Abstract
γ-Herpesviruses, Epstein–Barr virus, and Kaposi's sarcoma-associated herpesvirus are important human pathogens, because they are involved in tumor development. Murine γ-herpesvirus-68 (MHV-68 or γHV-68) has emerged as a small animal model system for the study of γ-herpesvirus pathogenesis and host–virus interactions. To identify the genes required for viral replication in vitro and in vivo, we generated 1,152 mutants using signature-tagged transposon mutagenesis on an infectious bacterial artificial chromosome of MHV-68. Almost every ORF was mutated by random insertion. For each ORF, a mutant with an insertion proximal to the N terminus of each ORF was examined for the ability to grow in fibroblasts. Our results indicate that 41 genes are essential for in vitro growth, whereas 26 are nonessential and 6 attenuated. Replication-competent mutants were pooled to infect mice, which led to the discovery of ORF 54 being important for MHV-68 to replicate in the lung. This genetic analysis of a tumor-associated herpesvirus at the whole genome level validates signature-tagged transposon mutagenesis screening as an effective genetic system to identify important virulent genes in vivo and define interactions with the host immune system.
Keywords: functional mapping, herpesvirus, bacterial artificial chromosome, deoxyuridine-triphosphatase, transposition
Because they are involved in tumor development, γ-herpesviruses are important human pathogens. The prototypic γ1-herpesvirus, Epstein–Barr virus (EBV), is associated with lymphomas, nasopharyngeal carcinoma, and other types of malignancies (1). Kaposi's sarcoma-associated herpesvirus (KSHV, also known as human herpesvirus 8), a γ2-herpesvirus, is associated with Kaposi's sarcoma and B cell lymphomas (2). In vivo studies of human γ-herpesvirus infection and pathogenesis have been limited because of restricted host range. With its genomic organization similar to that of human γ-herpesviruses, murine γ-herpesvirus-68 (MHV-68 or γHV-68) has emerged as a small animal model system to study γ-herpesvirus pathogenesis and host interaction (3–7). After intranasal infection, MHV-68 establishes productive infection in the epithelia of the respiratory tract and latent infection in splenocytes, macrophages, dendritic cells, and lung epithelial cells (8–11). Analysis of the complete nucleotide sequence of MHV-68 revealed a double-stranded DNA of ≈118 kb unique sequences with 1,213-bp terminal repeats, which is predicted to encode at least 80 genes (12). Despite recent progress in our understanding of the γ-herpesviruses, a substantial portion of the ORFs have not been studied in vitro or in vivo.
Signature-tagged mutagenesis (STM) is based on random transposon insertional mutagenesis and allows simultaneous screening of multiple mutants by negative selection (13–15). This approach is accompanied by tagging each mutant with a unique short DNA sequence so that it can subsequently be identified within a pool of mutants. A mutant that fails to establish infection in an animal is readily detected based on the absence of a unique tag signal in the output (in vivo) as compared with the input (in vitro). Hensel et al. (14) used random synthetic oligonucleotides [NK]20 as a variant tag, comprised of up to ≈1017 complexity of mutants. However, due to the problems involving cross-hybridization and reproducibility of [32P]-labeled PCR-amplified individual tags, STM technology has been modified by using preselected tagged transposons to construct a mutant library (16, 17). STM, a powerful genetic method to screen virulent genes, has been used in various pathogens (13, 15). However, the method using transposon-inserted mutagenesis has not been easily adapted to large viruses until the recent availability of their infectious bacterial artificial chromosome (BAC) clones.
Here, we took advantage of MHV-68 in the BAC plasmid and in vitro Mu transposition system as a rapid mutant generation tool and adapted the STM technique as an effective screening system. We conducted functional analysis of the MHV-68 genome for in vitro growth. Replication-competent mutant viruses were further examined for in vivo growth by infection of pooled mutant viruses with different signature tags.
Materials and Methods
Cells, Viruses, and Plasmids. BHK-21 and NIH3T12 cells were propagated in growth medium containing 10% FBS and antibiotics. The wild-type virus (WT), pBAC/MHV-68 virus, and its mutant derivatives were all propagated in BHK-21 cells for in vitro and in NIH3T12 cells for in vivo study. We generated an independent BAC clone of MHV-68 (pBAC/MHV-68) (T.-T.W., H.-I.L., L. Tong, H. Deng, N. Reyes, G. Smith, and R.S., unpublished results). BAC (pBeloBAC plasmid, chloramphenicol; CamR) was inserted at the left end of the viral genome (nucleotide 1892). pEntranceposon (kanamycin; KanR) containing Mu binding sites was used to clone a signature-tagged transposon by using XbaI and StuI sites. Twelve tags were designed for real-time PCR detection. The sequences of the STM tags and primers are provided in Table 2, which is published as supporting information on the PNAS web site.
Generation of STM Mutants. pEntranceposons tagged with 12 unique sequences were cut with BglII to release the STM-transposons. In vitro transposition was performed for 10–15 min with pBAC/MHV-68 and purified STM transposons (Finnzymes, Helsinki). The reaction mixtures were transformed into bacteria (DH10b), and colonies were recovered from LB plates containing kanamycin (20 μg/ml) and chloramphenicol (20 μg/ml). BAC DNAs of 1,152 STM mutants were subjected to sequencing analysis, by using the Seq1 primer (5′-cgtcgcttactaggatccg-3′) (Macrogen, Seoul, Republic of Korea) to determine the location and the orientation of inserted transposons. EcoRI digestion, followed by Southern analysis, was performed to confirm the correct genomic arrangement of STM mutants (Fig. 5, which is published as supporting information on the PNAS web site).
Analysis of STM Mutants in Vitro and in Vivo. BAC DNAs of the STM mutants with transposon inserted in the closest proximity to the N terminus of each ORF were transfected into BHK21 cells in 96-well plates, by using the Lipofectamine Plus reagent (Invitrogen). At 6–8 days posttransfection (p.t.), supernatant was transferred at 1:10 dilution to freshly seeded BHK21 cells for two rounds, and the cytopathic effects (CPE) of each mutant were scored to determine the essentialness of the disrupted gene for in vitro growth of the virus. Plaque assays and real-time PCR with ORF 56-specific primers (nucleotides 75598–75783; M56F, 5′-gtaactcgagactgaaacctcgcagaggtcc-3′; M56R, 5′-ccgaagcttgcacggtgcaatgtgtcacag-3′) were used to measure the titers of replication-competent viruses. Three pools of 11 replication-competent STM mutants and a control STM mutant with the transposon insertion at the BAC plasmid (STM BAC) with distinct tags were intranasally administered [45 plaque-forming units (pfu) per mutant, total 20 μl of 500 pfu] to infect BALB/c mice (≈6 weeks old, n = 6 per pool). The mice were killed at 7 days postinfection, and the total genomic DNA from the lung tissue was extracted by using the DNeasy kit (Qiagen, Valencia, CA), then subjected to real-time PCR by using STM tag-specific primers (B1-3, A1-4) and a common dual-labeled probe (TaqMan probe, Qiagen). The viral DNA extracted from the inocula and the total cellular DNA extracted from NIH3T12 cells infected with the inocula (multiplicity of infection = 0.05; 5,000 pfu per 105 cells) were also used for real-time PCR as an input control.
Real-Time PCR. DNA template (50 ng) and ORF 56-specific primers or STM tag-specific primers with STM TaqMan probe (sequences shown in Table 2) were mixed with 2× Master mix (Applied Biosystems). Real-time PCR with TaqMan probe was run at 95°C for 15 min and 45 cycles at 95°C for 30 s and 60°C for 10 s, with the results analyzed in Opticon II (MJ Research, Cambridge, MA). PCR with SYBR Green was run at 95°C for 15 min and 45 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 10 s, followed by melting curve analysis.
Results
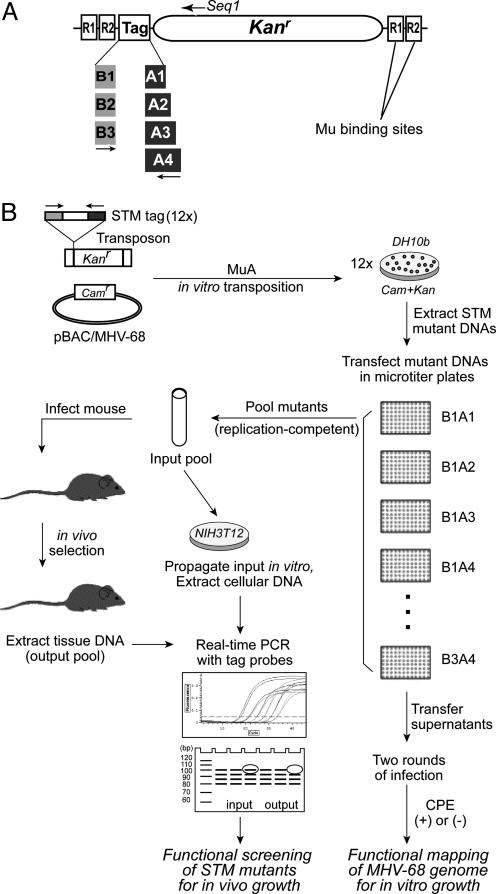
Generation of an MHV-68 STM Mutant Library. We generated a library of MHV-68 mutants labeled with unique tags for use in in vitro and in vivo screening (Fig. 1). To circumvent the problems in the use of random synthetic oligonucleotides as STM tags and to maximize the sensitivity in detection of the tags in a quantitative manner, we used a combination of three forward and four reverse primers suitable for real-time PCR. All 12 unique tag sequences of our STM transposons carry Mu binding sites with the kanamycin selection marker (Fig. 1 A). For random transposon insertion, we used an in vitro transposition system involving Mu A transposase (18, 19). Mu is known to be one of the most efficient transposon systems but has less target sequence preference. As a template for mutagenesis, we used our infectious BAC clone of MHV-68 carrying chloramphenicol resistance (20). With double selection of antibiotics, in vitro transposition of Mu into pBAC/MHV-68 resulted in >104 colonies. The insertion locus and the orientation of the transposon in each of 1,152 STM mutants (96 mutants per tag) were determined by direct sequencing of the insertion-junction. Whereas 131 mutants showed unreliable sequencing results, possibly due to double insertion of the transposon, high G + C content, and/or poor DNA quality, 988 insertions hit the MHV-68 genome and 33 hit the BAC plasmid sequence. Multiple hits were obtained for each ORF, except ORF 30 (which can potentially encode 80 aa). The number of mutants for each ORF correlated with the size of the corresponding ORF, except M10a, b, and c, suggesting random transposon insertion of STM mutants. The longest distance between determined insertions was 2,094 bp, encompassing M10a, b, and c (100-bp repeat regions, nucleotides 98981–101170). This result was probably due to difficulties in sequencing a region of high G + C content (86%) (12). Based on further characterization of STM mutants by analyzing restriction enzyme digestions and Southern hybridizations and on consideration of the coding capacity and the repetitive sequences within ORFs, mutants of ORF 47, M10a, b, and c, M13, and M14 were not further studied. It is not yet clear whether these viral genes, except ORF 47, are really expressed as functional proteins.
Fig. 1.
STM of MHV-68 for in vivo and in vitro screening. (A) A schematic diagram of STM transposons. Tag sequences are composed of 12 different combinations of three forward and four reverse sequences and a common sequences in the middle, which is designed for real-time PCR. (B) A schematic representation of the MHV-68 STM strategy. By using MuA transposase and an STM transposon with pBAC/MHV-68 as a template, in vitro transposition yields thousands of transposon-inserted mutants. BAC DNAs of STM mutants that have transposon insertion at each ORF of MHV-68 are extracted and transfected into fibroblasts in 96-well plates. After two rounds of infection with transfected supernatants, ORFs are categorized to be essential or nonessential, based on the growth phenotype of a mutant with an insertion proximal to the N terminus of each ORF. Replication-competent mutants are pooled and simultaneously used to infect mice. The inoculum is also propagated in NIH3T12 cells. DNAs from tissues of infected mice (output) and from infected NIH3T12 cells (input) were subjected to real-time PCR, by using STM tag-specific primer sets to determine the fate of each virus within the pool. The copy number of each tag in the input and the output is quantitatively compared to identify the attenuated mutants.
Determining the Viral Genes Essential for Replication in Vitro. Although to date 12 recombinant viruses with disrupted ORFs have been reported among 80 putative ORFs of MHV-68 (20–34), the majority of the viral genes have not yet been characterized. Using the STM mutant library, we took a systematic approach to determine the requirement of each ORF for in vitro growth of MHV-68. A mutant virus may produce a truncated form of a gene product that may or may not be functional, depending on its transposon insertion location. Considering the lack of available antibodies against most ORFs in MHV-68 and functional assay systems to determine the functionality of the truncated protein (if applicable), we used only a mutant with a transposon insertion proximal to the N terminus of each ORF for the following experiments. Mutant BAC DNAs were isolated and transfected into BHK21 cells in 96-well plates. An STM mutant with a transposon insertion at the BAC plasmid was used as a positive control, and a mutant generated by site-directed mutagenesis to disrupt ORF 31 was the negative control (20). At 6–8 days posttransfection, supernatants were transferred to freshly seeded BHK21 cells for two more rounds to score the cytopathic effects of mutants.
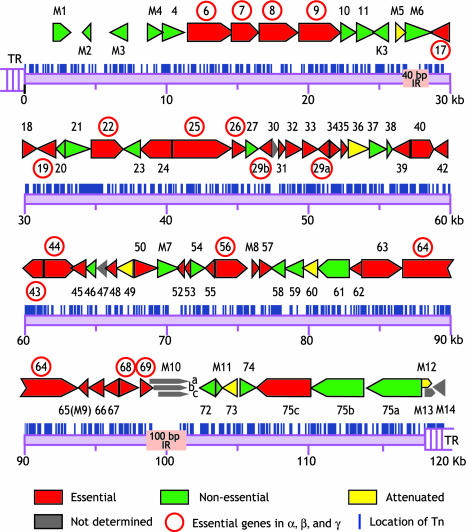
Thirty-two mutants were able to produce infectious viruses, and 41 showed no signs of cytopathic effects, even in multiple rounds of incubation with supernatants from transfected cells (Fig. 2and Table 3, which is published as supporting information on the PNAS web site). BAC DNAs of mutants that did not yield any infectious virions were independently prepared at two different laboratories and transfected to confirm the no-growth phenotype. Additional mutants with the disruption of the same ORF but with different insertion sites were also used to verify the results. Our results were consistent with earlier reports on two essential genes, and we newly identified 39 genes that are essential. Among 32 replication-competent viruses, 10 have been previously reported (21–34). All of the essential genes seem to be conserved in γ-herpesviruses (35), except M8, which is unique to MHV-68. The phenotype of M8null could be due to the facts that M8 locus is overlapped with spliced ORF 57 transcript and that ORF 57 is essential (36). In this study, we do not attempt to adjust any possible splicing events that may affect the phenotype, because transcripts for most ORFs of MHV-68 have not been characterized. The conserved essential ORFs are thought to encode proteins involved in DNA replication and virion assembly (Fig. 2 and Table 3) (37–39). Consistent with this hypothesis, most viral proteins that were previously identified in the virion of MHV-68 seem to be essential for virus replication in fibroblasts, except for ORF 20 and M7 (gp150) (40). Among ORF 75a, -b, and -c genes that encode tegument proteins highly homologous to one another, only ORF 75c, found in the virion preparation, was essential (40). The number of essential genes of MHV-68 for in vitro growth is comparable with the number of previously characterized herpesviruses such as herpes simplex virus-1 and human cytomegalovirus. We found that 17 essential ORFs (ORFs 6, 7, 8, 9, 17, 19, 22, 25, 26, 29b, 29a, 43, 44, 56, 64, 68, and 69) are common among α, β, and γ subfamilies (37–39).
Fig. 2.
Functional mapping of MHV-68 for in vitro growth. Putative ORFs of MHV-68 originally predicted by Virgin and colleagues (10) are color-coded, according to the growth properties of STM mutants with transposon insertion at the proximity to N terminus of each ORF in fibroblast. The ORFs with rightward direction are named in the upper lines, whereas those with leftward orientation in the lower lines. The red circles indicate essential genes conserved in α-, β-, and γ-herpesvirus subfamilies. The vertical lines represent transposon insertion sites, based on sequencing results of 988 mutants.
In vitro growth properties of 32 replication-competent mutants were further analyzed. Replication-competent mutants grown in NIH3T12 were titered by plaque assays and used for multiple-step growth curves. Results from multiple-step growth curves showed that most STM mutants for nonessential genes were indeed replication-competent, although there are six mutants showing various degrees of attenuation including M5, ORFs 36, 49, 60, and 73, and M12 (Fig. 2; see also Fig. 6A, which is published as supporting information on the PNAS web site). Attenuated phenotype of an ORF 73 mutant was weak and only apparent at later time points, which is consistent with published results (30). Real-time PCR analysis using STM tags in pooled infection of multiple mutant viruses also showed the similar phenotypes (Fig. 6B). The information from direct comparison of our results with previous publications is included in Table 3.
To make sure that the transposon insertion indeed disrupts the function of a specific gene, we examined viral gene expressions using a series of mutants that have transposon insertion at different locations of a viral gene (M7, as an example). The mutant viruses have disrupted expression of the transposon-inserted gene (M7), not of the others (Fig. 7, which is published as supporting information on the PNAS web site). Genomic stability of STM mutants was also tested by comparing dozens of BAC DNAs isolated from Escherichia coli with DNAs from cells infected with the same virus. Upon several rounds of infections before this experiment, no significant genomic alterations of our STM mutants, except the number of terminal repeats, were observed, suggesting that the genomes of STM mutants are stable in cultured cells (Fig. 8, which is published as supporting information on the PNAS web site).
Determining the Viral Genes That Are Important for Viral Replication in Vivo. STM mutants that can grow in vitro were further examined for their ability to establish acute infection in vivo. Our STM strategy allows us to simultaneously infect a mouse with multiple mutants, because they can be individually identified within a pool based on the unique tag sequences (41). Three pools of 11 replication-competent STM mutants with distinct tags (45 pfu per virus) were intranasally administered to infect BALB/c mice (six mice per pool). A mutant with a transposon insertion at the BAC plasmid, STM BAC (WT), was included in every pool as a positive control. STM BAC also served as a reference point for comparison among different pools as well as different mice. At 7 days postinfection, DNAs from lung tissues of infected mice were subjected to real-time PCR for maximal sensitivity to identify STM mutants that are defective in replicating in vivo. Our pBAC/MHV-68 virus replicated normally in vitro and showed near normal in vivo lytic replication in the lung. The variation (2- to 3-fold) does not reach statistical significance compared with the WT or its revertant, although its latency is clearly defective (T.-T.W., unpublished data). All of the STM mutants in our study carried the BAC sequences, and their relative abilities to grow in vivo were compared. DNAs were extracted from the inocula itself and from NIH3T12 cells that were infected with the same inocula, then subjected to real-time PCR, by using STM tags. Thus, as a control, we used viral DNAs propagated in fibroblasts to normalize the effect of variations in in vitro replication capacity of the viruses on our in vivo study.
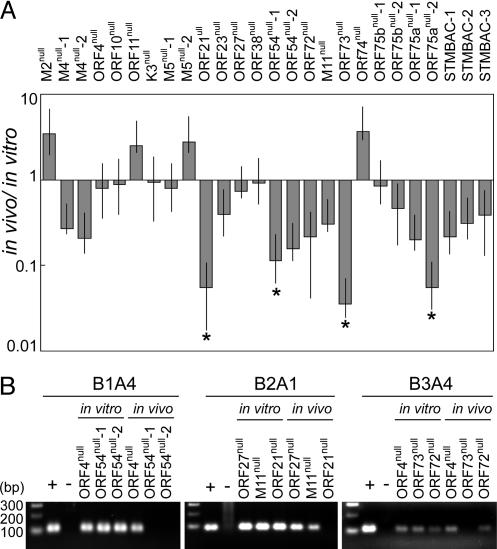
Comparison of the signals between in vitro and in vivo led to identification of four mutants that showed statistically significant defective growth in the lung as compared with STM BACs (P < 0.05) (Fig. 3A). These results were also confirmed when the PCR products were run on agarose gels (Fig. 3B). A mutant with a disrupted ORF 21 (thymidine kinase, ORF 21null) showed severe attenuation in the lung, which is consistent with an earlier study (26). In addition, a mutant with a disrupted ORF 73 (ORF 73null), a homologue of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen (LANA), had significantly reduced growth in acute infection at a low inoculation dose (<1,000 pfu) (30). Interestingly, ORF 75a, encoding a tegument protein homologous to ORF 75b and -c, seemed to be important in the lung infection, whereas ORF 75c was shown to be important for growth in vitro. It is not clear whether ORF 75b is also important. These ORFs warrant investigation in greater detail.
Fig. 3.
Identification of STM mutants likely to be critical for acute infection in vivo. (A) Quantitative analysis of STM mutant growth in the lung. Pooled STM mutants with distinct tags (45 pfu per virus) were used to intranasally infect BALB/c mice (six mice per pool) and also propagated in NIH3T12 cells. Relative copy numbers of STM mutants with specific tag primer sets using DNAs (50 ng) from infected cells (in vitro) and lung tissues of infected mice (in vivo), as determined by real-time PCR, were compared to identify STM mutants defective in establishing acute infection in mice. Mutants included in different pools are indicated with numbers. SEMs are shown as error bars. *, Mutants showing significant attenuation in the lung. (B) The PCR products of STM mutants were run on agarose gels. A set of representative data is shown. Compared with the in vitro data, mutants ORF 21null, ORF 73null, and ORF 54null failed to show detectable growth in vivo, whereas others replicated normally. The specific PCR products ranged from 80 to 100 bps. + control, STM plasmid mix; - control, lung DNA from an uninfected BALB/c mouse.
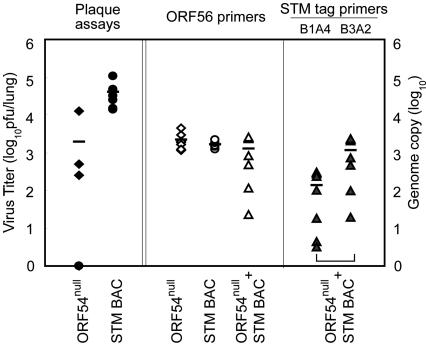
Furthermore, a mutant of ORF 54 (ORF 54null), putatively encoding deoxyuridine-triphosphatase (dUTPase), which is well-conserved among γ-herpesviruses (42), was shown to be reduced in growth in the lung. These data suggest that ORF 54 may be a novel candidate that is important for establishing acute infection in the lung. To further verify the defective growth of ORF 54null in the lung, six to seven mice were individually infected with ORF 54null or STM BAC alone (50 pfu), or coinfected with ORF 54null and STM BAC (50 pfu per virus). Mice were killed at 9 days postinfection. The titers of infectious virus in the lung were determined by plaque assays (Fig. 4). In accordance with our initial screening results, ORF 54null infection showed attenuated growth in the lung (P = 0.038). To distinguish in vivo growth of each mutant in mice coinfected with ORF 54null and STM BAC, real-time PCR was performed by using both virus-specific (ORF 56) and STM tag-specific primers. We calculated relative copy numbers of each virus. Because the PCR efficiencies differed depending on the primer sets used, copy numbers obtained from PCR with each STM tag primer set were normalized with a ratio of copy numbers from STM tags to those from ORF 56 obtained from individual infections. In dual infection of ORF 54null and STM BAC, growth of ORF 54null in the lung was also significantly reduced (P = 0.015) compared with the replication of STM BAC, confirming our screening results (Table 1). In vivo growth kinetic studies of ORF 54null demonstrated the significantly reduced growth of ORF 54null in the lung in both individual and dual infections (Fig. 9 A and B, which is published as supporting information on the PNAS web site). An independently isolated ORF 54null mutant also showed attenuated growth in the lung during the course of infection, whereas STM BAC and ORF 10null showed normal growth (Fig. 9C). Taken together, our results suggest that ORF 54, which encodes dUTPase, plays an important role in acute infection of MHV-68 in the lung.
Fig. 4.
Attenuation of ORF 54null growth in the lung. Mice were infected with ORF 54null (⋄), STM BAC (WT, ○), or ORF 54null + STM BAC (▵) at 50 pfu per virus and killed at 9 days postinfection. Infectious viruses in the lung were assayed by plaque assays (Left, filled symbols). Results from real-time PCR using viral genome-specific (ORF 56, Center, open symbols) or STM tag-specific (Right, gray symbols) primers with DNAs from lung tissues of infected mice are shown. ORF 54null is tagged with B1A4, and STM BAC with B3A2. Genome copies of individual viruses in mixed infection were normalized based on comparison of results from viral genome-specific primers with those from STM tags in individual infection. The average of each group is shown as a bar within the group.
Table 1. Relative genome copy numbers of individual viruses.
| Relative copy number
|
|||
|---|---|---|---|
| Mouse | ORF 54null | STM BAC | Ratio of ORF 54null/STM BAC,* % |
| 1 | 4.4 | 473.5 | 0.9 |
| 2 | 102.9 | 740.5 | 13.9 |
| 3 | 3.2 | 19.9 | 16.1 |
| 4 | 311.8 | 2,082.9 | 15.0 |
| 5 | 18.7 | 98.8 | 18.9 |
| 6 | 279.5 | 2,253.0 | 12.4 |
| 7 | 234.5 | 2,338.2 | 10.0 |
To determine growth of each virus in mice infected with both ORF 54null and WT, relative genome copy numbers of individual viruses were calculated for each mouse in a comparison with total viral genome (ORF 56). The ratios of ORF 54null/STM BAC provide the relative growth of ORF 54null in the presence of STM BAC.
In mice infected with ORF 54null + STM BAC
Discussion
Global analysis of STM mutants generated by in vitro transposition with MuA transposase demonstrated a highly effective random mutagenesis system. We have constructed an STM mutant library that disrupts nearly every ORF in MHV-68 and have established a functional map of MHV-68 for in vitro growth. Moreover, using distinct tag sequences, we conducted efficient genetic screening and confirmed previous studies with mutants of ORF 21 and ORF 73. Our screening identified ORF 54, which encodes viral dUTPase, as likely being important for acute infection in the lung. Taken together, our results represent a functional analysis of a tumor-associated herpesvirus at the whole genome level in the context of in vitro and in vivo infection. Therefore, our systematic approach by using the STM screening system to define defective phenotypes in vivo will enable us to address critical issues in γ-herpesvirus pathogenesis and immune interactions with hosts.
STM of MHV-68. We have adapted the STM technique as an effective screening system for MHV-68 replication with the following considerations (13). (i) As the complexity of the pool increases, it might lead to false identification of attenuated mutants because of the increased probability of individual mutants that fail to be recovered adequately to yield hybridization signals. Problems in cross-hybridization and reproducibility of 32P-labeled PCR-amplified individual tags from random synthetic oligonucleotides complicated a later screening process. (ii) If the inoculum dose is too low, there may be a random chance for individual mutants to fail to initiate successful infection; if the dose is too high, the host immune system may be overwhelmed, resulting in the growth of mutants that would otherwise be attenuated. (iii) We also considered the genomic size of MHV-68 to estimate the number of mutants to be analyzed in vivo. Taking these into account, we designed our 12 STM tags as a combination of three forward and four reverse primers with defined sequences suitable for highly sensitive real-time PCR. In our hands, an inoculum dose as low as 10 pfu of virus induced reproducible replication kinetics in mice, although lower doses still can initiate infection (T.-T.W., unpublished results). We used an inoculum dose of 45 pfu per virus and a total of 500 pfu for screening so as not to overwhelm the immune system, yet sufficient to initiate infection for every mutant. Lower inoculum doses may have advantages such as mimicking natural viral infection with less transcomplementation in an animal. As the results from in vivo screening indicated, our modified STM protocol works efficiently in MHV-68.
Essential Viral Genes for in Vitro Growth of MHV-68. Our functional mapping of the MHV-68 genome indicated 41 ORFs to be essential, 26 nonessential, and 6 attenuated. In our study, two rounds of infection with an extended incubation time were carried out to ensure that initially low titers of a mutant would have the chance to be amplified to a saturating level in subsequent incubation. However, a mutant that replicates in cells without being able to release infectious virion progeny may have been classified as essential. Due to defects in viral entry, a mutant that is cytopathic effect (+) in transfection but loses its infectivity in subsequent infection would also be categorized as essential. Many nonessential genes including attenuated ones are clustered at both ends of the viral genome rather than being randomly distributed. For example, an ≈9-kb region spanning M1 to ORF 4 and a 6-kb region from ORF 10 to M6 form blocks of nonessential genes at the left end of the genome. A 16-kb region from ORF 72 to M12, except ORF 75c, forms another block of nonessential genes at the right end of the genome.
Nevertheless, MHV-68 seems to carry the number of essential genes comparable with previously characterized herpesviruses such as herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV). The prototype herpesvirus, HSV-1, has been collectively shown to encode 37 essential genes among 85 genes (37). Recent studies on HCMV reported that 41–45 essential genes among 150–162 genes predicted ORFs (38, 39). Among the 41 ORFs essential for MHV-68 lytic replication in vitro, 10 ORFs are conserved only in γ-herpesviruses, and not found in α or β subfamilies (Table 3). Eleven essential ORFs are common between MHV-68 and HCMV, although two of them (ORF 33 and 39) showed nonessential phenotype in HSV-1. Two ORFs (ORF 42 and 57) displayed essential phenotype in MHV-68 and HSV-1, attenuated phenotype in HCMV. Comparing the data from MHV-68, HSV-1, and HCMV, we found that 17 conserved ORFs are essential for viral lytic replication. We proposed that these 17 ORFs, conserved among all herpesvirus subfamilies, form the core group of proteins required for herpesvirus lytic replication in cultured cells. Whereas the function of ORF 69 has not been clearly defined, most of the 17 conserved genes are thought to encode proteins involved in viral DNA replication and virion assembly (37–39). These 17 viral proteins might serve as targets of therapeutic agents that have the potential efficacy against virus replication in all three subfamilies.
ORF 54, a dUTPase, Is Important for MHV-68 Replication in the Lung in the Presence of Other Viruses. ORF 54 encodes dUTPase, an enzyme highly conserved among α- and γ-herpesviruses, but substantially diverged in β-herpesviruses (42). Found in a wide variety of organisms, including eukaryotic cells, Saccharomyces cerevisiae, E. coli, and viruses, dUTPase is an essential enzyme in the nucleotide metabolism involved in (i) providing dUMP, the direct precursor for biosynthesis of dTTP; and (ii) maintaining a low dUTP:dTTP ratio to minimize the misincorporation of uracil into DNA (42–44). In a number of retroviruses and DNA viruses, dUTPase may control local dUTP levels during replication, and also enhance viral replication in nondividing host tissues. Infection of WT HSV-1 resulted in down-regulation of cellular dUTPase expression, whereas cellular levels of dUTPase remained normal in the cells infected with dUTPase-deficient mutants (45). The growth of HSV-1-lacking dUTPase has also been reported to be severely attenuated in the central nervous system, but normal in other locations, in addition to its reactivation deficiency in neurons (46). Therefore, it is conceivable that there may be a limited amount of endogenous dUTPase for virus replication in cells that may be complemented by virally encoded dUTPase. A mutant lacking viral dUTPase may be disadvantaged in DNA metabolism, such as misincorporation of uracil into the viral genome, yielding fewer infectious viruses.
At first glance, our results on ORF 54null infection seemed confounding. Although the virus growth measured by plaque assays was severely reduced, viral DNA copy numbers of ORF 54null were comparable with that of WT (Fig. 4 and Table 1). This phenomenon may be due to the presence of defective viral DNA in ORF 54null, rendering it incapable of producing infectious viruses. However, the DNA copy number of ORF 54null was significantly lower than that of WT in dual infection, which may be due to competition with other viruses in early infection. Thus, these observations suggest that our screening conditions with multiple mutants in a single animal would be more stringent than with a single mutant alone. Kinetic studies of ORF 54null and 2nd ORF 54null replication in the lung consistently recapitulated the defective phenotype of ORF 54null growth in both individual and dual infections (Fig. 9). These results were also reproduced by using independent mutants such as ORF 54STOP with stop codons in all three ORFs engineered at the N terminus of ORF 54, whereas STM BAC and ORF 10null as well as a marker-rescued revertant of ORF 54STOP resulted in normal growth (S.H., unpublished data). These results reinforce our conclusion that ORF 54 plays an important role in acute infection in the lung. Detailed mechanisms of this defective growth of ORF 54null warrant further investigation and will elucidate an important role of dUTPase in acute infection of γ-herpesviruses.
In conclusion, our study of signature-tagged mutagenesis highlights the use of an unbiased systematic approach for identifying viral determinants important for viral replication in vitro and in vivo. The advantage of studying MHV-68 as a model system of human γ-herpesviruses includes the availability of an amenable genetic system in the host. Screening of the STM mutant library in other organs, latent infection, and transgenic or knockout mice will also facilitate identification of viral determinants interacting with a host factor of interest, e.g., interactions with the host immune system. Furthermore, identification of attenuated mutants in vivo may contribute to development of new strategies for vaccines and therapeutic targets.
Note. After this manuscript was submitted, an independent study that analyzed the role of 29 ORFs in MHV-68 replication in vitro was published (47).
Supplementary Material
Acknowledgments
We thank Nikki Reyes and Wendy Aft for editing the manuscript. We also thank Dr. Fenyong Liu (University of California, Berkeley) for initial suggestions and all members of the R.S. laboratory for critical discussions. This study was supported by National Institutes of Health Grants CA83525, CA91791, and DE14153, the Stop Cancer Foundation, the California Cancer Research Committee (to R.S.), Hallym University Research Fund 2004 Grant HRF-2004-45, and Korea Research Foundation Grant KRF-2004-015-C00416 (to M.J.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MHV-68, murine γ-herpesvirus-68; STM, signature-tagged mutagenesis; BAC, bacterial artificial chromosome; pfu, plaque-forming unit; dUTPase, deoxyuridine-triphosphatase; HSV-1, herpes simplex virus 1; HCMV, human cytomegalovirus.
References
- 1.Rickinson, A. B. & Kieff, E. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2575-2627. [Google Scholar]
- 2.Moore, P. S. & Chang, Y. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2803-2833. [Google Scholar]
- 3.Simas, J. P. & Efstathiou, S. (1998) Trends Microbiol. 6, 276-282. [DOI] [PubMed] [Google Scholar]
- 4.Virgin, H. W. & Speck, S. H. (1999) Curr. Opin. Immunol. 11, 371-379. [DOI] [PubMed] [Google Scholar]
- 5.Speck, S. H. & Virgin, H. W. (1999) Curr. Opin. Microbiol. 2, 403-409. [DOI] [PubMed] [Google Scholar]
- 6.Doherty, P. C., Christensen, J. P., Belz, G. T., Stevenson, P. G. & Sangster, M. Y. (2001) Philos. Trans. R. Soc. London B. Biol. Sci. 356, 581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nash, A. A., Dutia, B. M., Stewart, J. P. & Davison, A. J. (2001) Philos Trans. R. Soc. London B. Biol. Sci. 356, 569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart, J. P., Usherwood, E. J., Ross, A., Dyson, H. & Nash, T. (1998) J. Exp. Med. 187, 1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weck, K. E., Barkon, M. L., Yoo, L. I., Speck, S. H. & Virgin, H. I. (1996) J. Virol. 70, 6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weck, K. E., Kim, S. S., Virgin, H. I. & Speck, S. H. (1999) J. Virol. 73, 3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sunil-Chandra, N. P., Efstathiou, S. & Nash, A. A. (1992) J. Gen. Virol. 73, 3275-3279. [DOI] [PubMed] [Google Scholar]
- 12.Virgin, H. W., 4th, Latreille, P., Wamsley, P., Hallsworth, K., Weck, K. E., Dal Canto, A. J. & Speck, S. H. (1997) J. Virol. 71, 5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, F. (2003) Annu. Rev. Genet. 37, 3-29. [DOI] [PubMed] [Google Scholar]
- 14.Hensel, M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E. & Holden, D. W. (1995) Science 269, 400-403. [DOI] [PubMed] [Google Scholar]
- 15.Chiang, S. L., Mekalanos, J. J. & Holden, D. W. (1999) Annu. Rev. Microbiol. 53, 129-154. [DOI] [PubMed] [Google Scholar]
- 16.Mei, J. M., Nourbakhsh, F., Ford, C. W. & Holden, D. W. (1997) Mol. Microbiol. 26, 399-407. [DOI] [PubMed] [Google Scholar]
- 17.Lehoux, D. E., Sanschagrin, F. & Levesque, R. C. (1999) BioTechniques 26, 473-480. [DOI] [PubMed] [Google Scholar]
- 18.Haapa, S., Suomalainen, S., Eerikainen, S., Airaksinen, M., Paulin, L. & Savilahti, H. (1999) Genome Res. 9, 308-315. [PMC free article] [PubMed] [Google Scholar]
- 19.Haapa, S., Taira, S., Heikkinen, E. & Savilahti, H. (1999) Nucleic Acids Res. 27, 2777-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia, Q., Wu, T.-T., Liao, H.-I., Chernishof, V. & Sun, R. (2004) J. Virol. 78, 6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clambey, E. T., Virgin, H. W., 4th, & Speck, S. H. (2000) J. Virol. 74, 1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowler, P., Marques, S., Simas, J. P. & Efstathiou, S. (2003) J. Gen. Virol. 84, 3405-3416. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby, M. A., Virgin, H. W., 4th, & Speck, S. H. (2002) J. Virol. 76, 1790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macrae, A. I., Usherwood, E. J., Husain, S. M., Flano, E., Kim, I. J., Woodland, D. L., Nash, A. A., Blackman, M. A., Sample, J. T. & Stewart, J. P. (2003) J. Virol. 77, 9700-9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson, P. G., May, J. S., Smith, X. G., Marques, S., Adler, H., Koszinowski, U. H., Simas, J. P. & Efstathiou, S. (2002) Nat. Immunol. 3, 733-740. [DOI] [PubMed] [Google Scholar]
- 26.Coleman, H. M., de Lima, B., Morton, V. & Stevenson, P. G. (2003) J. Virol. 77, 2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lima, B. D., May, J. S. & Stevenson, P. G. (2004) J. Virol. 78, 5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simas, J. P., Bowden, R. J., Paige, V. & Efstathiou, S. (1998) J. Gen. Virol. 79, 149-153. [DOI] [PubMed] [Google Scholar]
- 29.Moorman, N. J., Virgin, H. W., 4th, & Speck, S. H. (2003) Virology 307, 179-190. [DOI] [PubMed] [Google Scholar]
- 30.Moorman, N. J., Willer, D. O. & Speck, S. H. (2003) J. Virol. 77, 10295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridgeman, A., Stevenson, P. G., Simas, J. P. & Efstathiou, S. (2001) J. Exp. Med. 194, 301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dyk, L. F., Virgin, H. W., 4th, & Speck, S. H. (2000) J. Virol. 74, 7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gangappa, S., van Dyk, L. F., Jewett, T. J., Speck, S. H. & Virgin, H. W., 4th (2002) J. Exp. Med. 195, 931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova, I. V., Virgin, H. W., 4th, & Speck, S. H. (2003) J. Virol. 77, 5731-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alba, M. M., Das, R., Orengo, C. A. & Kellam, P. (2001) Genome Res. 11, 43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackett, M., Stewart, J. P., Pepper, S. d. V., Chee, M., Efstathiou, S., Nash, A. A. & Arrand, J. R. (1997) J. Gen. Virol. 78, 1425-1433. [DOI] [PubMed] [Google Scholar]
- 37.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), Vol. 2, pp. 2399-2460. [Google Scholar]
- 38.Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., Zhu, H. & Liu, F. (2003) Proc. Natl. Acad. Sci. USA 100, 14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, D., Silva, M. C. & Shenk, T. (2003) Proc. Natl. Acad. Sci. USA 100, 12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bortz, E., Whitelegge, J. P., Jia, Q., Zhou, Z. H., Stewart, J. P., Wu, T. T. & Sun, R. (2003) J. Virol. 77, 13425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hengel, H., Lucin, P., Jonjic, S., Ruppert, T. & Koszinowski, U. H. (1994) J. Virol. 68, 289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldo, A. M. & McClure, M. A. (1999) J. Virol. 73, 7710-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, R., Wang, H. & Mansky, L. M. (2002) J. Gen. Virol. 83, 2339-2345. [DOI] [PubMed] [Google Scholar]
- 44.Payne, S. L. & Elder, J. H. (2001) Curr. Protein Pept. Sci. 2, 381-388. [DOI] [PubMed] [Google Scholar]
- 45.Lirette, R. & Caradonna, S. (1990) J. Cell Biochem. 43, 339-353. [DOI] [PubMed] [Google Scholar]
- 46.Pyles, R. B., Sawtell, N. M. & Thompson, R. L. (1992) J. Virol. 66, 6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorman, N. J., Lin, C. Y. & Speck, S. H. (2004) J. Virol. 78, 10282-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.