Abstract
Polyketide synthases (PKSs) represent a powerful catalytic platform capable of effecting multiple carbon-carbon bond forming reactions and oxidation state adjustments. We explored the functionality of two terminal PKS modules that produce the 16-membered tylosin macrocycle, using them as biocatalysts in the chemoenzymatic synthesis of tylactone and its subsequent elaboration to complete the first total synthesis of the juvenimicin, M-4365, and rosamicin classes of macrolide antibiotics via late-stage diversification. Synthetic chemistry was employed to generate the tylactone hexaketide chain elongation intermediate that was accepted by the juvenimicin (Juv) ketosynthase of the penultimate JuvEIV PKS module. The hexaketide is processed through two complete modules (JuvEIV and JuvEV) in vitro, which catalyze elongation and functionalization of two ketide units followed by cyclization of the resulting octaketide into tylactone. After macrolactonization, a combination of in vivo glycosylation, selective in vitro cytochrome P450-mediated oxidation, and chemical oxidation was used to complete the scalable construction of a series of macrolide natural products in as few as 15 linear steps (21 total) with an overall yield of 4.6%.
Introduction
Improved methods for creating novel chemical structures and their application toward developing effective antibiotics are of critical importance in the ongoing battle against drug-resistant microbes. Macrolide (macrolactone glycoside) antibiotics, such as pikromycin,1 erythromycin,2 and tylosin,3–4 have proven to be useful and adaptable tools against evolving pathogens by targeting the ribosome and are amenable to improvement through semisynthetic modification.5 In order to further improve production of this important class of antimicrobials, efforts have focused during the past two decades on investigating the biological sources of these potent natural products,6 with important studies probing in vivo usage7–8 and in vitro analysis.9–10 As secondary metabolites, macrolides presumably offer an evolutionary advantage to their producing organism (e.g., Streptomyces venezuelae, Saccharopolyspora erythraea, Streptomyces fradiae) through inhibition of competitors or by protective signaling to other symbiotic organisms. Their production is typically mediated by three major groups of enzymes: polyketide synthases (PKSs) that are responsible for construction and cyclization of the unmodified aglycone core, glycosylation enzymes that synthesize and append sugar moieties, and cytochrome P450 monooxygenases (P450s) that further functionalize the macrolactone via late-stage C-H bond oxidation. These final modification steps play a significant role in enhancing the potency of the antibiotic.
Type I polyketide synthases are large multidomain enzymes that select activated malonic acid derivatives, processing two carbons from each ketide unit and their various side chains into the final complex structure. Modular PKSs, such as those that produce macrolides, utilize discrete modules to sequentially effect the attachment and oxidation state adjustment of specific ketide units, with the entire cassette being engaged to complete the unique linear chain or macrocycle.
While a rich diversity of domains and modifications exist, the most common actions of a modular PKS are dependent upon the activities of up to six canonical domains and their distinct roles.11 Three of these domains are critical to extension: an acyl transferase (AT) is responsible for selection of an acyl-CoA subunit, an acyl carrier protein (ACP) shuttles the growing polyketide amongst domains,9–10 and a ketosynthase (KS) accepts the upstream polyketide intermediates and catalyzes a decarboxylative Claisen reaction resulting in chain elongation. The presence, absence, or null activity of three additional domains directs oxidation state adjustment on the nascent polyketide chain after extension. A ketoreductase (KR) domain reduces a β-keto group to a β-hydroxyl, a dehydratase (DH) eliminates the alcohol, typically forming an α/β double bond, and an enoyl reductase (ER) saturates the double bond to an alkane. These domains comprise a module, which expresses singly or within multimodular PKS megaenzymes. At the end of many PKS pathways, a terminal thioesterase (TE) domain offloads the completed polyketide as a linear acid or as a macrolactone product.
We have previously elucidated the structure9 and function10 of PikAIII, a PKS responsible for the formation of 12- and 14-membered macrolactones, as well as disclosed the actions and use of various glycosylation,12–13 P450,13–16 and methyltransferase17–18 tailoring enzymes. These studies resulted in the biocatalytic syntheses of the pikromycin aglycone (2005),19 the erythromycin aglycone (2009),20 and pikromycin (2013).21 Despite several years of effort, however, the PKSs responsible for the production of the 16-membered macrolide tylosin in Streptomyces fradiae22–23 have eluded analysis due to the inability to effectively express the key PKS enzymes. The terminal two monomodular gene products of this system, TylGIV and TylGV, were of high interest because they can accommodate larger polyketides, TylGV selects for a malonyl rather than a methylmalonyl extender unit, and both monomodules could serve as a potential platform for cryoEM investigations9–10 of complete PKS modules that include thioesterase domains.
The 16-membered macrolactone core of tylosin (tylactone, 1) is formed via a modular PKS pathway (Figure 1A) through the actions of five separate proteins (TylGI – TylGV) containing eight modules (including the loading module), each of which contributes two carbon atoms to the backbone of the final octaketide.23 The first protein, TylGI, contains three modules and is responsible for installing the carbons of 1 highlighted in purple. The AT of the loading module selectively loads the ACP using methylmalonyl-CoA, which is then decarboxylated by a specialized KS domain (KSQ). The ACP passes the resulting propionate to the KS of module 1, with the proximity for interaction ensured by a linker region between the two domains (denoted by a black bar in Figure 1A). The AT of module 1 also selects a methylmalonate and loads its ACP, which then moves to the KS active site and facilitates a decarboxylative Claisen reaction by interacting with the KS-bound propionate, resulting in an unreduced diketide. The KR of module 1 stereoselectively reduces the β-keto of the ACP-bound diketide to an R alcohol, and the ACP then delivers this substrate to the KS of module 2. Module 2 undertakes the same reaction sequence as module 1, with the addition of a DH-catalyzed dehydration. After this process is complete, the resulting ACP-bound triketide is passed from the terminal ACP of TylGI to the KS of TylGII via the interaction of docking domains24 (yellow chevrons) that enable the PKS proteins to recognize their downstream partners.25 Module 3 installs two additional carbon atoms (yellow) by selecting malonyl-CoA and, after oxidation-state adjustment, passes a tetraketide to TylGIII via the interaction of its C-terminal docking domain (blue chevron). TylGIII installs the carbons highlighted in blue (notably, the KR of module 4 is inactive and a β-ketone is maintained) with module 5 selecting for ethylmalonyl-CoA and containing an ER domain that saturates the KR/DH-processed alkene. The hexaketide product of TylGIII is subsequently extended and reduced twice by the monomodules TylGIV (green) and TylGV (red), finally being passed to the TE of TylGV, which cyclizes the linear octaketide into the 16-membered ring of tylactone (1).
Figure 1.
Figure 1A: The PKS system responsible for the production of the cyclized tylosin octaketide (1) and (inset) its subsequent tailoring to the juvenimicin, rosamicin, and M-4365 series of natural products (2–8).
Figure 1B: Gene clusters that code for biosynthesis of tylactone, tylosin21 and the juvenimicins (MF033535).
Beyond the PKSs responsible for assembling macrolactone cores, tailoring enzymes encoded in the gene cluster are responsible for processing them into macrolides and adding further selective functionality. Specifically, the interdependent action of glycosylation pathway enzymes to synthesize and transfer sugars, cytochrome P450 monooxygenases to install oxidative functionality, and methyltransferases eventually completes the final forms of the macrolide antibiotics reported from distinct bacterial strains. The tailoring steps responsible for tylosin biosynthesis were well established through a concerted effort by Eli Lilly in the 1980s using structural elucidation of metabolites from S. fradiae blocked-mutant strains and cross-feeding of accumulated intermediates with other mutants,22 with the complete biosynthetic cluster being reported later.23
To address the intransigence of the terminal TylGIV and TylGV monomodules toward soluble protein overexpression, we hypothesized that homologous polyketide synthases from a different macrolide-producing species sharing the same tylactone core (1) could enable us to effectively study 16-membered ring formation. The juvenimicins26 and M-4365,27 produced by Micromonospora chalcea (ssp. izumensis)28 and Micromonospora capillata,29 respectively, appeared to be likely candidates that might contain tailoring enzymes enabling the diverse oxidation patterns observed in the isolated natural products (2–8, Figure 1A, inset). We reasoned that after completion of tylactone (1) by the PKS, a glycosylation sub-cluster similar to des in S. venezuelae12 would create and append a desosamine sugar to give the macrolide (2). A P450 monooxygenase was predicted to selectively oxidize the C20 position (R1) to an alcohol (3) or aldehyde (5), and the action of another P450 could install the C23 alcohol at R2 (4) or the C12-C13 epoxide (6–8).30 Thus, in addition to confirming the predicted functions of these enzymes, we envisioned using a biocatalytic approach (Figure 2) involving late-stage polyketide assembly, tailoring, and C-H functionalization to achieve the first total synthesis of tylactone and several additional 16-membered macrolides.
Figure 2.
Proposed late-stage tailoring/C-H functionalization and retrosynthetic analysis of a hexaketide advanced intermediate (inset) suitable for interception and utilization of the tylosin PKS biosynthetic machinery.
Toward this goal, we imagined synthesizing the natural tylactone hexaketide chain elongation intermediate (Figure 2, inset) that we could suitably activate and employ as a substrate for a penultimate PKS module homologous to TylGIV (Figure 1A), reconstituting in vitro the natural processes that result in tylactone (1). Our retrosynthetic approach to the hexaketide suggested a disconnection through the α/β double bond via the Horner-Wadsworth-Emmons (HWE) version of the Wittig olefination to maximize the future synthesis of derivatives. The desired β-ketophosphonate could replace a chiral auxiliary, which would be used to install the desired stereochemistry along with a previously reported alkyl halide. We reasoned that the aldehyde portion could be achieved using extended-enolate chemistry, an approach further strengthened by a similar, recent synthesis.31 Upon successful processing of the hexaketide by the PKS modules to tylactone (1), we envisioned employing in vivo glycosylation and in vitro P450-mediated oxidation13 to achieve late-stage diversification, enabling facile access to the complete set of juvenimicin,26 M-4365,27 and rosamicin32 macrolides.
Results and Discussion
Synthesis of the Tylactone Hexaketide Chain Elongation Intermediate
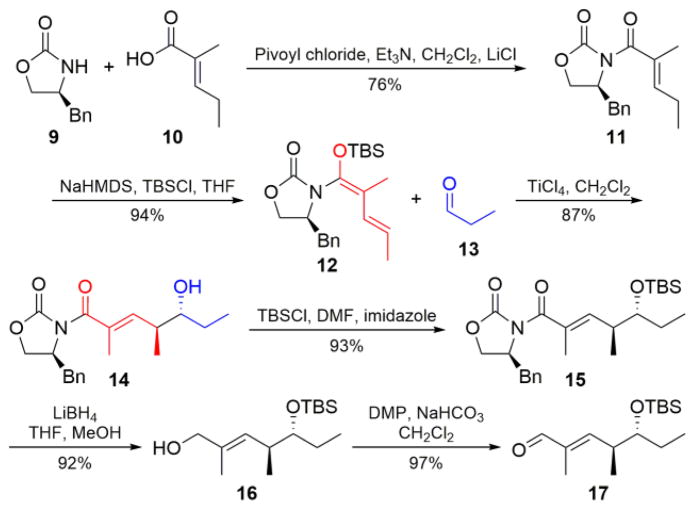
In order to generate a suitably activated natural tylactone hexaketide intermediate (Figure 2, inset), we undertook the synthesis of aldehyde 17 (Scheme 1). Our initial approach employed an iridium-based coupling strategy33 to realize the desired stereochemical configuration, followed by homologation of the resulting aldehyde according to chemistry originally developed for the synthesis of the rosamicin aglycone.34 While successful in generating 17, this strategy was burdened by complex reagents and beset by low yields, leading us to pursue a more conventional approach (Scheme 1). The unsaturated acid 10 was appended to (S)-4-benzyl-2-oxazolidinone (9) using the established mixed anhydride strategy.35 Compound 11 was activated as the silylated extended enol ether (12)35 and reacted36 with freshly distilled propionaldehyde (13), furnishing the completed carbon skeleton (14) in a high diastereomeric ratio (>20:1). After protection of alcohol 14 as the silyl ether (15), the chiral auxiliary was cleaved37 to the primary alcohol (16), the last stable intermediate in the aldehyde pathway. Oxidation to 17 was readily effected by treating 16 with Dess-Martin Periodinane (DMP),38 but extended storage of 17, especially without scrupulous protection from atmospheric oxygen, resulted in auto-oxidation to the corresponding α/β unsaturated acid. Despite the requisite pause at alcohol 16, the overall yield for this six-step process was 52%.
Scheme 1.
Synthesis of aldehyde partner 17.
With a robust and high yielding strategy for the aldehyde portion established, our attention next turned to the synthesis of an appropriate coupling partner. We envisioned using a HWE olefination strategy as our final convergent union (Figure 2), but required a route capable of accommodating the C6 ethyl group instead of resolution from a symmetric substrate.19 The combination of a chiral alkyl halide precursor with a Myers-type chiral auxiliary offered an improved strategy capable of providing the requisite desymmetrization while avoiding expensive reagents such as Roche ester derivatives.20–21
Experiments with enantioselective hydroxymethylation39 and lipase resolution furnished material for initial explorations, but not in an enantiomeric ratio (<84%) sufficient for reactions on scale. We ultimately employed a strategy reported for the enantiomer40 of 22 (Scheme 2) where butyric anhydride (18) was appended to 9 generating butanoic oxazolidinone 19.41 The stereoselective reaction of this material with benzyl chloromethyl ether as a carbon source yielded 20 in a high dr (>20:1).37 Subsequent cleavage of the chiral auxiliary37 to alcohol 21 and its activation as the iodide40 (22) effectively provided material suitable for further elucidation.
Scheme 2.
Synthesis of chiral alkyl iodide fragment 22.
The coupling of 22 (Scheme 3) with (+)-pseudoephedrine-activated propionate (23) prepared by the Myers method42 smoothly installed the second stereocenter (24) in greater than a 20:1 dr with a small amount of eliminated byproduct due to excess lithium diisopropyl amine. Reductive cleavage of the auxiliary37, 42 furnished alcohol 25, which could be efficiently converted to the methyl ether (26). The oxidation of diether 26 was attempted using ruthenium chloride,43 but the benzylic position was selectively oxidized and the resulting benzoyl ester would not hydrolyze or undergo further oxidation, precluding the direct formation of 28. Instead, the benzyl ether was first cleaved by hydrogenolysis, and 27 was subjected to oxidation.43 Initial conversions of 27 to 28 were hindered by formation of a diacid byproduct, which was subsequently suppressed by the replacement of water with aqueous phosphate buffer (pH 7), an adaptation that prevented the acid product from undergoing self-catalyzed hydrolysis. The final hexaketide carbon was installed with concomitant activation for HWE olefination by treatment with more than two equivalents of deprotonated dimethyl methylphosphonate,19 first neutralizing the acid of 28 before reacting with its ester to form 29 in a 24% yield over ten steps.
Scheme 3.
Completion of acid coupling partner 29.
Despite the testing of a number of established HWE olefination conditions, phosphonate 29 and aldehyde 17 (Scheme 4) were unreactive except with barium hydroxide,19 which epimerized dienone 30, presumably at the C8 position alpha to the ketone. Further screening revealed that cesium carbonate in dry isopropanol44 was also effective and gave a 2:1 diastereomeric ratio of the desired configuration in small-scale reactions and a 4:1 dr upon scale-up. Compound 30 was carried through thioesterification21 (31) as a mixture of epimers, as separation was most easily effected at this stage. Finally, fluoride-mediated cleavage of the silyl ether furnished 32, a fully formed and activated hexaketide advanced intermediate suitable for reaction with select PKSs.
Scheme 4.
HWE coupling and activation of the advanced hexaketide intermediate.
Isolation, Sequencing, and Cloning of Macrolide Genes
Initial efforts focused on TylGIV and TylGV PKS expression, the last two modules from the tylosin producer Streptomyces fradiae (Figure 1A). However, TylGV proved remarkably resistant to expression. Changes in buffer conditions, solubility modifiers, expression time, and use of a Streptomyces host provided only marginal improvements to a poor yield; the vast majority of expressed protein remained insoluble. To circumvent these problems, we sequenced other bacterial species whose secondary metabolites shared the tylactone core (1) with the goal of using homologous proteins from these systems. The juvenimicin producer Micromonospora chalcea subspecies izumensis (ATCC 21561)28 and Micromonospora sp. CNJ878, a recently disclosed species with variability of the C5 glycoside,45 showed the presence of a similar five-protein, eight-module PKS cluster after genome sequencing and bioinformatic gene cluster mining (Figure 1B). The predicted proteins, JuvEI–JuvEV and CnjHI–CnjHV, showed high homology to TylGI–TylGV (SI Table 2), and we expected them to serve as surrogate modules for chemoenzymatic construction of the tylactone core (1, Figure 1A). Further annotation of the juv cluster (accession number MF033535) revealed a desosaminylation cassette (juvFI - juvFV) similar to the des genes in Streptomyces venezuelae12 and P450 monooxygenases (juvC and juvD) that we hypothesized were responsible for introduction of alcohol and aldehyde functionality at C20 and epoxide functionality at C12/C13 because of their high sequence identity to recently reported rosamicin monooxygenases.30 This deeper analysis suggested that the juv gene cluster is not only responsible for production of the juvenimicins (2, 3, 7, 8), but could also yield the M-4365 family (5, 6) and possibly unreported natural products as well.
Preparation and screening of a fosmid library resulted in identification of a clone that contained both juvEIV and juvEV, the M. chalcea homologs to tylGIV and tylGV, as well as one containing P450 genes juvC and juvD (Figure 1B). These M. chalcea open reading frames were used to construct plasmids encoding 6x His-tagged PKSs JuvEIV and JuvEV and P450s JuvC and JuvD. The two P450 genes were cloned both as native monooxygenases and as the corresponding engineered single-component P450-RhFRED fusion proteins.46
Optimization of PKS Expression and in vitro Catalysis
With the desired constructs in hand, we focused on expression and optimal use of JuvEIV and JuvEV both to confirm their function and to employ them as biocatalysts in the total synthesis of tylactone and the juvenimicins. JuvEIV expressed well, but it resisted purification as a homogeneous polypeptide. Fortunately, the catalytic efficiency of this PKS monomodule was found to be high in reactions as an E. coli cell-free lysate. JuvEV was readily obtained as a pure protein using fast protein liquid chromatography (FPLC) and enabled the isolation of up to 50 mg of the terminal monomodule per liter of culture (SI Figure 1).
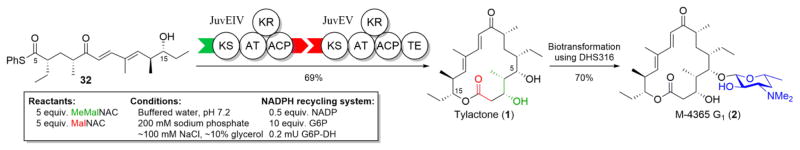
Batch analysis of JuvEV indicated a high degree of consistency between observed activity and protein concentration, but experimentation with JuvEIV indicated significant variability per unit volume due to batch-to-batch differences in concentration of the monomodule within the clarified E. coli lysate. By titrating JuvEIV against known concentrations of JuvEV, the activity of a particular batch could be assessed (SI Figure 2). Additionally, the relative amount of protein needed to complete the conversion of 32 to 1 (Scheme 5) within three hours was determined (SI Figure 3) to ensure rapid, complete conversion.
Scheme 5.
Chemoenzymatic synthesis of tylactone (1) and glycosylation to 2 via biotransformation.
Optimization of the biocatalytic extension reaction of 32 (Scheme 5) revealed that keeping its concentration lower than 0.5 mM was optimal for its high conversion to 1. The ratio of JuvEIV to JuvEV was assumed to be lower than the 4 μM (0.8% mole) JuvEV concentration and confirmed by titration (SI Figures 2 and 3) to be at a threshold sufficient to suppress unproductive side reactions such as hydrolysis.47 Assessing other reaction parameters, including malonyl and methylmalonyl N-acetyl cysteamine (NAC) equivalents, ratios of buffer components, pH, dimethyl sulfoxide concentration, and the NADP+ recycling system,48 indicated that variation was tolerated, albeit with slightly reduced yields. These experiments were performed using stock solutions, the combination of which enabled testing at known molarities of each component in a final reaction volume of 50 μL. The resulting optimized ratio was linearly scalable to a level where 32 could be accurately massed, dissolved in DMSO, and added to the reaction mixture (SI Figure 4). These optimizations enabled the coupled use of JuvEIV and JuvEV as biocatalysts capable of 1) accepting hexaketide 32 as a substrate, 2) installing the final four carbon atoms in the macrocycle (Scheme 5, red and green), 3) stereoselective introduction of three chiral centers, and 4) completing macrolactonization of the resulting octaketide to tylactone (1), the unoxidized aglycone core of tylosin, in high yield.
In vivo Glycosylation
With the success of this chemoenzymatic approach to the complete macrocycle, our attention turned to the final glycosylation and oxidative tailoring steps to complete the total syntheses of the juvenimicins, the M-4365 series, and rosamicin (2–8, Figure 1A, inset). We previously described a mutated S. venezuelae strain, DHS316,13 lacking the pikromycin PKS and P450 genes, which renders it incapable of producing its own macrolides. However, the des genes12 are intact, which enables the recombinant strain to biotransform exogenous macrolactones into their corresponding macrolides. S. venezuelae strain DHS316 is extraordinarily effective at converting 1 (Scheme 5) into M-4365 G1 (2)27 compared to standard synthetic methodologies used for the desoaminylation of the aglycones of pikromycin,49 methymycin,50 and mycinamicin.51 None of these syntheses were able to obtain the activated sugar in greater than 50% yield despite the facile semi-synthetic approach that includes harvesting desosamine from erythromycin,52 let alone the additional steps required for attachment to the macrolactone and deprotection. The DHS316 biotransformation strain enabled us to routinely achieve greater than a 60% yield of 2 by adding 100 milligrams of 1 to an actively growing one liter culture (SI Figure 5); the product was readily purified by flash chromatography,53 though care needed to be taken to exclude light to prevent photoisomerization of the γ-δ double bond.
Structural Diversification via Late-stage P450 Oxidation
With the natural product 2 in hand, we assessed the potential for late-stage diversification to the remaining family members via cytochrome P450-catalyzed oxidation (Scheme 6 and SI Figure 6). The P450s JuvC and JuvD exhibited 87% and 84% identity to the P450s RosC and RosD,30 respectively, which are involved in the biosynthesis of rosamicin (juvenimicin A3, 8) in Micromonospora rosaria. As the functions of these enzymes have been established by genetic and whole cell biotransformation studies,30 we surmised that JuvC could iteratively oxidize C20 to produce M-4365 G2 (5) from 2 via juvenimicin B1 (3) while JuvD could install an epoxide across the C12-C13 double bond of each of these compounds to produce known natural products 6–8.
Scheme 6.
Selective oxidation state adjustment by in vitro P450 oxidation.
In order to test the ability of the JuvC and JuvD P450s to catalyze their predicted oxidation reactions, we cloned and expressed the corresponding genes as fusions to the FMN/[2Fe-2S]-containing reductase domain of P450RhF (RhFRED) as previously described,13, 46 generating catalytically self-sufficient biocatalysts in each case. While JuvC-RhFRED expressed well and exhibited the characteristic P450 CO-reduced spectrum, its activity toward 2 was rather modest, producing 3 with ~20% conversion and trace amounts of 5 under standard analytical-scale reaction conditions. Due to its low activity, we tested the homologous enzyme from the tylosin biosynthetic pathway, TylI (72% sequence identity to JuvC), and found that the TylI-RhFRED fusion protein expressed at a high level and readily accepted 2 as a substrate, producing a mixture of C20 primary alcohol product 3 (~80%) and C20 aldehyde 5 (~5%). Upon scale-up, TylI-RhFRED effectively converted 2 into 3 with an isolated yield of 61% plus trace amounts of 5 and recovered 2. Further analytical-scale investigation of TylI-RhFRED reactivity revealed that despite its ability to readily convert 2 into 3, it was much less effective generating 5 from 3 (10–15%). Therefore, we opted employ a copper(I)/TEMPO catalyst system54–55 to selectively convert the C20 primary alcohol of 3 into aldehyde 5 in a 57% yield.
With 2, 3, and 5 in hand, we tested the proposed epoxidase JuvD-RhFRED on each compound and determined that this enzyme effectively converted each substrate to a single product. Thus, JuvD-RhFRED was employed in large-scale reactions to readily obtain 6, 7, and 8 in isolated yields of 55%, 63%, and 43%, respectively. Finally, based on the unusual substrate flexibility of the P450 MycCI from the mycinamicin biosynthetic pathway in Micromonospora griseorubida,13 we employed the MycCI-RhFRED fusion protein to selectively convert 2, 3, and 5 to 23-hydroxy-5-O-desosaminyl-tylactone (33), juvenimicin B3 (4), and 5-O-desosaminyl-tylonolide (34), respectively. Although only 4 has been previously reported from M. chalcea,26 33 and 34 are potentially produced as well as evidenced by the hybrid biosynthesis of 33,56 and the semisynthesis57 and eventual isolation and identification of 34 from Micromonospora sp. YS-02930K.58
Antimicrobial Testing
The macrolides were tested against a panel of human bacterial pathogens (Table 1 and SI Figure 7) to determine their relative minimum inhibitory concentrations (MICs). The activities of 5 and 8 were comparable to commercial antibiotics tylosin and erythromycin, showing strong activity against Gram positive strains and enhanced activity against Gram negative pathogens, results that are consistent with those described previously.28–29, 59 The aldehyde functionality of 5, 34, and 8 (Scheme 6 and SI Figure 7) is critical to high activity showing ~10–100-fold improvement over the corresponding hydroxyl (3, 4, 7) and unoxidized (2, 33, 6) counterparts. Testing of the photoisomer (2-iso, SI Figure 8) revealed that the Z geometry at C12/C13 dramatically reduced activity relative to 2. This result suggests that the epoxide potentially serves as a protective mechanism to retain activity rather than a critical pharmacophore, as evidenced by the similar level of activity regardless of the oxidation at C12/C13 (8) or C23 (34) compared to the parent alkene (5). A similar decrease in activity was also observed in photoisomeric tylosin degradation products.60
Table 1.
Select MIC values (see also SI Figure 7)
| Bacterial Strain | Macrolide | |||||||
|---|---|---|---|---|---|---|---|---|
| Tylosin | Erythromycin | 2-iso | 2 | 3 | 5 | 34 | 8 | |
| MIC (μM) | ||||||||
| B. subtilis DHS 5333 | 1.6 | 0.781 | >100 | >100 | >100 | 3.1 | 3.1 | 1.6 |
| B. subtilis 168 | 1.6 | 0.781 | >100 | >100 | >100 | 3.1 | 12.5 | 0.781 |
| B. anthracis 34f2 | 0.781 | 3.1 | >100 | 50 | 100 | 1.6 | 3.1 | 0.781 |
| B. cereus | 6.3 | 6.3 | >100 | >100 | 100 | 6.3 | 25 | 3.1 |
| S. aureus ATCC 6538P | 1.6 | 0.781 | >100 | 50 | 50 | 1.6 | 3.1 | 0.781 |
| S. aureus NorA | 6.3 | 1.6 | >100 | 12.5 | 50 | 3.1 | 3.1 | 1.6 |
| E. coli TolC | 1.6 | 0.781 | 100 | 1.6 | 1.6 | 0.391 | 0.391 | 0.391 |
| E. coli MC1061 | 25 | 12.5 | 50 | 1.6 | 1.6 | 1.6 | 1.6 | 1.6 |
Conclusions
We describe for the first time an efficient biocatalytic approach to the 16-membered macrolactone core of tylosin and the first total synthesis of the juvenimicin, M-4365, and rosamicin family of macrolide antibiotics. Using this approach, natural product 2 was generated in 15 linear steps (21 total) with an overall yield of 4.6%. Our approach employed the native monomodular polyketide synthases responsible for completing polyketide chain elongation, processing, and cyclization of the core macrolactone followed by an engineered biotransformation strain to append the desosamine glycoside to the macrolactone in the absence of protecting groups. By understanding the activities of the P450 monooxygenases required for the native production of several different macrolides, we were able to effectuate late-stage oxidation to obtain a series of highly modified antibiotics based on the tylactone core, a unique divergent strategy that enables efficient, regio- and stereoselective C-H activation and/or epoxidation.
Our work has also experimentally confirmed that the purported juv gene cluster is indeed responsible for the production of the juvenimicins in Micromonospora chalcea. This outcome was achieved by synthesis of the fully elaborated and thiophenyl-activated hexaketide chain elongation intermediate 32, which was used as a substrate for penultimate PKS module JuvEIV. Following chain elongation and keto-group reduction by JuvEIV, the heptaketide was transferred to JuvEV where it was elongated, reduced, and cyclized to efficiently generate tylactone (1). Moreover, we have demonstrated that the inherent substrate flexibility of several distinct cytochromes P450 provides access to the plethora of antibiotics produced by a single gene cluster via late-stage diversification. These unique P450 reactions enabled access to macrolides 33 and 34, compounds that have not been previously reported from M. chalcea.
These efforts reveal the versatility of polyketide biosynthetic pathway enzymes in creating complex natural products and new derivatives, and suggest that judicious implementation of biocatalytic methods offers a compelling approach for modern synthetic methodology, both as an effective catalytic tool and as a resource for structural diversification. Future efforts will be directed toward determining the limits of flexibility in these systems, using them as a platform to create additional novel functionality, and attempting to incorporate them into synthetic biology approaches to create new, bioactive molecules. Moreover, these tools will be essential for on-going structural studies of biosynthetic enzymes, particularly megasynthases, in order to fully understand their functional limits and develop rational strategies for protein engineering.
Supplementary Material
Acknowledgments
The authors thank the NIH (R35 GM118101, R01 GM076477, T32 GM008353), the National Science Foundation under the CCI Center for Selective C–H Functionalization (CHE-1205646), the University of Michigan Rackham Predoctoral Fellowship program (M.D.D. II), and the Hans W. Vahlteich Professorship (to D.H.S.) for financial support. The authors are grateful to Professor Brian Murphy for supplying Micromonospora sp. CNJ878 cell mass and Eli Lilly and Company for providing media components for S. fradiae fermentation. We thank Dr. Ashootosh Tripathi for assistance with HPLC separations, the University of Michigan Sequencing Core for acquisition of all Sanger, Illumina, and SMRT data, the University of Michigan Center for Chemical Genomics for assistance preparing antimicrobial assay plates, and the laboratory of Professor Pavel Nagorny for the use of their polarimeter.
Footnotes
Supporting Information: Synthetic experimental; M. chalcea gDNA isolation, fosmid library preparation, screening and sequencing methodology, and juv cluster annotation; biochemistry experimental for Juv, Tyl, and Myc proteins; antimicrobial assay protocol and full inhibition table; characterization data and spectra.
References
- 1.Muxfeldt H, Shrader S, Hansen P, Brockmann H. J Am Chem Soc. 1968;90:4748. doi: 10.1021/ja01019a055. [DOI] [PubMed] [Google Scholar]
- 2.Flynn EH, Sigal MV, Jr, Wiley PF, Gerzon K. J Am Chem Soc. 1954;76:3121. [Google Scholar]
- 3.Hamill RL, Haney ME, Jr, Stamper M, Wiley PF. Antibiot Chemother (Washington, D C) 1961;11:328. [PubMed] [Google Scholar]
- 4.McGuire JM, Boniece WS, Higgens CE, Hoehn MM, Stark WM, Westhead J, Wolfe RN. Antibiot Chemother (Washington, D C) 1961;11:320. [Google Scholar]
- 5.Henninger TC. Expert Opin Ther Pat. 2003;13:787. [Google Scholar]
- 6.Pletz J, Breinbauer R. From Biosynthesis to Total Synthesis. John Wiley & Sons, Inc; 2016. p. 519. [Google Scholar]
- 7.Kinoshita K, Williard PG, Khosla C, Cane DE. J Am Chem Soc. 2001;123:2495. doi: 10.1021/ja004139l. [DOI] [PubMed] [Google Scholar]
- 8.Harvey CJB, Puglisi JD, Pande VS, Cane DE, Khosla C. J Am Chem Soc. 2012;134:12259. doi: 10.1021/ja304682q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan ARH, Hakansson K, Sherman DH, Smith JL, Skiniotis G. Nature (London, U K) 2014;510:512. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whicher JR, Dutta S, Hansen DA, Hale WA, Chemler JA, Dosey AM, Narayan ARH, Hakansson K, Sherman DH, Smith JL, Skiniotis G. Nature (London, U K) 2014;510:560. doi: 10.1038/nature13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keatinge-Clay AT. Nat Prod Rep. 2012;29:1050. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 12.Borisova SA, Zhao L, Sherman DH, Liu H-w. Org Lett. 1999;1:133. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- 13.DeMars MD, Sheng F, Park SR, Lowell AN, Podust LM, Sherman DH. ACS Chem Biol. 2016;11:2642. doi: 10.1021/acschembio.6b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Chaulagain MR, Knauff AR, Podust LM, Montgomery J, Sherman DH. Proc Natl Acad Sci U S A. 2009;106:18463. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzai Y, Tsukada S-i, Sakai A, Masuda R, Harada C, Domeki A, Li S, Kinoshita K, Sherman DH, Kato F. Antimicrob Agents Chemother. 2012;56:3648. doi: 10.1128/AAC.06063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negretti S, Narayan ARH, Chiou KC, Kells PM, Stachowski JL, Hansen DA, Podust LM, Montgomery J, Sherman DH. J Am Chem Soc. 2014;136:4901. doi: 10.1021/ja5016052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Anzai Y, Kinoshita K, Kato F, Sherman DH. ChemBioChem. 2009;10:1297. doi: 10.1002/cbic.200900088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard SM, Akey DL, Tripathi A, Park SR, Konwerski JR, Anzai Y, Li S, Kato F, Sherman DH, Smith JL. ACS Chem Biol. 2015;10:1340. doi: 10.1021/cb5009348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aldrich CC, Beck BJ, Fecik RA, Sherman DH. J Am Chem Soc. 2005;127:8441. doi: 10.1021/ja042592h. [DOI] [PubMed] [Google Scholar]
- 20.Mortison JD, Kittendorf JD, Sherman DH. J Am Chem Soc. 2009;131:15784. doi: 10.1021/ja9060596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen DA, Rath CM, Eisman EB, Narayan ARH, Kittendorf JD, Mortison JD, Yoon YJ, Sherman DH. J Am Chem Soc. 2013;135:11232. doi: 10.1021/ja404134f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltz RH, Seno ET. Annu Rev Microbiol. 1988;42:547. doi: 10.1146/annurev.mi.42.100188.002555. [DOI] [PubMed] [Google Scholar]
- 23.Cundliffe E. Biosynthesis of the macrolide antibiotic, tylosin. Birkhaeuser Verlag; 2002. p. 177. [Google Scholar]
- 24.Buchholz TJ, Geders TW, Bartley FE, Reynolds KA, Smith JL, Sherman DH. ACS Chem Biol. 2009;4:41. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whicher JR, Smaga SS, Hansen DA, Brown WC, Gerwick WH, Sherman DH, Smith JL. Chem Biol (Oxford, U K) 2013;20:1340. doi: 10.1016/j.chembiol.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishi T, Harada S, Yamana H, Miyake A. J Antibiot. 1976;29:1171. doi: 10.7164/antibiotics.29.1171. [DOI] [PubMed] [Google Scholar]
- 27.Kinumaki A, Harada K, Suzuki T, Suzuki M, Okuda T. J Antibiot. 1977;30:450. doi: 10.7164/antibiotics.30.450. [DOI] [PubMed] [Google Scholar]
- 28.Hatano K, Higashide E, Shibata M. J Antibiot. 1976;29:1163. doi: 10.7164/antibiotics.29.1163. [DOI] [PubMed] [Google Scholar]
- 29.Furumai T, Maezawa I, Matsuzawa N, Yano S, Yamaguchi T, Takeda K, Okuda T. J Antibiot. 1977;30:443. doi: 10.7164/antibiotics.30.443. [DOI] [PubMed] [Google Scholar]
- 30.Iizaka Y, Higashi N, Ishida M, Oiwa R, Ichikawa Y, Takeda M, Anzai Y, Kato F. Antimicrob Agents Chemother. 2013;57:1529. doi: 10.1128/AAC.02092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiers WD, Dodge GJ, Li Y, Smith JL, Fecik RA, Aldrich CC. Chem Sci. 2015;6:5027. doi: 10.1039/c5sc01505g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimann H, Jaret RS. J Chem Soc, Chem Commun. 1972:1270. [Google Scholar]
- 33.Gao X, Townsend IA, Krische MJ. J Org Chem. 2011;76:2350. doi: 10.1021/jo200068q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlessinger RH, Poss MA, Richardson S. J Am Chem Soc. 1986;108:3112. [Google Scholar]
- 35.Nicolaou KC, Sun YP, Guduru R, Banerji B, Chen DYK. J Am Chem Soc. 2008;130:3633. doi: 10.1021/ja710485n. [DOI] [PubMed] [Google Scholar]
- 36.Shirokawa S-i, Shinoyama M, Ooi I, Hosokawa S, Nakazaki A, Kobayashi S. Org Lett. 2007;9:849. doi: 10.1021/ol0630191. [DOI] [PubMed] [Google Scholar]
- 37.Guo X, Liu T, Valenzano CR, Deng Z, Cane DE. J Am Chem Soc. 2010;132:14694. doi: 10.1021/ja1073432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dess DB, Martin JC. J Org Chem. 1983;48:4155. [Google Scholar]
- 39.Boeckman RK, Miller JR. Org Lett. 2009;11:4544. doi: 10.1021/ol9017479. [DOI] [PubMed] [Google Scholar]
- 40.Sefkow M, Neidlein A, Sommerfeld T, Sternfeld F, Maestro MA, Seebach D. Liebigs Ann Chem. 1994:719. [Google Scholar]
- 41.Ager DJ, Allen DR, Schaad DR. Synthesis. 1996:1283. [Google Scholar]
- 42.Myers AG, Yang BH, Chen H, McKinstry L, Kopecky DJ, Gleason JL. J Am Chem Soc. 1997;119:6496. [Google Scholar]
- 43.Carlsen PHJ, Katsuki T, Martin VS, Sharpless KB. J Org Chem. 1981;46:3936. [Google Scholar]
- 44.Roy S, Spilling CD. Org Lett. 2010;12:5326. doi: 10.1021/ol102345v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlson S, Marler L, Nam SJ, Santarsiero BD, Pezzuto JM, Murphy BT. Mar Drugs. 2013;11:1152. doi: 10.3390/md11041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Podust LM, Sherman DH. J Am Chem Soc. 2007;129:12940. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonnett SA, Rath CM, Shareef AR, Joels JR, Chemler JA, Hakansson K, Reynolds K, Sherman DH. Chem Biol (Cambridge, MA, U S) 2011;18:1075. doi: 10.1016/j.chembiol.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong CH, Whitesides GM. J Am Chem Soc. 1981;103:4890. [Google Scholar]
- 49.Oh HS, Kang HY. J Org Chem. 2012;77:1125. doi: 10.1021/jo201158q. [DOI] [PubMed] [Google Scholar]
- 50.Oh HS, Xuan R, Kang HY. Org Biomol Chem. 2009;7:4458. doi: 10.1039/b911200f. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto T, Maeta H, Suzuki K, Tsuchihashi G. Tetrahedron Lett. 1988;29:3575. [Google Scholar]
- 52.Suzuki K, Maeta H, Matsumoto T, Tsuchihashi G. Tetrahedron Lett. 1988;29:3571. [Google Scholar]
- 53.Still WC, Kahn M, Mitra A. J Org Chem. 1978;43:2923. [Google Scholar]
- 54.Hoover JM, Stahl SS. J Am Chem Soc. 2011;133:16901. doi: 10.1021/ja206230h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoover JM, Steves JE, Stahl SS. Nat Protoc. 2012;7:1161. doi: 10.1038/nprot.2012.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadakane N, Tanaka Y, Omura S. J Antibiot. 1983;36:921. doi: 10.7164/antibiotics.36.921. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka A, Tsuchiya T, Umezawa S, Umezawa H. J Antibiot (Tokyo) 1981;34:1374. doi: 10.7164/antibiotics.34.1374. [DOI] [PubMed] [Google Scholar]
- 58.Imai H, Suzuki K, Morioka M, Sasaki T, Tanaka K, Kadota S, Iwanami M, Saito T. J Antibiot. 1989;42:1000. doi: 10.7164/antibiotics.42.1000. [DOI] [PubMed] [Google Scholar]
- 59.Wagman GH, Waitz JA, Marquez J, Murawski A, Oden EM, Testa RT, Weinstein MJ. J Antibiot (Tokyo) 1972;25:641. doi: 10.7164/antibiotics.25.641. [DOI] [PubMed] [Google Scholar]
- 60.Werner JJ, Chintapalli M, Lundeen RA, Wammer KH, Arnold WA, McNeill K. J Agric Food Chem. 2007;55:7062. doi: 10.1021/jf070101h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.