 and BRD4 amplification data from the TCGA data portal via the cBio Cancer Genomics portal (http://cbioportal.org) (1,20,21). Somatic amplification of at least one member of the NSD3-CHD8-BRD4 axis was observed in 18% (88/489) of the cases (Fig. 1A). Survival analysis of TCGA patients with ovarian HGSC (n=489), revealed that patients with somatic amplification of at least one member of the NSD3-CHD8-BRD4 pathway had significantly worse overall and progression-free survival compared with those with non-amplified pathway members, with log-rank test P-values of 0.00078 and 0.00085, respectively (Fig. 1B and C). The median number of months until relapse was 14.0 for cases with pathway amplification, compared with 17.6 months for cases without amplification. Patients with amplification had a median survival of 35.8 months compared with a median survival of 45.0 months for patients without amplification.
Figure 1.
Amplification of the NSD3-CHD8-BRD4 pathway and survival in ovarian HGSC. (A) Amplification of the NSD3-CHD8-BRD4 pathway in ovarian HGSC (only tumors exhibiting amplification of at least one member of the pathway are shown). (B and C) Survival Kaplan-Meier curves for patients whose tumor exhibited amplification of at least one member of the NSD3-CHD8-BRD4 pathway: (B) Overall and (C) disease-free survival of TCGA cases with copy number alteration data (n=489) and NSD3, CHD8 and/or BRD4 amplification. HGSC, high-grade serous carcinoma; TCGA, The Cancer Genome Atlas.
Our analysis focused on the ovarian TCGA cohort of cases with copy number alterations (n=489), as this cohort maximizes the number of cases with gene amplifications. However, significant worse overall survival and progression-free survival effects (P<0.05) with NSD3, BRD4 and CHD8 amplification were also observed in the following ovarian HGSC TCGA cohorts (data not shown): i) All tumors (n=557), ii) all complete tumors (n=316), iii) platinum-sensitive tumors (n=197) and iv) tumors with complete response to primary therapy (n=276). In addition, amplification of BRD4-associated genes in the TCGA ovarian carcinoma cohort appeared to be specific to amplification of NSD3, CHD8 and BRD4, since other known BRD4-associated genes were not significantly amplified in the TCGA study. For example, amplification of the following known BRD4-associated genes occurred at <1% of total cases for each gene: CDK9, CCNT1, HEXIM1, MLLT1, AFF1, KAT5, MED12 and MED24 (data not shown).
Co-occurrence of NSD3 and CHD8 amplification in ovarian HGSC
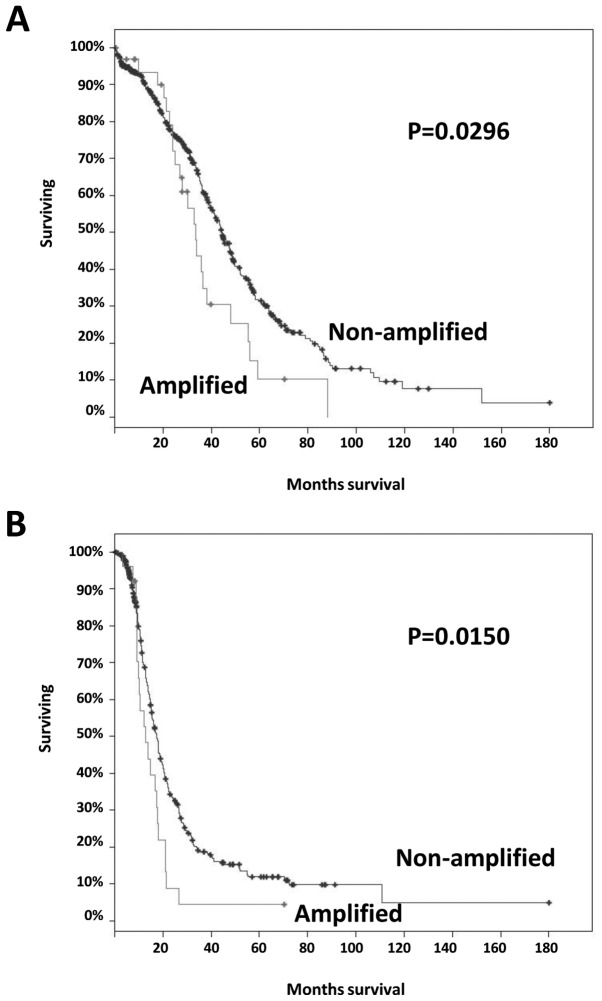
It has been previously demonstrated that BRD4 amplification alone in ovarian HGSC (observed in 12% of the cases) is associated with worse survival (9). Furthermore, in the ovarian HGSC TCGA cohort, there was a significant tendency towards co-occurrence of NSD3 and CHD8 amplification (P<0.001, log odds ratio=2.383), while there was a trend for mutual exclusivity of NSD3 and BRD4 amplification (P=0.091, log odds ratio <-3). For these reasons, we assessed whether amplifications of NSD3 and/or CHD8 were independently associated with worse survival. Somatic amplification of NSD3 and/or CHD8 was observed in 7% (34/489) of the cases, and amplification of NSD3 and/or CHD8 was significantly associated with worse overall and progression-free survival compared with those in non-amplified cases; log-rank test P=0.0296 and P=0.0150, respectively (Fig. 2A and B). The median number of months to relapse was 12.8 for cases with NSD3 and/or CHD8 amplification compared with 17.5 months for cases without amplification. Patients with NSD3 and/or CHD8 amplification had a median survival of 33.4 months compared with a median overall survival of 44.5 months in patients without amplification. These results suggest that the survival effects of NSD3/CHD8 amplification are mutually exclusive to BRD4 amplification in ovarian HGSC.
Figure 2.
Survival Kaplan-Meier curves for patients with ovarian high-grade serous carcinoma with tumors exhibiting amplification of NSD3 and/or CHD8. (A) Overall and (B) disease-free survival of TCGA cases with copy number alteration data (n=489), NSD3 and/or CHD8 amplification. TCGA, The Cancer Genome Atlas.
Overexpression of NSD3 and CHD8 mRNA in ovarian HGSC
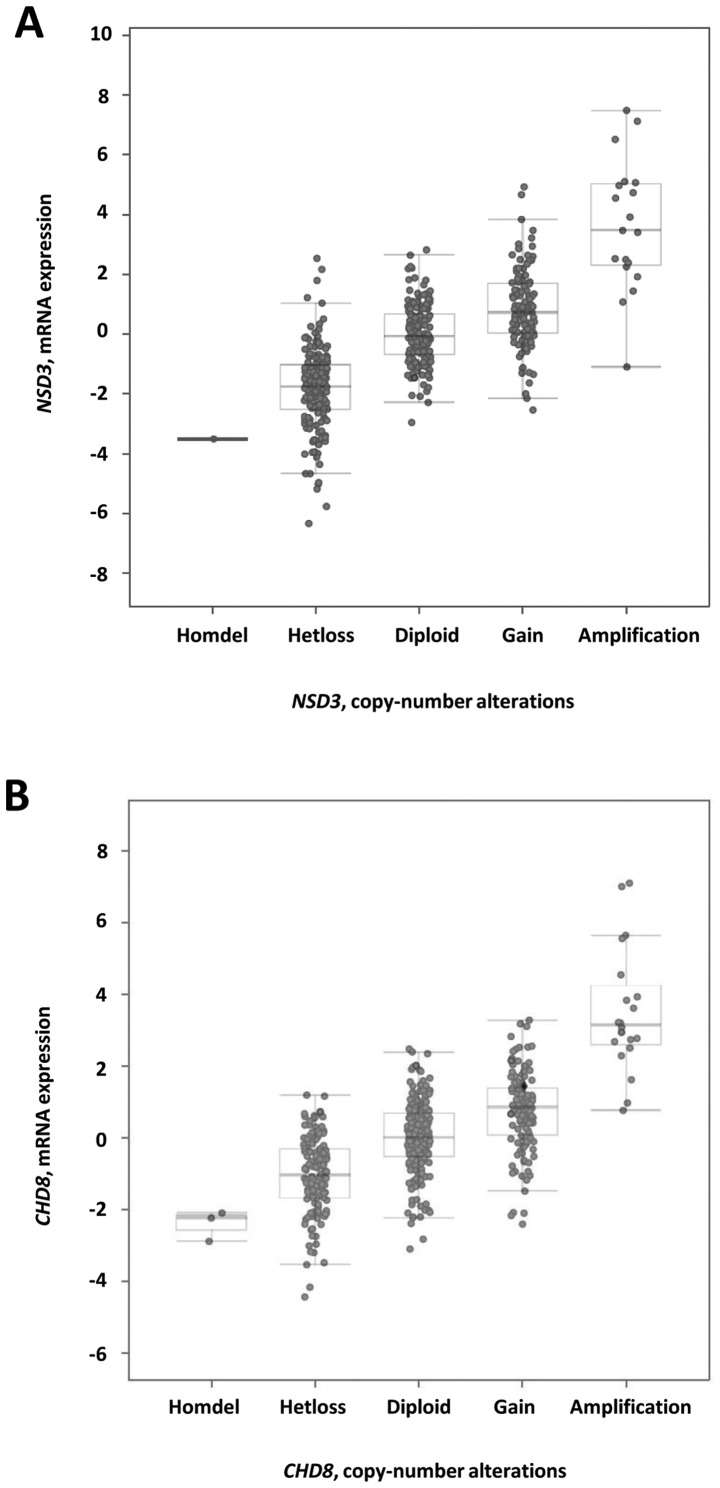
The NSD3 and CHD8 mRNA data for the ovarian carcinoma HGSC TCGA cohort were next examined. Similar to previously reported data for BRD4 (9), NSD3 and CHD8 mRNA levels tended to increase with gene copy number increases (Fig. 3A and B). To define significant NSD3 and CHD8 mRNA upregulation, Z score thresholds of either +2.0 or +3.0 were used via the cBio portal, specifically in the cohort of cases with copy number alterations (n=489). By using a Z score threshold of +2.0, NSD3 mRNA expression levels were increased in 9% (43/489) of the cases, and CHD8 mRNA expression levels were increased in 7% (36/489) of the cases. By contrast, by using a more stringent Z score threshold of +3.0, NSD3 mRNA expression levels were increased in 3.5% (17/489) of the cases, and CHD8 mRNA expression levels were increased in 2.9% (14/489) of the cases.
Figure 3.
NSD3 and CHD8 mRNA expression vs. copy number data for ovarian high-grade serous carcinoma. (A) NSD3 and (B) CHD8 mRNA levels tended to increase with increased copy number.
Using a Z score threshold of +2.0, in 78.9% of NSD3-amplified cases (15/19), NSD3 amplification resulted in NSD3 mRNA upregulation. A similar analysis revealed that, in 85% of CHD8-amplified cases (17/20), CHD8 amplification resulted in CHD8 mRNA upregulation. Using a more stringent Z score threshold of +3.0, in 57.9% of NSD3-amplified cases (11/19), NSD3 amplification resulted in NSD3 mRNA upregulation, while in 55.0% of CHD8-amplified cases (11/20), CHD8 amplification resulted in CHD8 mRNA upregulation.
mRNA overexpression of NSD3 and/or CHD8 was associated with worse overall survival, with Z scores of either +2.0 (log-rank test P=0.0247, Fig. 4A), or with a more stringent Z score of +3.0 (log-rank test P=0.0064, Fig. 4B). A smaller P-value with a more stringent Z score of +3.0 compared with a score of +2.0 suggests a dose-response type overall survival effect with increasing levels of NSD3 and CHD8 mRNA. However, there was no significant difference in progression-free survival in cases with NSD3 and/or CHD8 mRNA upregulation, with Z scores of either +2.0 or +3.0 (log-rank test P=0.38 for either Z score).
Figure 4.
Overall survival with NSD3 and/or CHD8 mRNA upregulation. (A) Kaplan-Meier curve depicting overall survival with NSD3 and/or CHD8 mRNA upregulation (Z score threshold of +2.0), of NSD3 and/or CHD8 mRNA levels in ovarian high-grade serous carcinoma. (B) Kaplan-Meier curve depicting overall survival with NSD3 and/or CHD8 mRNA upregulation (Z score threshold of +3.0), of NSD3 and/or CHD8 mRNA levels in ovarian high-grade serous carcinoma.
Amplification of NSD3, CHD8 and BRD4 in endometrial carcinoma
We next investigated the endometrial carcinoma TGCA cohort in order to determine whether amplification of the NSD3-CHD8-BRD4 axis occurred in endometrial carcinomas, and whether amplification in endometrial cancer was associated with specific survival outcomes and specific histological or molecular subgroups. Initial analysis of 232 endometriod-type tumors with complete molecular data revealed amplification of at least one member of the pathway (NSD3, CHD8 and/or BRD4), in 9% (21/232) of the overall endometrioid cancer TCGA cases (data not shown). Of note, amplification of at least one pathway member was significantly associated with worse overall patient survival; log-rank test P=0.015 (Fig. 5A), while no significant difference in progression-free survival was observed; log-rank test P=0.15 (progression-free survival curve not shown).
Figure 5.
Amplification of the NSD3-CHD8-BRD4 pathway in the endometrial endometrioid and serous carcinoma TCGA cohorts. (A) Overall survival Kaplan-Meier curve for patients with tumors exhibiting amplification of at least one member of the NSD3-CHD8-BRD4 pathway, regardless of subtype. Amplification of the NSD3-CHD8-BRD4 pathway in the (B) endometrioid ‘serous-like’ carcinoma, (C) endometrioid MSI high and (D) endometrioid low copy number molecular subgroups, and (E) in tumors with specific serous-type histology. TCGA, The Cancer Genome Atlas; MSI, microsatellite instability.
The TCGA endometrial endometrioid carcinoma cohort has been stratified into four different molecular subgroups, each with distinct survival outcomes: i) ‘Serous-like’ copy number high alterations (worst prognosis); ii) MSI hypermutated; iii) copy number low alterations; and iv) POLE ultramutated (best prognosis) (5). For this reason, it was assessed whether amplification of the NSD3-CHD8-BRD4 axis in endometrioid adenocarcinoma was associated with specific molecular subgroups. In the TCGA cohort, amplification of at least one member of the NSD3-CHD8-BRD4 pathway occurred specifically in the ‘serous-like’ copy number high endometrial endometrioid carcinoma (33%, 20/60) subgroup (Fig. 5B), compared with the MSI high (0%, 0/65) (Fig. 5C), low copy number (1%, 1/90) (Fig. 5D) or POLE ultramutated (0%, 0/17, data not shown) molecular subgroups.
Finally, it was investigated whether amplification of the NSD3-CHD8-BRD4 pathway also occurred in endometrial carcinomas of serous histological subtype. The TGCA study has additionally characterized the genomic landscape of 53 endometrial serous carcinomas, of which 25% (13/53) harbored NSD3, CHD8 and/or BRD4 amplification (Fig. 5E). In endometrial serous and in endometrioid ‘serous-like’ carcinoma, amplification of the NSD3-CHD8-BRD4 axis was not associated with worse overall or progression-free survival (data not shown). However, the overall findings support the notion that amplification of members of the NSD3-BRD4-CDH8 pathway occurs frequently in a subset of pelvic HGSCs of both ovarian and endometrial origin.
Discussion
Recently, the chromatin regulators have become attractive targets for cancer therapy. BRD4 is a chromatin reader, which binds acetylated chromatin at specific promoters/enhancers and recruits additional factors to specific genomic sites. The BRD4 chromatin-remodeling complex is partly composed of BRD4-NSD3-CHD8 protein complexes, which all co-localize in the genome to promote transcription and expression of target genes such as CMYC, CD274 (encoding PD-L1) or ALDH1A1 (18,22–24). Importantly, small-molecule inhibitors of BRD4, such as JQ1 and I-BET151, have been developed and may hold promise for the treatment of cancer, including ovarian HGSC and endometrial adenocarcinoma (7,10,25).
We used the publicly available data of the TCGA project on ovarian and endometrial carcinomas to investigate the extent of genomic alterations of the NSD3-CHD8-BRD4 chromatin-remodeling complex in pelvic HGSCs. We herein propose that amplification of the NSD3-CDH8-BRD4 axis frequently occurs in a subset of pelvic HGSC of tubo-ovarian as well as endometrial origin. In patients with tubo-ovarian HGSCs, amplification of this pathway may be associated with unfavorable prognosis and survival. In endometrial cancers, amplification occurred more specifically in endometrial serous and in endometrioid ‘serous-like’ carcinomas, which are intrinsically more aggressive compared with the more commonly encountered low-grade endometrioid endometrial adenocarcinoma. The percentage of cases exhibiting NSD3-CHD8-BRD4 pathway amplification appears to be similar among ovarian HGSC (18%), endometrial serous carcinoma (25%) and endometrial serous-like carcinoma (33%), suggesting a shared mechanism of tumor progression in a subset of pelvic HGSCs. These findings support the hypothesis that the NSD3-BRD4-CHD8 pathway is potentially targetable with the newly developed BRD4-specific small-molecule inhibitors in a subset of HGSC patients of either tubo-ovarian or endometrial origin.
NSD3, CHD8 and BRD4 are located in 3 different amplicons in the genome, and within these amplicons, there is evidence to suggest that NSD3, CHD8 and/or BRD4 may be putative cancer driver genes rather than passenger genes. First, amplification was identified in two different TCGA datasets of pelvic HGSCs of i) ovarian and ii) endometrial origin. Second, amplification of NSD3, CHD8 and BRD4 correlated with increased mRNA expression, and in ovarian HGSC, worse patient survival is also observed with increased NSD3, CHD8 and BRD4 mRNA levels (9). Third, the oncogenic activity of NSD3 and BRD4 in ovarian HGSC has been previously validated by inhibition of NSD3 via siRNA/shRNA knockdown or of BRD4 via BRD4-specific small-molecule inhibitors, which resulted in decreased proliferation of ovarian cancer cells (6). Similar cytotoxic results were also seen in endometrial cancer cell culture and xenograft systems, wherein the BRD4-specific inhibitor, JQ1, suppressed growth of phosphatase and tensin homolog-positive endometrial cancer cells (25). Additionally, it has been demonstrated that knockdown of NSD3 (also referred to as WHSC1L1) in breast cancers with amplification of the 8p11-12 region resulted in significant decreases in cell proliferation (26).
However, it should be noted that certain nearby amplified genes within the amplicons also displayed increased mRNA expression in the TCGA cohort. For example, RAB2B is located in the same amplicon as CHD8, NOTCH3 is located in the same amplicon as BRD4, and BAG4 is located in the same amplicon as NSD3, all of which exhibited gene amplification and increased mRNA expression in a subset of ovarian HGSC patients from the TCGA cohort (data not shown). In addition, based on our overall and progression-free survival curve analyses, prognosis appears to be better correlated with amplification rather than mRNA expression, suggesting a potential collaborative role of co-amplified genes in the amplicons. Despite being a part of amplicons, there are several studies demonstrating the importance of NSD3, CHD8 and BRD4 in oncological processes (27–31), and it is noteworthy that amplicons may not necessarily contain only one oncogene, but may act as a unit with several genes of importance (26). Future knockdown of NSD3, BRD4 and CHD8 or treatment of HGSC displaying pathway amplification with BRD4-specific small-molecule inhibitors are required to confirm the growth-promoting properties of specific gene amplification in patients with pelvic HGSC.
Possible therapeutic downstream effector genes that are regulated by the NSD3-BRD4-CHD8 complex in pelvic HGSC include CMYC, CD274 (encoding PD-L1) and/or ALDH1A1. In a human xenograft model, BRD4 inhibition was effective in a subset of ovarian HGSCs that exhibited high CMYC or MYCN levels (7). In another ovarian cancer mouse model, treatment with BRD4-specific inhibitors limited tumor progression via immunotherapeutic effects by significantly reducing PD-L1 expression on tumor cells, dendritic cells and macrophages, and by increasing the activity of antitumor cytotoxic T cells (23). In addition, a third independent group has demonstrated that combination treatment of ovarian cancer cells with both cisplatin and the BRD4-specific inhibitor, JQ1, suppressed the outgrowth of cisplatin-resistant cells and improved the survival of ovarian cancer-bearing mice by suppressing ALDH1A1 levels and stem cell-like characteristics (24). In the ovarian HGSC TCGA cohort, our finding that worse overall survival is also observed with increased NSD3, BRD4 and CHD8 mRNA levels, in addition to gene amplification, suggests that this survival effect may be specific to NSD3-BRD4-CHD8 function by regulating the gene expression of potential downstream target genes.
Due to the limitations of the TCGA studies (such as relatively short follow-up time, lack of comorbidities and clinical covariables), these findings are exploratory and hypothesis-generating rather than definitive. For this reason, future independent studies with multivariate analyses and long-term follow-up to determine the effect of other clinical covariables (including age, stage, comorbidities and treatment regimens) on survival outcome are required to confirm our TCGA findings. However, it is worth highlighting that the vast majority of pelvic HGSCs from the TCGA study were high-stage (stage III or IV) as well as high-grade (1,5). In addition, our analysis also revealed significant survival effects with pathway amplification, specifically in a platinum-sensitive ovarian TCGA cohort and in an ovarian TCGA cohort with complete response to primary therapy.
Finally, we hypothesized that NSD3-BRD4-CHD8-amplified pelvic HGSCs may exhibit increased sensitivity to newly developed BRD4 small-molecule inhibitors. Future studies, including animal xenografts or cell culture models, are required to further elucidate the mechanisms by which amplification and overexpression of the pathway may affect tumor progression and patient survival in patients with HGSC. In summary, this study presents evidence that the NSD3-BRD4-CHD8 pathway is amplified and overexpressed in a subset of pelvic HGSCs of both tubo-ovarian and endometrial origin, and that amplification and overexpression may be associated with worse prognosis and survival. Our findings suggest that target inhibition of the BRD4 axis with specific inhibitors should be tested in a subset of patients with pelvic HGSC of gynecological (endometrial or tubo-ovarian) origin.
Acknowledgements
The present study protocol was in accordance with the TCGA publication policy for ovarian and endometrial carcinomas, which may be found at http://cancergenome.nih.gov. This study was also approved by the corresponding author's Institutional Review Board. In addition, the present study has been previously published as an abstract for the 2017 United States and Canadian Academy of Pathology annual meeting (www.nature.com/modpathol/journal/v30/n2s/pdf/modpathol2016251a.pdf).
References
- 1.Cancer Genome Atlas Research Network, corp-author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bast RC, Hennessy B, Mills GB. The biology of ovarian cancer: New opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pal T, Permuth-Wey J, Betts JA, Krischer JP, Fiorica J, Arango H, LaPolla J, Hoffman M, Martino MA, Wakeley K, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 4.Etemadmoghadam D, Au-Yeung G, Wall M, Mitchell C, Kansara M, Loehrer E, Batzios C, George J, Ftouni S, Weir BA, et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin Cancer Res. 2013;19:5960–5971. doi: 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, McGee J, Chen X, Doman TN, Gong X, Zhang Y, Hamm N, Ma X, Higgs RE, Bhagwat SV, et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS One. 2014;9:e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baratta MG, Schinzel AC, Zwang Y, Bandopadhayay P, Bowman-Colin C, Kutt J, Curtis J, Piao H, Wong LC, Kung AL, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma; Proc Natl Acad Sci USA; 2014; pp. 232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goundiam O, Gestraud P, Popova T, De la Motte Rouge T, Fourchotte V, Gentien D, Hupé P, Becette V, Houdayer C, Roman-Roman S, et al. Histo-genomic stratification reveals the frequent amplification/overexpression of CCNE1 and BRD4 genes in non-BRCAness high grade ovarian carcinoma. Int J Cancer. 2015;137:1890–1900. doi: 10.1002/ijc.29568. [DOI] [PubMed] [Google Scholar]
- 9.Ucar D, Lin DI. Amplification of the bromodomain-containing protein 4 gene in ovarian high-grade serous carcinoma is associated with worse prognosis and survival. Mol Clin Oncol. 2015;3:1291–1294. doi: 10.3892/mco.2015.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Ma P, Jing Y, Yan Y, Cai MC, Zhang M, Zhang S, Peng H, Ji ZL, Di W, et al. BET bromodomain inhibition as a therapeutic strategy in ovarian cancer by downregulating FoxM1. Theranostics. 2016;6:219–230. doi: 10.7150/thno.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3:e196–e204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 12.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 13.Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol. 2016;19:45–50. doi: 10.1016/j.ddtec.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., III Targeting MYC dependence in cancer by inhibiting BET bromodomains; Proc Natl Acad Sci USA; 2011; pp. 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-Short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu LL, Tian M, Li X, Li JJ, Huang J, Ouyang L, Zhang Y, Liu B. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, Zhang R. BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 2016;16:2829–2837. doi: 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kossenkov AV, Wu SR, Wickramasinghe JM, Yin X, Palozola KC, Gardini A, Showe LC, Yokoyama Y, Zhu H, Lee JH, et al. BET inhibitors suppress ALDH activity by targeting ALDH1A1 super-enhancer in ovarian cancer. Cancer Res. 2016;76:6320–6330. doi: 10.1158/0008-5472.CAN-16-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu H, Li J, Clark LH, Jackson AL, Zhang L, Guo H, Kilgore JE, Gehrig PA, Zhou C, Bae-Jump VL. JQ1 suppresses tumor growth via PTEN/PI3K/AKT pathway in endometrial cancer. Oncotarget. 2016;7:66809–66821. doi: 10.18632/oncotarget.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010;70:8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shingleton JR, Hemann MT. The chromatin regulator CHD8 is a context-dependent mediator of cell survival in murine hematopoietic malignancies. PLoS One. 2015;10:e0143275. doi: 10.1371/journal.pone.0143275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subtil-Rodríguez A, Vázquez-Chávez E, Ceballos-Chávez M, Rodríguez-Paredes M, Martín-Subero JI, Esteller M, Reyes JC. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res. 2014;42:2185–2196. doi: 10.1093/nar/gkt1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Zeng L, Shen C, Ju Y, Konuma T, Zhao C, Vakoc CR, Zhou MM. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure. 2016;24:1201–1208. doi: 10.1016/j.str.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung M, Gelato KA, Fernández-Montalván A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]