Abstract
HOXA distal transcript antisense RNA (HOTTIP), a critical oncogenic long non-coding RNA, has been reported to be aberrantly regulated in various cancer types. The present meta-analysis aimed to investigate HOTTIP as a potential clinical applicable prognostic biomarker in malignant neoplasms. Literature collections were performed by searching the electronic databases, PubMed and Web of Science (up to July 20, 2016). All the relevant searches were conducted to identify the association of HOTTIP with the overall survival (OS) rate. A total of six articles consisting of 508 patients were included in the present meta-analysis. The results suggested that the overexpression of HOTTIP is closely correlated with poor OS (hazard ratio=2.28; 95% confidence interval=1.71–3.04; P=0.000). In conclusion, the present meta-analysis has demonstrated that an increased expression level of HOTTIP is correlated with poor OS in different types of cancer, suggesting that HOTTIP potentially serves as a reliable prognostic biomarker in different types of cancer.
Keywords: long non-coding RNA, HOXA distal transcript antisense RNA, HOTTIP, cancer, prognosis, meta-analysis
Introduction
Long non-coding RNAs (lncRNAs) are a class of transcripts >200 nucleotides in length with limited protein-coding potential (1). Recently, several studies have revealed that lncRNAs are dysregulated in numerous types of human cancers, including gastric, colorectal, gastric, ovarian, lung and breast cancer (2–7). Certain lncRNAs have multiple functions in a wide range of biological processes, including proliferation, apoptosis, or cell migration (2,3,6).
HOXA distal transcript antisense RNA is an antisense non-coding transcript located at the 5′-end of the Homeobox A (HOXA) cluster (8). An increased expression level of HOTTIP has been observed in various types of tumor (9–14). Furthermore, the overexpression of HOTTIP has been previously shown to be significantly correlated with poor prognosis (9–14). Therefore, HOTTIP may be a candidate to serve as a feasible prognostic factor. Therefore, the aim of the present study was to perform a quantitative meta-analysis to further determine the correlation of HOTTIP with tumor prognosis.
Data collection methods
Search strategy
Electronic databases were searched, including PubMed and Web of Science (up to July 20, 2016). The following keywords were used for the search: ‘long non-coding RNA HOTTIP’, ‘lncRNA HOTTIP’, ‘cancer’, ‘prognosis’, and ‘survival’.
Selection criteria
Inclusion criteria were as follows: i) patients were divided into high and low groups, according to the expression levels of HOTTIP; ii) the correlation between HOTTIP expression quantity and overall survival (OS) was investigated. Exclusion criteria were as follows: i) editorials, expert opinions, letters, case reports and reviews were excluded, as were ii) studies lacking helpful data on OS and iii) duplicate publications.
Data extraction
Two investigators (LY.Y. and N.J.) independently extracted data from the eligible studies, and disagreements were resolved by discussion with a third investigator (Z-P.X.). The extracted data included the first author, year of publication, the country in which the study participants were enrolled, number of patients, tumor type, detection method of HOTTIP, cut-off values, survival analysis method, the sources of hazard ratios (HRs) [95% CIs (confidence intervals)], and the numbers in the high and low HOTTIP expression groups. The HRs were obtained using two methods. First, the HRs were obtained directly from publications; secondly, the HRs were extrapolated using Engauge Digitizer version 4.1 software from the Kaplan-Meier survival curve (15). The strength of the link between HOTTIP and survival outcome were evaluated by the HRs with corresponding 95% CIs. The I2 statistic was used to evaluate the heterogeneity of the data (16). The random-effects model was used if there was significant heterogeneity between studies (I2 >50% or P<0.05), whereas the fixed-effects model was applied for the present meta-analysis. The meta-analysis results were displayed using Forest plots. The stability of the results was evaluated using sensitivity analysis. Begg's funnel plots were used to evaluate publication bias (17). All the statistical analyses were performed using Stata 12.0 (Stata Corporation, College Station, TX, USA).
Results
As shown in Fig. 1, the searches with key terms disclosed that 64 articles that had been published up to July 20, 2016. After screening the title and abstract carefully, 56 references were excluded due to duplicate publications and irrelevant content. Upon further review of the full articles, 2 were excluded since they were records lacking information concerning the survival outcomes. Finally, 6 articles consisting of 508 patients were included in the present meta-analysis. Among these 6 studies, 5 came from China and 1 was from Switzerland. Five different types of cancer were evaluated, with 2 cases of hepatocellular carcinoma, 1 case of tongue squamous cell carcinoma, 1 of colorectal cancer, 1 of osteosarcoma, and 1 of gastric cancer. All these characteristics are summarized in Table I.
Figure 1.
Flow diagram of publication selection.
Table I.
Characteristics of HOTTIP studies included in the meta-analysis.
| Authors | Year | Country | Cancer type | Total no. | HOTTIPP detection method | Cut-off | Outcome | Survival analysis | HR estimate | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|
| Quagliata et al | 2014 | Switzerland | HCC | 52 | RT-qPCR | Median | OS | Univariate | Survival curve | (12) |
| Ge et al | 2015 | China | HCC | 48 | RT-qPCR | Median | OS | Univariate | Survival curve | (14) |
| Zhang et al | 2015 | China | TSCC | 86 | RT-qPCR | Median | OS | Multivariate, univariate | Reported | (9) |
| Ren et al | 2015 | China | CRC | 156 | RT-qPCR | Median | OS | Multivariate, univariate | Reported | (11) |
| Li et al | 2015 | China | OSA | 68 | RT-qPCR | Mean | OS | Multivariate, univariate | Reported | (13) |
| Ye et al | 2016 | China | GC | 98 | RT-qPCR | Median | OS | Univariate | Survival curve | (10) |
HOTTIP, HOXA distal transcript antisense RNA; HR, hazard ratio; OS, overall survival; HCC, hepatocellular carcinoma; TSCC, tongue squamous cell carcinoma; CRC, colorectal cancer; OSA, osteosarcoma; GC, gastric cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Association between HOTTIP and OS
To investigate the association between HOTTIP expression and OS, 6 studies consisting of 508 patients were included. Heterogeneity analysis revealed that no evidence of statistically significant heterogeneity across these 6 studies was identified (P=0.983 for heterogeneity test; I2=0%). Subsequently, a fixed-effect model was used to calculate the HR with corresponding 95% CI. According to the meta-analysis results (HR=2.28; 95% C I=1.71–3.04; P<0.0001, Fig. 2), it may be concluded that patients for whom a high expression of HOTTIP in cancerous tissues was detected were more prone to a poor outcome.
Figure 2.
Forest plots showing the association between HOTTIP expression and the OS. OS, overall survival; HR, hazard ratio.
Sensitivity analysis and publication bias
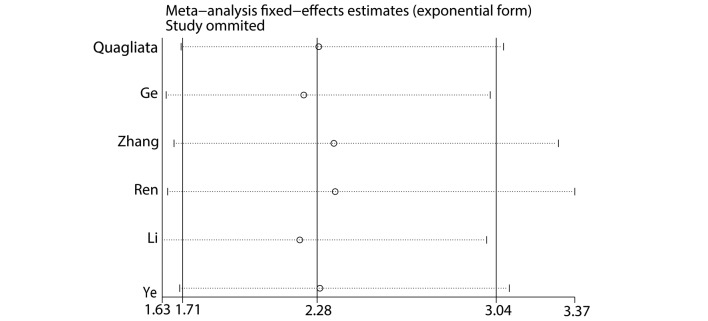
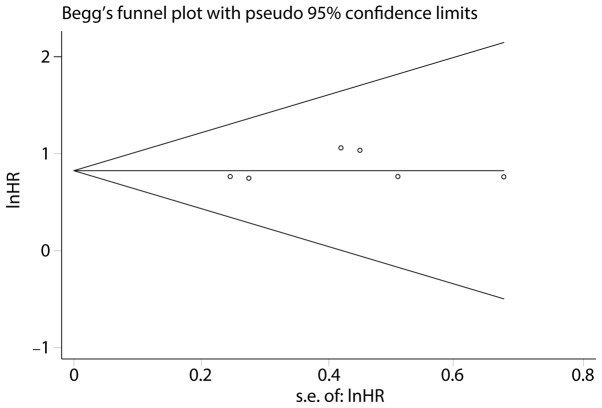
Sensitivity analysis indicated that the exclusion of any individual study did not change the significance of HR (Fig. 3). This demonstrated that the HR of the OS was reliable. A Begg's funnel plot and Egger's linear regression test were performed to evaluate publication bias. The Egger's and Begg's tests suggested the publication bias was not significant (P=0.151 for Egger's test; P=0.06 for Begg's test) (Fig. 4).
Figure 3.
Sensitivity analysis of the HRs of HOTTIP expression and OS for the included studies.
Figure 4.
Begg's funnel plot to explore publication bias of the included studies between HOTTIP and OS. lnHR, logarithm of hazard ratio; s.e., standard error.
Discussion
HOTTIP is an lncRNA transcribed from the 5′-end of the HOXA. It is able to bind to and target WD repeat-containing protein 5 (WDR5)/mixed lineage leukemia (MLL) complexes to the 5′-HOXA locus, driving histone H3 lysine 4 trimethylation and gene transcription (8). The most recently published studies have identified that HOTTIP is a critical oncogenic lncRNA that is highly expressed in numerous types of cancers, including hepatocellular carcinoma and colorectal, gastric, and lung cancer. At the same time, these studies have demonstrated that the HOTTIP/HOXA13 axis is associated with cell growth and the cell cycle, which serves an important role in the genesis and progression of cancer (18,19). Although it has already been demonstrated that HOTTIP is closely associated with multiple tumors, the prognostic role of HOTTIP expression in different types of cancer has yet to be fully elucidated. Therefore, a meta-analysis was performed to explore the connection between a high expression level of HOTTIP and the OS rate for cancer patients. The present meta-analysis has highlighted the tumor prognostic role of HOTTIP, and provided sufficient evidence to establish an association between HOTTIP expression and the prognosis of different human cancer types, suggesting that overexpression of HOTTIP is significantly corrected with poor OS. However, it should be emphasized that there were several limitations in the present study. First, the cut-off definition of HOTTIP expression varied in each study. Secondly, only 6 studies with 508 patients were included in the present meta-analysis, which may influence the reliability of our results. Thirdly, three HRs in our study were calculated according to the survival curve, which may have affected the accuracy of the research results. Fourthly, only English language papers were included in the present study.
Taken together, the present meta-analysis has demonstrated that elevated levels of HOTTIP expression are significantly associated with a poor OS (HR=2.28; 95% CI=1.71–3.04; P<0.0001). Therefore, HOTTIP may be used as a negative, unfavorable prognostic marker for most types of cancer.
References
- 1.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 2.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, Liu Y, Ou J, Wang D, Yao L, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 4.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Fan Y, Wang YF, Su HF, Fang N, Zou C, Li WF, Fei ZH. Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial-mesenchymal transition. J Hematol Oncol. 2016;9:57. doi: 10.1186/s13045-016-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, Yuan J, Shan W, Li C, Hu X, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Zhao L, Wang YX, Xi M, Liu SL, Luo LL. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36:8805–8809. doi: 10.1007/s13277-015-3645-2. [DOI] [PubMed] [Google Scholar]
- 10.Ye H, Liu K, Qian K. Overexpression of long noncoding RNA HOTTIP promotes tumor invasion and predicts poor prognosis in gastric cancer. Onco Targets Ther. 2016;9:2081–2088. doi: 10.2147/OTT.S95414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren YK, Xiao Y, Wan XB, Zhao YZ, Li J, Li Y, Han GS, Chen XB, Zou QY, Wang GC, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8:11458–11463. [PMC free article] [PubMed] [Google Scholar]
- 12.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Cao L, Hang D, Wang F, Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8:11414–11420. [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 corrected and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger M, Smith G Davey, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SS, Wuputra K, Liu CJ, Lin YC, Chen YT, Chai CY, Lin CS, Kuo KK, Tsai MH, Wang SW, et al. Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget. 2016;7:36049–36064. doi: 10.18632/oncotarget.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang SR, Yang JK, Xie JK, Zhao LC. Long noncoding RNA HOTTIP contributes to the progression of prostate cancer by regulating HOXA13. Cell Mol Biol (Noisy-le-grand) 2016;62:84–88. [PubMed] [Google Scholar]