Abstract
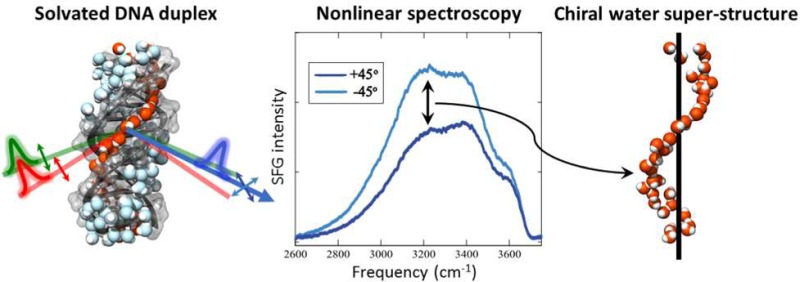
The iconic helical structure of DNA is stabilized by the solvation environment, where a change in the hydration state can lead to dramatic changes to the DNA structure. X-ray diffraction experiments at cryogenic temperatures have shown crystallographic water molecules in the minor groove of DNA, which has led to the notion of a spine of hydration of DNA. Here, chiral nonlinear vibrational spectroscopy of two DNA sequences shows that not only do such structural water molecules exist in solution at ambient conditions but that they form a chiral superstructure: a chiral spine of hydration. This is the first observation of a chiral water superstructure templated by a biomolecule. While the biological relevance of a chiral spine of hydration is unknown, the method provides a direct way to interrogate the properties of the hydration environment of DNA and water in biological settings without the use of labels.
Short abstract
Using chiral nonlinear spectroscopy, we discover a chiral water superstructure surrounding DNA at ambient conditions. This chiral spine of hydration is templated by the helical structure and provides a direct label-free probe of the solvation shell.
Introduction
The first few layers of water around DNA are critical to its structure and biological function. Dehydration can change the structure from the classic right-handed B-form to the shorter and wider A-form and the left-handed Z-form.1 Furthermore, interactions of DNA strands with small molecules and proteins are mediated by the solvation structure. As such, the hydrating water molecules form an activation barrier for molecular drugs targeting the major and minor grooves of DNA, and essential biological processes such as DNA transcription involving biomolecules binding to DNA displacing the solvation shell.2−5 Consequently, DNA’s hydration shell has been studied extensively with X-ray crystallography,6−9 neutron scattering,10 NMR spectroscopy,11−13 electronic14 and vibrational spectroscopy15 and molecular dynamics (MD) simulations.16,17 X-ray experiments performed at cryogenic temperatures have shown the existence of structural water molecules in particular minor groove sequences, giving notion to the presence of a spine of hydration in the DNA minor groove. NMR experiments have measured different time scales concerning the dynamics of water in the solvation structure of DNA,12,13 and time-resolved fluorescence measurements have suggested evidence of slow water molecules in the hydration shell,14 but the interpretation of the latter measurements has been questioned.16 Recent MD simulations have shown a broad range of water dynamics in the DNA solvation structure and identified very slow water molecules in the minor groove of DNA consistent with the structural waters observed in the X-ray experiments.17 To differentiate the bulk water response from the response of the relatively few hydration waters, many of the previous experimental methods have relied on cryogenic temperatures, indirect probes, dehydrated DNA, or other methods that likely disrupt the biologically relevant equilibrium hydration structure. Recently third-order vibrational spectroscopy has been used to investigate the phosphate vibrations of DNA, indirectly probing the hydration structure.18 The present study focuses on directly targeting the solvation structure using second-order nonlinear vibrational spectroscopy, which is capable of probing interfacial and chiral structures in situ without relying on labels.19−21 The main idea is that if structural waters persist at room temperature, they should form an ordered, chiral, helical structure surrounding DNA. Using chiral sum-frequency generation (SFG) spectroscopy, we examine the vibrational frequencies of the hydration water molecules of two DNA sequences bound to a self-assembled monolayer under near-physiological conditions (room temperature and 100 mM NaCl solution). We indeed observe that DNA imprints its chirality on the surrounding water molecules, generating a chiral SFG water response. This confirms the existence of a DNA minor groove spine of hydration at room temperature and further shows that the chiral structure of biomolecules can be imprinted on the surrounding solvation structure.
Approach
Chiral SFG has emerged as a powerful tool for studying chiral biomolecules.21−28 SFG is a nonlinear optical method in which an infrared and a visible photon interact with the sample producing a photon with the sum of the incident frequencies (energies). Second-order nonlinear optical techniques, like SFG, require a lack of inversion symmetry under the electric dipole approximation.19,20 Chiral-specific SFG spectroscopy is possible because chirality, by definition, breaks inversion symmetry. Where the chiral response in linear vibrational circular dichroism is on the order of 0.1%, the chiral SFG response can be on the same order of magnitude as the achiral SFG response.26 However, as a nonlinear optical method, chiral SFG typically suffers from a relatively poor signal-to-noise ratio, making it very difficult to detect a subtle chiral solvation structure. In linear spectroscopy, the small chiral response is separated from the large achiral response by taking the difference between two large signals containing both contributions, e.g., right- and left-handed polarized light. Similarly, chiral signatures can be detected in SFG spectroscopy by taking a difference between polarization combinations, but pure chiral signals can also be obtained. SFG spectroscopy involves three light fields that can be polarized either parallel (p) or perpendicular (s) to the plane of incidence, giving rise to eight distinct polarization combinations. Polarizations are listed in descending order of photon frequency, i.e., spp indicated s-polarized SFG, p-polarized visible, and p-polarized IR beams. For a surface that is isotropic with respect to rotations within the plane (C∞), polarization combinations containing one or three p-polarized fields (ssp, sps, pss, and ppp) give rise to a signal for both chiral and achiral samples.21,26 However, polarization combinations containing two p-polarized fields (spp, psp, and pps) only produce a SFG signal for chiral samples. The polarization combination sss cannot produce a SFG signal. A full description of achiral and chiral SFG is given in the Supporting Information. Accordingly, pure chiral signals can be obtained in SFG spectroscopy, but given limitations to signal-to-noise, it is often advantageous to interfere a pure chiral with an achiral signal. This can be done by having any of the light fields linearly polarized at an odd angle, e.g., ±45°, containing both p and s polarizations. A chiral signature is then obtained by taking the difference between the +45° and −45° signals, analogously to circular dichroism.22,24,25,27,28
We have recently made technical advancements improving the sensitivity and reliability of chiral SFG spectroscopy.28 The chiral SFG method, illustrated in Figure 1, relies on simultaneously measuring two polarization components and rigorously calibrating the alignment through self-referencing. Here either the pure chiral and achiral SFG responses, or alternatively, the +45° and −45° polarized SFG responses, can be measured simultaneously. The blue and red arrows represent the orthogonally polarized achiral and chiral SFG signals, respectively, for a given set of incident polarizations. In the interference scheme, the orthogonal signals are first rotated by 45° by a waveplate. A beam displacer then splits the SFG signals into vertical and horizontal polarizations, containing the positively and negatively interfered achiral and chiral signals, which are then collected simultaneously. By measuring the +45° and −45° polarized SFG responses simultaneously, the difference signal is immune to laser fluctuations. Furthermore, the self-referencing approach described in the Supporting Information corrects for any potential false chiral responses from alignment imperfections. Both these improvements facilitate measuring the weak chiral imprint onto the solvation structure of DNA.
Figure 1.
Incident visible (green) and infrared (red) beams produce a second-order SFG signal (blue) at the buried surface-bound DNA/water interface. The inset shows click chemistry linking the DNA to a SiO2-coated CaF2 prism. The SFG beam’s polarization is rotated 45° by an achromatic waveplate (AW). The beam is then split into vertical and horizontal polarizations by a beam displacer (BD). Finally, the +45° and −45° polarized SFG responses are collected simultaneously (CCD). The difference between the measured +45° and −45° polarized SFG responses across the OH stretch region shown in the graph proves the existence of a chiral water superstructure surrounding DNA.
Results
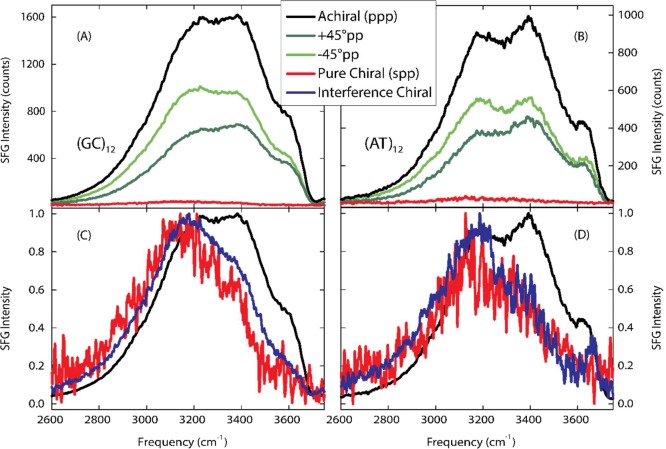
The results presented here are the first SFG experiments focused on the water OH stretch region surrounding DNA. Previous experiments examined the CH stretch region of surface bound DNA in contact with air using chiral SFG,25 showing that SFG is a sensitive probe of the structural chirality of DNA, and in contact with water using achiral SFG.29 The water OH stretch vibrational frequency probed here is very sensitive to the local hydrogen-bonded network with lower vibrational frequencies corresponding to stronger hydrogen-bonded configurations.30 The OH stretch of the water–air surface spans the frequency range of 3000–3800 cm–1, but strongly hydrogen-bonded water can exhibit lower vibrational frequencies. Within the same frequency range, the NH stretch vibrations between DNA base pairs appear as two narrower features at 3200 and 3350 cm–1,15 but since these are orientated perpendicular to the DNA strain, they do not contribute to a net chiral superstructure. The strong difference between the +45°pp and –45°pp spectra shown in Figure 1 across the water OH stretch vibrational frequency region demonstrates the existence of a chiral water superstructure templated by the DNA helical structure. Chiral ordering of individual molecules in contact with small biomolecules has previously been observed in circular vibrational dichroism experiments on the water bend vibration and theoretical calculations.31−33 The present finding is the first discovery of a chiral water superstructure. Figure 2A,B compares the pure chiral spp SFG signal and the difference spectrum taken between the +45°pp and –45°pp polarized SFG responses for two DNA duplexes with 24 alternating GC or AT base pairs. Both sequences display a chiral water structure showing that the chiral structure is not sequence specific. The chiral signatures measured by the two methods agree well, where the interference method exhibits improved signal strength.
Figure 2.
Chiral SFG responses for 24-base pair double-stranded DNA sequences in aqueous 100 mM NaCl. (A) and (C) show results for 24 base pair double-stranded DNA consisting of alternating guanine and cytosine bases (GCGC). (B) and (D) show results for 24 base pair double-stranded DNA strands consisting of alternating thymine and adenine bases (ATAT). (A) and (B) demonstrate the chirality of the water around both sequences through the nonzero intensity of the pure chiral spp (red) and in the difference between the of the +45°pp (black), and –45°pp (green) spectra. (C) and (D) compare the normalized pure achiral ppp (gray), pure chiral spp (red), and interference +45°pp - –45°pp (blue) spectra. The interference chiral spectra have greater signal-to-noise ratios than the pure chiral results, but have similar spectral shapes. Compared to the achiral spectra, the pure and interference chiral spectra are red-shifted, indicating stronger hydrogen-bonded waters. In addition, a peak at 3660 cm–1 in (D) indicates non-hydrogen bonded chiral water population, attributed to non-hydrogen-bonded OH’s in contact with a hydrophobic surface in the ATAT minor groove.
In addition to establishing the existence of a chiral water structure surrounding DNA at ambient conditions, our method facilitates characterizing the hydrogen-bonding strength of this chiral water superstructure. Figure 2C,D compares the achiral and interference chiral SFG response for the GCGC and ATAT duplexes. The achiral ppp spectra display a broad OH stretch band reflecting the broad range of hydrogen-bonding interactions in the solvation shell. The achiral signals contain three main features at 3200, 3400, and 3600 cm–1. While the 3200 and 3400 cm–1 features could result from inter- and intramolecular coupling between the OH vibrations as suggested for the air–water interface,34−37 for this complex system, they likely correspond to specific hydrogen-bonding interactions within the general solvation structure: the 3200 cm–1 band is due to water molecules orientated in the electric field from the DNA backbone charges, the 3400 cm–1 band is due to medium-strength hydrogen-bonded water molecules in the major and minor grooves, and the 3600 cm–1 band is due to weakly hydrogen-bonded water molecules in the grooves. The achiral SFG spectrum is consistent with the linear spectrum of DNA in weakly hydrated films.15
Compared to the achiral spectra, the interference chiral spectra for both the (GC)12 and (AT)12 duplexes are shifted to lower frequencies while still exhibiting a broad frequency distribution. This red shift reveals that the chiral water structure around DNA is more strongly hydrogen-bonded than the general achiral solvation shell as expected due to the strong interaction of localized waters with the DNA base pairs. The chiral water structure around the (AT)12 duplex further displays a population of non-hydrogen-bonded OH stretch vibrations in contact with a hydrophobic structure at 3660 cm–1. Such non-hydrogen-bonded OH vibrations have previously been observed both at macroscopic flat hydrophobic surfaces,38,39 and in the solvation shell around hydrophobic residues.40−42 The observation of non-hydrogen-bonded OH vibrations in the solvation shell of DNA is initially surprising. However, it is known that the minor groove at AT base pairs is particularly narrow.43 An analysis of the sequence dependence of the minor groove width of 88 DNA crystal structures found that 82% of tetranucleotides with especially narrow minor groove widths (<5 Å) contain three or more AT pairs.44 The (AT)12 duplex likely has a narrower minor groove than the (GC)12 duplex and therefore may have more restricted waters in its minor groove. Accordingly, we attribute the non-hydrogen-bonded OH vibrations to water molecules that donate a single hydrogen-bond to a neighboring water molecule or DNA base but, due to the narrow width of the minor groove in the (AT)12 duplex, are not able to find another hydrogen-bonding partner. Our results thus show that the chiral water structure around DNA is a general phenomenon, while some sequence-specific fine structure exists.
Discussion
Having observed the existence of a chiral water structure surrounding DNA, the question arises as to its origin. While the intrinsic chiral response of stereocenters dominates linear chiroptical signals, structural chirality of achiral chromophores arranged in ordered chiral structures dominates chiral SFG.26 Chiral SFG has thus the unique capability of being able to isolate the response of the water molecules that adopt an overall chiral structure through the interaction with DNA.
The chiral water structure observed here is consistent with structural minor groove waters observed by other experiments. The X-ray experiments at cryogenic temperatures showed the existence of structural waters inside the minor groove of DNA around the central AATT region of the Drew–Dickerson dodecamer (CGCGAATTCGCG).6,7 An idealized strand of B-DNA has a minor groove width of 5.7 Å and a major groove width of 11.7 Å. The relative size of the minor groove compared to water (3.2 Å) is consistent with the crystallographic observation of a single-file spine of hydration in the minor groove of B-DNA.6 In contrast, the major groove is about twice as large as the minor groove and can accommodate 2–3 water molecules across its width. Molecular dynamics simulations have established anomalous water dynamics slowed by a factor of 10 in the minor groove, but the dynamics of major groove water is only modestly perturbed by a factor of 2.17,45 These observations suggest that water in the major groove is substantially more disordered, which is likely to disrupt the chirality of the water in the major groove compared to the minor groove. Therefore, we conclude that the measured chiral signal is dominated by the water molecules in immediate contact with the DNA strands, likely in the minor groove, but a small contribution from the net alignment of major groove waters cannot be excluded experimentally. For illustrative purposes, the water structure within the minor and major grooves of the double-stranded CGCAAATTTGCG oligonucleotide from a MD simulation46 are highlighted in Figure 3. Our results confirm the existence of structural water molecules under ambient conditions and that this is a general effect occurring for both (GC)12 and (AT)12 duplexes.
Figure 3.

Water structure in the minor groove (red) and major groove (blue) of the double-stranded CGCAAATTTGCG oligonucleotide in a MD simulation.46 Water in the minor groove is substantially more ordered with respect to the DNA-axis than water in the major groove, which suggests that the chiral SFG measurements are selectively interrogating the minor groove spine of hydration without the use of labels.
Conclusions
The present study provides the first observation of a chiral water superstructure templated by a large biomolecule and affords a direct label-free characterization of the hydration shell structure of DNA under ambient solvated conditions. The study confirms the existence of structural water molecules and a spine of hydration in the minor groove of DNA under near-physiological conditions that has previously been observed at cryogenic temperatures. The spectroscopic results presented here further reveal that the chiral structure of DNA is imprinted on the solvation structure resulting in a chiral spine of hydration surrounding DNA. The chiral water superstructure exhibits a broad range of hydrogen-bonding strengths reflecting the large heterogeneity of the water structure and dynamics in the solvation shell of DNA. In comparison to the general achiral solvation structure, however, the chiral water structure is more strongly hydrogen-bonded.. The chiral water structure is observed for both the (GC)12 and (AT)12 duplexes, showing that the chiral spine of hydration is a general phenomenon not confined to a specific sequence. However, sequence-specific differences occur, where non-hydrogen-bonded waters are observed for the (AT)12 duplex, which exhibits a narrower minor groove compared to (GC)12. This is consistent with the idea that a narrower minor groove would confine the water more and limit availability of hydrogen-bonding partners.
The discovered chiral nature of a water superstructure provides a means for directly targeting and characterizing the hydrogen-bonding strength of water molecules in biological settings more generally. While it is not clear whether or not a chiral water superstructure could have a biological function, such as providing a nonspecific binding motif, it is known that the hydration state affects the structure and function of DNA. Changes in the hydration state can change the structure from the classic right-handed B-form to the shorter and wider A-form and the left-handed Z-form that are thought to be involved in DNA packing and transcription. Chiral SFG spectroscopy can now be used to examine how the solvation structure of DNA might transform under different hydration conditions and under interactions with salts, biomolecules, and drugs. For example, since water molecules in the minor groove of DNA can present a barrier to drug binding, the strongly hydrogen-bonded water can be monitored as the binding event occurs, thereby providing a means of elucidating a mechanism. Given the abundance of chirality in biology, this technique could provide a general label-free method for studying the local water structure in biological systems.
Methods
10 mm × 10 mm 60° equilateral CaF2 prisms (Crystran) were used to improve the SFG signal strength from the buried solid–water interface compared to flat windows. A homemade Teflon flow cell housed the prisms for the SFG experiments. The CaF2 prisms were coated with a 125 nm thick SiO2 layer using an atomic layer deposition system with 110 °C plasma (Oxford Instruments ALD FlexAL). The SiO2 coatings were annealed at 800 °C for 30 min in a high vacuum furnace (Lindberg). A gold band was added to a section of each sample prism for normalizing the SFG signal. First, a 5 nm thick titanium adhesion layer was applied, followed by a 150 nm thick gold layer by magnetron argon sputtering through a stainless steel mask. With a skin depth of 16.2 nm for 792.5 nm light and 33.5 nm for 3400 nm light, the titanium adhesion layer does not influence the gold nonresonant SFG response. The nonresonant gold response allows collection of the infrared envelope of the SFG spectrometer.
SiO2-coated CaF2 prisms were reacted with azidomethyltrimethoxytrichlorosilane (Gelest) using a modified version of a literature gas-phase silanization procedure.47 Briefly, a 2 L desiccator was dried at 140 °C for 4 h, then cooled for less than 10 min. Silane (200 μL) was added to filter paper. The filter paper and 800 mg of MgSO4 were placed in the desiccator. A home-built steel prism holder was placed in the desiccator above the reactants. The desiccator was evacuated for 60 s and placed in a 110 °C oven for 1 h. Deposition was verified by Fourier transform infrared spectroscopy of the azide absorbance at 2100 cm–1.
DNA was attached to the azide SAM using dibenzocyclooctyl copper-free click chemistry. Dibenzocyclooctyl-modified 24-base pair DNA sequences were purchased from the Keck Oligonucleotide Synthesis Resource at Yale University. The purchased sequences were alternating guanines and cytosines (XCGCGCGCGCGCGCGCGCGCGCGCG, where X = 5′-DBCO-TEG Phosphoramidite) and alternating thymines and adenines (XATATATATATATATATATATATAT) along with the unmodified complementary strands. The strained octyne-modified DNA sequences were dissolved to 60 μM concentration in phosphate buffered saline (PBS) and placed in contact with the azide SAM-modified SiO2-coated CaF2 prisms for 1 day at room temperature. The DNA-modified prisms were then rinsed with 250 mM NaCl solution to remove any nonbonded DNA. The presence of the DNA was verified by FTIR spectroscopy of the CH-stretches of the DNA and the presence of phosphate by X-ray photoelectron spectroscopy. The complementary DNA strands were dissolved in PBS solution and paired with the immobilized DNA by exposure for 1 h at room temperature. Nonbonded DNA was removed by rinsing with 250 mM NaCl aqueous solution. Azide SAMs and DNA samples were subjected to chiral SFG experiments both dry and in contact with 100 mM NaCl aqueous solution.
For these SFG experiments, a 3 mJ beam from a Ti:sapphire amplifier (Coherent, Legend Elite Duo, 793 nm, 1 kHz, 25 fs, 38 nm fwhm) pumps an OPA (OPerA Solo, Coherent) to generate mid-infrared laser pulses. To cover the breadth of the OH stretch region, the OPA is centered at eight positions (2500, 2600, 2650, 2700, 3100, 3300, 3500, and 3700 nm) with approximately 250 cm–1 fwhm and 30 μJ pulse energy. To combine into one spectrum, each of the eight spectra was cut off when the infrared envelope intensity (measured by gold nonresonant spectrum) was below 5% of the maximum and the intensities were added.
The infrared laser pulses were filtered (quartz broadband filter, Spectrogon) and focused at the sample at an incident angle of 65° with a pulse energy at the sample of 5 μJ. A separate 793 nm beam from the amplifier was filtered by an etalon (TecOptics, 795 nm, 0.6 nm, 10 cm–1). Higher and lower order Fabry-Pérot transmission fringes were filtered out with a bandpass filter (Thorlabs, 10 nm fwhm) angle-tuned to maximize throughput. The narrowband 793 nm pulse was then focused on the sample at an incident angle of 60° with a pulse energy of 5 μJ. The incident photons induce a second-order polarization that emits a photon at the sum of the incident frequencies. In order to normalize SFG signals, the infrared envelope is obtained from the shape of the nonresonant gold SFG signals in the ppp polarization (the only polarization with a strong nonresonant signal). The gold spectra were normalized 0 to 1 and the sample spectra were divided by the normalized gold spectra to produce normalized sample spectra.
In the interference scheme, the polarization of the SFG signal was rotated by 45° by the achromatic λ/2 waveplate (air-spaced, Eksma 467–4205). A birefringent calcite beam displacer (Thorlabs BD27) displaced the vertically polarized projection of the SFG signal in the vertical direction, while the horizontal polarization component was transmitted unaffected. The horizontal and vertical projections were then directed into a monochromator (Acton SP-2500i, Princeton Instruments, 1800 grooves/mm blazed at 500 mm) and collected simultaneously on a liquid nitrogen-cooled CCD array (Spec-10, Princeton Instruments, 1340 × 400 pixels).
Acknowledgments
This work was supported by The Arnold and Mabel Beckman Foundation under a Young Investigator Award and the National Science Foundation under an NSF CAREER Award (CHE-1151079). Sample preparation made use of the Cornell Center for Materials Research Shared Facilities, which are supported through the NSF MRSEC program (DMR-1120296), and the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (ECCS-0335765).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00100.
Theoretical background for chiral and achiral SFG spectroscopy and self-referencing results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Saenger W.; Hunter W. N.; Kennard O. DNA Conformation Is Determined by Economics in the Hydration of Phosphate Groups. Nature 1986, 324 (6095), 385–388. 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W. The Role of Water in Protein-DNA Interactions. Curr. Opin. Struct. Biol. 1997, 7 (1), 126–134. 10.1016/S0959-440X(97)80016-4. [DOI] [PubMed] [Google Scholar]
- Jayaram B.; Jain T. The Role of Water in Protein-DNA Recognition. Annu. Rev. Biophys. Biomol. Struct. 2004, 33 (1), 343–361. 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- Privalov P. L.; Dragan A. I.; Crane-Robinson C.; Breslauer K. J.; Remeta D. P.; Minetti C. A. S. A. What Drives Proteins into the Major or Minor Grooves of DNA?. J. Mol. Biol. 2007, 365 (1), 1–9. 10.1016/j.jmb.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer G. M.; Fuchs J. E.; Markt P.; Kirchmair J.; Wellenzohn B.; Langer T.; Liedl K. R. Sequence-Specific Positions of Water Molecules at the Interface between DNA and Minor Groove Binders. ChemPhysChem 2008, 9 (18), 2766–2771. 10.1002/cphc.200800647. [DOI] [PubMed] [Google Scholar]
- Drew H. R.; Dickerson R. E. Structure of a B-DNA Dodecamer. III. Geometry of Hydration. J. Mol. Biol. 1981, 151 (3), 535–556. 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Kopka M. L.; Fratini A. V.; Drew H. R.; Dickerson R. E. Ordered Water Structure around a B-DNA Dodecamer. A Quantitative Study. J. Mol. Biol. 1983, 163 (1), 129–146. 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Schneider B.; Cohen D.; Berman H. M. Hydration of DNA Bases: Analysis of Crystallographic Data. Biopolymers 1992, 32 (7), 725–750. 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- Egli M.; Tereshko V.; Teplova M.; Minasov G.; Joachimiak A.; Sanishvili R.; Weeks C. M.; Miller R.; Maier M. A.; An H.; Cook P. D.; Manoharan M. X-Ray Crystallographic Analysis of the Hydration of A- and B-Form DNA at Atomic Resolution. Biopolymers 1998, 48 (4), 234–252. . [DOI] [PubMed] [Google Scholar]
- Chen S. H.; Liu L.; Chu X.; Zhang Y.; Fratini E.; Baglioni P.; Faraone A.; Mamontov E. Experimental Evidence of Fragile-to-Strong Dynamic Crossover in DNA Hydration Water. J. Chem. Phys. 2006, 125, 171103. 10.1063/1.2372491. [DOI] [PubMed] [Google Scholar]
- Denisov V. P.; Carlström G.; Venu K.; Halle B. Kinetics of DNA Hydration. J. Mol. Biol. 1997, 268, 118–136. 10.1006/jmbi.1996.0862. [DOI] [PubMed] [Google Scholar]
- Liepinsh E.; Otting G.; Wüthrich K. NMR Observation of Individual Molecules of Hydration Water Bound to DNA Duplexes: Direct Evidence for a Spine of Hydration Water Present in Aqueous Solution. Nucleic Acids Res. 1992, 20 (24), 6549–6553. 10.1093/nar/20.24.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck J. M.; Ding Y.; Stone K.; Qin P. Z.; Han S. Anomalously Rapid Hydration Water Diffusion Dynamics Near DNA Surfaces. J. Am. Chem. Soc. 2015, 137 (37), 12013–12023. 10.1021/jacs.5b05813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K.; Zhao L.; Zewail A. H. Water at DNA Surfaces: Ultrafast Dynamics in Minor Groove Recognition. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (14), 8113–8118. 10.1073/pnas.1433066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyc Ł.; Yang M.; Nibbering E. T. J.; Elsaesser T. Ultrafast Vibrational Dynamics and Local Interactions of Hydrated DNA. Angew. Chem., Int. Ed. 2010, 49 (21), 3598–3610. 10.1002/anie.200905693. [DOI] [PubMed] [Google Scholar]
- Furse K. E.; Corcelli S. a. The Dynamics of Water at DNA Interfaces: Computational Studies of Hoechst 33258 Bound to DNA. J. Am. Chem. Soc. 2008, 130 (39), 13103–13109. 10.1021/ja803728g. [DOI] [PubMed] [Google Scholar]
- Duboué-Dijon E.; Fogarty A. C.; Hynes J. T.; Laage D. Dynamical Disorder in the DNA Hydration Shell. J. Am. Chem. Soc. 2016, 138 (24), 7610–7620. 10.1021/jacs.6b02715. [DOI] [PubMed] [Google Scholar]
- Guchhait B.; Liu Y.; Siebert T.; Elsaesser T. Ultrafast Vibrational Dynamics of the DNA Backbone at Different Hydration Levels Mapped by Two-Dimensional Infrared Spectroscopy. Struct. Dyn. 2016, 3, 43202 10.1063/1.4936567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. R.The Principles of Nonlinear Optics; Wiley-Interscience: New York, 1984. [Google Scholar]
- Eisenthal K. B. Liquid Interfaces Probed by Second-Harmonic and Sum-Frequency Spectroscopy. Chem. Rev. 1996, 96 (4), 1343–1360. 10.1021/cr9502211. [DOI] [PubMed] [Google Scholar]
- Yan E. C. Y.; Fu L.; Wang Z.; Liu W. Biological Macromolecules at Interfaces Probed by Chiral Vibrational Sum Frequency Generation Spectroscopy. Chem. Rev. 2014, 114 (17), 8471–8498. 10.1021/cr4006044. [DOI] [PubMed] [Google Scholar]
- Belkin M. a; Kulakov T. a; Ernst K. H.; Yan L.; Shen Y. R. Sum-Frequency Vibrational Spectroscopy on Chiral Liquids: A Novel Technique to Probe Molecular Chirality. Phys. Rev. Lett. 2000, 85 (21), 4474–4477. 10.1103/PhysRevLett.85.4474. [DOI] [PubMed] [Google Scholar]
- Belkin M. A.; Shen Y. R. Non-Linear Optical Spectroscopy as a Novel Probe for Molecular Chirality. Int. Rev. Phys. Chem. 2005, 24 (2), 257–299. 10.1080/01442350500270601. [DOI] [Google Scholar]
- Wang J.; Chen X.; Clarke M. L.; Chen Z. Detection of Chiral Sum Frequency Generation Vibrational Spectra of Proteins and Peptides at Interfaces in Situ. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (14), 4978–4983. 10.1073/pnas.0501206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes G. Y.; Gibbs-Davis J. M.; Boman F. C.; Stepp B. R.; Condie A. G.; Nguyen S. T.; Geiger F. M. Making “sense” of DNA. J. Am. Chem. Soc. 2007, 129 (24), 7492–7493. 10.1021/ja071848r. [DOI] [PubMed] [Google Scholar]
- Haupert L. M.; Simpson G. J. Chirality in Nonlinear Optics. Annu. Rev. Phys. Chem. 2009, 60, 345–365. 10.1146/annurev.physchem.59.032607.093712. [DOI] [PubMed] [Google Scholar]
- Fu Li; Zhang Y.; Wei; Zhe-Hao; Wang H.-F. Intrinsic Chirality and Prochirality at Air/R-(+)- and S-(−)-Limonene Interfaces: Spectral Signatures With Interference Chiral Sum-Frequency Generation Vibrational Spectroscopy. Chirality 2014, 26 (9), 509–520. 10.1002/chir.22337. [DOI] [PubMed] [Google Scholar]
- McDermott M. L.; Petersen P. B. Robust Self-Referencing Method for Chiral Sum Frequency Generation Spectroscopy. J. Phys. Chem. B 2015, 119 (38), 12417–12423. 10.1021/acs.jpcb.5b08176. [DOI] [PubMed] [Google Scholar]
- Li Z.; Weeraman C. N.; Azam M. S.; Osman E.; Gibbs-Davis J. M. The Thermal Reorganization of DNA Immobilized at the Silica/buffer Interface: A Vibrational Sum Frequency Generation Investigation. Phys. Chem. Chem. Phys. 2015, 17 (19), 12452–12457. 10.1039/C5CP00781J. [DOI] [PubMed] [Google Scholar]
- Auer B. M.; Skinner J. L. Water: Hydrogen Bonding and Vibrational Spectroscopy, in the Bulk Liquid and at the Liquid/vapor Interface. Chem. Phys. Lett. 2009, 470 (1–3), 13–20. 10.1016/j.cplett.2009.01.010. [DOI] [Google Scholar]
- Merten C.; Xu Y. Chirality Transfer in a Methyl Lactate-Ammonia Complex Observed by Matrix-Isolation Vibrational Circular Dichroism Spectroscopy. Angew. Chem., Int. Ed. 2013, 52 (7), 2073–2076. 10.1002/anie.201208685. [DOI] [PubMed] [Google Scholar]
- Yang G.; Xu Y. Probing Chiral Solute-Water Hydrogen Bonding Networks by Chirality Transfer Effects: A Vibrational Circular Dichroism Study of Glycidol in Water. J. Chem. Phys. 2009, 130, 164506. 10.1063/1.3116582. [DOI] [PubMed] [Google Scholar]
- Losada M.; Xu Y. Chirality Transfer through Hydrogen-Bonding: Experimental and Ab Initio Analyses of Vibrational Circular Dichroism Spectra of Methyl Lactate in Water. Phys. Chem. Chem. Phys. 2007, 9 (24), 3127. 10.1039/b703368k. [DOI] [PubMed] [Google Scholar]
- Raymond E. A.; Tarbuck T. L.; Brown M. G.; Richmond G. L. Hydrogen-Bonding Interactions at the Vapor/water Interface Investigated by Vibrational Sum-Frequency Spectroscopy of HOD/H2O/D2O Mixtures and Molecular Dynamics Simulations. J. Phys. Chem. B 2003, 107, 546–556. 10.1021/jp021366w. [DOI] [Google Scholar]
- Sovago M.; Campen R.; Wurpel G.; Müller M.; Bakker H. J.; Bonn M. Vibrational Response of Hydrogen-Bonded Interfacial Water Is Dominated by Intramolecular Coupling. Phys. Rev. Lett. 2008, 100, 173901. 10.1103/PhysRevLett.100.173901. [DOI] [PubMed] [Google Scholar]
- Myalitsin A.; Urashima S.-H.; Nihonyanagi S.; Yamaguchi S.; Tahara T. Water Structure at the Buried Silica/Aqueous Interface Studied by Heterodyne-Detected Vibrational Sum-Frequency Generation. J. Phys. Chem. C 2016, 120, 9357–9363. 10.1021/acs.jpcc.6b03275. [DOI] [Google Scholar]
- Schaefer J.; Backus E. H. G.; Nagata Y.; Bonn M. Both Inter- and Intramolecular Coupling of O–H Groups Determine the Vibrational Response of the Water/Air Interface. J. Phys. Chem. Lett. 2016, 7, 4591–4595. 10.1021/acs.jpclett.6b02513. [DOI] [PubMed] [Google Scholar]
- Du Q.; Freysz E.; Shen Y. R. Surface Vibrational Spectroscopic Studies of Hydrogen Bonding and Hydrophobicity. Science (Washington, DC, U. S.) 1994, 264, 826–828. 10.1126/science.264.5160.826. [DOI] [PubMed] [Google Scholar]
- Scatena L. F.; Brown M. G.; Richmond G. L. Water at Hydrophobic Surfaces: Weak Hydrogen Bonding and Strong Orientation Effects. Science (Washington, DC, U. S.) 2001, 292 (5518), 908–912. 10.1126/science.1059514. [DOI] [PubMed] [Google Scholar]
- Davis J. G.; Gierszal K. P.; Wang P.; Ben-Amotz D. Water Structural Transformation at Molecular Hydrophobic Interfaces. Nature 2012, 491 (7425), 582–585. 10.1038/nature11570. [DOI] [PubMed] [Google Scholar]
- Perera P. N.; Fega K. R.; Lawrence C.; Sundstrom E. J.; Tomlinson-Phillips J.; Ben-Amotz D. Observation of Water Dangling OH Bonds around Dissolved Nonpolar Groups. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (30), 12230–12234. 10.1073/pnas.0903675106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Petersen P. B. Solvation Shell Structure of Small Molecules and Proteins by IR-MCR Spectroscopy. J. Phys. Chem. Lett. 2017, 8 (3), 611–614. 10.1021/acs.jpclett.6b02925. [DOI] [PubMed] [Google Scholar]
- Yoon C.; Privé G. G.; Goodsell D. S.; Dickerson R. E. Structure of an Alternating B-DNA Helix and Its Relationship to A-Tract DNA. Proc. Natl. Acad. Sci. U. S. A. 1988, 85 (17), 6332–6336. 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; West S. M.; Sosinsky A.; Liu P.; Mann R. S.; Honig B. The Role of DNA Shape in Protein – DNA Recognition. Nature 2009, 461 (7268), 1248–1253. 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furse K. E.; Corcelli S. a. Effects of an Unnatural Base Pair Replacement on the Structure and Dynamics of DNA and Neighboring Water and Ions. J. Phys. Chem. B 2010, 114 (30), 9934–9945. 10.1021/jp105761b. [DOI] [PubMed] [Google Scholar]
- Furse K. E.; Corcelli S. A. Molecular Dynamics Simulations of DNA Solvation Dynamics. J. Phys. Chem. Lett. 2010, 1 (12), 1813–1820. 10.1021/jz100485e. [DOI] [Google Scholar]
- Lowe R. D.; Pellow M. A.; Stack T. D. P.; Chidsey C. E. D. Deposition of Dense Siloxane Monolayers from Water and Trimethoxyorganosilane Vapor. Langmuir 2011, 27 (16), 9928–9935. 10.1021/la201333y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.