Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Droplet digital PCR is an affordable and user-friendly method for the detection of F8 and F9 sequence variants in maternal plasma.
Targeted MPS accurately determines fetal inheritance of F8 int22h-related inversions, using maternal plasma of pregnant hemophilia carriers.
Abstract
Direct detection of F8 and F9 sequence variants in maternal plasma of hemophilia carriers has been demonstrated by microfluidics digital PCR. Noninvasive prenatal assessment of the most clinically relevant group of sequence variants among patients with hemophilia, namely, those involving int22h-related inversions disrupting the F8 gene, poses additional challenges because of its molecular complexity. We investigated the use of droplet digital PCR (ddPCR) and targeted massively parallel sequencing (MPS) for maternal plasma DNA analysis to noninvasively determine fetal mutational status in pregnancies at risk for hemophilia. We designed family-specific ddPCR assays to detect causative sequence variants scattered across the F8 and F9 genes. A haplotype-based approach coupled with targeted MPS was applied to deduce fetal genotype by capturing a 7.6-Mb region spanning the F8 gene in carriers with int22h-related inversions. The ddPCR analysis correctly determined fetal hemophilia status in 15 at-risk pregnancies in samples obtained from 8 to 42 weeks of gestation. There were 3 unclassified samples, but no misclassification. Detailed fetal haplotype maps of the F8 gene region involving int22h-related inversions obtained through targeted MPS enabled correct diagnoses of fetal mutational status in 3 hemophilia families. Our data suggest it is feasible to apply targeted MPS to interrogate maternally inherited F8 int22h-related inversions, whereas ddPCR represents an affordable approach for the identification of F8 and F9 sequence variants in maternal plasma. These advancements may bring benefits for the pregnancy management for carriers of hemophilia sequence variants; in particular, the common F8 int22h-related inversions, associated with the most severe clinical phenotype.
Introduction
Hemophilia is one of the most common inherited bleeding disorders and is caused by a reduction in plasma levels of the coagulation factor VIII (coded by the F8 gene) or coagulation factor IX (coded by the F9 gene), clinically manifesting as spontaneous or injury-associated joint or deep-muscle bleeding episodes.1 Prenatal diagnosis (PND) is an integral part of antenatal care for pregnant carriers of hemophilia. The aim is to diagnose fetal sex and, in the case of a male fetus, to assess its hemophilia status. So far, the available options for prenatal diagnosis are through invasive testing, with an uptake of 25% being reported in the United Kingdom.2 The purposes of PND in hemophilia are twofold. First, it provides the option of termination of pregnancy. The attitudes toward termination of affected pregnancies among carriers of hemophilia vary widely between countries, depending on the availability and quality of medical care for hemophilia.3,4 Second, PND can optimize obstetric care during labor and delivery to ensure newborn safety.5 Noninvasive prenatal testing (NIPT) for fetal sex determination is now considered a “pretest” to invasive PND, avoiding the risk of invasive procedures in female pregnancies. However, invasive testing is still required for half of pregnant carriers to establish the hemophilia status of a male fetus.6 We have previously reported that it is feasible to perform NIPT for fetal genotype assessment by maternal plasma DNA analysis of F8 and F9 sequence variants by microfluidics digital PCR (dPCR).7 The European Hematology Association Roadmap for European Hematology Research has identified development in NIPT for hemophilia as a research priority, in particular for severe and common sequence variants such as F8 int22h-related inversions.8
The well-established hemophilia A and hemophilia B databases gather detailed descriptions of more than 2000 sequence variants occurring in the F8 gene and approximately 1000 sequence variants in the F9 gene, respectively.9-11 Notably, approximately half of the patients with severe hemophilia A harbor the inversion mutations associated with intron 22 in the F8 gene, the int22h-related inversions, located on chromosome X.12 The mutations stem from an intrachromosomal homologous recombination between the intragenic int22h-1 region and 1 of its 2 extragenic copies (int22h-2 and int22h-3), positioned more telomerically.13-15 However, because of technical obstacles in characterization of the inversion breakpoints, its direct detection is arduous, and so far has not been shown for NIPT of hemophilia carriers. This can potentially be resolved through a haplotype-based analysis examining mutation-linked polymorphic sites.16-18
The protocol described by Tsui et al7 for NIPT of hemophilia assesses the allelic imbalance from the proportion of the mutant (M) and normal/wild-type (N) allele in maternal plasma of pregnant hemophilia carriers. As for any NIPT-based protocol using a molecule counting strategy, the fetal DNA fraction governs the degree of such allelic imbalance, and thus requires precise molecular tools for its measurement.19 The lower is the fetal DNA fraction in a particular sample, the higher is the number of plasma DNA molecules that would need to be counted. The chip-based microfluidics platform adopted by Tsui et al7 has less flexibility in terms of scaling the number of digital reactions achieved. At this time, droplet dPCR (ddPCR) and massively parallel sequencing (MPS) technologies offer the highest sensitivity and precision for quantification of maternal plasma DNA.20 Such advantageous features are attained by a simultaneous analysis of either plasma DNA molecules distributed into more than 104 reaction partitions (droplets) in ddPCR or millions or more sequence reads of plasma DNA libraries in a sequencing flow cell for MPS.
Given that more affordable and user-friendly alternatives to the costly MPS technologies are currently available, we first investigated the use of ddPCR to establish fetal inheritance of maternal F8 and F9 sequence variants for NIPT of hemophilia as an extension of the previous study.7 We further focused on the detection of more complex sequence variants, namely, int22h-related inversions associated with the F8 gene, through the targeted MPS analysis of maternal plasma in pregnant hemophilia carriers.
Methods
Patient recruitment
We recruited 18 pregnant women with singleton male fetuses who were either hemophilia A carriers (7 carriers of F8 sequence variants and 3 carriers of F8 int22h-related inversions) or hemophilia B carriers (8 carriers of F9 sequence variants) from the Royal Free Hospital, London, United Kingdom (Table 1). Peripheral blood was collected on several occasions during the pregnancy. Samples collected at different points during the same pregnancy were labeled using the same sample number followed by a subscript A, B, or C to indicate the chronological order of the samples for that pregnancy. We further obtained proband’s peripheral blood in 2 families with F8 int22h-related inversions (family 16 and family 17) and the placental tissue (family 16 and family 18) for the haplotype-based analysis. The hemophilia status of the newborn was determined from the cord blood sample based on the factor VIII and factor IX assays, as appropriate. The study was approved by the institutional ethical board at the Royal Free Hospital, London, United Kingdom, and conducted in accordance with the Helsinki Declaration. All participants agreed to participate in the study by providing an informed written consent.
Table 1.
Clinical information and noninvasive detection of fetal F8 and F9 sequence variants by ddPCR
| Plasma sample* | Affected gene | Sequence variant† | Gestation, wk | Droplet digital PCR results | SPRT classification | Fetal status§ | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of reaction droplets | Total M allele count‡ | Total N allele count‡ | Fetal DNA fraction, %¶ | No. of reaction wells to reach SPRT classification | No. of positive droplets to reach SPRT classification | ||||||
| 1A | F8 | c.6278A>G (p.Asp2093Gly) | 18 | 35 941 | 192 | 130 | 10.9 | 2 | 157 | Affected | Affected |
| 1B | 34 | 49 337 | 259 | 188 | 21.2 | 1 | 77 | Affected | |||
| 2A | F8 | c.6278A>G (p.Asp2093Gly) | 18 | 365 097 | 734 | 817 | 3.7 | 16 | 1144 | Unaffected | Unaffected |
| 2B | 36 | 74 389 | 221 | 297 | 23.5 | 3 | 325 | Unaffected | |||
| 3A | F8 | c.6532C>T (p.Arg2178Cys) | 40 | 54 396 | 348 | 437 | 24.3 | 1 | 127 | Unaffected | Unaffected |
| 4A | F8 | c.6532C>T (p.Arg2178Cys) | 24 | 83 891 | 252 | 200 | 9.8 | 1 | 93 | Affected | Affected |
| 4B | 34 | 83 535 | 271 | 185 | 15.2 | 1 | 92 | Affected | |||
| 5A | F8 | c.2417delC (p.Ser806fs) | 8 | 138 458 | 233 | 309 | 4.8 | 7 | 243 | Unaffected | Unaffected |
| 6A | F8 | c.1171C>T (p.Arg391Cys) | 11 | 43 266 | 463 | 604 | 4.2 | 2 | 502 | Unaffected | Unaffected |
| 6B | 23 | 59 215 | 552 | 647 | 8.1 | 1 | 212 | Unaffected | |||
| 7A | F8 | c.826G>A (p.Val276Met) | 28 | 55 390 | 289 | 335 | 12.2 | 4 | 409 | Unaffected | Unaffected |
| 8A | F9 | c.520+13A>G (Splice site) | 10 | 57 734 | 174 | 210 | 17.0 | 3 | 250 | Unaffected | Unaffected |
| 8B | 34 | 55 860 | 88 | 102 | 25.6 | 1 | 28 | Unaffected | |||
| 9A | F9 | c.520+13A>G (Splice site) | 36 | 85 166 | 157 | 96 | 33.0 | 1 | 58 | Affected | Affected |
| 9B | 42 | 85 400 | 778 | 610 | 25.5 | 1 | 285 | Affected | |||
| 10A | F9 | c.316G>A (p.Gly106Ser) | 8 | 56 146 | 150 | 112 | 0.8 | NA | NA | Unclassified | Affected |
| 10B | 23 | 59 716 | 193 | 156 | 10.0 | 2 | 165 | Affected | |||
| 11A | F9 | c.874delC (p.Gln292fs) | 36 | 48 538 | 909 | 726 | 17.1 | 1 | 374 | Affected | Affected |
| 12A | F9 | c.1144T>C (p.Cys382Arg) | 34 | 63 669 | 564 | 434 | 12.8 | 1 | 177 | Affected | Affected |
| 12B | 38 | 53 549 | 3056 | 2668 | 10.5 | 1 | 843 | Affected | |||
| 13A | F9 | c.1069G>A (p.Gly357Arg) | 32 | 58 627 | 470 | 636 | 23.1 | 1 | 212 | Unaffected | Unaffected |
| 13B | 40 | 38 782 | 2763 | 3299 | 11.9 | 1 | 1551 | Unaffected | |||
| 14A | F9 | c.802T>A (p.Cys268Ser) | 23 | 50 616 | 577 | 505 | 3.0 | 4 | 895 | Affected | Affected |
| 14B | 32 | 49 366 | 138 | 136 | 18.4 | NA | NA | Unclassified | |||
| 15A | F9 | c.-35G>A (Promoter, 5′ UTR) | 12 | 140 647 | 674 | 671 | 4.0 | NA | NA | Unclassified | Unaffected |
| 15B | 25 | 134 135 | 620 | 784 | 6.9 | 6 | 620 | Unaffected | |||
| 15C | 34 | 34 347 | 594 | 654 | 5.8 | 2 | 578 | Unaffected | |||
ddPCR analyses were performed to test each maternal plasma sample until reaching the fetal genotype classification by the SPRT, or until the maternal plasma was exhausted. Each maternal plasma sample was analyzed by at least 4 reaction wells and evaluated based on the accumulated ddPCR data. The number of reaction wells required for SPRT classification was influenced by the fetal DNA fraction and the total number of allele counts. Total number of positive droplets equals the sum of the M and the N alleles.
NA, not applicable.
Maternal blood samples were taken on 1, 2, or 3 occasions from the same pregnancy, denoted as a sample with subscript A, B, or C, respectively.
Nomenclature of sequence variants is based on the guidelines of the Human Genome Variation Society.21 GenBank accession NM_000132.3 (F8 mRNA variant 1) and NM_000133.3 (F9 mRNA) were used as the reference sequences for hemophilia A and B sequence variants, respectively.
M allele count, the number of droplets positive for the mutant allele. N allele count, the number of droplets positive for the normal/wild-type allele.
Fetal DNA fractions were determined by the digital ZFY/ZFX assay.
Hemophilia status of the newborn was determined from the cord blood sample based on the factor VIII and factor IX assays, as appropriate.
Sample processing
Peripheral blood collected into EDTA-containing tubes was processed by a double-centrifugation protocol7 and stored frozen locally, and all samples were shipped on dry ice to Hong Kong. We extracted maternal plasma DNA with the QIAamp DSP DNA Blood Mini Kit (Qiagen) and buffy coat DNA, using the QIAamp DNA Blood Mini Kit (Qiagen) according to manufacturer’s instructions. Placental DNA was extracted from the tissue with the QIAamp DNA Mini Kit (Qiagen) following the “DNA Purification from Tissues Spin” Protocol.
ddPCR and relative mutation dosage analysis for detecting F8 and F9 sequence variants
We designed ddPCR assays for detecting F8 and F9 sequence variants based on the maternal mutational status, using the Primer Express software (Life Technologies). The mutation assays were performed in a duplex format to discriminate between the N and M alleles, using 2 allele-specific TaqMan probes and 1 pair of primers. We measured fetal DNA fractions in maternal plasma by digital ZFY/ZFX assay.22 All ddPCR analyses were carried out on the QX100/QX200 Droplet Digital PCR System (Bio-Rad). Using the system’s droplet generator, up to 20 000 reaction droplets were generated within a single reaction well, consisting of 10 μL of 2× ddPCR Supermix for Probes (Bio-Rad), relevant forward and reverse primers and probes (supplemental Tables 1 and 2, available on the Blood Web site), 0.5 μL of uracil N-glycosylase, 5 μL of plasma DNA, and deionized water in a final volume of 20 μL. Reaction droplets with positive fluorescent emission from 1 or both probes were considered positive, whereas those without fluorescent emission were assigned as negative. Conventional, nondigital PCRs were carried out in a 96-well foil-sealed PCR plate on the C1000 Thermal Cycler or iCycler (Bio-Rad), following the thermal profile in the manufacturer’s protocol. We analyzed the data with the QuantaSoft Software (version 1.7.4.0917, Bio-Rad) with a manual threshold setting.
We adopted the relative mutation dosage (RMD) approach for X-linked inheritance.7 The sequential probability ratio test (SPRT) statistically evaluated the dosage imbalance between the M and N alleles to infer the hemophilia status of fetuses carried by pregnant women who were carriers of F8 or F9 sequence variants. Briefly, if the fetus had inherited the M allele, there would be overrepresentation of the M allele, indicating the presence of an affected fetus. In contrast, if the fetus had inherited the N allele, the M allele count would be underrepresented, suggesting the presence of an unaffected fetus.
Targeted massively parallel sequencing of F8 int22h-related inversions
Target enrichment and library preparation.
A custom-designed SeqCap EZ Choice Library (NimbleGen) protocol was used for the in-solution target enrichment of DNA libraries. The custom probe set covered regions on chromosomes 3-9, 11-17, 19, 22, and X, encompassing a total length of 27.3 Mb of genomic DNA. A region analyzed for the detection of F8 int22h-related inversions spanned 7.58 Mb on chromosome X, representing 0.7% of the whole target capture design. It comprised 795 targeted segments in the centromeric region (denoted as CEN) and 27 targeted segments in the telomeric region (denoted as TEL) (Figure 1). Targeted regions on other chromosomes were used to estimate fetal DNA fractions with the FetalQuant algorithm.23 The SeqCap probes were designed to preferentially target single nucleotide polymorphisms (SNPs) with a minimum heterozygosity rate of 20%, with the GC content of the targeted region in a range of 38% to 42%, and excluded SNPs that were separated by less than 200 bp.
Figure 1.
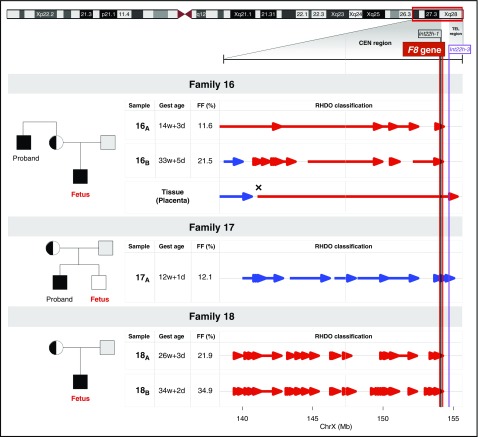
Noninvasive prenatal testing for hemophilia families with F8 int22h-related inversions. Pedigree for 3 families with a history of hemophilia examined by a targeted MPS and RHDO approach. Genotypes of the mother and proband in family 16 and family 17, or the mother and the placental tissue in family 18, were used in a haplotype analysis to infer fetal inheritance of F8 int22h-related inversions. The informative SNPs that were heterozygous in the mother (AB) and hemizygous in proband (A-) were selected. The SNP alleles found in proband belonged to the haplotype linked with the F8 int22h-related inversion (Hap I, red). The SNP alleles absent from the proband belonged to the haplotype not linked with the F8 int22h-related inversion (Hap II, blue). The tail and tip of an arrow denote the start and end, respectively, of a RHDO block. Vertical red, black, and purple lines correspond to the location of the F8 gene and int22h-1 and int22h-3 homologs on chromosome X, respectively. The vertical dashed gray line illustrates a centromere-region boundary. For family 16 and family 18, the fetus had inherited Hap I from the mother, where the haplotype blocks in the F8 region were found to be linked to the proband’s int22h-related inversion. The cross symbol in the placental tissue in family 16 indicates a recombination event in the chromosome position chrX: 141,078,676. In family 17, haplotype blocks in the F8 region were found to be linked to Hap II, suggesting the presence of an unaffected fetus. CEN region and TEL region, centromeric and telomeric region, respectively; FF, fetal DNA fraction; Gest age, gestational age.
DNA libraries were constructed using the KAPA Library Preparation Kit (Kapa Biosystems) according to manufacturer’s recommendations. The plasma and genomic DNA libraries were enriched by 13 and 8 cycles of precapture ligation-mediated PCR, respectively, to obtain a minimum of 1 μg of DNA libraries further subjected to target capture (NimbleGen) according to the manufacturer’s protocol. Twelve cycles of PCR were used for a postcapture ligation-mediated PCR amplification, and captured PCR products were purified with the MinElute PCR Purification Kit (Qiagen). The pre- and postcapture amplified DNA libraries were measured by the nondigital real-time ZFX assay to estimate the relative fold enrichment by calculating the ΔCt values of the pre- and postcapture samples.22 Maternal plasma DNA samples were prepared as a single sample targeted capture, whereas genomic DNA was subjected to a 4-plex DNA library pooling. Each plasma DNA library was sequenced with 1 lane of a flow cell on the HiSeq 2500 System with a paired-end format (75 bp × 2). Genomic DNA library was sequenced in a 4-plex sequencing protocol per flow cell. Paired-end reads were then aligned to the reference human genome (hg19; non-repeat-masked), using the SOAP2 program (http://soap.genomics.org.cn/). Up to 2 nucleotide mismatches for each member of the paired-end reads were allowed.
Selection of informative SNPs in the F8 region for relative haplotype dosage analysis.
To establish fetal inheritance of maternal haplotypes by targeted MPS, we identified informative SNPs based on the genotypes of the mother and proband (family 16 and family 17), or the mother and the placental tissue (family 18). We genotyped DNA by microarray analysis, using the HumanOmni2.5-8 BeadChip Kit (Illumina) on the iScan System (Illumina) and by targeted MPS. We explored the F8 region located up to 7 Mb downstream from the int22h-1 homolog in the direction of the F8 gene transcription start site (chrX: 147,109,089-154,109,089; hg19), denoted as the CEN region. The TEL region spanning a 577-kb region downstream from the int22h-3 covered a region between chrX: 154,693,870 and chrX: 155,270,560. Informative SNPs were those found heterozygous in the mother’s DNA (AB). Hemizygous SNPs identified in the proband’s DNA or the placental DNA were assigned to Hap I, as they belonged to the haplotype linked to the F8 int22h-related inversion (M allele). The SNPs absent from the proband’s DNA were classified as Hap II, as they belonged to the haplotype linked to the N allele. Relative haplotype dosage (RHDO)16 and SPRT analyses were used for the assessment of haplotype dosage imbalance in maternal plasma from the targeted MPS data. Briefly, the null hypothesis for the SPRT classification represents a balance in the dosage of 2 maternal haplotypes. An odds ratio of 100 was used to evaluate whether an overrepresentation or underrepresentation of Hap I or Hap II haplotype in the maternal plasma is present. An overrepresentation of the Hap I indicated the presence of an affected fetus, whereas an overrepresentation of the Hap II indicated the presence of an unaffected fetus.
Results
Detection of F8 and F9 sequence variants in maternal plasma by ddPCR
We first analyzed maternal plasma of 15 pregnant women who were carriers of sequence variants either in the F8 or the F9 gene to assess the fetal hemophilia status by ddPCR and RMD analyses. Among the whole cohort of studied pregnancies at the gestational age between 8 and 42 weeks, the fetal DNA fractions ranged from 0.8% to 33.0%. We performed SPRT analyses based on 2 parameters; namely, the proportion of the M allele and the fetal DNA fraction in maternal plasma, as measured by ddPCR, and predicted fetal genotype consistent with the clinical outcome for the studied pregnancies (Table 1). The number of ddPCR reaction wells required for a conclusive SPRT classification was influenced by the fetal DNA fraction and the number of plasma DNA molecules analyzed. On average, 3 reaction wells in ddPCR (range, 28-1551 positive droplets) were sufficient to establish the fetal hemophilia status among the classified samples, with fetal DNA fractions ranging from 3.0% to 33.0%. In the whole cohort, we detected 2 unclassified results with low fetal DNA fractions (0.8% and 4.0%), and 1 unclassified third-trimester case. The second-trimester sample from this pregnancy (sample 14A) correctly indicated the presence of an affected fetus, but no classification was obtained from a sample collected 9 weeks later (sample 14B), perhaps because the total DNA was found to be 4 times lower for the unclassified sample.
Computer simulations on diagnostic accuracy of ddPCR for detecting F8 and F9 sequence variants
We further conducted computer simulation analyses to estimate the accuracy of performing noninvasive prenatal testing for F8 and F9 sequence variants by ddPCR. The simulations considered fetal DNA fractions ranging from 5% to 30%, and the number of reaction wells in a range of 1 to 10 (corresponding to 200-2000 positive droplets). The analysis assumed the average template concentration of the reference allele per droplet, mr, to be 0.01 and the number of droplets per well in ddPCR to be 20 000. Table 2 shows that for a ddPCR assay examining at least 5 reaction wells and testing a maternal plasma sample with fetal DNA fraction of at least 15%, one would reach more than 99% accuracy at a call rate of more than 98% by digital RMD and SPRT analyses.
Table 2.
Diagnostic accuracies of genotype classification by RMD and SPRT assessed by computer simulations
| Fetal DNA fraction, % | No. of positive droplets | Call rate, %* | Accuracy, %† |
|---|---|---|---|
| 5 | 200 | 0.7 | 84.8 |
| 400 | 6.5 | 93.5 | |
| 600 | 16.1 | 93.2 | |
| 800 | 24.4 | 95.4 | |
| 1000 | 33.5 | 94.8 | |
| 1200 | 40.3 | 96.0 | |
| 1400 | 47.3 | 95.9 | |
| 1600 | 52.1 | 96.2 | |
| 1800 | 57.2 | 96.7 | |
| 2000 | 60.7 | 97.1 | |
| 10 | 200 | 28.8 | 93.6 |
| 400 | 56.9 | 95.8 | |
| 600 | 72.2 | 97.2 | |
| 800 | 80.0 | 98.1 | |
| 1000 | 86.9 | 98.6 | |
| 1200 | 90.8 | 99.0 | |
| 1400 | 93.5 | 99.4 | |
| 1600 | 95.6 | 99.5 | |
| 1800 | 96.5 | 99.6 | |
| 2000 | 97.5 | 99.8 | |
| 15 | 200 | 60.9 | 96.9 |
| 400 | 85.1 | 98.5 | |
| 600 | 93.2 | 99.4 | |
| 800 | 97.4 | 99.7 | |
| 1000 | 98.3 | 99.9 | |
| 1200 | 99.4 | 99.9 | |
| 1400 | 99.7 | 100.0 | |
| 1600 | 99.9 | 100.0 | |
| 1800 | 100.0 | 100.0 | |
| 2000 | 100.0 | 100.0 | |
| 20 | 200 | 84.1 | 98.2 |
| 400 | 96.4 | 99.7 | |
| 600 | 99.2 | 99.9 | |
| 800 | 99.8 | 100.0 | |
| 1000 | 99.9 | 100.0 | |
| 1200 | 100.0 | 100.0 | |
| 1400 | 100.0 | 100.0 | |
| 1600 | 100.0 | 100.0 | |
| 1800 | 100.0 | 100.0 | |
| 2000 | 100.0 | 100.0 | |
| 30 | 200 | 98.6 | 99.8 |
| 400 | 100.0 | 100.0 | |
| 600 | 100.0 | 100.0 | |
| 800 | 100.0 | 100.0 | |
| 1000 | 100.0 | 100.0 | |
| 1200 | 100.0 | 100.0 | |
| 1400 | 100.0 | 100.0 | |
| 1600 | 100.0 | 100.0 | |
| 1800 | 100.0 | 100.0 | |
| 2000 | 100.0 | 100.0 |
Call rate (%) corresponds to the percentage of samples that achieves the SPRT threshold to make a call.
Accuracy (%) refers to the percentage of samples showing a correct call.
Detection of F8 int22h-related inversions in maternal plasma by targeted MPS
With the aim to noninvasively detect int22h-related inversions associated with the F8 gene in maternal plasma of hemophilia carriers, we investigated the applicability of targeted MPS in 3 hemophilia families. A mean of 111 million paired-end reads covered almost the entire target spanning 27.3 Mb, yielding a mean sequencing depth of 149-fold per base. Table 3 summarizes the statistics for the sequencing quality parameters for each studied case.
Table 3.
Targeted massively parallel sequencing of maternal plasma DNA for detection of F8 int22h-related inversions
| Plasma sample | Raw reads, M | Mapped reads, M | No. of reads/ on target, M | No. of reads/ off target, M | Coverage, %* | PCR duplication rate, %† | Sequencing depth per base, -fold‡ | No. of SNPs in CEN region¶ | No. of RHDO classifications in CEN region¶ | No. of SNPs in TEL region¶ | No. of RHDO classifications in TEL region¶ | RHDO analysis | Fetal status§ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16A | 161.8 | 153.8 | 147.1 | 16.8 | 99.96 | 70.4 | 129.2 | 15 | 4 | 1 | 0 | Affected | Affected |
| 16B | 70.6 | 67.2 | 65.0 | 7.8 | 99.67 | 39.7 | 130.6 | 13 | 5 | 1 | 0 | Affected | |
| 17A | 172.0 | 163.3 | 158.4 | 17.8 | 99.67 | 63.7 | 177.1 | 20 | 4 | 1 | 1 | Unaffected | Unaffected |
| 18A | 75.0 | 71.5 | 68.7 | 8.6 | 99.66 | 39.9 | 135.6 | 19 | 9 | 0 | 0 | Affected | Affected |
| 18B | 130.0 | 123.1 | 117.4 | 15.8 | 99.67 | 52.1 | 174.4 | 22 | 21 | 0 | 0 | Affected |
CEN, centromeric region; RHDO, relative haplotype dosage; TEL, telomeric region.
Coverage corresponds to the length of the covered nucleotides in the targeted region divided by the length of the targeted region.
PCR duplication rate corresponds to the proportion of PE reads with the same sequences and start-end coordinates.
Sequencing depth per base is defined as the sum of the bases sequenced within the targeted region divided by the length of the targeted region.
CEN corresponds to the region located up to 7 Mb downstream from the int22h-1 homolog in the direction of the F8 gene transcription start site (chrX: 147,109,089-154,109,089). TEL corresponds to a telomeric region spanning a 577-kb region downstream from the int22h-3 and covering chrX: 154,693,870-155,270,560.
Hemophilia status of the newborn was determined from the cord blood sample based on the factor VIII and factor IX assays, as appropriate.
RHDO analysis assessed whether the fetus had inherited the maternal haplotype associated with the F8 int22h-related inversion or the normal/wild-type haplotype in 5 maternal plasma samples from pregnant hemophilia carriers at gestational ages between 12 and 34 weeks (Figure 1). SPRT classifications identified an affected fetus in family 16 and family 18, and 1 unaffected fetus in family 17, thus accurately predicting the fetal genotype (Table 3; Figure 1). Families were found to be informative for, on average, 18 polymorphic markers located in the 7-Mb CEN region, with a limited number of SNPs identified in the 577-kb TEL region (Table 3).
Specifically, we analyzed 2 maternal plasma samples collected in the early second and the third trimesters in family 16, where fetal DNA fractions were found to be 11.6% and 21.5%, respectively (Figure 1). The genotyping data identified 15 and 13 informative SNPs in the CEN region for the second- and the third-trimester samples, respectively. In contrast, only 1 informative SNP was detected in the TEL region in both samples, not contributing to any SPRT classification. RHDO analyses identified 4 and 5 haplotype blocks linked to the F8 int22h-related inversion (Hap I) spanning the CEN region in the second- and the third-trimester samples, respectively. A detailed haplotype map obtained from the sequencing data revealed a haplotype switch in the chromosome position chrX: 142,484,121 for the sample 16A and in the chromosome position chrX: 141,082,684 for the sample 16B. The results obtained through targeted MPS of the placental tissue confirmed the occurrence of a recombination event in the position chrX: 141,078,676.
In family 17, RHDO analysis correctly determined the presence of an unaffected fetus that had inherited haplotype blocks linked to the normal/wild-type allele (Hap II). The fetal DNA fraction of the studied first-trimester plasma sample in this family was 12.1%. According to the genotyping information of the mother and the proband, 20 informative SNPs were identified in the CEN region, and 1 informative SNP was found in the TEL region, providing a correct classification. Four haplotype blocks spanning the CEN region indicated presence of an unaffected fetus consistent with the hemophilia status of the newborn. We noted no recombination event in the selected region on the long arm of chromosome X.
Informative SNPs in family 18 were identified on the basis of the genotypes of the mother and the placental tissue. We analyzed 2 plasma samples collected from the second and the third trimesters with fetal DNA fractions of 21.9% and 34.9%, respectively. In total, we detected 19 and 22 informative SNPs in the CEN region for the second- and the third-trimester samples, respectively, with no SNP identified in the TEL region. As a result of high fetal DNA fractions found in these samples, SPRT classifications were obtained from a higher proportion of interrogated SNPs. RHDO analyses identified 9 and 21 haplotype blocks spanning the CEN region for the second- and the third-trimester samples, respectively. Haplotype blocks were found to be linked to the proband’s F8 int22h-related inversion (Hap I), indicating the presence of an affected fetus consistent with the clinical phenotype.
Discussion
In this study, among a cohort of 15 pregnant carriers of F8 and F9 sequence variants, ddPCR enabled noninvasive assessment of fetal hemophilia status inferred from the maternal mutational profile in samples obtained from 8 to 42 weeks of gestation. We have further provided the first demonstration of NIPT of F8 int22h-related inversions by establishing the linkage of the SNPs within the F8 region in 3 hemophilia families.
Recent developments in dPCR have brought alternatives to relatively costly sequencing methods for its application in NIPT. Previously, Tsui et al7 recruited 7, mostly third-trimester pregnancies at risk for hemophilia, whose maternal plasma was tested by microfluidics dPCR. The microfluidics system analyzes samples in a microfluidics chip capable of accommodating approximately 9000 reaction wells. In contrast, the whole reaction plate used in ddPCR can generate more than 1.9 M droplets distributed among 96 reaction wells, which represents a more scalable and affordable approach for plasma DNA quantification. Here, we performed an in silico analysis demonstrating the important effect of the fetal DNA fraction in affecting the accuracy of noninvasive prenatal testing for hemophilia using ddPCR and SPRT (Table 2). Clinically, samples with low levels of fetal DNA fractions, typically collected at earlier gestational ages, would require more plasma DNA molecules to be counted. Because of the scalability of ddPCR, the protocol performed reliably even in cases with fetal DNA fraction lower than 10%. ddPCR analyses carried out on samples with insufficient fetal DNA fractions or a low number of analyzed molecules might not reach the statistical confidence required for fetal genotype classification, as represented by the 3 unclassified plasma samples detected in our cohort. However, as shown in the data of this studied cohort, the test either provided a correct classification or no classification. There had been no misclassification. When an unclassified result is encountered, one either performs more digital analyses on the sample to accumulate more data points, or when the sample is consumed, one may resort to an additional blood draw, possibly at a later gestational age with higher fetal DNA fraction. Further validation in a larger cohort would certainly be useful in understanding the performance of this approach.
The targeted MPS strategy has been shown in other studies to be a robust method for NIPT of monogenic diseases based on selective sequencing of the genome.17,18 This approach is able to achieve a higher than 99% diagnostic sensitivity at a 200-fold sequencing depth for 500 analyzed SNPs in maternal plasma samples with fetal DNA fractions as low as 3%.18 We particularly applied this approach for the noninvasive prenatal assessment of F8 int22h-related inversions because the use of RHDO analysis bypasses the need to determine the precise breakpoints of the inversion in each case. In the present study, we performed target enrichment of the F8 region and used RHDO analysis to noninvasively establish fetal inheritance using SNP markers linked to maternal int22h-related inversions. This approach had enabled us to construct a haplotype map of the fetus in the vicinity of the F8 region, which had led to a correct prediction of fetal hemophilia status in all tested families.
In our cohort, RHDO analysis identified 1 recombination event in family 16, located relatively close to the Xq27 band, being consistent with previous observations.24,25 These data, as well as those from our earlier study, show that the RHDO approach could pinpoint the site of recombination fairly precisely.18,26
Approximately one third of all hemophilia A sequence variants occur de novo.27 Until recently, maternal plasma DNA sequencing of de novo sequence variants faced technical limitations because of sequencing error rates of MPS platforms. Kitzman et al28 showed that fetal de novo sequence variants could be detected from maternal plasma at a sensitivity of 88.6%, but with a positive predictive value of only 0.000156%. Further improvements in sequencing technologies reducing the sequencing error rate,16 development of optimized protocols for DNA library preparation that minimize the PCR error rate,29 and introduction of more sophisticated bioinformatics algorithms has increased the specificity of such an approach. Chan et al30 reported the ability to detect fetal de novo sequence variants from maternal plasma at about 80% sensitivity with at least 62% positive predictive value. Another practical challenge represents the family-based setting of this study, requiring construction of the maternal haplotype inferred from the sequencing data of the mother and the proband. This requirement has limited the applicability of the approach in certain cases, as no proband might be available in some families. However, linked-read haplotype sequencing technology can establish mutation haplotype signature in the carrier sample directly, and thus permits PND without the availability of the proband.26 Alternative methods for maternal haplotyping thus hold promise in resolving this issue and open up new potential targets for further refinements of NIPT-based protocols for other monogenic diseases.26
The present developments in NIPT of hemophilia by ddPCR offer an affordable method to detect a spectrum of predefined sequence variants in the F8 and F9 genes in maternal plasma. In a clinical setting, ddPCR analysis would first assess the fetal sex through chromosome Y or other paternally inherited SNP markers. If a male fetus was identified, the mutation-specific ddPCR assay could be used to determine the fetal hemophilia status. Diagnosis could be readily achieved for samples with fetal DNA fractions above a certain threshold, and for samples with low fetal DNA fractions, an additional number of digital analyses would further enhance the detection. If a set of predesigned ddPCR assays for testing hemophilia sequence variants was developed, the sample processing and data analysis could be performed within 1 day at reagent costs of approximately US$3 per reaction well. Such an approach would be impractical in routine practice to detect more complex hemophilia sequence variants, such as F8 int22h-related inversions, which requires analyzing a higher number of polymorphic DNA sites. To this end, we show that targeted MPS strategy facilitates a fine mapping of the F8 region by performing an accurate and robust measurement of maternally inherited fetal haplotypes. As the costs of sequencing continue to decrease and larger cohorts of pregnant carriers of F8 int22h-related inversions will be examined, targeted MPS approaches, such as the one presented in our study, are envisaged to become an essential part in NIPT of hemophilia carriers.
Acknowledgments
The authors thank Keith Gomez, Bilal Jradeh, and Debra Pollard at the Katharine Dormandy Haemophilia and Thrombosis Centre for their assistance in conducting this study.
This work was supported by the Research Grants Council of the Hong Kong SAR Government under the theme-based research scheme (T12-403/15-N) and the Vice Chancellor’s One-Off Discretionary Fund of The Chinese University of Hong Kong (VCF2014021).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: I.H., P.J., Y.M.D.L. and R.W.K.C. designed the research; I.H., P.J., J.D., Y.M.D.L., R.A.K., and R.W.K.C. performed the research; I.H. wrote the first draft of the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: P.J., R.W.K.C., and Y.M.D.L. have granted patents and filed patent applications for plasma nucleic acid analysis. Part of the portfolio has been licensed to Sequenom, Illumina, Xcelom, and Cirina. R.W.K.C. and Y.M.D.L are on the board of directors of Xcelom and Cirina and hold equities in Sequenom, Xcelom, and Cirina. P.J. and R.W.K.C. are consultants to Xcelom. P.J. is a consultant to Cirina. The remaining authors declare no competing financial interests.
Correspondence: Rossa W. K. Chiu, Department of Chemical Pathology, The Chinese University of Hong Kong, Prince of Wales Hospital, 30-32 Ngan Shing St, Shatin, New Territories, Hong Kong SAR, China; e-mail: rossachiu@cuhk.edu.hk.
References
- 1.Bowen DJ. Haemophilia A and haemophilia B: molecular insights. Mol Pathol. 2002;55(2):127-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi C, Lee CA, Shiltagh N, Khan A, Pollard D, Kadir RA. Pregnancy in carriers of haemophilia. Haemophilia. 2008;14(1):56-64. [DOI] [PubMed] [Google Scholar]
- 3.Varekamp I, Suurmeijer TP, Bröcker-Vriends AH, et al. . Carrier testing and prenatal diagnosis for hemophilia: experiences and attitudes of 549 potential and obligate carriers. Am J Med Genet. 1990;37(1):147-154. [DOI] [PubMed] [Google Scholar]
- 4.Karimi M, Peyvandi F, Siboni S, Ardeshiri R, Gringeri A, Mannucci PM. Comparison of attitudes towards prenatal diagnosis and termination of pregnancy for haemophilia in Iran and Italy. Haemophilia. 2004;10(4):367-369. [DOI] [PubMed] [Google Scholar]
- 5.Huq FY, Kadir RA. Management of pregnancy, labour and delivery in women with inherited bleeding disorders. Haemophilia. 2011;17(1):20-30. [DOI] [PubMed] [Google Scholar]
- 6.Kadir RA, Davies J, Winikoff R, et al. . Pregnancy complications and obstetric care in women with inherited bleeding disorders. Haemophilia. 2013;19(4):1-10. [DOI] [PubMed] [Google Scholar]
- 7.Tsui NBY, Kadir RA, Chan KCA, et al. . Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117(13):3684-3691. [DOI] [PubMed] [Google Scholar]
- 8.Engert A, Balduini C, Brand A, et al. ; EHA Roadmap for European Hematology Research. The European Hematology Association Roadmap for European Hematology Research: a consensus document. Haematologica. 2016;101(2):115-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne AB, Miller CH, Kelly FM, Michael Soucie J, Craig Hooper W. The CDC Hemophilia A Mutation Project (CHAMP) mutation list: a new online resource. Hum Mutat. 2013;34(2):E2382-E2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Association for Haemophilia and Allied Disorders Coagulation Factor Variant Databases. http://www.eahad-db.org. Accessed 6 November 2016. [DOI] [PubMed]
- 11.Rallapalli PM, Kemball-Cook G, Tuddenham EG, Gomez K, Perkins SJ. An interactive mutation database for human coagulation factor IX provides novel insights into the phenotypes and genetics of hemophilia B. J Thromb Haemost. 2013;11(7):1329-1340. [DOI] [PubMed] [Google Scholar]
- 12.Bagnall RD, Giannelli F, Green PM. Int22h-related inversions causing hemophilia A: a novel insight into their origin and a new more discriminant PCR test for their detection. J Thromb Haemost. 2006;4(3):591-598. [DOI] [PubMed] [Google Scholar]
- 13.Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5(3):236-241. [DOI] [PubMed] [Google Scholar]
- 14.Naylor JA, Buck D, Green P, Williamson H, Bentley D, Giannelli F. Investigation of the factor VIII intron 22 repeated region (int22h) and the associated inversion junctions. Hum Mol Genet. 1995;4(7):1217-1224. [DOI] [PubMed] [Google Scholar]
- 15.Naylor J, Brinke A, Hassock S, Green PM, Giannelli F. Characteristic mRNA abnormality found in half the patients with severe haemophilia A is due to large DNA inversions. Hum Mol Genet. 1993;2(11):1773-1778. [DOI] [PubMed] [Google Scholar]
- 16.Lo YMD, Chan KCA, Sun H, et al. . Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. [DOI] [PubMed] [Google Scholar]
- 17.Lam KWG, Jiang P, Liao GJW, et al. . Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to β-thalassemia. Clin Chem. 2012;58(10):1467-1475. [DOI] [PubMed] [Google Scholar]
- 18.New MI, Tong YK, Yuen T, et al. . Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab. 2014;99(6):E1022-E1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lun FMF, Tsui NBY, Chan KCA, et al. . Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(50):19920-19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu RWK, Cantor CR, Lo YMD. Non-invasive prenatal diagnosis by single molecule counting technologies. Trends Genet. 2009;25(7):324-331. [DOI] [PubMed] [Google Scholar]
- 21.den Dunnen JT, Dalgleish R, Maglott DR, et al. . HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564-569. [DOI] [PubMed] [Google Scholar]
- 22.Lun FMF, Chiu RWK, Chan KCA, Leung TY, Lau TK, Lo YMD. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008;54(10):1664-1672. [DOI] [PubMed] [Google Scholar]
- 23.Jiang P, Chan KCA, Liao GJW, et al. . FetalQuant: deducing fractional fetal DNA concentration from massively parallel sequencing of DNA in maternal plasma. Bioinformatics. 2012;28(22):2883-2890. [DOI] [PubMed] [Google Scholar]
- 24.Drayna D, Davies K, Hartley D, et al. . Genetic mapping of the human X chromosome by using restriction fragment length polymorphisms. Proc Natl Acad Sci USA. 1984;81(9):2836-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drayna D, White R. The genetic linkage map of the human X chromosome. Science. 1985;230(4727):753-758. [DOI] [PubMed] [Google Scholar]
- 26.Hui WWI, Jiang P, Tong YK, et al. . Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clin Chem. 2017;63(2):513-524. [DOI] [PubMed] [Google Scholar]
- 27.Ljung RC, Sjörin E. Origin of mutation in sporadic cases of haemophilia A. Br J Haematol. 1999;106(4):870-874. [DOI] [PubMed] [Google Scholar]
- 28.Kitzman JO, Snyder MW, Ventura M, et al. . Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4(137):137ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlsson K, Sahlin E, Iwarsson E, Westgren M, Nordenskjöld M, Linnarsson S. Amplification-free sequencing of cell-free DNA for prenatal non-invasive diagnosis of chromosomal aberrations. Genomics. 2015;105(3):150-158. [DOI] [PubMed] [Google Scholar]
- 30.Chan KCA, Jiang P, Sun K, et al. . Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci USA. 2016;113(50):E8159-E8168. [DOI] [PMC free article] [PubMed] [Google Scholar]