Abstract
A method to genetically program mouse hematopoietic stem cells to develop into functional CD8 or CD4 T cells of defined specificity in vivo is described. For this purpose, a bicistronic retroviral vector was engineered that efficiently delivers genes for both α and β chains of T cell receptor (TCR) to hematopoietic stem cells. When modified cell populations were used to reconstruct the hematopoietic lineages of recipient mice, significant percentages of antigen-specific CD8 or CD4 T cells were observed. These cells expressed normal surface markers and responded to peptide antigen stimulation by proliferation and cytokine production. Moreover, they could mature into memory cells after peptide stimulation. Using TCRs specific for a model tumor antigen, we found that the recipient mice were able to partially resist a challenge with tumor cells carrying the antigen. By combining cells modified with CD8- and CD4-specific TCRs, and boosting with dendritic cells pulsed with cognate peptides, complete suppression of tumor could be achieved and even tumors that had become established would regress and be eliminated after dendritic cell/peptide immunization. This methodology of “instructive immunotherapy” could be developed for controlling the growth of human tumors and attacking established pathogens.
Although the immune system handles most pathogens well, it does a poor job of suppressing the growth of tumors. This phenomenon is not totally understood, but much evidence suggests that limited numbers of T cells capable of responding to tumor cells, insufficient avidity of these T cells for tumor antigens, and tolerogenic attenuation by the tumor contribute to this immunological failure (1–5). Therefore, a major goal of cancer immunotherapy has been to generate a large number of highly avid, tumor-specific T cells that can last in vivo for a long time and resist tolerization (6). Existing methods focus on reshaping the normal T cell repertoire and fall into two categories: active immunotherapy, which involves activating the effectors in the host immune system to inhibit cancer cell growth and reject tumor (e.g., cancer vaccination) (7, 8), and passive immunotherapy, which directly provides the host with effectors to react against cancer (e.g., adoptive transfer of in vitro expanded or modified anti-tumor T cells) (9–14).
We have developed a different approach to anti-tumor immunotherapy that involves programming hematopoietic stem cells (HSCs) to make T cells of a desired specificity in a mouse. Conventional T cell receptor (TCR) transgenic mice can be made via pronuclear injection of prerearranged TCR genes (15). However, this method offers no opportunity for therapeutic application in humans. In contrast, HSCs are attractive targets for TCR gene transfer because of their roles as T cell progenitors, enormous regeneration capacity, and availability for modification in humans (16). In this article we describe a method to impart the desired anti-tumor specificities to the mouse T cell repertoire by delivering tumor-specific TCR genes into HSCs, followed by adaptive transfer to generate a continuous stream of anti-tumor T cells in host mice.
Materials and Methods
Mice. C57BL/6J(B6) female mice were purchased from Charles River Breeding Laboratories. RAG1–/– mice, OT1 TCR transgenic mice, and OT2 TCR transgenic mice were purchased from The Jackson Laboratory. RAG1/OT2 Tg mice were generated by breeding OT2 Tg mice with RAG1–/– mice. All mice were housed in the California Institute of Technology animal facility in accordance with institute regulations.
MOT1 and MOT2 Retrovirus. The MOT1 and MOT2 constructs were generated from the MIG retrovirus (17). OT1 and OT2 TCR cDNAs were kind gifts of F. R. Carbone (University of Melbourne, Melbourne) and W. R. Heath (The Walter and Eliza Hall Institute of Medical Research, Melbourne). Retroviruses were made in HEK293.T cells as described (17).
HSC Isolation, Infection, and Transfer. B6 or RAG1–/– female mice 6–8 weeks old were treated with 250 μg of 5-fluorouracil per gram of body weight (Sigma). Five days later, bone marrow (BM) cells were harvested and cultured for 4 days in RPMI medium 1640 containing 10% FBS with 20 ng/ml recombinant murine IL-3, 50 ng/ml recombinant murine IL-6, and 50 ng/ml recombinant murine stem cell factor (all from Biosource International, Camarillo, CA). On days 2 and 3, the cells were spin-infected with MOT1 or MOT2 retroviruses supplemented with 8 μg/ml polybrene for 90 min at 770 × g at 30°C. On day 4 of culture, BM cells were collected and transferred by tail vein injection into B6 female hosts or RAG1–/– female hosts that had received 1,200 or 360 rads of whole-body radiation, respectively. Each host received 2–3 × 106 infected BM cells. BM recipient mice were maintained on the mixed antibiotic sulfmethoxazole and trimethoprim oral suspension (HiTech Pharmacal, Amityville, NY) in a sterile environment for 6–8 weeks until analysis or usage for further experiments.
In Vitro T Cell Stimulation and Functional Assays. For antigen dosage response, cells were cultured at 2 × 105 cells per well in T cell culture medium containing ovalbumin (OVA)p257–269 (designated as OVAp1) at 0–1 μg/ml or OVAp329–337 (designated as OVAp2) at 0–10 μg/ml. Three days later, culture supernatants were collected and assayed for IL-2, IL-4, or IFN-γ production by ELISA, and proliferation was assessed by [3H]thymidine incorporation (17). For time-course response, cells were stimulated with 0.1 μg/ml OVAp1 or 1 μg/ml OVAp2, and the culture supernatants were collected and assayed for IL-2, IL-4, or IFN-γ production by ELISA on days 1.5, 2.5, and 3.5. In cytokine proliferation response, cells were cultured with 10 ng/ml recombinant murine IL-2, 10 ng/ml IL-4, or 10 ng/ml recombinant murine IL-15 (all from BioSource International) for 4 days in the absence of antigen, and proliferation was assessed by [3H]thymidine incorporation.
Antibodies and FACS Analysis. Fluorochrome-conjugated antibodies specific for mouse CD4, CD8, CD25, CD69, CD62L, CD44, TCRVα2, and TCRVβ5.1,5.2 were purchased from BD Pharmingen. Surface staining was performed by blocking with anti-CD16/CD32 (mouse Fc receptor, BD Pharmingen) followed by staining with fluorochrome-conjugated antibodies. Intracellular staining of TCR was preformed by using the Cytofix/Cytoperm Kit from BD Pharmingen. A FACScan flow cytometer was used for detailed analysis.
Tumor Challenge of Mice. The tumor cell lines EL.4 (C57BL/6, H-2b, thymoma) and E.G7 (EL.4 cells transfected with the chicken OVA cDNA) (18) were used for tumor challenge of mice. A total of 5 × 106 EL.4 or E.G7 cells were injected s.c. into the left flank of the mice. Tumor size was measured every other day by using fine calipers (Manostat, Merenschwand, Switzerland) and is shown as the product of the two largest perpendicular diameters a × b (mm2). Mice were killed when the tumors reached 400 mm2.
Dendritic Cell (DC) Generation, Antigen Pulsing, and Mouse Immunization. DCs were generated from BM cultures as described by Lutz et al. (19), with some minor modifications. Briefly, BM cells were harvested from B6 female mice (6–8 weeks old) and cultured in 10-cm diameter Petri dishes at 2 × 106 cells per dish in medium containing granulocyte–macrophage colony-stimulating factor, with fresh medium feeding on days 3, 6, and 8. On day 9, nonadherent cells were collected and plated into new 10-cm diameter Petri dishes at 4–6 × 106 cells per dish in the presence of 1 μg/ml LPS (Sigma). On day 10, nonadherent cells (usually >80% are mature DCs) were collected and washed once with Iscove's modified Dulbecco's media/50 mM 2-mercaptoethanol and resuspended in 0.8 ml of the same medium containing 100 μg of OVAp1 (or 100 μg of OVAp1 plus 100 μg of OVAp2). The cells were then incubated at 37°C for 3 h with gentle shaking every 30 min. Three hours later, the OVAp1 or OVAp1+2-loaded DCs were washed twice with PBS and used to immunize mice by tail vein injection. Each mouse received ≈0.5 × 106 OVAp-loaded DCs.
Results
Tumor Model. We chose chicken OVA as the model antigen for our study. OVA is the well characterized target antigen for both a CD8 TCR, OT1 (recognizing OVAp257–269, designated as OVAp1) (20), and a CD4 TCR, OT2 (recognizing OVAp329–337, designated as OVAp2) (21), offering us the opportunity to study both the CD8 cytotoxic and CD4 helper arms of antigen-specific T cell immunity.
Construction of Retroviral Vector to Deliver TCR Genes into HSCs. We used a mouse stem cell virus-based retroviral vector (17) to deliver TCR genes into HSCs. Coexpression of the two TCR subunits from the same vector has proved critical because no antigen-specific T cells were detected when we used a two-vector system reported previously (17) to infect WT HSCs (data not shown). We achieved the coexpression by linking cDNAs encoding TCR α and β chains with an internal ribosome entry site and inserting this into the retroviral vector under control of the viral LTR promoter (Fig. 1a). The vectors expressing OT1 or OT2 TCR were designated MOT1 or MOT2 (Fig. 1a). In addition to using HSCs as the target for gene transfer, this approach has other features that facilitate clinical application: single-vector gene delivery, small genetic elements (TCR α and β cDNAs), and a convenient viral promoter to drive the expression of transgenes.
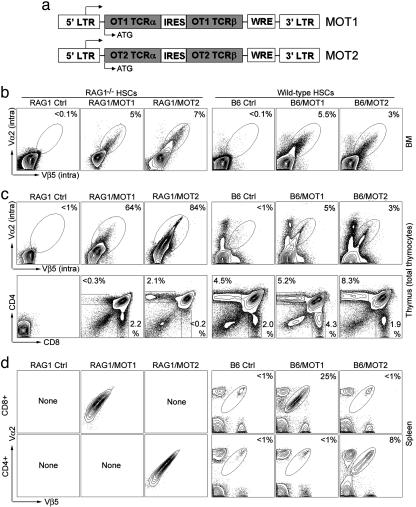
Fig. 1.
Imparting the desired CD8 cytotoxic or CD4 helper T cell specificity to the mouse T cell repertoire by genetic modification of HSCs. HSCs from RAG1–/– or B6 mice were infected with MOT1 or MOT2 retroviruses and transferred into either RAG1–/– (denoted as RAG1/MOT1 or RAG1/MOT2) or B6 (denoted as B6/MOT1 or B6/MOT2) recipient mice, respectively. RAG1–/– (denoted as RAG1) or B6 mice were included as controls. OT1 and OT2 TCRs were detected by costaining of TCR Vα2 and Vβ5. (a) Schematic representation of the MOT1 and MOT2 retrovirus constructs. IRES, internal ribosomal entry site; WRE, woodchuck responsive element. (b) Expression of OT1 or OT2 TCRs in BM detected by intracellular staining. (c) Thymic development of OT1 or OT2 T cells. (Upper) Thymocyte expression of OT1 or OT2 TCRs was detected by intracellular staining. (Lower) Distribution of developmental markers CD4 and CD8 is shown. (d) Detection of mature OT1 CD8 or OT2 CD4 T cells in periphery.
Imparting the Desired CD8 or CD4 T Cell Specificity to the Mouse T Cell Repertoire by Genetic Modification of HSCs. In our first step to test MOT1 and MOT2, we found that both retroviruses efficiently mediated functional TCR expression upon infecting peripheral mouse T cells (Supporting Methods and Fig. 5, which are published as supporting information on the PNAS web site). We then tested whether MOT1- and MOT2-mediated expression of TCRs could program WT HSCs to develop into OT1 and OT2 T cells in vivo. RAG1–/– HSCs were included as a control. B6 WT and RAG1–/– HSCs were infected with MOT1 or MOT2 viruses and then transferred into WT mice (denoted B6/MOT1 or B6/MOT2) or RAG1–/– recipients (denoted RAG1/MOT1 or RAG1/MOT2). The recipients were allowed to reconstitute their T cell compartments for 7 weeks before analysis. Some 3–7% of the total BM cells were transduced (Fig. 1b), including long-term HSCs that expressed c-kit and Sca-1 (16) (data not shown). The percentage of transduced HSCs remained constant to 8-month posttransfer and persisted through a secondary BM transfer (data not shown), indicating that they maintained their stem cell features of longevity and self-renewal. Therefore, this genetic method is sufficiently robust to ensure the maintenance of a transduced HSC population for the lifetime of the host.
We then analyzed T cell development in thymus. Owing to a lack of endogenous TCRs, unmodified RAG1–/– thymocytes cannot progress to the CD4+CD8+ double-positive (DP) stage (22). As shown in Fig. 1c, thymocytes from RAG1/MOT1 and RAG1/MOT2 mice expressed transgenic TCRs and gained the ability to develop into DP cells, and further into either CD8 or CD4 single-positive T cells. This system displayed a high specificity in the guidance of T cell development to the expected fate, which was confirmed by the observation that in the periphery there were only OT1 monospecific CD8 T cells in RAG1/MOT1 mice and only OT2 monospecific CD4 T cells in RAG1/MOT2 mice (Fig. 1d).
In the thymi of B6/MOT1 and B6/MOT2 mice, we detected thymocytes expressing transgenic TCRs and observed an augmentation of the CD8 or CD4 single-positive T cell compartment in comparison with WT animals (Fig. 1c). Further analysis of peripheral T cells showed that in a representative experiment 25% of the total peripheral CD8 T cells in B6/MOT1 expressed OT1 TCR and 8% of the total peripheral CD4 T cells in B6/MOT2 mice expressed OT2 TCR, with no leakage into the other T cell compartment (Fig. 1d). In numerous experiments, we achieved an average of ≈20% (ranging from 15% to 30%) of the total peripheral CD8 T cells in B6/MOT1 mice carrying the OT1 TCR specificity, and an average of ≈8% (ranging from 5% to 10%) of the total peripheral CD4 T cells carrying the OT2 TCR specificity. This high percentage of antigen-specific T cells remained constant for 8 months posttransfer. Therefore, retrovirus-mediated expression of TCR cDNAs in WT HSCs can stably generate a significant population of T cells with the desired features and specificity. In natural conditions, this percentage can be achieved only by clonal expansion of a few high-affinity T cell clones after strong immune stimulation (23).
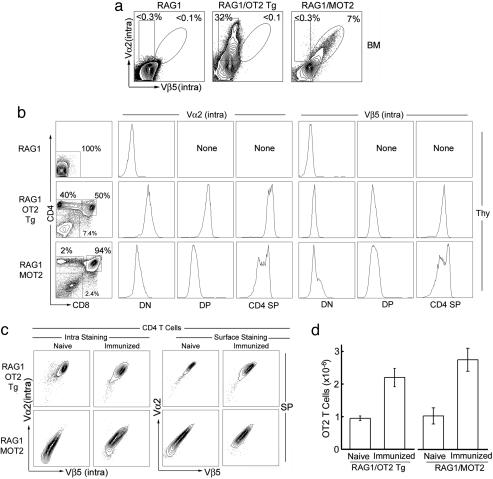
Comparison of the Efficacy of Generating OT2 T Cells in RAG1/MOT2 Mice and the Conventional RAG1/OT2 Tg Mice. To evaluate the efficacy of our method for instructing the host to generate antigen-specific T cells in vivo, we performed a detailed comparison between RAG1/MOT2 mice and conventional RAG1/OT2 Tg mice (Fig. 2). The RAG1–/– background offered a clean system for our intended study, because of the lack of endogenous TCR expression (22). Surprisingly, TCR α and β chain expression patterns in the thymocytes of RAG1/MOT2 mice closely resembled the β chain expression in RAG1/OT2 Tg mice that is under control of the endogenous TCR promoter and enhancers (21) (Fig. 2b). The mature OT2 T cells recovered from the periphery of the two kinds of mice were comparable, although there was more heterogeneity of TCR expression in the OT2 T cells from RAG1/MOT2 mice (Fig. 2c). A similar magnitude of antigenic response was observed when we immunized the animals with OVAp2 antigen (Fig. 2d).
Fig. 2.
Comparison of the efficacy of generating OT2 T cells in RAG1/MOT2 mice and the conventional RAG1/OT2 Tg mice. OT2 TCRs were detected by costaining of TCR Vα2 and Vβ5. (a) OT2 TCR expression in BM as measured by intracellular staining. (b) OT2 TCR expression and T cell development in thymus (Thy). T cell development was assessed by distribution of developmental markers CD4 and CD8. OT2 TCR expression at each development stage is shown. DN, double negative; DP, double positive; SP, single positive. (c) TCR expression in peripheral OT2 T cells. Mice were immunized with OVAp2 antigen and complete Freund's adjuvant for 6 days. Both intracellular and surface expressions of OT2 TCR was measured. (d) Total OT2 T cells in unchallenged (naïve) and immunized mice.
Characterization of the Antigen-Specific CD8 and CD4 T Cells Generated by Genetic Programming of WT HSCs. This methodology of programming of HSCs could, with high efficacy, continuously generate in the host a large population of T cells directed against a desired antigen. We then focused our attention on the functionality of the T cells generated by this method. As shown in Fig. 3a, OT1 T cells harvested from B6/MOT1 mice displayed the canonical naïve phenotype of CD8 T cells: CD25–CD69–CD62LhighCD44low. Upon stimulation with OVAp1 in vitro, these OT1 T cells responded vigorously as measured by proliferation and IFN-γ production, with a magnitude comparable to the response of transgenic OT1 T cells obtained from conventional OT1 TCR transgenic mice (20) (Fig. 3b). FACS analysis showed that the activated OT1 T cells exhibited the typical effector cytotoxic T cells phenotype: CD25+CD69+CD62LlowCD44high (Fig. 3a).
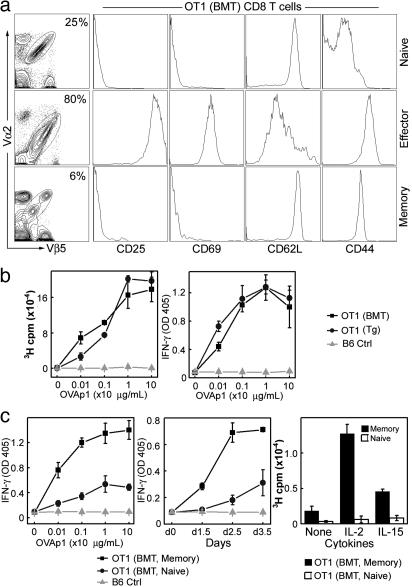
Fig. 3.
Characterization of the OT1 CD8 T cells generated by genetic programming of WT HSCs. OT1 T cells harvested from B6/MOT1 mice [denoted as OT1(BMT), where BMT is BM transfer] 8 weeks after HSCs transfer were considered to be naïve. They were stimulated with OVAp1 in vitro for 3 days to generate effector OT1 T cells, which were then transferred into host mice for memory study. (a) Patterns of surface activation markers on OT1(BMT) T cells at the naïve, effector, or memory stages. (b) Functional analysis of the naïve OT1(BMT) T cells (▪). Proliferation and IFN-γ production in response to OVAp1 stimulation are shown. The responses were compared with OT1 T cells from the conventional OT1 TCR transgenic mice, denoted as OT1(Tg) (•). B6 spleen cells were included as negative control (B6 Ctrl, ▴). Equal numbers of OT1(BMT) and OT1(Tg) T cells were used in all of the experiments. (c) Functional analysis of memory OT1(BMT) T cells [denoted as OT1(BMT, Memory), ▪]. Dosage response (Left) and time-course response (Center) to OVAp1 stimulation as measured by IFN-γ production and proliferation response to cytokine stimulation (Right) are shown. The responses were compared with those of the naïve OT1(BMT) T cells [denoted as OT1(BMT, Naïve), •). B6 spleen cells were included as a negative control (B6 Ctrl, ▴). Equal numbers of OT1(BMT, Memory) and OT1(BMT, Naive) T cells were used in all of the experiments.
We then tested the ability of these OT1 T cells to generate and maintain CD8 T cell memory by adoptive transfer of in vitro-activated OT1 cytotoxic T cell to host mice; we used this approach because continuous generation of naïve OT1 T cells makes it difficult to directly analyze memory formation in B6/MOT1 mice. Sixteen weeks after transfer, long-lived OT1 T cells were detected in the hosts, which displayed the memory phenotype (CD25–CD69–CD62LhighCD44high) (Fig. 3a). These cells showed a stronger and faster response to antigen stimulation when compared with naïve OT1 T cells (Fig. 3c). Furthermore, they proliferated upon cytokine stimulation, which is a unique feature of memory T cells (24) (Fig. 3c).
Similar results were obtained for OT2 CD4 T cells (Fig. 6, which is published as supporting information on the PNAS web site). Thus, OT1 and OT2 T cells generated by our method are fully functional and normal in all aspects we evaluated. In particular, the ability of these T cells to generate and maintain long-term memory makes the method attractive for immunotherapy.
In Vivo Constitution of Anti-Tumor T Cell Immunity. The therapeutic potential of this method was tested in the E.G7 mouse tumor model (18). OVA serves as the tumor antigen in this model, allowing us to study either anti-tumor cytotoxic T cell immunity in B6/MOT1 mice, anti-tumor helper T cell immunity in B6/MOT2 mice, or the combination of both arms in B6/MOT1+MOT2 mice (B6 mice receiving both MOT1- and MOT2-transduced WT HSCs) (Fig. 4a) (25). Suppression of syngenic tumor growth was studied by using the following protocol: B6/MOT1, B6/MOT2, and B6/MOT1+MOT2 mice were allowed to reconstitute their T cell compartment over 8–10 weeks after receiving retroviral transduced HSCs. E.G7 tumor cells were then injected s.c., and EL.4 tumor cells were used as a control (18). To evaluate the effects of peptide immunization, half of the experimental groups were immunized by using DCs loaded with OVAp1 (DC/OVAp1) 4 days after tumor injection. Four mice were used in each treatment group, and the experiments were performed three times.
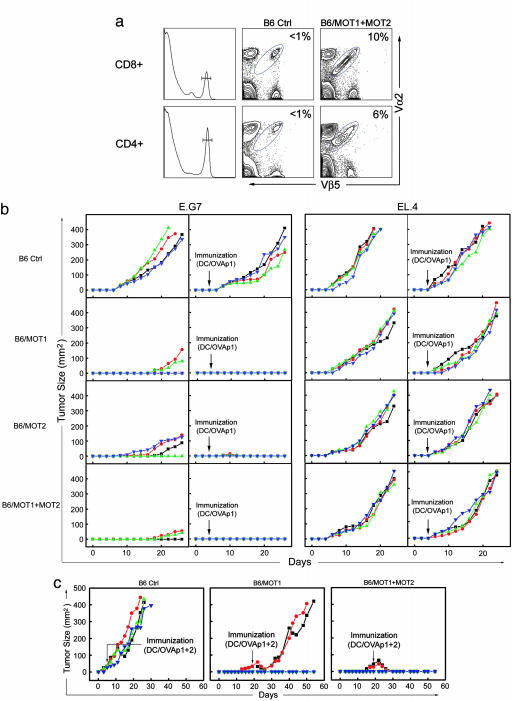
Fig. 4.
In vivo constitution of anti-tumor T cell immunity. E.G7 mouse tumor model was used. Tumor size is shown as the product of the two largest perpendicular diameters a × b (mm2). Each line represents one mouse. Four mice were included in each group, and experiments were performed three times. Results from one representative experiment are shown. (a) Constitution in mouse of both arms of anti-tumor T cell immunity. FACS staining showed the detection of mature OT1 CD8 and OT2 CD4 T cells in the spleen of B6/MOT1+MOT2 mice (B6 mice receiving both MOT1- and MOT2-transduced WT HSCs). (b) Suppression of syngenic tumor growth by in vivo constitution of anti-tumor T cell immunity. EL.4 and its OVA-expressing derivative, E.G7, were the two mouse tumor types examined. DC/OVAp1, DCs loaded with OVAp1. (c) Eradication of established solid tumors by in vivo constitution of both arms of anti-tumor T cell immunity. E.G7 tumor cells were used. DC/OVAp1+2, DCs loaded with both OVAp1 and OVAp2.
The results of one representative experiment are shown in Fig. 4b. Compared with the B6 control, significant tumor suppression was observed in B6/MOT1 mice, with complete suppression in 50% of the mice. No recurrence of tumors was observed as long as the experiments ran (up to 200 days). For the other 50%, tumor growth was suppressed up to 18 days but eventually progressed. We recovered E.G7 tumor cells from these mice and found they were still recognized by naïve OT1 T cells (Fig. 7, which is published as supporting information on the PNAS web site), excluding the possibility of “epitope escape” by OVA antigen mutation. When we analyzed the OT1 cells recovered from the tumor-bearing mice, we found they could no longer respond to OVAp1 antigen stimulation in vitro, as measured by IFN-γ production (Supporting Methods and Fig. 8 a and b, which is published as supporting information on the PNAS web site). These anergic OT1 T cells displayed a surface phenotype that resembled naïve T cells but with higher CD44 expression (Fig. 8c), indicating that they had encountered tumor antigen. Interestingly, total T cells still responded to anti-CD3 stimulation (data not shown), suggesting the existence of an active antigen-specific tumor tolerogenic mechanism (26). Similar results were obtained for B6/MOT2 tumor-bearing mice (Fig. 9, which is published as supporting information on the PNAS web site), suggesting that tolerogenic mechanisms act on both anti-tumor cytotoxic and helper T cells.
We tested whether DC-based peptide immunization could enhance OT1 T cell function and thus improve their anti-tumor activity (27). As hoped, this strategy thoroughly suppressed tumor growth in all of the B6/MOT1 mice (Fig. 4b).
Interestingly, we also observed significant tumor suppression in B6/MOT2 mice (Fig. 4b) despite the fact that unlike OT1 T cells OT2 T cells cannot directly recognize E.G7 tumor cells because of their lack of MHC II expression (Fig. 10, which is published as supporting information on the PNAS web site). The possible mechanism in this case would be cross-presentation of tumor antigens by the host-derived antigen-presenting cells (25). Even more strikingly, when we immunized B6/MOT2 mice with DC/OVAp1 complete suppression of tumor growth was observed in all of the mice (some tumors grew up to a barely detectable size but soon regressed) (Fig. 4b). This series of experiments indicated that the CD4 anti-tumor T cells could provide essential help for the endogenous cytotoxic T cell to mount an anti-tumor response, regardless of whether they can recognize the tumor directly or not.
As expected, we observed a combinatorial tumor suppression effect in B6/MOT1+MOT2 mice, as exhibited by a slower tumor growth in mice even when the tumor broke through (Fig. 4b). DC/OVAp1 immunization totally suppressed tumor growth in all of the mice (Fig. 4b).
Importantly, peptide immunization did not work for B6 control mice, and in all experimental groups challenged with control tumor EL.4, the tumors grew up at a similar rate regardless of immunization, confirming that suppression of E.G7 tumor growth was tumor antigen-specific and mediated by the engineered anti-tumor CD8 and/or CD4 T cells (Fig. 4b).
We further extended the idea of combining the two arms of anti-tumor T cell immunity with the aim of eradicating an established large vascularized solid tumor. Physiologically, tumors achieve growth by initially overcoming the host's anti-tumor immuno-surveillance and then constantly tolerizing the anti-tumor T cells in the host (5). We devised a protocol to mimic these processes: mice in which E.G7 tumors grew despite the presence of anti-tumor CD8 (B6/MOT1) or both anti-tumor CD8 and CD4 T cells (B6/MOT1+MOT2) were immunized with one dose of DCs loaded with both OVAp1 and OVAp2 (denoted as DC/OVAp1+2), at the time when their tumors reached 30 mm2 [18 days after tumor challenge, tumors extant for this duration and with this size are considered to be long-term established tumors (26)]. As depicted in Fig. 4c, the tumors in B6/MOT1 mice receiving immunization were suppressed for 10 days but then grew, reaching 400 mm2 in ≈50 days. In contrast, tumors in B6/MOT1+MOT2 mice shrank after the immunization and were not longer detectable within ≈12 days; no tumor recurrence was observed for as long as the experiments ran (>200 days). These results demonstrated that established solid vascularized tumors could be eradicated by providing both the CD8 and CD4 arms of anti-tumor T cell immunity combined with appropriate peptide immunization to boost the responses of both arms.
Discussion
We have developed an effective method representing a unique direction for T cell immunotherapy. It combines stem cell therapy, gene therapy, and immunotherapy to guide the host to develop, in vivo, a large population of antigen-specific T cells and thus could be called instructive immunotherapy. Previous T cell immunotherapy methods focused on manipulating the existing T cell repertoire and fell into two categories: active expansion of the antigen-specific T cells in vivo by immunization and passive (adoptive) transfer of in vitro expanded T cells (8, 11). These methods rely on massive expansion of the few antigen-specific T cells, which is usually a demanding task and not always successful (5, 11). Furthermore, antigen-specific T cells expanded from an individual's T cell repertoire cannot be readily transferred to others because of the risk of severe graft-versus-host disease (28). TCR gene transfer avoids these issues. Recently, several groups showed that retrovirus-mediated expression of TCR genes in peripheral T cells could endow them with the desired specificity (29, 30). However, such T cells present certain challenges: they are activated mature cells that have already expressed an endogenous TCR of the unknown specificity; their effector function may be restricted by the conditions under which they are activated in vitro; and it is unclear how long they persist in vivo (31).
Our method shows promising results in the E.G7 mouse tumor model, implying its therapeutic potential for treating cancers. The OVA tumor antigen in this model represents a category of “non-self” tumor antigen including cancer testes antigens and antigens encoded after genetic alterations or encoded by alternative/atypical transcripts (32). “Self” tumor antigens are more common (32). Despite the concern that T cells recognizing these antigens may be subjected to self-tolerance mechanisms, peripheral high-affinity T cell clones specific for such tumor antigens are observed and have proved their therapeutic potential in various animal models and clinical trials (9, 32). TCR genes of such T cell clones could be handled as we describe here to generate tumor-specific T cell immunity in vivo.
Our method can be easily extended to generate T cells targeting multiple epitopes of the antigen, thus countering “epitope escape” by antigen mutations (33). It might also be extended to engineering the B cell receptor on B cells, allowing the cells to secrete antibodies of predefined specificity. Therefore, instructive immunotherapy has the potential to establish in vivo a lifelong “complete” adaptive immunity (including CD8 cytotoxic T cell, CD4 helper T cell, and B cell arms) to target any pathogens where antigens and the cognate TCR/B cell receptors are known (6, 34). In addition, this method could be used for rapid and efficient analysis of such problems as the development and life history of antigen-specific T cells and in vivo anti-tumor T cell response and tolerance, providing a useful tool for basic T cell studies.
Supplementary Material
Acknowledgments
We thank F. R. Carbone and W. R. Heath for the OT1 and OT2 TCR cDNAs; M. Bevan (University of Washington, Seattle) for the E.G7 tumor cell line; S. Kovats (City of Hope, Duarate, CA) for cell proliferation assays; E. Santiestevan for critical reading of the manuscript; and members of the D.B. laboratory for useful discussions. This work was supported by National Institute of Health Grant R01 GM39458.
Author contributions: L.Y. and D.B. designed research; L.Y. performed research; L.Y. and D.B. analyzed data; and L.Y. and D.B. wrote the paper.
Abbreviations: HSC, hematopoietic stem cell; TCR, T cell receptor; BM, bone marrow; BMT, BM transfer; DC, dendritic cell; OVA, ovalbumin.
References
- 1.Chambers, C. A., Kuhns, M. S., Egen, J. G. & Allison, J. P. (2001) Annu. Rev. Immunol. 19, 565–594. [DOI] [PubMed] [Google Scholar]
- 2.Smyth, M. J., Godfrey, D. I. & Trapani, J. A. (2001) Nat. Immunol. 2, 293–299. [DOI] [PubMed] [Google Scholar]
- 3.Matzinger, P. (2002) Science 296, 301–305. [DOI] [PubMed] [Google Scholar]
- 4.Ochsenbein, A. F. (2002) Cancer Gene. Ther. 9, 1043–1055. [DOI] [PubMed] [Google Scholar]
- 5.Marincola, F. M., Wang, E., Herlyn, M., Seliger, B. & Ferrone, S. (2003) Trends Immunol. 24, 335–342. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg, S. A. (2001) Nature 411, 380–384. [DOI] [PubMed] [Google Scholar]
- 7.Finn, O. J. (2003) Nat. Rev. Immunol. 3, 630–641. [DOI] [PubMed] [Google Scholar]
- 8.Berzofsky, J. A., Ahlers, J. D. & Belyakov, I. M. (2001) Nat. Rev. Immunol. 1, 209–219. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, M. E., Wunderlich, J. R., Robbins, P. F., Yang, J. C., Hwu, P., Schwartzentruber, D. J., Topalian, S. L., Sherry, R., Restifo, N. P., Hubicki, A. M., et al. (2002) Science 298, 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee, C., Thompson, J. A., Byrd, D., Riddell, S. R., Roche, P., Celis, E. & Greenberg, P. D. (2002) Proc. Natl. Acad. Sci. USA 99, 16168–16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudley, M. E. & Rosenberg, S. A. (2003) Nat. Rev. Cancer 3, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, W. Y., Blattman, J. N., Dossett, M. L., Yee, C. & Greenberg, P. D. (2003) Cancer Cell 3, 431–437. [DOI] [PubMed] [Google Scholar]
- 13.Pardoll, D. M. (2002) Nat. Rev. Immunol. 2, 227–238. [DOI] [PubMed] [Google Scholar]
- 14.Sadelain, M., Riviere, I. & Brentjens, R. (2003) Nat. Rev. Cancer 3, 35–45. [DOI] [PubMed] [Google Scholar]
- 15.Vonboehmer, H. (1990) Annu. Rev. Immunol. 8, 531–556. [DOI] [PubMed] [Google Scholar]
- 16.Morrison, S. J., Uchida, N. & Weissman, I. L. (1995) Annu. Rev. Cell Dev. Biol. 11, 35–71. [DOI] [PubMed] [Google Scholar]
- 17.Yang, L., Qin, X., Baltimore, D. & Van Parijs, L. (2002) Proc. Natl. Acad. Sci. USA 99, 6204–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, M. W., Carbone, F. R. & Bevan, M. J. (1988) Cell 54, 777–785. [DOI] [PubMed] [Google Scholar]
- 19.Lutz, M. B., Kukutsch, N., Ogilvie, A. L. J., Rossner, S., Koch, F., Romani, N. & Schuler, G. (1999) J. Immunol. Methods 223, 77–92. [DOI] [PubMed] [Google Scholar]
- 20.Hogquist, K. A., Jameson, S. C., Heath, W. R., Howard, J. L., Bevan, M. J. & Carbone, F. R. (1994) Cell 76, 17–27. [DOI] [PubMed] [Google Scholar]
- 21.Barnden, M. J., Allison, J., Heath, W. R. & Carbone, F. R. (1998) Immunol. Cell Biol. 76, 34–40. [DOI] [PubMed] [Google Scholar]
- 22.Mombaerts, P., Iacomini, J., Johnson, R. S., Herrup, K., Tonegawa, S. & Papaioannou, V. E. (1992) Cell 68, 869–877. [DOI] [PubMed] [Google Scholar]
- 23.Butz, E. A. & Bevan, M. J. (1998) Immunity 8, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanchot, C., Lemonnier, F. A., Perarnau, B., Freitas, A. A. & Rocha, B. (1997) Science 276, 2057–2062. [DOI] [PubMed] [Google Scholar]
- 25.Pardoll, D. M. & Topalian, S. L. (1998) Curr. Opin. Immunol. 10, 588–594. [DOI] [PubMed] [Google Scholar]
- 26.Overwijk, W. W., Theoret, M. R., Finkelstein, S. E., Surman, D. R., de Jong, L. A., Vyth-Dreese, F. A., Dellemijn, T. A., Antony, P. A., Spiess, P. J., Palmer, D. C., et al. (2003) J. Exp. Med. 198, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerundolo, V., Hermans, I. F. & Salio, M. (2004) Nat. Immunol. 5, 7–10. [DOI] [PubMed] [Google Scholar]
- 28.Goulmy, E., Schipper, R., Pool, J., Blokland, E., Falkenburg, J. H. F., Vossen, J., Gratwohl, A., Vogelsang, G. B., vanHouwelingen, H. C. & vanRood, J. J. (1996) N. Eng. J. Med. 334, 281–285. [DOI] [PubMed] [Google Scholar]
- 29.Kessels, H., Wolkers, M. C., van den Boom, M. D., van der Valk, M. A. & Schumacher, T. N. M. (2001) Nat. Immunol. 2, 957–961. [DOI] [PubMed] [Google Scholar]
- 30.Stanislawski, T., Voss, R. H., Lotz, C., Sadovnikova, E., Willemsen, R. A., Kuball, J., Ruppert, T., Bolhuis, R. L. H., Melief, C. J., Huber, C., et al. (2001) Nat. Immunol. 2, 962–970. [DOI] [PubMed] [Google Scholar]
- 31.Opferman, J. T., Ober, B. T. & Ashton-Rickardt, P. G. (1999) Science 283, 1745–1748. [DOI] [PubMed] [Google Scholar]
- 32.Houghton, A. N., Gold, J. S. & Blachere, N. E. (2001) Curr. Opin. Immunol. 13, 134–140. [DOI] [PubMed] [Google Scholar]
- 33.Khong, H. T. & Restifo, N. P. (2002) Nat. Immunol. 3, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riddell, S. R. & Greenberg, P. D. (2000) J. Antimicrob. Chemother. 45, 35–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.