Abstract
Mutations in BRAT1, encoding BRCA1-associated ATM activator 1, are associated with a severe phenotype known as rigidity and multifocal seizure syndrome, lethal neonatal (RMFSL; OMIM # 614498), characterized by intractable seizures, hypertonia, autonomic instability, and early death. We expand the phenotypic spectrum of BRAT1 related disorders by reporting on four individuals with various BRAT1 mutations resulting in clinical severity that is either mild or moderate compared to the severe phenotype seen in RMFSL. Representing mild severity are three individuals (Patients 1–3), who are girls (including two sisters, Patients 1–2) between 4–10 years old, with subtle dysmorphisms, intellectual disability, ataxia or dyspraxia, and cerebellar atrophy on brain MRI; additionally, Patient 3 has well-controlled epilepsy and microcephaly. Representing moderate severity is a 15 month old boy (Patient 4) with severe global developmental delay, refractory epilepsy, microcephaly, spasticity, hyperkinetic movements, dysautonomia, and chronic lung disease. In contrast to RMFSL, his seizure onset occurred later at 4 months of age, and he is still alive. All four of the individuals have compound heterozygous BRAT1 mutations discovered via whole exome sequencing: c.638dupA (p.Val214Glyfs*189); c.803+1G>C (splice site mutation) in Patients 1–2; c.638dupA (p.Val214Glyfs*189); c.419T>C (p.Leu140Pro) in Patient 3; and c.171delG (p.Glu57Aspfs*7); c.419T>C (p.Leu140Pro) in Patient 4. Only the c.638dupA (p.Val214Glyfs*189) mutation has been previously reported in association with RMFSL. These patients illustrate that, compared with RMFSL, BRAT1 mutations can result in both moderately severe presentations evident by later-onset epilepsy and survival past infancy, as well as milder presentations that include intellectual disability, ataxia/dyspraxia, and cerebellar atrophy.
Keywords: BRAT1, clinical severity, RMFSL, cerebellar atrophy, intellectual disability, epilepsy
INTRODUCTION
Mutations in BRAT1 (MIM# 614506), encoding BRCA1-associated ATM activator 1, are associated with a severe phenotype known as rigidity and multifocal seizure syndrome, lethal neonatal (RMFSL; MIM# 614498), which is inherited in an autosomal recessive manner. RMFSL is characterized by microcephaly, rigidity, intractable focal seizures, apnea, and bradycardia. Death can occur in RMFSL within several months after birth due to cardiopulmonary arrest. EEG can reveal bilateral temporal and central spike activity, multifocal seizures, background slowing, and absent posterior dominant rhythm [Puffenberger et al., 2012]. Brain MRIs can be normal or reveal frontal lobe hypoplasia or global atrophy. Neuropathology may show neuronal loss and gliosis in the corticobasal region [Puffenberger et al., 2012; Saitsu et al., 2014].
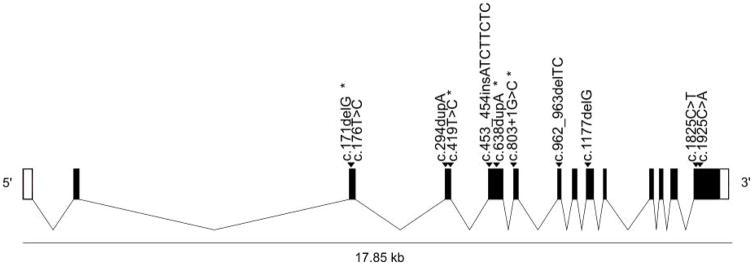
To date, several distinct homozygous and compound heterozygous mutations in BRAT1 (NM_152743.3) have been identified among 15 patients with RMFSL or an RMFSL-like phenotype (Figure 1, Table I). The homozygous mutations are as follows: c.638dupA (p.Val214Glyfs*189), identified in Amish infants from Pennsylvania, Wisconsin, and Kentucky [Puffenberger et al., 2012] and Moroccan siblings [van de Pol et al., 2015]; c.453_454insATCTTCTC (p.Leu152Ilefs*70), identified in a Mexican neonate [Saunders et al., 2012]; and c.1177delG (p.Ala393Leufs*3) [described in the publication as c.1173delG (p.Leu391fs)], found in a brother and sister sib-pair born to consanguineous parents [Straussberg et al., 2015]. Two BRAT1 mutations in the trans configuration [c.176T>C (p.Leu59Pro); c.962_963delTC (p.Leu321Profs*81)] were present in a Japanese child with a phenotype consistent with RMFSL including dysmorphic features, hypertonia, progressive microcephaly, optic atrophy, apnea, and neonatal-onset intractable seizures with burst suppression pattern on EEG (Ohtahara syndrome), who also had cerebral and cerebellar atrophy on brain MRI [Saitsu et al., 2014]. This child died at age 3 months from pneumonia. She had a sister who was similarly affected and died at 1 year of age, but from whom DNA was unavailable for confirmatory testing. Interestingly, there are two reported patients with compound heterozygous BRAT1 mutations who have an initial presentation similar to RMFSL with the exception of long term survival. One is 6 years old [c.294dupA (p.Leu99Thrfs*92); c.1925C>A (p.Ala642Glu)][Mundy et al., 2015], and the other is almost 4 years old [c.294dupA (p.Leu99Thrfs*92); c.1825C>T (p.Arg609Trp)][Hanes et al., 2015].
Figure 1.
Table I.
Characteristics of individuals with previously reported BRAT1 mutations in addition to our patients.
| Previously Reported Patients | Patients 1 and 2 | Patient 3 | Patient 4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | c.638dupA (p.Val214Glyfs*189); c.638dupA (p.Val214Glyfs*189) | c.453_454insATCTTCTC (p.Leu152Ilefs*70); c.453_454insATCTTCTC (p.Leu152Ilefs*70) | c.1177delG (p.Ala393Leufs*3)A; c.1177delG (p.Ala393Leufs*3)A | c.176T>C (p.Leu59Pro); c.962_963delTC (p.Leu321Profs*81) | c.294dupA (p.Leu99Thrfs*92); c.1925C>A (p.Ala642Glu) | c.294dupA (p.Leu99Thrfs*92); c.1825C>T (p.Arg609Trp) | c.638dupA (p.Val214Glyfs*189); c.638dupA (p.Val214Glyfs*189) | c.638dupA (p.Val214Glyfs*189); c.803+1G>C (splice site mutation) | c.638dupA (p.Val214Glyfs*189); c.419T>C (p.Leu140Pro) | c.171delG (p.Glu57Aspfs*7); c.419T>C (p.Leu140Pro) |
| Reference | [Puffenberger et al., 2012] | [Saunders et al., 2012] | [Straussberg et al., 2015] | [Saitsu et al., 2014] | [Mundy et al., 2015] | [Hanes et al., 2015] | [van de Pol et al., 2015] | |||
| Number of patients described | 5 (different sibships) | 1 (female) | 2 (sister, brother) | 2 (sisters); 1 with confirmed mutation | 1 (female) | 1 (female) | 3 (2 brothers; 1 sister); 2 with confirmed mutation | 2 (sisters) | 1 (female) | 1 (male) |
| Ethnicity | Pennsylvania, Wisconsin, and Kentucky Amish | Mexican | Arab | Japanese | Not specified | Caucasian | Moroccan | Mixed European | Mixed European | West Indian/mixed European |
| Consanguinity | No | Not reported | Yes | No | No | No | Yes | No | No | No |
| Clinical Phenotype | RMFSL | RMFSL | RMFSL | RMFSL | RMFSL | RMFSL | RMFSL | Intellectual disability, ataxia | Intellectual disability, epilepsy, rigidity, microcephaly | RMFS, not neonatal lethal |
| Brain MRI | Normal or mild frontal hypoplasia | Normal | Normal | Cerebral and cerebellar atrophy; delayed white matter myelination | Decreased myelination, thin corpus callosum, cerebellar hypoplasia, right temporal encephalomalacia | Cerebellar atrophy, brainstem atrophy, dysmyelination | Generalized atrophy over time in 2 patients, one of whom also had “hardly any myelination”; not performed in 1 patient | Cerebellar atrophy | Cerebellar atrophy, mild myelination delay | Mild diffuse cerebral atrophy |
| EEG | Background slowing; bilateral spikes | Focal epileptiform activity; multifocal sharp waves | Sharp waves; bilateral spikes | Burst-suppression | Epileptiform activity; absent posterior dominant rhythm | Background slowing | Multifocal epileptiform activity and electrographic seizures in all 3 patients, one of whom also had burst-suppression | Normal in Patient 2 (performed due to rapid mood swings, staring spells) | Spike and wave complexes; background slowing | Generalized and focal slowing; multifocal epileptiform activity; electrographic seizures |
| Age of presentation | Day 1 of life | Day 1 of life | Day 1 of life | Day 1 of life | 3 months | First month of life | First month of life in 2 patients; day 1 of life in 1 patient | Infancy (developmental delay) | 5 months (microcephaly, developmental delay) | Day 1 of life |
| Age at Death | 4 weeks (in one infant) and < 4 months in others (all from cardiopulmonary arrest) | >5 weeks (withdrawal of support) | 5 months; 6 months (both from cardiopulmonary arrest) | 21 months; 3 months (both from pneumonia) | Alive at age 6 years | Alive at age 3 years 8 months | 3 months (from respiratory failure); 17 months (from respiratory failure); 2 months (from prematurity-related complications) | Alive at ages 10 and 6 years | Alive at age 4 years 4 months | Alive at age 15 months |
Described in the publication as c.1173delG (p.Leu391fs).
In this report, we expand the clinical spectrum of BRAT1 related disorders by presenting four patients with compound heterozygous BRAT1 mutations who, in contrast to the severe phenotype of RMFSL, have a mild (in three of the patients) and moderate (in one of the patients) clinical course. The mild clinical course is characterized by intellectual disability (ID), ataxia, and cerebellar atrophy, sometimes in combination with microcephaly and epilepsy. The moderate clinical course is characterized by features similar to RMFSL but with survival past infancy, later onset epilepsy, and continued development. We propose that BRAT1 mutations confer a spectrum of disease severity ranging from mild to severe.
CLINICAL REPORT
Patient 1 and Patient 2
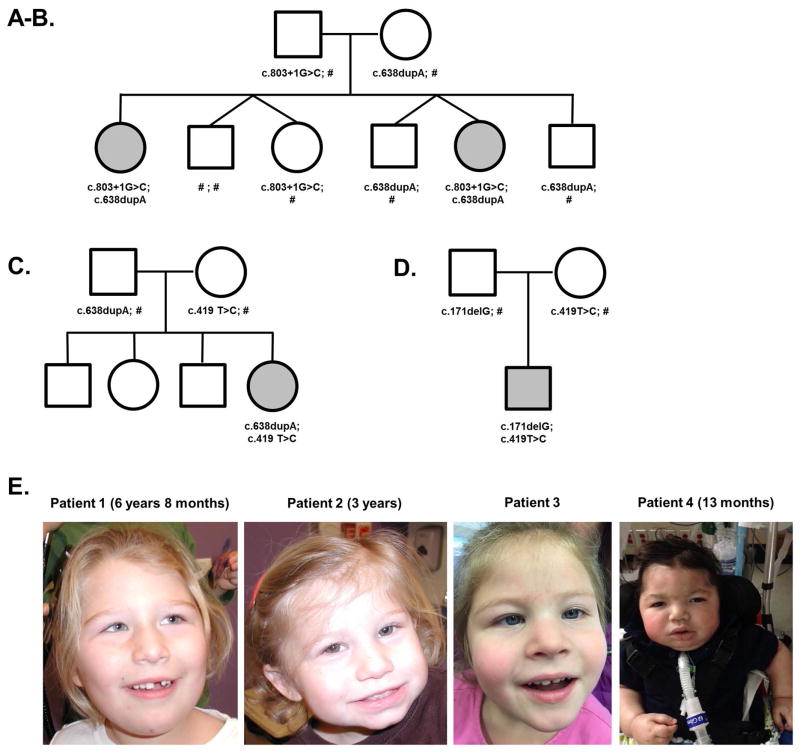
Patient 1 and Patient 2 are sisters who presented for evaluation of ataxia and developmental delay. The family history is negative for other individuals with neurodevelopmental disorders. Full siblings include an unaffected sister and three unaffected brothers as shown in Figure 2A–B.
Figure 2.
Patient 1
Patient 1 is currently 10 years old and presented initially between the ages of 1–2 years with global developmental delay. Her prenatal and neonatal course was uncomplicated. In terms of motor development, she started crawling at 16 months of age. By 2 years 3 months of age, she was able to pull to stand, cruise, and climb on to the couch; she could not walk but was able to stand with support. By 3 years of age, she could stand independently for 10 seconds. By 5 years 5 months of age, she could walk with one hand held, and 5 months later, she could walk with a walker. In terms of language and cognition, at 2 years 3 months, she was nonverbal but smiling and interactive. She communicated her needs by whining and pointing, demonstrated 10–15 signs but no words, understood “no”, and pointed to at least 8 body parts. By 3 years of age, she could say several single words that were dysarthric. By 5 years 10 months of age, she could also say all the letters of the alphabet and knew some colors. At 7 years 1 month of age, she could write out her name and draw a circle. From a behavioral standpoint, she developed disruptive behaviors since around 5 years of age, including defiance and hitting/biting others when upset. She did not have episodes concerning for seizures, so EEG was not performed.
On general examination, her growth parameters have been normal including head circumference (25th centile). Her dysmorphisms include epicanthal folds, slightly high arched palate, tented upper lip, and bilateral 5th finger clinodactyly (Figure 2E). Her cranial nerves showed no deficits apart from pendular nystagmus. She exhibited early hypotonia which improved with age. There was no evidence of rigidity. She had normal strength, reflexes, and sensation. Since 2 years 3 months of age, she has had dysmetria and truncal titubation. At 5 years 10 months of age, her gait was wide-based and ataxic.
Patient 2
Patient 2 is currently 6 years old and presented initially between the ages of 1–2 years with developmental delay after an unremarkable prenatal and neonatal course. Her motor development was as follows: at 15 months of age, she was able to cruise, and at 3 years 4 months of age, she could walk with a walker. Her language was also delayed. She could babble and say several words specifically at age 15 months. At 2 years 1 month of age, she had a 15-word vocabulary in addition to possessing some sign language. At 3 years 4 months of age, she could point to body parts, identify one color, and recite letters of the alphabet. By this point, she also had spells of extreme anger, requiring greater than expected amounts of time to settle down after becoming agitated. By 5 years 3 months of age, she had developed staring spells and rapid mood swings; three hour EEG was normal with no epileptiform discharges or lateralizing signs.
On general examination, her growth parameters have been normal, except for microcephaly (2nd centile). She has epicanthal folds, slightly high arched palate, tented upper lip, bilateral 5th finger clinodactyly, and a wide face (Figure 2E). At 4 years 3 months of age, she had developed dysarthric speech and pendular nystagmus; otherwise her cranial nerves were intact. She was diffusely hypotonic from an early age, but her tone improved in the lower extremities with time. She did not have rigidity. She had normal strength, reflexes, and sensation. She exhibited the following cerebellar findings: At 1 year 8 months of age, she had dysmetria in her upper extremities. By 2 years 1 months of age, she had developed a slight activity-induced tremor. She had poor balance but could stand on her toes. At 4 years 3 months of age, she continued to have a fine tremor and dysmetria and also exhibited truncal titubation. She could walk with support, with a tendency to toe walk.
Patient 3
Patient 3 is currently 4 years 4 months old and presented to Neurology at age 5 months of age for evaluation of microcephaly, delayed gross motor skills, and delayed visual maturation, subsequently diagnosed as cortical visual impairment. She was born at term to non-consanguineous parents, and she has 3 older healthy siblings (Figure 2C). In terms of her motor development, she rolled at 7 months, cruised at 18 months, and walked independently at 2 years 4 months. She could make consonant sounds, babble, and follow some 1-step commands, but at 3.5 years of age, she was using only 1–2 words, signing “more”, and indicating desires with gaze or by pointing with her hand. She is very happy, social, and interactive with peers and family.
At age 3 years 2 months, she developed episodes of staring unresponsively for 10 to 15 seconds with associated autonomic changes of pallor, blotchiness, and perioral cyanosis. Her EEG showed frequent 3–4 Hz generalized spike and wave complexes (without clinical correlate) that resolved together with her clinical seizures when treated with ~60 mg/kg/day of levetiracetam.
On physical exam, her height and weight have been normal while her head circumference has grown consistently close to 2nd centile. She has a flat facial profile, no true epicanthal folds, and bilateral 5th finger clinodactyly (Figure 2E). At her most recent visit at 4 years 4 months, she was very social and visually interactive; she followed simple commands; and she was able to bring a person by the hand to obtain objects she wanted. She has a persistent right esotropia, with mild amblyopia by formal ophthalmology exam treated with part time patching. She has mild optic nerve hypoplasia, with decreased visual acuity bilaterally. She had a relative paucity of facial expression/movement. Her motor exam has shown moderate appendicular rigidity, unchanged since 5 months of age, but tendon reflexes were normal. She demonstrated dyspraxia with fine motor skills but did not have true ataxia. She used a near mature pincer to grasp small objects. She had a rigid and wide-based gait with flexed arm posture and no arm swing, and she could walk well but not run.
Patient 4
Patient 4 is currently 15 months old and was first evaluated for hypertonia and mild microcephaly at birth (HC 32 cm). His parents are non-consanguineous. Pregnancy was complicated by oligohydramnios, but he was born at term. At 2.5 months, he was hospitalized due to increased work of breathing. By 1 year of age, he developed head control, could briefly sit, and started reaching intermittently. In terms of language, he smiles spontaneously and was cooing by 5 months of age. More recently, he has stopped vocalizing because of tracheostomy and continuous ventilation.
He first presented with tonic seizures at 4 months of age, and he had episodes of focal electrographic status epilepticus on EEG. His epilepsy has remained refractory with seizure semiology predominantly apneas, at times with subtle stiffening. He has required multiple anti-epileptic drugs and ketogenic diet, which triggered protein losing enteropathy and was discontinued. EEGs over time have shown seizures, generalized and focal bi-posterior quadrant slowing, and multi-focal epileptiform activity.
He has cortical visual impairment and dysphagia requiring G-tube feeding. He has significant intermittent asymptomatic bradycardia and hypothermia. Non-epileptic apnea/hypoventilation and chronic lung disease led to tracheostomy and chronic mechanical ventilation.
His growth parameters have shown progressive microcephaly (most recently −3.5 SD), normal weight, and normal height. He has borderline downslanting palpebral fissures, supraorbital fullness, slightly flattened nose, and low-set and posteriorly rotated ears with mildly cupped upper helices (Figure 2E). He has severe dry skin. He is alert at times and attempts to vocalize. He does not consistently track and has intermittent dysconjugate gaze. He has axial hypotonia and symmetric hypertonia with spasticity and variable rigidity in all extremities. Tendon reflexes are brisk and toes upgoing. He had uncoordinated hyperkinetic movements at approximately 4–6 months of age, which resolved over time. Videos have shown him sitting with minimal support.
Evaluations
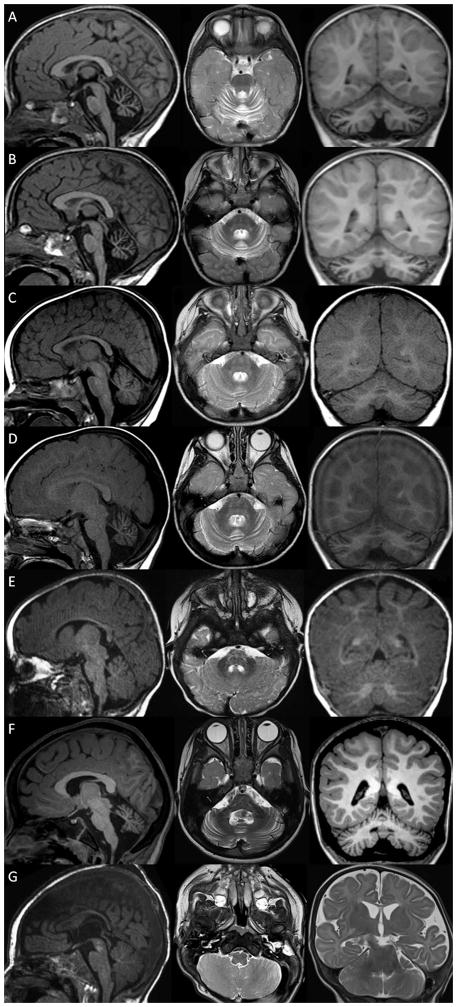
Neuroimaging in patient 1 showed prominent cerebellar folia which remained unchanged from 2 to 3 years (Figure 3A–B). Neuroimaging in patient 2 showed progressive enlargement of the cerebellar interfolial spaces from 8 months to 1 year and 6 months compatible with cerebellar atrophy (Figure 3C–D). Neuroimaging in Patient 3 was reported as normal at 5 months, but showed enlargement of the cerebellar interfolial spaces compatible with cerebellar atrophy and mildly delayed myelination on repeat brain MRI scans at 21 months and 4 years 3 months (Figure 3E–F). Neuroimaging in patient 4 at 1 day of life showed normal structures but subtle non-specific T2 hyperintensities. Repeat MRI at 4.5 months of age was notable for mild global cerebral volume loss with prominence of the sulci and secondary enlargement of the lateral ventricle, but normal cerebellar structures (Figure 3G).
Figure 3.
Extensive genetic and metabolic testing prior to WES was normal for all the patients with two exceptions. In patient 1, CGH array showed a de novo 26 kilobase duplication on chromosome 17p13.3 that encompasses exons 2–5 of the YWHAE gene. This duplication was not present in patient 2 and hence does not segregate with the sisters’ phenotype. In addition, Patient 4 had an inherited deletion at 17p13.2, not thought to explain his clinical presentation.
METHODS
For Patients 1 and 2, WES on the affected sisters and their mother was performed by Ambry Genetics on genomic DNA (gDNA) isolated from whole blood. Exome library preparation, sequencing, bioinformatics, and data analysis were performed as previously described [Farwell Gonzalez et al., 2015; Gandomi et al., 2014; Farwell et al., 2015]. Briefly, samples prepared using either the SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA) [Gnirke et al., 2009] and sequenced using paired-end, 100-cycle chemistry on the Illumina HiSeq 2000 (Illumina, San Diego, CA). The sequence data were aligned to the reference human genome (GRCh37), and variant calls were generated using CASAVA and Pindel [Ye et al., 2009]. Stepwise filtering included the removal of common SNPs, intergenic and 3′/5′ UTR variants, non-splice-related intronic variants, and lastly synonymous variants. Variants were then filtered further based on family history and possible inheritance models. Data are annotated with the Ambry Variant Analyzer tool (AVA). Candidate alterations were confirmed using automated fluorescence dideoxy sequencing. Co-segregation analysis was performed using each available family member.
For Patient 3, WES on the affected girl and both parents was performed by GeneDx on gDNA isolated from whole blood. Samples were prepared using Agilent Clinical Research Exome kit for sequence capture, and sequenced using 100bp paired-end reads on an Illumina HiSeq 2000 (Illumina, San Diego, CA). Sequence data were aligned to the reference human genome (GRCh37), and analyzed for sequence variants using a custom-developed analysis tool (Xome Analyzer). Capillary sequencing was used to confirm all potentially pathogenic variants.
For Patient 4, sequencing of the affected child was performed by Claritas Genomics, using genomic DNA isolated from whole blood. Two orthogonal sequencing approaches were undertaken: (1) whole exome sequence capture using the Ion Ampliseq Exome Kit with HiQ chemistry, followed by sequencing on the Ion Proton (Thermo Fisher), and (2) large-scale capture of 4,813 genes of established medical significance using the TruSight One kit (Illumina), followed by 150 bp paired-end sequencing on the MiSeq (Illumina, San Diego, CA). Ion Proton sequence data underwent alignment and variant calling using Torrent Suite software v4.4 (Thermo Fisher). Illumina MiSeq sequence data underwent alignment and variant calling using MiSeq Reporter (Illumina). Mean target coverage achieved for patient 4 was 134X (Proton Ampliseq) and 310X (MiSeq TruSight One), respectively. Variant data for the two platforms was combined using tools custom-developed by Claritas Genomics (Combinator, manuscript in preparation). Variant annotation, filtering, and interpretation was conducted in collaboration with the clinical team using an interpretive pipeline provided by Claritas and WuXi NextCODE. Sanger sequencing was used to confirm and phase all potentially pathogenic variants.
RESULTS
In each of the four patients, WES identified compound heterozygous mutations in BRAT1 (NM_152743.3). Patients 1 and 2 have the following BRAT1 mutations: c.638dupA (p.Val214Glyfs*189) and c.803+1G>C (splice site mutation). The mother was heterozygous for the frameshift mutation only. Sanger sequencing in the other family members demonstrated that the father was heterozygous for the splice site mutation. The unaffected sister was heterozygous for the splice site mutation, two unaffected brothers were heterozygous for the frameshift mutation, and the third unaffected brother had neither mutation. The c.638dupA (p.Val214Glyfs*189) variant is seen at a very low frequency in the general population, particularly among those with European ancestry: 0.00051 allele frequency in non-Finnish Europeans in Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org) and 0.00085 allele frequency in European Americans in the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/). The c.803+1G>C variant is not observed in ExAC or EVS.
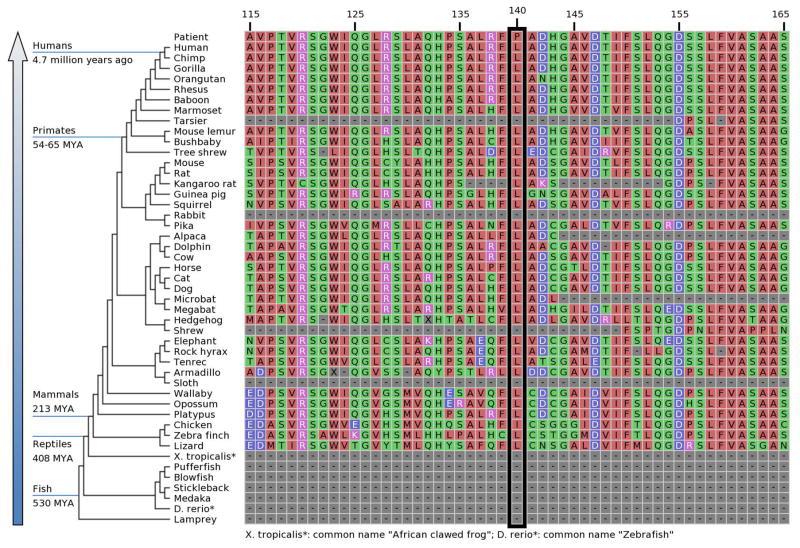
Patient 3 is compound heterozygous for c.638dupA (p.Val214Glyfs*189) and c.419T>C (p.Leu140Pro). Parental analysis was performed to determine that the variants are in trans configuration in the proband, with the frameshift mutation being paternally inherited, and the missense mutation maternally inherited. The proband’s siblings are asymptomatic and have not been tested. The c.419T>C (p.Leu140Pro) is not observed in ExAC or EVS. Leu140 lies within a domain of the BRAT1 protein that is predicted to form an Armadillo-type fold (InterPro; http://www.ebi.ac.uk/interpro/protein/Q6PJG6), and Leu140 is evolutionarily conserved across multiple species (Figure 4). Alterations to proline are often structurally impactful due to imposition of backbone constraints, and consistent with this, p.Leu140Pro is predicted to be probably damaging by PolyPhen2 (score of 1.000) [http://genetics.bwh.harvard.edu/ggi/pph2], deleterious by SIFT (score of 0) [http://sift.jcvi.org/www/SIFT_enst_submit.html], and disease-causing by MutationTaster [http://www.mutationtaster.org/].
Figure 4.
Patient 4 is compound heterozygous for c.171delG (p.Glu57Aspfs*7) and c.419T>C (p.Leu140Pro). Parental analysis was performed to determine that the variants are in trans configuration in the proband, with the frameshift mutation paternally inherited and the missense mutation maternally inherited. The c.171delG (p.Glu57Aspfs*7) variant is not seen in ExAC or EVS.
DISCUSSION
The BRAT1 variants in these four patients are likely pathogenic. For Patients 1–2, the c.803+1G>C alteration has not been previously reported but is expected to be deleterious, since it was thought to alter a canonical splice donor site. The c.638dupA (p.Val214Glyfs*189) alteration has been previously reported in the homozygous state in patients with RMFSL among Amish and Moroccan families. Patient 3 has the c.638dupA (p.Val214Glyfs*189) mutation on one allele, but the other allele is a novel missense variant. Patient 4 has this same novel missense mutation, in addition to a novel frameshift mutation. None of the novel variants have been reported in ExAC or EVS.
Our patients expand the clinical spectrum of BRAT1-related disorders. Collectively, the phenotypes of Patients 1–3 differ from that of RMFSL and appear to represent a mild form of BRAT1 related disorders. All three of these patients have shared dysmorphisms, ID, ataxia or dyspraxia, and cerebellar atrophy. However, the cardiopulmonary problems or dysautonomia originally reported in the Amish subjects was prominently absent. Notably, all three are alive, well into childhood, and acquiring new developmental skills over time.
Patient 4 manifests in part some features of RMFSL, but in contrast to the earliest reports of RMFSL, there is slightly increased survival, slowed development, and later onset of epilepsy (4 months of age). Thus, it would be reasonable to categorize his presentation as moderate in severity relative to the severe deteriorating clinical course reported originally with RMFSL. The presentation of Patient 4 is similar to two other patients with BRAT1 mutations and RMFSL-like phenotype who were reported alive in childhood [Mundy et al., 2015; Hanes et al., 2015].
There are a few possibilities why there may be a spectrum of severity associated with BRAT1 mutations. First, our patients may have inherited certain variants upstream or downstream of BRAT1-related pathways that are modulating their phenotype. Second, with respect to Patients 1 and 2, while the c.803+1G>C mutation likely alters splicing patterns, it is not known to what extent this alteration leads to aberrant expression of mRNA from that allele. For example, splice site mutations in the ATP7A gene may result in Menkes disease or a milder form known as Occipital Horn Syndrome (OHS) [Skjørringe et al., 2011]. Thus, in the two sisters, it is possible the BRAT1 allele affected by the splicing variant may be hypomorphic which ameliorates the phenotype. Third, with respect to Patients 3 and 4, who share a common missense mutation [c.419T>C (p.Leu140Pro)], the mutation on the other allele may be modulating the severity. Phenotypic variability is common in epilepsy genetics and neurodevelopmental disorders [Olson et al., 2014], and it is likely that such may be the case for BRAT1 as well.
The presence of cerebellar atrophy may be a hallmark of mild (Patients 1–3) but not moderate (Patient 4) or severe BRAT1 related disease. In support of this notion, Patients 1–3 presented with, or eventually developed evidence of, cerebellar atrophy. Moreover, in the literature, patients with BRAT1 mutations who were reported to be alive in childhood also showed signs of cerebellar atrophy [Mundy et al., 2015; Hanes et al., 2015]. In contrast, with previously reported patients of severe RMFSL associated with neonatal or infantile death, brain MRI showed either normal findings [Saunders et al., 2012; Straussberg et al., 2015], frontal lobe hypoplasia [Puffenberger et al., 2012], or global atrophy [Saitsu et al., 2014; van de Pol et al., 2015]. Given the period of rapid growth in postnatal cortical and cerebellar development during the first two years of life [Knickmeyer et al., 2008], it is reasonable to suggest that those with severe RMFSL may die before the cerebellar changes induced by their BRAT1 defect become more prominent.
The interaction between BRAT1 and BRCA1 may have theoretical implications for heterozygous carriers of BRAT1 mutations. For Patients 1 and 2, the family history is notable for cancer in several maternal relatives: maternal grandmother with ovarian cancer at age 49 who had negative BRCA1/2 mutation testing; two sisters of this maternal grandmother (maternal great-aunts) with ovarian cancer diagnosed in their 30’s and 50’s respectively, both now deceased; maternal great-grandmother (mother of maternal grandmother) with breast cancer diagnosed in her 80’s, now deceased; one maternal great-aunt (sister of maternal grandfather) with unspecified cancers (possibly ovarian or others) who died in her 40’s. For patient 4, the family history is notable for maternal grandmother with vulvar cancer. Autosomal dominant mutations in BRCA1 are associated with increased risks of breast, ovarian, and other cancers [Antoniou et al., 2003; Tai et al., 2007]. For parental carriers of BRAT1 variants, it is unclear whether their risk of developing malignancy is increased. However, in light of the fact that tumor suppression by BRCA1 depends on its binding to BRAT1, a single defective copy of BRAT1 may not be sufficient to result in full-fledged disease but may lead to reduced tumor suppression. There is precedent for this phenomenon in ataxia-telangiectasia, an autosomal recessive disorder caused by ATM mutations, which confer a moderately increased risk of breast cancer for female carriers [Easton, 1994]. We were particularly concerned about this possibility in Patients 1–2 because of striking family history of early-onset ovarian cancer in the maternal grandmother and two of her sisters. The maternal grandmother tested negative for the frameshift mutation, indicating that our patient’s mother likely inherited the mutation from her father, who had a sister die of cancer (possibly ovarian) in her 40’s. DNA was not available from this individual, and further carrier testing was not performed in the family. Thus, this possible relationship between BRAT1 mutation in heterozygous state and cancer susceptibility needs to be studied further before a definite conclusion is made.
In summary, our patients demonstrate biallelic BRAT1 mutations causing mild and moderate phenotypes. Patients 1–3 have a mild phenotype of non-lethal ID, non-progressive ataxia or dyspraxia, and cerebellar atrophy, with well-controlled epilepsy in Patient 3. Patient 4 has a moderate phenotype that resembles RMFSL except for longer survival, continued development, and later onset of epilepsy. A BRAT1-related disorder should be considered, with increased awareness of the phenotypic variability, in children with ID, non-progressive cerebellar ataxia or dyspraxia, and cerebellar atrophy. We suggest renaming this spectrum BRAT1-related neurodevelopmental disorders.
Acknowledgments
We would like to thank the families for their involvement. David Miller receives funding support from NHGRI 1U41HG006834. Heather Olson receives support from the NINDS (K12 NS079414-02).
Footnotes
CONFLICTS OF INTEREST
Dr. Davis and Ms. Shahmirzadi are paid employees of Ambry Genetics. David Miller and Tim Yu serve as part-time consultant to Claritas Genomics, a provider of genetic testing services (non-equity professional services agreement).
References
- Antoniou A, Pharoah PDP, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi O-P, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF. Cancer risks in A-T heterozygotes. Int J Radiat Biol. 1994;66:S177–182. doi: 10.1080/09553009414552011. [DOI] [PubMed] [Google Scholar]
- Farwell Gonzalez KD, Li X, Lu H-M, Lu H, Pellegrino JE, Miller RT, Zeng W, Chao EC. Diagnostic Exome Sequencing and Tailored Bioinformatics of the Parents of a Deceased Child with Cobalamin Deficiency Suggests Digenic Inheritance of the MTR and LMBRD1 Genes. JIMD Rep. 2015;15:29–37. doi: 10.1007/8904_2014_294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, Baxter RM, Zeng W, Mroske C, Parra MC, Gandomi SK, Lu I, Li X, Lu H, Lu H-M, Salvador D, Ruble D, Lao M, Fischbach S, Wen J, Lee S, Elliott A, Dunlop CLM, Tang S. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- Gandomi SK, Farwell Gonzalez KD, Parra M, Shahmirzadi L, Mancuso J, Pichurin P, Temme R, Dugan S, Zeng W, Tang S. Diagnostic exome sequencing identifies two novel IQSEC2 mutations associated with X-linked intellectual disability with seizures: implications for genetic counseling and clinical diagnosis. J Genet Couns. 2014;23:289–298. doi: 10.1007/s10897-013-9671-6. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes I, Kozenko M, Callen DJA. Lethal Neonatal Rigidity and Multifocal Seizure Syndrome-A Misnamed Disorder? Pediatr Neurol. 2015 doi: 10.1016/j.pediatrneurol.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH. A Structural MRI Study of Human Brain Development from Birth to 2 Years. J Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy SA, Krock BL, Mao R, Shen JJ. BRAT1-related disease-identification of a patient without early lethality. Am J Med Genet A. 2015 doi: 10.1002/ajmg.a.37434. [DOI] [PubMed] [Google Scholar]
- Olson HE, Poduri A, Pearl PL. Genetic forms of epilepsies and other paroxysmal disorders. Semin Neurol. 2014;34:266–279. doi: 10.1055/s-0034-1386765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pol LA, Wolf NI, van Weissenbruch MM, Stam CJ, Weiss JM, Waisfisz Q, Kevelam SH, Bugiani M, van de Kamp JM, van der Knaap MS. Early-Onset Severe Encephalopathy with Epilepsy: The BRAT1 Gene Should Be Added to the List of Causes. Neuropediatrics. 2015;46:392–400. doi: 10.1055/s-0035-1564791. [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, Cassidy RP, Fiorentini CJ, Heiken KF, Lawrence JJ, Mahoney MH, Miller CJ, Nair DT, Politi KA, Worcester KN, Setton RA, Dipiazza R, Sherman EA, Eastman JT, Francklyn C, Robey-Bond S, Rider NL, Gabriel S, Morton DH, Strauss KA. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Yamashita S, Tanaka Y, Tsurusaki Y, Nakashima M, Miyake N, Matsumoto N. Compound heterozygous BRAT1 mutations cause familial Ohtahara syndrome with hypertonia and microcephaly. J Hum Genet. 2014;59:687–690. doi: 10.1038/jhg.2014.91. [DOI] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjørringe T, Tümer Z, Møller LB. Splice Site Mutations in the ATP7A Gene. PLoS One. 2011:6. doi: 10.1371/journal.pone.0018599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussberg R, Ganelin-Cohen E, Goldberg-Stern H, Tzur S, Behar DM, Smirin-Yosef P, Salmon-Divon M, Basel-Vanagaite L. Lethal neonatal rigidity and multifocal seizure syndrome--report of another family with a BRAT1 mutation. Eur J Paediatr Neurol. 2015;19:240–242. doi: 10.1016/j.ejpn.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99:1811–1814. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]