Abstract
Five new isoprenoids, 3,4,8,16-tetra-epi-lobocrasol (1), 1,15β-epoxy-deoxysarcophine (2), 3,4-dihydro-4α,7β,8α-trihydroxy-Δ2-sarcophine (3), ent-sarcophyolide E (4), and 16-deacetyl- halicrasterol B (5) and ten known compounds 6‒15, were characterized from the marine soft coral Sarcophyton glaucum, collected off Taitung coastline. Their structures were defined by analyzing spectra data, especially 2D NMR and electronic circular dichroism (ECD). The structure of the known compound lobocrasol (7) was revised. Cytotoxicity potential of the isolated compounds was reported, too.
Keywords: sarcophyton glaucum, lobocrasol, ent-sarcophyolide E, sarcophine
1. Introduction
Soft corals classified in the genus Sarcophyton are the predominant species in many coral reefs. They are endowed with diverse secondary metabolites, including cembranoids [1,2], biscembranoids [3] and steroids [4]. For pharmacological research, some of the metabolites are reported to possess cytotoxic [5,6], antimicrobial [7], and neuroprotective [8] activities. Ecologically, sarcophytoxide, usually found in sarocophyton species, is an allelopathic chemical used in competition for space with scleractinian corals [9]; while another well-known metabolite, sarcophine, is known to have a toxic effect on fishes [10]. In our previous work, we had isolated novel biscembranoids with cytotoxic activity and anti-inflammatory properties from the cultured soft coral S. glaucum [3]. Our present investigation disclosed the purification and structural elucidation of five new and ten known isoprenoid-derived compounds from the same wild-type species, collected off the coastline of Taitung County. The structure of 7 was also revised. Cytotoxic activity of the isolates was assayed by the inhibition of cancer cell proliferation.
2. Results and Discussion
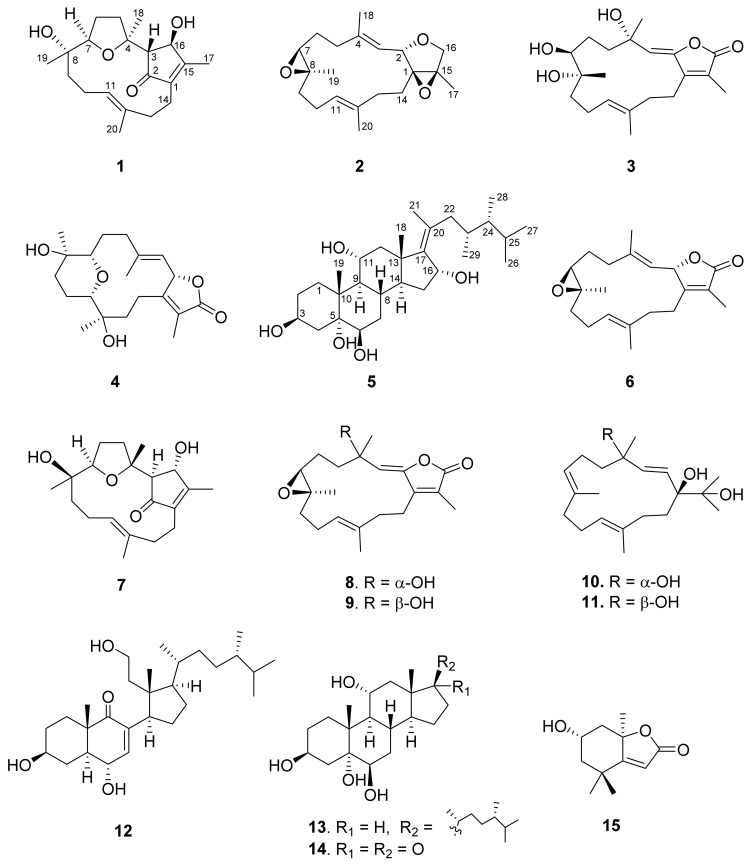
The EtOAc extract of Sarcophyton glaucum was repeatedly separated by column chromatography and HPLC to obtain five new isoprenoids 1‒5 and ten known compounds 6‒15 (Figure 1). By comparing with data in the literature, the known compounds were identified as sarcophine (6) [11], lobocrasol (7) [12], 3,4-dihydro-4α-hydroxy-Δ2-sarcophine (8) [11,13], 3,4-dihydro-4β-hydroxy-Δ2-sarcophine (9) [11,13], crassumol A (10) [14], klyflaccicembranol F (11) [15], sarcomilasterol (12) [16], sarcoaldesterol B (13) [17], sarglaucsterol (14) [18], loliolide (15) [19].
Figure 1.
Structures of compounds 1–15.
The molecular formula of 3,4,8,16-tetra-epi-lobocrasol (1), a colorless oil, was determined as C20H30O4 based on the [M + Na]+ ion peak obtained by (+)-HRESIMS. The 13C NMR data showed 20 carbon signals, including evidence of an α,β-unsaturated enone (δC 204.3, 167.0, and 142.8), an additional double bond (δC 133.3 and 128.4), and four oxygenated carbons (δC 83.8, 83.2, 75.9, and 73.6) (Table 1). The 1H NMR, in conjunction with the HSQC spectra, revealed that the structure of 1 possessed four methyl groups [δH 2.08 (3H, s), 1.69 (3H, s), 1.35 (3H, s), 1.03 (3H, s)], an olefinic methine [δH 5.15 (1H, t, J = 7.6 Hz)], and two oxygenated methines [δH 4.90 (1H, s); 3.80 (1H, dd, J = 9.2, 5.6 Hz)] (Table 2). The above spectral data were similar to those of lobocrasol (7) [12], suggesting that 1 might be an epimer of 7.
Table 1.
13C NMR spectroscopic data of compounds 1‒3 (100 MHz, CDCl3) and 4 (100 MHz, DMSO-d6).
| No. | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 142.8 (C) | 71.9 (C) | 152.2 (C) | 165.3 (C) |
| 2 | 204.3 (C) | 76.6 (CH) | 148.2 (C) | 79.6 (CH) |
| 3 | 59.4 (CH) | 122.4 (CH) | 116.5 (CH) | 119.5 (CH) |
| 4 | 83.2 (C) | 140.0 (C) | 74.2 (C) | 143.8 (C) |
| 5 | 32.4 (CH2) | 37.9 (CH2) | 38.8 (CH2) | 35.7 (CH2) |
| 6 | 25.0 (CH2) | 25.4 (CH2) | 26.9 (CH2) | 24.2 (CH2) |
| 7 | 83.8 (CH) | 61.9 (CH) | 74.6 (CH) | 83.4 (CH) |
| 8 | 73.6 (C) | 59.8 (C) | 74.9 (C) | 68.3 (C) |
| 9 | 34.0 (CH2) | 40.2 (CH2) | 38.0 (CH2) | 40.3 (CH2) |
| 10 | 21.5 (CH2) | 23.7 (CH2) | 22.1 (CH2) | 23.0 (CH2) |
| 11 | 128.4 (CH) | 123.9 (CH) | 128.5 (CH) | 80.0 (CH) |
| 12 | 133.3 (C) | 136.1 (C) | 131.4 (C) | 71.3 (C) |
| 13 | 39.1 (CH2) | 35.2 (CH2) | 38.3 (CH2) | 36.8 (CH2) |
| 14 | 20.0 (CH2) | 27.5 (CH2) | 22.3 (CH2) | 20.1 (CH2) |
| 15 | 167.0 (C) | 67.4 (C) | 123.7 (C) | 121.2 (C) |
| 16 | 75.9 (CH) | 69.4 (CH2) | 170.3 (C) | 174.5 (C) |
| 17 | 13.2 (CH3) | 12.1 (CH3) | 8.8 (CH3) | 8.5 (CH3) |
| 18 | 25.2 (CH3) | 15.6 (CH3) | 29.5 (CH3) | 16.2 (CH3) |
| 19 | 25.8 (CH3) | 16.7 (CH3) | 25.5 (CH3) | 20.0 (CH3) |
| 20 | 15.3 (CH3) | 14.9 (CH3) | 15.9 (CH3) | 23.4 (CH3) |
Table 2.
1H NMR spectroscopic data of compounds 1‒3 (400 MHz, CDCl3) and 4 (400 MHz, DMSO-d6).
| No. | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 2 | - | 4.87 d (11.2) | - | 5.53 d (10.4) |
| 3 | 2.16 br s | 5.26 dd (10.8, 0.8) | 5.21 s | 4.99 d (10.4) |
| 5 | 3.04 q (10.8) | 2.34 m | 2.09 m | 2.18 m |
| 1.67 m | - | 1.78 m | - | |
| 6 | 2.00 dd (9.6, 2.4) | 1.92 m | 1.54 m | 1.97 m |
| 1.54 m | 1.63 m | 1.51 m | 1.42 m | |
| 7 | 3.80 dd (9.2, 5.6) | 2.65 t (4.0) | 3.52 dd (9.2, 3.2) | 3.01 dd (10.0, 2.8) |
| 9 | 1.50 m | 2.13 m | 1.64 m | 1.73 m |
| 1.25 m | 0.93 m | 1.47 m | 1.49 m | |
| 10 | 2.05 m | 2.27 m | 2.17 m | 1.62 m |
| 1.90 m | 1.87 m | 1.86 m | 1.34 m | |
| 11 | 5.15 t (7.6) | 5.11 dd (9.6, 5.6) | 4.99 t (6.8) | 3.13 d (10.4) |
| 13 | 2.28 br d (13.2) | 2.22 m | 2.28 m | 1.70 m |
| 1.84 m | 1.95 m | - | 1.49 m | |
| 14 | 2.61 td (13.2, 2.1) | 1.69 m | 2.56 m | 2.45 m |
| 2.12 m | - | - | 1.91 m | |
| 16 | 4.90 br s | 3.89 d (10.4) | - | - |
| - | 3.75 d (10.4) | - | - | |
| 17 | 2.08 s | 1.43 s | 1.94 s | 1.73 s |
| 18 | 1.35 s | 1.84 s | 1.40 s | 1.77 s |
| 19 | 1.03 s | 1.27 s | 1.61 s | 0.98 s |
| 20 | 1.69 s | 1.57 s | 1.67 s | 0.98 s |
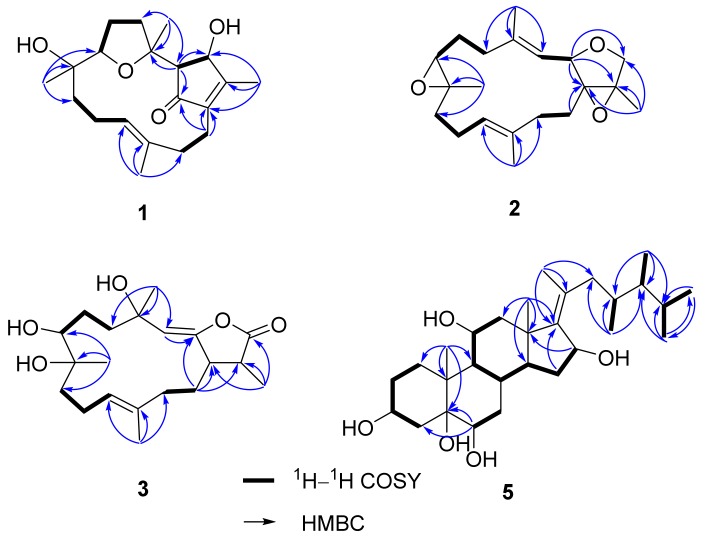
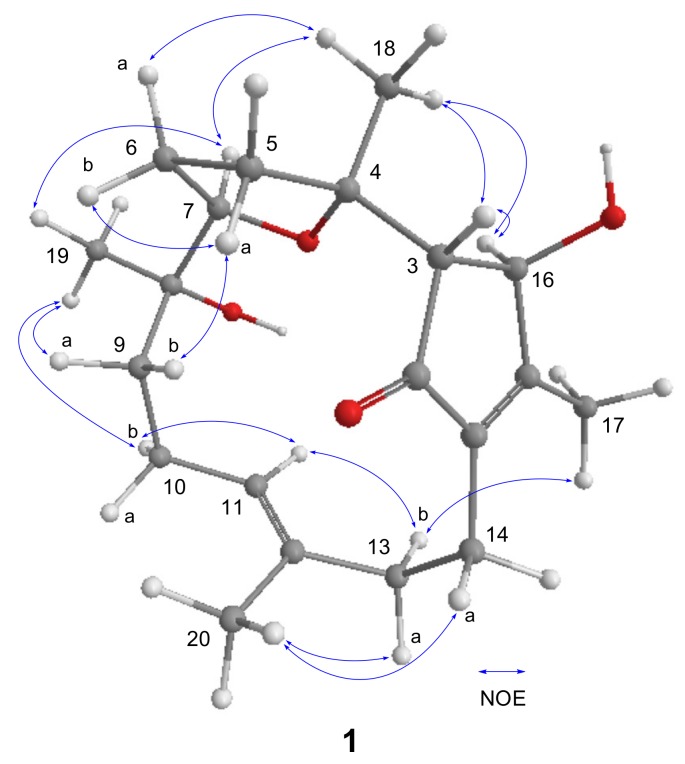
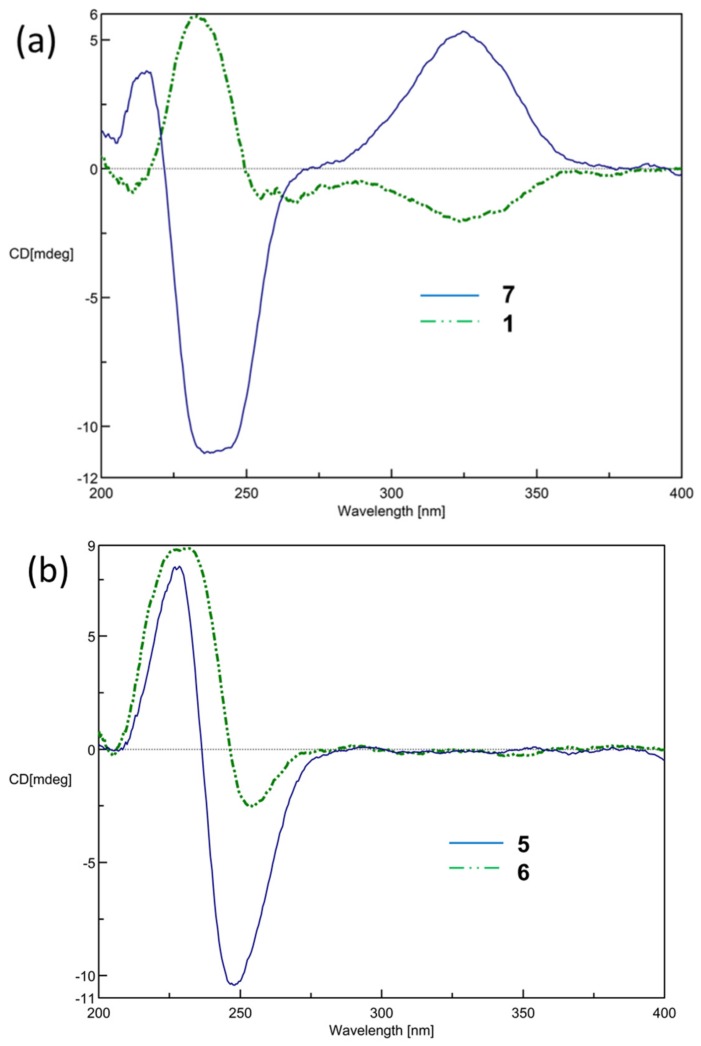
Analysis of COSY spectra established four proton spin systems: H-3/H-16, H2-5/H2-6/H-7, H2-9/H2-10/H-11, and H2-13/H2-14 (Figure 2). The HMBC correlations from H3-18 to C-3, C-4, and C-5 made the connection of the tetrahydrofuran ring and the 4-hydroxy-3-methylcyclopent-2-enone moiety. The HMBC correlations from H3-19 to C-7, C-8, and C-9; from H3-20 to C-11, C-12, and C-13; and from H3-17 to C-1, C-15, and C-16 combined with correlations from H2-14 to C-1, C-2, and C-15 constructed the remaining fragments. Consequently, the planar structures of 1 and 7 were found to be the same. The relative configurations of all the chiral centers in 1 were deduced by interpretation of nuclear Overhauser effect (NOE) data, analysis of 3JH-H values, and comparison of carbon chemical shifts. As depicted in Figure 3, the NOE cross-peaks of H3-18/H-7 supported that they oriented on the same face, and casually assigned these protons to be α-oriented. The downfield-shifted proton H-5a (δH 3.04), deshielded by the C-2 carbonyl group, showed an NOE correlation with H-9b (δH 1.25), while H3-19 showed correlations with both H-7 and H-9a (δH 1.50), suggesting that H-7 and C-9 were located in anti-orientation, thus suggesting a β-orientation for H3-19. The strong NOE cross-peaks of H3-18/H-16 suggested the α-orientation of H-16. A weak NOE correlation was observed between H-16 and H-3, while H-3 also showed a correlation with H3-18, and suggested H-3 should be located anti to H-16. This was confirmed by comparing the small 3JH-H values with those of cyclopent-2-enone analogues, in which adjacent protons with a trans configuration showed small coupling constants [20,21]. The E geometry for Δ11 double bond was deduced by NOE cross-peaks of H-11/H-13b (δH 1.84), as well as by comparing carbon chemical shift for the C-20 at δC 15.3 (<20 ppm) [22]. Thus, the chemical shift for C-20 of 7 (δC 14.8) revealed the Δ11 double bond should possess the E, not Z geometry, as assigned in the literature [12]. This was further confirmed by our observation of NOE correlation between H-11 and H-13 (δH 2.48) (Supplementary Materials, Figure S6-3). The absolute configuration at C-16 of 7 was determined by Lin et al. using Mosher’s method [12]. The electronic circular dichroism (ECD) spectrum of 1 showed Cotton effects with approximately opposite signs compared to that of 7 (Figure 4a), suggesting different configurations at C-3 and C-16 for both 1 and 7. Thus, the absolute configuration of 1 was determined as shown in Figure 1.
Figure 2.
Selected 1H–1H COSY and HMBC correlations of 1–3 and 5.
Figure 3.
Selected NOE correlations of compound 1.
Figure 4.
Electronic circular dichroism (ECD) curves of (a) compounds 1 and 7; (b) compounds 5 and 6.
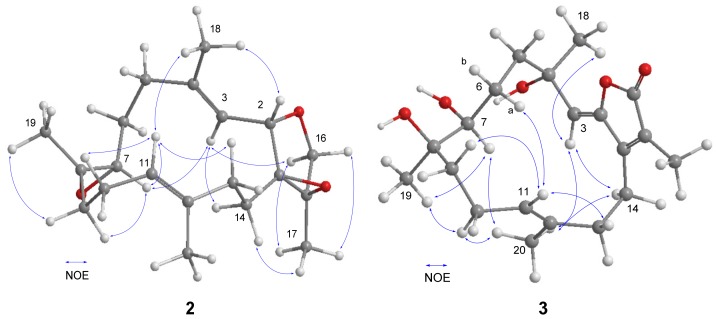
The 1,15β-Epoxy-deoxysarcophine (2) was isolated as a colorless oil and had a molecular formula of C20H30O3, determined by HRESIMS analysis. The NMR spectra were quite similar to those of 1,15β-epoxy-2-epi-deoxysarcophine [23] with the exception of the signals around the tetrahydrofuran (THF) ring. The 1H–1H COSY and HMBC correlations, as depicted in Figure 2, confirmed that 2 and 1,15β-epoxy-2-epi-deoxysarcophine shared the same planar structure. The 14-membered ring of 2 is suggested to be identical to that of sarcophine (6), of which the C-7 to C-11 fragment adopted a half-chair conformation [24], due to the fact that both compounds possessed similar key NOE correlation data: H-3/H-7 and H-7/H-11 (Figure 5). The NOE cross-peaks of H-3/H2-14, H2-14/H3-17, as well as H-2/H3-18, implied that both the 1,15-epoxy group and H-2 were β-oriented.
Figure 5.
Selected NOE correlations of compounds 2 and 3.
The molecular formula of 3,4-dihydro-4α,7β,8α-trihydroxy-Δ2-sarcophine (3), C20H30O5, was established by HRESIMS, exceeding that of 8 or 9 by 18 mass units. A series of characteristic absorption bands due to hydroxy (3444 cm−1) and carbonyl (1747 cm−1) groups were assigned from the IR spectrum of 3. The NMR data of 3 resembled those of 8 and 9, with significant differences in carbon chemical shifts at C-7 (δC 74.6) and C-8 (δC 74.9), suggesting that 3 is a 7,8-dihydroxy analogue of 8 or 9. The Δ2 and Δ11 double bonds were both determined as E geometry according to NOE cross-peaks of H-3/H2-14 and H-10/H3-20, respectively (Figure 5). Moreover, the correlations of H-7/H3-19, H-7/H3-20, and H-3/H3-20 disclosed that these protons located on the same face and subjectively designated as α protons. The above correlations, as well as that of H-6a/H-11, restricted the envenlop conformation for the C-7 to C-11 segment (Figure 5). Accordingly, H3-18 was correlated with H-3, but not with H-7, suggesting that this methyl group is likely β-oriented.
ent-Sarcophyolide E (4), a colorless oil, was found to have a molecular formula of C20H30O5, based on its HRESIMS. Its NMR spectroscopic data (Table 1 and Table 2) were superimposed to that of sarcophyolide E [11]. The ECD spectra of 4 showed a negative Cotton effect for n→π* (254 nm) and positive π→π* (230 nm) transitions, which are quite the same as those of the co-isolated sarcophine (6) (Figure 4b), but are in opposite signs to those reported for sarcophyolide E [11]. Accordingly, 4 was elucidated as an enantiomer of sarcophyolide E. Surprisingly, the specific optical rotation of sarcophyolide E was reported as [α]D +4, that has the same positive sign as the specific optical rotation value of 4 ([α]D +14). The [α]D of 4 was repeatedly measured and the positive sign was always obtained. The reason for this discrepancy with literature might need to be studied further.
The HRESIMS of 16-deacetyl-halicrasterol B (5) showed a sodiated adduct ion peak at m/z 501.3552, implying a molecular formula of C29H50O5. Its 1H NMR spectrum disclosed three methyl singlets (one assignable to be olefinic methyl), four methyl doublets, four oxygenated-methine protons [4.00 (m, H-3); 3.47 (br s, H-6); 3.88 (td, J = 10.4, 5.2 Hz); 4.62 (d, J = 5.2 Hz, H-16)], as well as a complex array of aliphatic methine and methylene protons (Table 3). The 13C NMR spectrum showed a tetra-substituted double bond at δC 148.2 (C-17) and 134.2 (C-20) and five oxygenated carbons at δC 68.2 (C-3), 77.4 (C-5), 76.6 (C-6), 69.6 (C-11), and 73.0 (C-16). The above spectroscopic data were very similar to those of a Δ17(20) sterol, halicrasterol B, with the acetyl signals disappeared in 5, suggesting that 5 is a deacetyl derivative of halicrasterol B. This was secured by analyzing the 1H–1H COSY correlations and the HMBC experiments (Figure 2). Compound 5 and halicrasterol B were found to share the same configurations at the side chain segment based on their identical 13C NMR data. Accordingly, the structure of 5 was characterized as depicted in Figure 1.
Table 3.
1H and 13C NMR spectroscopic data of compound 5.
| No. | δH (J in Hz) a | δC (mult.) b | No. | δH (J in Hz) a | δC (mult.) b |
|---|---|---|---|---|---|
| 1 | 1.99 m | 35.2 (CH2) | 15 | 1.62 m | 36.5 (CH2) |
| 1.54 m | - | - | 1.45 m | - | |
| 2 | 1.75 m | 32.0 (CH2) | 16 | 4.62 d (5.2) | 73.0 (CH) |
| 1.51 m | - | 17 | - | 148.2 (C) | |
| 3 | 4.00 m | 68.2 (CH) | 18 | 0.91 s | 18.8 (CH3) |
| 4 | 2.09 dd (13.2, 11.6) | 42.0 (CH2) | 19 | 1.28 s | 17.5 (CH3) |
| 1.54 m | - | 20 | - | 134.2 (C) | |
| 5 | - | 77.4 (C) | 21 | 1.77 s | 17.5 (CH3) |
| 6 | 3.47 br s | 76.6 (CH) | 22 | 2.41 dd (13.2, 10.8) | 43.0 (CH2) |
| 7 | 1.84 m | 35.3 (CH2) | - | 1.91 m | - |
| 8 | 1.88 m | 29.1 (CH) | 23 | 1.86 m | 34.0 (CH) |
| 9 | 1.57 m | 53.3 (CH) | 24 | 1.09 m | 45.9 (CH) |
| 10 | - | 41.1 (C) | 25 | 1.64 m | 31.3 (CH) |
| 11 | 3.88 td (10.4, 5.2) | 69.6 (CH) | 26 | 0.87 d (6.8) | 19.6 (CH3) |
| 12 | 2.62 dd (12.0, 5.2) | 50.2 (CH2) | 27 | 0.93 d (6.8) | 22.0 (CH3) |
| 1.58 m | - | 28 | 0.82 d (6.8) | 11.9 (CH3) | |
| 13 | - | 46.0 (C) | 29 | 0.72 d (6.8) | 13.9 (CH3) |
| 14 | 1.78 m | 52.5 (CH) | - | - | - |
a Spectra recorded at 400 MHz in CD3OD; b spectra recorded at 100 MHz in CD3OD.
The cytotoxicity of the isolates 1–15 against HepG2 (human hepatocellular liver carcinoma), MDA-MB-231 (human breast adenocarcinoma), and A-549 (human lung epithelial cells) cancer cells were assayed. Furthermore, compounds 6, 8, 9, and 12 were also assayed for cytotoxicity against MOLT-4 (human acute lymphoblastic leukemia), SUP-T1 (human T-cell lymphoblastic lymphoma), and U-937 (human histiocytic lymphoma) cell lines. The results showed that compound 12 exhibited cytotoxicity effect against MDA-MB-231, MOLT-4, SUP-T, and U-937 cell lines with IC50 values of 13.8, 6.7, 10.5, and 17.7 μg/mL, respectively, while compound 13 was found to possess cytotoxicity against HepG2, MDA-MB-231, and A-549 cell lines with IC50 values of 9.7, 14.0, and 15.8 μg/mL. The remaining compounds were found to be not cytotoxic against the above cancer cell lines, with IC50 values higher than 20 μg/mL.
3. Experimental Section
3.1. General Experimental Procedures
The optical rotation values and IR spectra were recorded on a JASCO P-1020 digital polarimeter (JASCO Corporation, Tokyo, Japan) and JASCO J-815 spectrophotometer (JASCO Corporation), respectively. A Varian 400 NMR instrument was used to record the 1H NMR and 13C NMR spectra with the chemical shifts shown as ppm referenced to the solvent residue of CDCl3 (δH 7.26 ppm and δC 77.0 ppm), DMSO-d6 (δH 2.50 ppm and δC 39.5 ppm), and CD3OD (δH 3.31 ppm and δC 49.0 ppm). A Bruker APEX II mass spectrometer equipped with an ESI ionization source (Bruker, Bremen, Germany) was used for acquiring high-resolution mass data. The HPLC system was composed of a Shimadzu LC-10ATVP series pump, a UV detector (Shimadzu, Milan, Italy), and an ODS column (5 µm, 250 × 10 mm, Inertsil ODS-3, GL Science Inc., Tokyo, Japan).
3.2. Animal Material
The collection of S. glaucum was performed off the coast of Jihui Fishing Port, Taitung county, Taiwan, in March 2013. A −20 °C freezer was used for specimen storage until extraction. Prof. Chang-Feng Dai performed the species identification, and a voucher specimen (JiH-201304) of this soft coral has been deposited in National Sun Yat-sen University.
3.3. Extraction and Isolation
The lyophilized samples of S. glaucum (195 g) were chopped and soaked (4 × 24 h) with EtOAc (4 × 2 L). After the solvent was evaporated, a residue (17.35 g) was obtained. The residue of the EtOAc layer was chromatographed by a silica gel column, using a stepwise gradient system composed of EtOAc/n-hexane (0:100 to 100:0, each four column volumes) and acetone/EtOAc (0:100 to 100:0, each four column volumes), and eventually washed by MeOH to afford 25 fractions according to TLC analysis. Fraction 19 was subjected to repeated column chromatography (CC) by Sephadex LH-20 (isocratic, acetone), C18 gel (MeOH-H2O, 75%), and silica gel (acetone-n-hexane, 10%) to obtain subfractions (SFr.) 19-1 and 19-2. In turn, SFr.19-1 was subjected to semipreparative HPLC (MeOH-H2O, 73%) to give compound 6 (23.4 mg), while compound 2 (1.3 mg) was obtained from SFr.19-2 using a CH3CN-H2O (53%) solvent system. Fraction 20 was subjected to an open column chromatography using Sephadex LH-20 (isocratic, acetone) and subsequently purified by C18 column chromatography (MeOH-H2O, 90%) to yield two subfractions (SFr. 20-1 and 20-2). Compounds 11 (2.7 mg) and 10 (2.0 mg) were purified from SFr. 20-1 using HPLC (MeOH-H2O, 79%). SFr.20-2 was fractionated into two subfractions (SFr. 20-2-1 and SFr. 20-2-2) using C18 CC (MeOH-H2O, 64%). Compounds 8 (1.7 mg) and 9 (6.9 mg) were yielded from SFr. 20-2-1 using HPLC (MeOH-H2O, 65%), and compounds 3 (2.7 mg) and 15 (2.1 mg) were obtained from SFr. 20-2-2 using eluent of CH3CN-H2O (33%). Four subfractions (SFr. 22-1–SFr.22-4) were obtained from fraction 22 by C18 CC (MeOH-H2O, 80%). Compounds 4 (4.9 mg) and 7 (4.7 mg) were purified from SFr.22-1 using HPLC (MeOH-H2O, 64%). Compounds 1 (2.2 mg), 12 (33.6 mg), and 13 (4.8 mg) were isolated from SFr. 22-2, SFr. 22-3, and SFr.22-4, respectively, using HPLC (MeOH-H2O: 71% for SFr. 22-2, 79% for SFr. 22-3, and 85% for SFr. 22-4). Fraction 23 was subjected to CC using a C18 column (MeOH-H2O, 80%) and subsequently by HPLC (MeOH-H2O, 66%) to give compounds 14 (3.6 mg) and 5 (1.4 mg).
3,4,8,16-Tetra-epi-lobocrasol (1): colorless oil; –98 (c 1.00, CHCl3); ECD (MeOH) λmax (Δε) 211 (−0.62), 233 (+1.79), 324 (−0.62); IR (KBr) νmax 3419, 2925, 2855, 1698, 1651, 1455, 1380, 1332, 1246, 1080, and 1049 cm−1; 13C and 1H NMR data, see Table 1 and Table 2; ESIMS m/z 357 [M + Na]+; HRESIMS m/z 357.2034 [M + Na]+ (calculated (calcd.) for C20H30O4Na, 357.2036).
1,15β-Epoxy-deoxysarcophine (2): colorless oil; −154 (c 0.63, CHCl3); IR (KBr) νmax 2926, 2855, 1653, 1514, 1455, 1381, 1239, 1164, and 1039 cm−1; 13C and 1H NMR data, see Table 1 and Table 2; ESIMS m/z 341 [M + Na]+; HRESIMS m/z 341.2085 [M + Na]+ (calcd. for C20H30O3Na, 341.2087).
3,4-Dihydro-4α,7β,8α-trihydroxy-Δ2-sarcophine (3): colorless oil; −84 (c 1.28, CHCl3); IR (KBr) νmax 3444, 2965, 2929, 2857, 1747, 1668, 1638, 1455, 1373, 1317, 1260, 1127, 1062, and 1018 cm−1; 13C and 1H NMR data, see Table 1 and Table 2; ESIMS m/z 373 [M + Na]+; HRESIMS m/z 373.1986 [M + Na]+ (calcd. for C20H30O5Na, 373.1986).
ent-Sarcophyolide E (4): colorless oil; +14 (c 1.70, CHCl3); ECD (MeOH) λmax (Δε) 230 (+1.41), 254 (−0.40); 13C and 1H NMR data, see Table 1 and Table 2; ESIMS m/z 373 [M + Na]+; HRESIMS m/z 373.1984 [M + Na]+ (calcd. for C20H30O5Na, 373.1986).
16-Deacetyl-halicrasterol B (5): white powder; −111 (c 0.78, CHCl3); IR (KBr) νmax 3330, 2922, 2854,1652, 1598, 1455, 1374, and 1023 cm−1; 13C and 1H NMR data, see Table 3; ESIMS m/z 501 [M + Na]+; HRESIMS m/z 501.3552 [M + Na]+ (calcd. for C29H50O5Na, 501.3551).
Sarcophine (6): colorless oil; +139 (c 4.98, CHCl3); lit. +97 (c 0.01, CHCl3); ECD (MeOH) λmax (Δε) 229 (+1.22), 248 (−1.58); MS, 1H and 13C NMR data were found to be in full agreement with literature data [11].
Lobocrasol (7): colorless oil; −212 (c 0.05, CHCl3); lit. −186 (c 0.10, CHCl3); ECD (MeOH) λmax (Δε) 215 (+0.57), 240 (−1.66), 325 (+0.81); MS, 1H and 13C NMR data were found to be in full agreement with literature data [12].
3,4-Dihydro-4α-hydroxy-Δ2-sarcophine (8): colorless oil; −17 (c 0.50, CHCl3); lit. −5 (c 0.10, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [11,13].
3,4-Dihydro-4β-hydroxy-Δ2-sarcophine (9): colorless oil; −1 (c 1.53, CHCl3); lit. −1 (c 0.10, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [11,13].
Srassumol A (10): colorless oil; −30 (c 0.49, CHCl3); lit. −20 (c 0.10, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [14].
Klyflaccicembranol F (11): colorless oil; −76 (c 1.28, CHCl3); lit. −32 (c 0.60, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [15].
Sarcomilasterol (12): amorphous solid; +30 (c 0.67, CHCl3); lit. +5 (c 0.08, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [16].
Sarcoaldesterol B (13): amorphous solid; −21 (c 1.50, MeOH); lit. −20 (c 0.23, MeOH); MS, 1H and 13C NMR data were found to be in full agreement with literature data [17].
Sarglaucsterol (14): amorphous solid; −15 (c 0.45, MeOH); lit. −25 (c 0.11, MeOH); MS, 1H and 13C NMR data were found to be in full agreement with literature data [18].
Loliolide (15): colorless oil; −11 (c 0.73, CHCl3); lit. −92 (c 1.10, CHCl3); MS, 1H and 13C NMR data were found to be in full agreement with literature data [19].
3.4. Cytotoxicity Assay
HepG2, MDA-MB-231, A-549, MOLT-4, SUP-T1, and U-937 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The MTT assays were performed as previous reported [25]. After a 15 h culture for the above cancer cell lines, the isolated compounds prepared in different concentrations of DMSO were added for an additional 72 h culture [25]. The cytotoxic potential of the isolated compounds was evaluated by means of a MTT [3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide] cell proliferation assay and the absorbance was measured using an ELISA reader and monitored at 570 and 620 nm, which allowed the calculation of IC50 values.
4. Conclusions
Our present study discovered four new diterpenoids 1–4 and a new steroid 5 as well as ten known compounds 6–15. The 3,4,8,16-tetra-epi-lobocrasol (1) represent the second example of lobocrasol-type diterpenoid, while the Δ11 double in lobocrasol (7) was revised as E geometry in this study. It is interesting that, similar to sarcophine and ent-sarcophine [26], sarcophyolide E and ent-sarcophyolide E (4) were also isolated from the same genus.
Acknowledgments
This work was supported by grants from Ministry of Science and Technology (MOST104-2113-M-110-006, and 105-2113-M-110-002), NSYSU-KMU Joint Research Project (NSYSUKMU 06C0301709) and Aim for the TOP University Program (05C030205) from Ministry of Education of Taiwan awarded to J.-H. S.
Supplementary Materials
The 1H and 13C NMR spectra of compounds 1–5 and 7 (Figures S1–S6), and partial NOESY spectrum of 7 (Figure S6-3) are available online at http://www.mdpi.com/1660-3397/15/7/202/s1.
Author Contributions
Jyh-Horng Sheu designed the whole experiment. Chih-Hua Chao contributed to structural elucidation and manuscript preparation. Wen-Liang Li performed purification, structural elucidation, and data acquisition. Chiung-Yao Huang, Atallah F. Ahmed and Chih-Chuang Liaw also performed data acquisition. Yang-Chang Wu and Mei-Chin Lu performed the cytotoxicity assay. Chang-Feng Dai contributed to the collection of soft coral and species identification.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Lin W.Y., Chen B.W., Huang C.Y., Wen Z.H., Sung P.J., Su J.H., Dai C.F., Sheu J.H. Bioactive cembranoids, sarcocrassocolides P−R, from the Dongsha Atoll soft coral Sarcophyton crassocaule. Mar. Drugs. 2014;12:840–850. doi: 10.3390/md12020840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin W.Y., Lu Y., Su J.H., Wen Z.H., Dai C.F., Kuo Y.H., Sheu J.H. Bioactive cembranoids from the dongsha atoll soft coral Sarcophyton crassocaule. Mar. Drugs. 2011;9:994–1006. doi: 10.3390/md9060994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C.Y., Sung P.J., Uvarani C., Su J.H., Lu M.C., Hwang T.L., Dai C.F., Wu S.L., Sheu J.H. Glaucumolides A and B, biscembranoids with new structural type from a cultured soft coral Sarcophyton glaucum. Sci. Rep. 2015;5:15624. doi: 10.1038/srep15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duh C.Y., Wang S.K., Chung S.G., Chou G.C., Dai C.F. Cytotoxic cembrenolides and steroids from the formosan soft coral Sarcophyton crassocaule. J. Nat. Prod. 2000;63:1634–1637. doi: 10.1021/np0002381. [DOI] [PubMed] [Google Scholar]
- 5.Gong K.K., Tang X.L., Zhang G., Cheng C.L., Zhang X.W., Li P.L., Li G.Q. Polyhydroxylated steroids from the South China Sea soft coral Sarcophyton sp. and their cytotoxic and antiviral activities. Mar. Drugs. 2013;11:4788–4798. doi: 10.3390/md11124788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C., Li J., Su J., Liang Y., Yang X., Zheng K., Zeng L. Cytotoxic diterpenoids from the soft coral Sarcophyton crassocaule. J. Nat. Prod. 2006;69:1476–1480. doi: 10.1021/np050499g. [DOI] [PubMed] [Google Scholar]
- 7.ElAhwany A.M.D., Ghozlan H.A., ElSharif H.A., Sabry S.A. Phylogenetic diversity and antimicrobial activity of marine bacteria associated with the soft coral Sarcophyton glaucum. J. Basic Microbiol. 2015;55:2–10. doi: 10.1002/jobm.201300195. [DOI] [PubMed] [Google Scholar]
- 8.Badria F.A., Guirguis A.N., Perovic S., Steffen R., Müller W.E.G., Schröder H.C. Sarcophytolide: A new neuroprotective compound from the soft coral Sarcophyton glaucum. Toxicology. 1998;131:133–143. doi: 10.1016/S0300-483X(98)00124-3. [DOI] [PubMed] [Google Scholar]
- 9.Fleury B.G., Coll J.C., Sammarco P.W. Complementary (secondary) metabolites in a soft coral: Sex-specific variability, inter-clonal variability, and competition. Mar. Ecol. 2006;27:204–218. doi: 10.1111/j.1439-0485.2006.00106.x. [DOI] [Google Scholar]
- 10.Néeman I., Fishelson L., Kashman Y. Sarcophine—A new toxin from the soft coral Sarcophyton glaucum (Alcyonaria) Toxicon. 1974;12:593–598. doi: 10.1016/0041-0101(74)90192-5. [DOI] [PubMed] [Google Scholar]
- 11.Xi Z., Bie W., Chen W., Liu D., van Ofwegen L., Proksch P., Lin W. Sarcophyolides B–E, new cembranoids from the soft coral Sarcophyton elegans. Mar. Drugs. 2013;11:3186–3196. doi: 10.3390/md11093186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S.T., Wang S.K., Cheng S.Y., Duh C.Y. Lobocrasol, a new diterpenoid from the soft coral Lobophytum crassum. Org. Lett. 2009;11:3012–3014. doi: 10.1021/ol901070e. [DOI] [PubMed] [Google Scholar]
- 13.Sayed K.A.E., Hamann M.T. Structurally novel bioconversion products of the marine natural product sarcophine effectively inhibit JB6 cell transformation. J. Org. Chem. 1998;63:7449–7455. doi: 10.1021/jo9813134. [DOI] [PubMed] [Google Scholar]
- 14.Lin S.T., Wang S.K., Duh C.Y. Cembranoids from the Dongsha Atoll soft coral Lobophytum crassum. Mar. Drugs. 2011;9:2705–2716. doi: 10.3390/md9122705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed A.F., Tsai C.R., Huang C.Y., Wang S.Y., Sheu J.H. Klyflaccicembranols A–I, new cembranoids from the soft coral Klyxum flaccidum. Mar. Drugs. 2017;15:23. doi: 10.3390/md15010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W.T., Gong H.L., Yao L.G., Guo Y.W. 9,11-Secosteroids and polyhydroxylated steroids from two South China Sea soft corals Sarcophyton trocheliophorum and Sinularia flexibilis. Steroids. 2014;92:56–61. doi: 10.1016/j.steroids.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z.B., Xiao H., Fan C.Q., Lu Y.N., Zhang G., Yin S. Bioactive polyhydroxylated sterols from the marine sponge Haliclona crassiloba. Steroids. 2013;78:1353–1358. doi: 10.1016/j.steroids.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C.X., He X.X., Zhang J., Guo Q., Lei L.F., Su J.Y., Zeng L.M. New precursor of tetraterpenoids from the soft coral Sarcophyton glaucum. Nat. Prod. Res. 2013;27:782–786. doi: 10.1080/14786419.2012.701211. [DOI] [PubMed] [Google Scholar]
- 19.Hodges R., Porte A.L. The structure of loliolide: A terpene from lolium perenne. Tetrahedron. 1964;20:1463–1467. doi: 10.1016/S0040-4020(01)99140-9. [DOI] [Google Scholar]
- 20.Lin S., Shi T., Chen K.Y., Zhang Z.X., Shan L., Shen Y.H., Zhang W.D. Cyclopenicillone, a unique cyclopentenone from the cultures of Penicillium decumbens. Chem. Comm. 2011;47:10413–10415. doi: 10.1039/c1cc12079d. [DOI] [PubMed] [Google Scholar]
- 21.Jung M., Jang K.H., Kim B., Lee B.H., Choi B.W., Oh K.B., Shin J. Meroditerpenoids from the Brown Alga Sargassum siliquastrum. J. Nat. Prod. 2008;71:1714–1719. doi: 10.1021/np800321y. [DOI] [PubMed] [Google Scholar]
- 22.Davis R.A., Carroll A.R., Quinn R.J. Longithorols C–E, Three new macrocyclic sesquiterpene hydroquinone metabolites from the Australian ascidian, Aplidium longithorax. J. Nat. Prod. 1999;62:1405–1409. doi: 10.1021/np990217a. [DOI] [PubMed] [Google Scholar]
- 23.Sawant S.S., Youssef D.T.A., Reiland J., Ferniz M., Marchetti D., El Sayed K.A. Biocatalytic and antimetastatic studies of the marine cembranoids sarcophine and 2-epi-16-deoxysarcophine. J. Nat. Prod. 2006;69:1010–1013. doi: 10.1021/np050527v. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein J. Sarcophine, a new epoxy cembranolide from Marine origin. Tetrahedron. 1974;30:2817–2824. doi: 10.1016/S0040-4020(01)97451-4. [DOI] [Google Scholar]
- 25.Hsiao T.H., Sung C.S., Lan Y.H., Wang Y.C., Lu M.C., Wen Z.H., Wu Y.C., Sung P.J. New anti-inflammatory cembranes from the cultured soft coral Nephthea columnaris. Mar. Drugs. 2015;13:3443–3453. doi: 10.3390/md13063443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L.G., Liu H.L., Guo Y.W., Mollo E. New cembranoids from the Hainan soft coral Sarcophyton glaucum. Helv. Chim. Acta. 2009;92:1085–1091. doi: 10.1002/hlca.200800417. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.