Abstract
Cytoplasmic Hsp70s of SSA family, especially Ssa1p, are involved in the degradation of a variety of misfolded proteins in yeast. However the importance of other Ssa proteins in this process is unclear. To clarify the role(s) of individual Ssa proteins in proteolysis, we measured the breakdown of various cell proteins in mutants lacking different Ssa proteins. In mutants lacking Ssa1p and Ssa2p, the proteasomal degradation of short-lived proteins was reduced, which was not restored fully by the over-expression of Ssa1p. By contrast, the degradation of stable cellular proteins did not require Ssa proteins. The degradation of the cytosolic model substrates (Ub-P-β-gal and R-β-gal) and their ubiquitylation were inhibited by the inactivation of Ssa proteins. In addition, Ssa1p and the co-chaperone Ydj1p are indispensable for the intracellular degradation of a mutant secretory protein, Siiyama variant of human antitrypsin. Our findings indicate that both Ssa1p and Ssa2p are essential for the ubiquitin-dependent degradation of short-lived proteins and the requirements of Ssa proteins and the co-chaperones widely vary depending on the conformations and folding status of the substrates.
Keywords: ubiquitin, degradation, chaperone, hsp70, co-chaperone
INTRODUCTION
Eukaryotic cells contain multiple species of Hsp70s; for example, human cells contain six cytosolic Hsp70s including the constitutively expressed Hsc70 and multiple Hsp70 orthologs exist in the yeast [1, 2]. In yeast cytosol, closely related but differentially expressed Ssa proteins (Ssa1-4p) function in the protein folding and the protein trafficking into the ER while the ribosome-associated Ssb proteins (Ssb1-2p) are involved in the stabilization of nascent polypeptides [3, 4]. Functions of Hsp70s are usually regulated by their co-chaperones of DnaJ homologs and nucleotide-exchange factors, which are often regarded as the mediators for functional diversity of Hsp70s [5].
With their ability to bind selectively to unfolded proteins, Hsp70s also play important roles in the selective degradation of proteins by the ubiquitin-proteasome system and the lysosomal pathways through chaperone-mediated autophagy [6, 7]. In these processes, Hsp70s aid in the recognition of substrates by the proteolytic machinery. Alternatively, Hsp70s function as a cofactor facilitating the unfolding of proteins prior to the ubiquitylation or simply maintain them in soluble conformations that are more susceptible to proteolytic enzymes [6]. Ssa proteins (mostly Ssa1p) and the co-chaperone Ydj1p are required for the ER-associated degradation (ERAD) of several misfolded proteins [8–10]. We also reported that Ydj1p is involved in the degradation of cytosolic short-lived and abnormal proteins in general [11].
Functional studies of individual Ssa proteins in protein breakdown are difficult due to the presence of multiple orthologs. Previous reports mostly focused on the involvement of Ssa1p for the degradation of ERAD substrates [8–10]. However, it is not yet clear whether other Ssa proteins are dispensable for protein degradation. Although constitutively expressed Ssa1p and Ssa2p are 97% identical and compensate each other, they possibly have distinct functions, e.g., in their ability to propagate yeast prions and in resistance to heat shock. Moreover, yeast strains expressing individual Ssa proteins exhibit distinctive transcriptional profiles [12, 13].
The present studies were undertaken to study the roles of individual Ssa proteins in the breakdown of different classes of proteins. In addition, we compared the effects of Ssa proteins and co-chaperones on the proteasomal degradation of cytosolic and ERAD substrates. Here we present evidence that both Ssa1p and Ssa2p are important for the degradation of short-lived proteins generally and demonstrated that the requirements of Ssa proteins and Ydj1p for the protein degradation vary widely depending on the conformations and folding status of the client proteins.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Yeast strains used in this study are as follows; wild-type (MATa his3-11,15, leu2-3,112, ura3-52, trp1-1, lys2), ssa1ssa2 (WT, ssa1::HIS3 ssa2::LEU2), ssa1ssa3 (WT, ssa1::HIS3 ssa3::TRP1), ssa1ssa4 (WT, ssa1::HIS3 ssa4::LYS2), ssa2ssa3ssa4 (MATα his3-11,15, leu2-3,112, ura3-52, trp1-Δ1, lys2 SSA1 ssa2-1 ssa3-1 ssa4-2), ssa1-45 ts (MATα his3-11,15, leu2-3,112, ura3-52, trp1-Δ1, lys2 ssa1-45 ssa2-1 ssa3-1 ssa4-2) (provided by Dr. E.A. Craig, Univ. of Wisconsin); ydj1-151 (MATa ade2-1, his3-11,15, leu2-3,112, ura3-1, trp1-1, can1-100 Δydj1-2::HIS3, LEU2::ydj1-151), sis1-85 (MATa ade2-1, his3-11,15, leu2-3,112, ura3-1, trp1-1, can1-100 Δsis1::HIS3, sis1-85 on LEU2/CEN plasmid) [11]. The expression plasmids for Ssa1p (pGAL-SSA1), β-galactosidase fusion constructs (Ub-P-gal and R-gal), and the Siiyama variant of human antitrypsin were provided by Dr. S. Lindquist (MIT), Dr. S. Jentsch (Max-Plank Institute) and Dr. M.-H. Yu (KIST), respectively.
Measurement of Protein Breakdown
Yeast cells exponentially growing in SD medium supplemented with amino acids and nucleotides at 28°C were collected by centrifugation and re-suspended in the same medium without methionine. For temperature-sensitive mutants, cells were pre-incubated at 38°C for 30 min prior to labeling. To measure the breakdown of short-lived proteins, cells were labeled for 10 min with 100 μCi of 35S-methionine (ICN). After labeling, the cells were washed and re-suspended in fresh SD medium containing methionine (0.5 mg/ml) and cycloheximide (0.5 mg/ml) (chase period). At given intervals, aliquots of cells were taken and mixed with 100% TCA to give a final concentration of 10%. After incubation at 4°C for 1 h, the samples were centrifuged and the radioactivity in the TCA-soluble material was measured. The rate of protein degradation is expressed as the percentage of incorporated radioactivity converted into acid-soluble material during chase period. To measure the breakdown of long-lived proteins, the cells were labeled with 20 μCi of [14C]-leucine (ICN) for 2 h and incubated in SD medium containing excess leucine (and no cycloheximide) for 12 h to allow the degradation of the short-lived proteins. The release of radioactivity from remaining cell proteins was then measured in the presence of excess leucine and cycloheximde.
Pulse-Chase Experiments and Immunoprecipitation
Yeast cells carrying a β-gal or Siiyama variant of α1-antitrypsin were grown overnight at 28°C in raffinose-containing minimal medium. Cells were then incubated in galactose-containing, methionine-free medium for protein induction for 6–8 h. After the induction, cells were pre-incubated at 38°C for 30 min and pulse-labeled with 100 μCi of 35S-methionine for 10 min. For the chase, cells were transferred to the medium containing methionine (0.5 mg/ml) and cycloheximide (0.5 mg/ml) and then incubated at 38°C. At given intervals, aliquots of cells were collected and re-suspended in IP buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100) containing protease inhibitors. Cells were lysed by glass beads and the radioactivity incorporated into proteins (supernatant) was measured by TCA precipitation. For immunoprecipitation, cell lysate was incubated with 1 μl of anti-β-gal antibody (Promega) or anti-human AT antibody (Sigma) for 2–3 h at 4°C. The supernatants were then mixed with 20 μl protein A-Trisacryl beads (Pierce) for 2 h at 4°C. The beads were washed with IP buffer containing 0.1 % SDS and boiled in 1× SDS sampling buffer. The proteins were subjected to 10% SDS-PAGE and subsequent autoradiography. The relative amounts of β-gal and antitrypsin were determined with a PhosphorImager (Molecular Dynamics).
Affinity Chromatography of β-galactosidase
To isolate β-gal and the associated proteins, yeast cells expressing Ub-P-gal or M-gal were grown in YP-galactose medium for 12 h for protein induction. Cells were collected and re-suspended in buffer (50 mM Tris-HCl, pH 7.3; 200 mM NaCl; 5 mM MgCl2; 1 mM EDTA; 1 mM PMSF; protease inhibitors) and then lysed by glass beads. Equal amounts of proteins (3–4 mg) were applied to a 1 ml column of p-aminobenzyl-1-thio-β-galactoside crosslinked to agarose beads at 4°C and the bound β-gal was eluted with 8% lactose. The presence of Ssa1p or β-gal in the eluent was determined by western-blot with antibodies against human Hsp70 (StressGen) or β-gal (Promega). Enzymatic activity of β-gal was assayed as described in ‘Yeast Protocol Handbook’ (Clonetech).
RESULTS
Effects of individual Ssa proteins on the breakdown of different types of cell proteins
Yeast SSA proteins are functionally redundant and can compensate for each other [13, 14]. To examine the role(s) of individual Ssa proteins in the breakdown of different types of proteins, we utilized double mutant strains lacking different Ssa proteins. Among these strains, ssa1,2 mutant was of particular interest because Ssa1p and Ssa2p are constitutively expressed and more closely related to mammalian Hsp70s. At 28°C, 10-min pulse-labeled proteins were degraded at an initial rate of 8%/h in the wild-type whereas the degradation of such proteins was reduced to 5%/h in ssa1,2 mutant (Fig. 1A). Exposure of cells to increased temperatures causes structural alterations of cellular proteins and increases the rate of proteolysis. Upon switch to 38°C, the rate of protein breakdown increased about 2-fold (8%/h to 15%/h) in the wild-type, but failed to increase significantly (5%/h to 6%/h) in ssa1,2 mutant (Fig. 1B). By contrast, ssa1,3 and ssa1,4 mutants did not show any clear defect in the degradation of short-lived normal proteins (Fig. 1A and B). A possible explanation for these results is that Ssa2p is more important for degradation of short-lived proteins than Ssa1p. To test whether the reduced proteolysis is due to the lack of Ssa2p or both Ssa proteins are necessary for protein breakdown, we over-expressed Ssa1p in ssa1,2 mutant and then measured the degradation of pulse-labeled proteins. Although over-expressed Ssa1p increased the rate of proteolysis to some extent (10%/h vs. 6%/h), it was still insufficient to fully restore to that of wild-type (18%/h) (Fig. 1C). These findings argue that both Ssa1p and Ssa2p are involved in the breakdown of short-lived proteins and support that Ssa2p is indispensable for the degradation of certain proteins. Interestingly Ssa2p, but not Ssa1p, is required for vacuolar import and degradation of fructose-1,6-bisphosphatase [15]. For further experiments, we then employed ssa2,3,4 triple mutant and ssa1-45 ts mutant. The ssa1-45 ts mutant has been widely used because it allows rapid inactivation of all Ssa proteins upon shift to the non-permissive temperature. In these mutants, the breakdown of short-lived proteins at 38°C was reduced to a similar extent as seen in ssa1,2 strain (Fig. 1D). Notably the defect in proteolysis in ssa2,3,4 strain was similar to that in ssa1-45 ts strain in which all Ssa proteins are inactivated, which again suggests that Ssa2p is necessary for the degradation of certain short-lived proteins and it cannot be fully complemented by other Ssa proteins.
Fig. 1. The degradation of short-lived and long-lived proteins in mutants lacking different Ssa proteins.
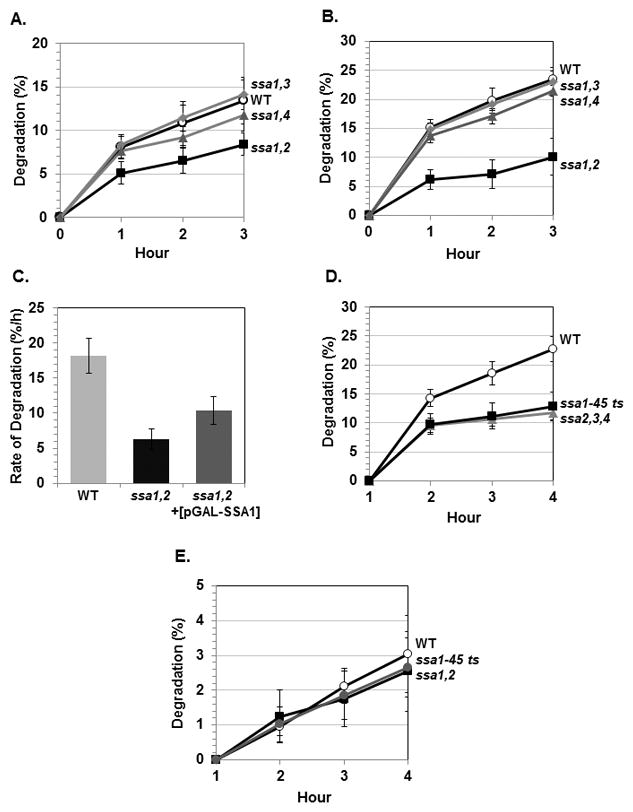
(A–B) Wild-type (WT) and the ssa1,2, ssa1,3, ssa1,4 mutants were labeled with 35S-methionine for 10 min and the breakdown of pulse-labeled proteins were measured at 28°C (A) or 38°C (B). (C) The plasmid carrying SSA1 gene (pGAL-SSA1) was introduced into ssa1,2 mutant and the rate of degradation of short-lived proteins was compared with those of WT and ssa1,2 mutant. (D) Degradation of short-lived proteins in WT, ssa2,3,4 triple mutant and ssa1-45 ts mutant at 38°C. (E) Degradation of long-lived proteins in WT, ssa1,2 and ssa1-45 ts mutants at 28°C. Data represent means ± S. D. from three independent experiments.
A 10-min pulse labels not only short-lived proteins but also the stable proteins, whose degradation occurs mainly in yeast vacuoles [16]. In mammals, Hsp70 is involved in lysosomal protein degradation through chaperone-mediated autophagy [7]. Unlike the short-lived proteins, breakdown of the stable components was not reduced in any of the ssa mutants. The degradation rate of long-lived proteins at 38°C was similar in the wild-type, ssa1,2 mutant and ssa1-45 ts mutant (about 2%/h) (Fig. 1E). Ydj1p, the co-chaperone of Ssa proteins, is not required for the degradation of long-lived proteins either [11]. Thus it seems that Ssa proteins and Ydj1p are required specifically for the proteasomal degradation of short-lived and abnormal proteins.
Effects of Ssa proteins on the degradation of β-gal fusion polypeptides
To further assess the requirement of Ssa proteins in the ubiquitin-dependent degradation of cytosolic proteins, we chose the UFD substrate ubiquitin-proline-β-galactosidase (Ub-P-gal) whose degradation requires Ubc4/5 and Ydj1p [11]. Although the degradation of short-lived and abnormal proteins was defective, the breakdown of Ub-P-gal was not reduced in ssa1,2 mutant or ssa2,3,4 mutant (data not shown). However, this polypeptide became stable (t1/2 = 25–30 min) in ssa1-45 ts mutant while it was rapidly degraded in the wild-type (t1/2 = 25–30 min) (Fig. 2A). Presumably, the residual Ssa proteins in ssa1,2 and ssa2,3,4 mutants support the degradation of particular substrates although they are not sufficient to fully restore the rate of overall proteolysis. Interestingly, a mutant form of yeast membrane ATPase also showed a similar pattern of dependence on different Ssa proteins for its degradation [17]. The degradation of the “N-end rule” substrate arginine-β-galactosidase (R-gal) was also reduced in ssa1-45 ts mutant (Fig. 2A). Interestingly, co-chaperone Ydj1p is required for the degradation of Ub-P-gal but not for that of R-gal, suggesting that Ssa1p and Ydj1p do not always cooperate in the protein degradation [11].
Fig. 2. The degradation of Ub-P-gal and R-gal was reduced in ssa1-45 ts mutant.
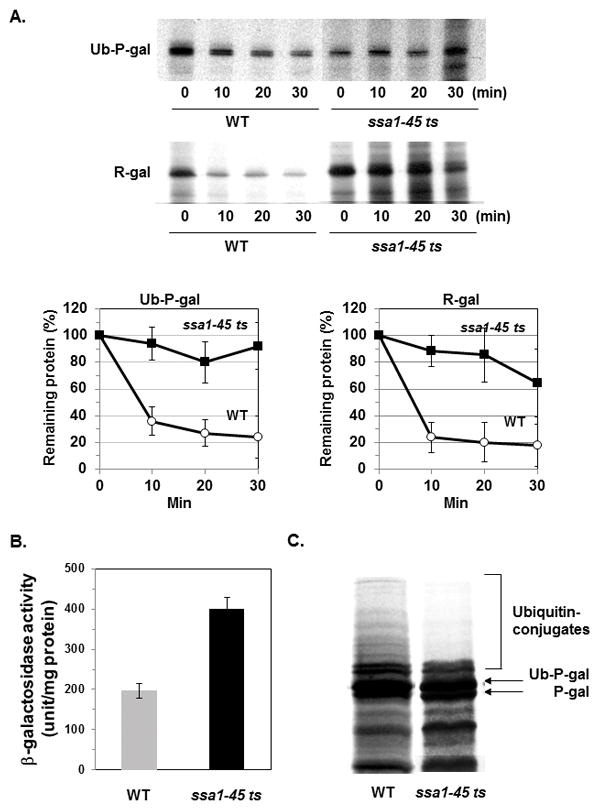
(A) At 38°C, wild-type and ssa1-45 ts mutant cells carrying the β-gal constructs were labeled with 35S-methionine for 10 min and their degradation was measured by the immunoprecipitation with an anti-β-gal antibody. Data represent means ± S. D. from three independent experiments. (B) To measure the β-gal activity in WT and ssa1-45 ts mutant at 38°C, yeast cells expressing Ub-P-gal were incubated at 38°C for 1 h, lysed and the β-gal activity was then assayed. (C) To compare the extent of the ubiquitylation of Ub-P-gal in WT and ssa1-45 ts mutant, cells were incubated and then labeled with 35S-methionine for 30 min at 38°C. The cells were collected and the β-gal fusion proteins were isolated by immunoprecipitation. The proteins were resolved in 8% SDS-PAGE gels and then subjected to autoradiography.
To determine if the reduced proteolysis by the inactivation of Ssa1p is due to a defect in protein folding or solubility, we measured the enzymatic activity of the β-gal in the soluble fraction from ssa1-45 ts mutant and wild-type. At 38°C, ssa1-45 ts mutant carrying Ub-P-gal showed much higher β-gal activity than the wild-type (Fig. 2B), confirming that non-degraded β-gal are enzymatically active and that the reduced degradation of the cytosolic substrates is caused by a specific defect in proteolysis.
Previously it was shown that Ssa1p is involved in the ubiquitylation of several ERAD substrates [9, 10]. To determine if Ssa proteins are also involved in the ubiquitylation of β-gal, we pulse-labeled cells with 35S-met for 30 min and examined the ubiquitylation of Ub-P-gal. In ssa1-45 ts mutant, poly-ubiquitylated forms of Ub-P-gal were not readily detected (Fig. 2C) confirming that Ssa proteins promote the ubiquitylation of substrates.
Association of Ssa1p with the substrate
Ssa proteins could promote proteolysis either by affecting the folding of ubiquitylation machinery or by facilitating its association of substrates. To determine if Ssa proteins associate with Ub-P-gal, we isolated Ub-P-gal or wild-type M-gal together with their associated proteins using affinity chromatography. Ssa1p was found in appreciable amounts in the fraction containing Ub-P-gal whereas much smaller amount of Ssa1p was detected in the fraction containing M-gal (Fig 3). Although much less of Ssa1p was isolated with the stable M-gal, these fractions contained at least 3 times more β-gal than the short-lived Ub-P-gal. Such preferential binding of Ssa1p to short-lived Ub-P-gal suggests that the direct association with chaperones facilitates the ubiquitylation and subsequent degradation of client proteins.
Fig. 3. Ssa1p was preferentially associated with the rapidly degraded Ub-P-gal.
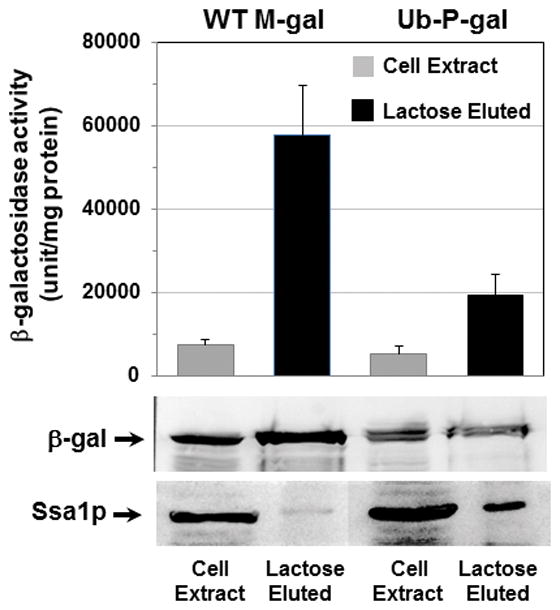
Wild-type cells expressing M-gal or Ub-P-gal were grown at 28°C in the presence of 2% galactose for 12–16 h. 3–4 mg of cell proteins were prepared and applied to a 1-ml column of p-aminobenzyl-1-thio-β-galactoside crosslinked to agarose beads at 4°C. The bound β-gal were eluted with lactose and the presence of Ssa1p or β-gal fusion proteins was determined by Western-blot with anti-human Hsp70 or anti-β-gal antibodies and also the β-gal activity was also measured.
Requirement of Ssa1p and Ydj1p for the degradation of Siiyama variant of antitrypsin
To compare the chaperone requirements for the degradation of cytosolic vs. ERAD substrates, we tested if a mutant of human α1-antirypsin (α1-AT), Siiyama variant, also requires Ssa-Ydj1p chaperones for its degradation. The Z variant of α1-AT causes AT deficiency by impeding the secretion and α1-AT proteins accumulated in the ER are subsequently degraded by ERAD pathway [18]. In yeast, the proteasomal degradation of Z variant requires ER Hsp70, BiP, but not Ssa1p [19]. By contrast, Siiyama variant was stabilized in ssa1-45 ts mutant (Fig 4A). We also examined the requirement of Ydj1p and Sis1p for its degradation. Inactivation of Ydj1p but not that of Sis1p significantly reduced the degradation of Siiyama variant (Fig 4B), indicating that Ssa1p and Ydj1p are both required for the degradation of this variant. Interestingly, in ydj1-151 mutant, the core-glycosylated forms of Siiyama variant (indicated by an asterisk) were barely detected while the inactivation of Ssa1p did not affect the core-glycosylation (Fig 4A) suggesting that only Ydj1p is involved in both the proteasomal degradation and the translocation of Siiyama variant into the ER.
Fig. 4. Degradation of Siiyama variant of human α1-AT requires both Ssa1p and Ydj1p.
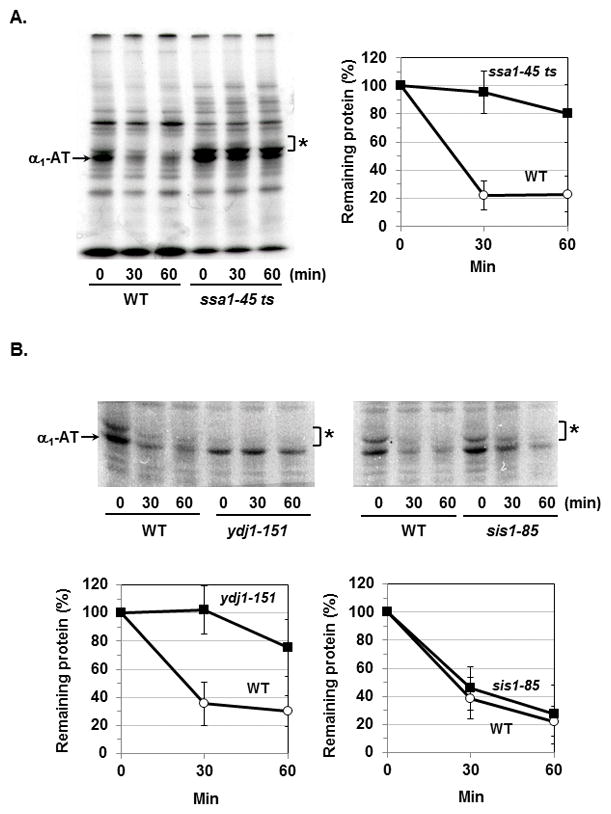
Wild-type and ssa1-45 ts mutant (A) or ydj1-151 and sis1-85 mutants (B) expressing Siiyama variant were labeled with 35S-methionine for 20 min at 38°C. The degradation of Siiyama variant during chase period was measured by the immunoprecipitation. Asterisk (*) denotes the core-glycosylated forms of α1-AT. Data represent means ± S. D. from three independent experiments.
DISCUSSION
Ssa proteins have overlapping functions and the lack of a certain member can be functionally replaced by another Ssa protein. However, recent findings suggested functional specificity among Ssa proteins [13]. Interestingly we found that all the mutants defective in proteolysis lack Ssa2p. In ssa1,2 mutant, heat-damaged proteins are degraded at less than a half of the rate seen in the wild-type. Moreover, ssa2,3,4 and ssa1-45 ts strains exhibited similar defects in the degradation of heat-damaged proteins. Despite these observations, the requirements of Ssa1p and Ssa2p for the degradation of individual substrates seem to be complex. Although the breakdown of the cytosolic substrate Ub-P-gal and several ERAD substrates requires Ssa1p [8–10], the failure of over-expressed Ssa1p in the ssa1,2 mutant to restore fully the ability to degrade short-lived proteins and the involvement of Ssa2p in the degradation of fructose-1,6-bisphosphatase [15] suggest essential roles of Ssa2p in the degradation of certain proteins. Presumably some client proteins require Ssa1p while others need Ssa2p for their degradation.
Moreover, unlike cytosolic substrates, the involvement of Ssa1p in ERAD pathway may vary depending on the conformations and the degree of misfolding of substrates. For example, Ssa1p and Ydj1p are both required for the degradation of the Siiyama variant of α1-AT while Ssa1p is not involved in the degradation of the Z variant [18]. Although both variants cause a secretion blockage and antitrypsin deficiency, the underlying mechanisms are different. The aggregation-prone Z variant (Glu342Lys) is defective in folding whereas Siiyama variant (Ser53Phe) causes the loss of stability [20]. On the other hand, Ssa1p is necessary for the ERAD of several transmembrane proteins [9, 21]. Interestingly Ssa1p facilitates the binding of misfolded proteins to the ubiquitin ligases San1p and Ubr1p [19]. Perhaps the requirements of Ssa proteins for ERAD substrates are determined by the specificity of the E3 ligase(s).
It has been believed that Ssa1p invariably functions with Ydj1p in protein degradation and the folding process. However, both chaperones can bind to substrates independently and they exhibit distinct substrate specificities. For example, DnaJ homologs can bind to certain proteins at different sites than Hsp70 [22]. Moreover, the degradation of CFTR, which requires Ssa1p, was not significantly reduced by inactivation of Ydj1p [23]. Degradation of the tumor suppressor VHL was affected by neither Ydj1p nor Sis1p; instead the inactivation of the Hsp70 cofactor Sti1 stabilized VHL [24]. We also observed that the degradation of Ub-P-gal requires both Ssa1p and Ydj1p while that of R-gal only needs Ssa1p [11]. Ub-P-gal is targeted for ubiquitylation by UFD pathway and R-gal is ubiquitylated by the N-end rule pathway. Ufd4p, the Ub ligase for UFD pathway, recognizes an N-terminal Ub moiety on substrates whereas the Ub ligase Ubr1p binds to destabilizing N-terminal residues of the N-end rule substrates [25]. Depending on the nature of client proteins and the associated Ub ligases, Ssa proteins may cooperate with distinct DnaJ homologs. Findings that the core-glycosylated forms of Siiyama variant were absent in ydj1-151 mutant while no such a defect was observed in ssa1-45 ts mutant suggest that Ydj1p is required for the translocation of Siiyama variant into the ER. Previous reports also demonstrated that inactivation of Ssa1p caused the accumulation of the non-glycosylated form of prepro-α-factor, while Ydj1p was not necessary for this process [26, 27]. Probably the requirements of Hsp70-Hsp40 for the degradation and the translocation of the individual substrate are much more complex than we initially believed.
The finding that Ssa1p and Ydj1p stably associate with Ub-P-gal implicates that this binding is critical for ubiquitylation. Chaperones may enhance the Ub-dependent degradation of proteins by preventing the aggregation of substrates or maintaining them in an unfolded conformation, which is essential for the recognition by ubiquitination machinery. Although the major function of chaperones in proteolysis seems to maintain substrates in a soluble form, such an explanation cannot fully account for findings that the non-degraded β-gal purified from ssa1-45 ts mutant was not only soluble but also enzymatically active. Presumably the association of Ssa proteins-Ydj1p with substrates helps maintain certain domains in an extended conformation that promotes modification by the ubiquitylation machinery. The chaperones may function either as factors that promote substrate recognition by the E3s or as cofactors that facilitate their catalytic action.
In this report, we demonstrate that both Ssa1p and Ssa2p are essential for the degradation of short-lived and abnormal proteins. Whether a certain substrate requires Ssa1p or Ssa2p for its degradation could be determined by its conformation and folding status. Moreover, DnaJ homologs other than Ydj1p may be involved in the breakdown of certain ERAD substrates. Since the E3s are presumed to physically associate with Hsp70-Hsp40, certain E3 ligases may bind to their substrates with the aid of substrate-specific chaperone complex. Future studies, e.g., degradome analysis for the requirement of Ssa proteins and DnaJ homologs, will allow us to gain insights into the mechanism by which cytosolic chaperones participate in the ubiquitin-proteasome system.
Acknowledgments
We are very grateful to Dr. Elizabeth Craig, Dr. Stefan Jentsch and Dr. Myeong-Hee Yu for the yeast strains and plasmids. We thank the members of Goldberg lab for their assistance in experiments. This study was supported by the grant from Korea Research Foundation (grant number 2013R1A1A2011628) to Do Hee Lee and the grant from NIGMS to Alfred L. Goldberg.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Kabani M, Martineau CN. Multiple Hsp70 isoforms in the eukaryotic cytosol: mere redundancy or functional specificity? Curr Genomics. 2008;9:338–348. doi: 10.2174/138920208785133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Schilke B, Craig EA, Horwich AL. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc Natl Acad Sci USA. 1998;95:12860–12865. doi: 10.1073/pnas.95.22.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peisker K, Chiabudini M, Rospert S. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1803:662–672. doi: 10.1016/j.bbamcr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Kampinga HH, Craig EA. The Hsp70 chaperone machinery: J-proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiber A, Ravid T. Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co-chaperones in targeting misfolded proteins to the proteasome. Biomolecules. 2014;4:704–724. doi: 10.3390/biom4030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S-H, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin–proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- 10.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal protein in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma D, Martineau CN, Le Dall MT, Reidy M, Masison DC, Kabani M. Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS ONE. 2009;4(8):e6644. doi: 10.1371/journal.pone.0006644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasin N, Cusack SA, Ali SS, Fitzpatrick DA, Jones GW. Global transcript and phenotypic analysis of yeast cells expressing Ssa1, Ssa2. Ssa3 or Ssa4 as sole source of cytosolic Hsp70-Ssa chaperone activity. BMC Genomics. 2014;15:194. doi: 10.1186/1471-2164-15-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of Hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1,6-bisphosphatase into vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teter SA, Klionsky DJ. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Sem Cell Develop Biol. 2000;11:173–179. doi: 10.1006/scdb.2000.0163. [DOI] [PubMed] [Google Scholar]
- 17.Gelling CL, Brodsky JL. Mechanisms underlying the cellular clearance of antitrypsin Z: Lessons from yeast expression systems. Proc Am Thorac Soc. 2010;7:363–367. doi: 10.1513/pats.201001-007AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer EA, Kruse KB, Fewell SW, Buchanan SM, Brodsky JL, McCracken AA. Differential requirements of novel A1PiZ degradation deficient (ADD) genes in ER-associated protein degradation. J Cell Sci. 2003;116:2361–2373. doi: 10.1242/jcs.00439. [DOI] [PubMed] [Google Scholar]
- 19.Guerriero CJ, Weiberth KF, Brodsky JL. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem. 2013;288:18506–18520. doi: 10.1074/jbc.M113.475905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang HA, Lee KN, Yu MH. Folding and stability of the Z and Siiyama genetic variants of human α1-antitrypsin. J Biol Chem. 1997;272:510–516. [PubMed] [Google Scholar]
- 21.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 23.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Hwang C-S, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic Hsp70 and a Dnaj-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:43784386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngosuwan J, Wang NM, Fung KL, Chirico WJ. Roles of cytosolic Hsp70 and Hsp40 molecular chaperones in post-translational translocation of presecretory proteins into the endoplasmic reticulum. J Biol Chem. 2003;278:7034–7042. doi: 10.1074/jbc.M210544200. [DOI] [PubMed] [Google Scholar]


