Abstract
CD8+ T lymphocytes play a major role in cellular-mediated immune responses to foreign antigen. We have previously demonstrated that costimulation of purified human CD8+ T cells induces de novo expression of the CD4 molecule and that ligation of CD4 on this cell type modulates CD8+ T cell activity in vitro. Herein, we investigate how the CD4 molecule expressed on murine CD8+ T cells contributes to CD8+ cell responses in vivo by employing adoptive transfer of CD8 cells from CD4 knockout mice into severe combined immunodeficient (SCID) recipients. Transfer of these cells into syngeneic SCID mice resulted in a decreased immune response to infection by lymphocytic choriomeningitis virus. These decreased responses occurred even in the presence of CD4+ T cells, indicating that this was truly a CD8-cell defect. Similarly, transfer of CD8+ T cells incapable of expressing CD4 into allogeneic SCID mice resulted in a decreased response to alloantigens compared with that of normal CD8+ T cells. Therefore, CD4 expression on CD8 T lymphocytes modulates cytotoxic T lymphocyte function and is critical in vivo for optimal cell-mediated immunity to viral and alloantigens.
Keywords: antiviral response, CD8+ T cells, alloantigen response, cytotoxic T lymphocytes
T cell development in the thymus is characterized by the appearance and disappearance of cell-surface CD4 and CD8, which are classic markers of ontogeny. It was previously thought that after synchronous expression of CD4 and CD8, the developing thymocyte becomes CD4 or CD8 single positive, permanently turning off expression of one of these molecules. These single-positive cells are then exported to the periphery to perform their respective functions: CD4+ cells serving primarily as T helper (Th) cells, and CD8+ cells serving primarily as cytotoxic T lymphocytes (CTLs). Our current investigation focuses on the CD8+ T cell population. CD8+ T cells become activated to perform their function when they encounter a specific antigen presented in the context of MHC class I and are costimulated by one of many molecules present on antigen-presenting cells (APCs) (1). A well described series of events is triggered after activation of CD8+ T cells, primarily resulting in CTLs that kill antigen-expressing target cells.
In contrast to the notion that commitment to the CD4 or CD8 lineage is complete or permanent after thymopoiesis, extrathymic CD8+CD4+ cells have been identified in many organisms, including humans, monkeys, mice, rats, swine, and chickens (reviewed in ref. 2). Various disease conditions appear to affect the levels of CD8+CD4+ T cells. In humans, infection with HIV, human T cell leukemia virus, Epstein–Barr virus, human herpesvirus 6, and increasing age have been linked with increased percentages of CD8+CD4+ T cells in the peripheral blood (3–6). In mice, increases in the CD8+CD4+ population have been observed after inoculation with reovirus (7), or recombinant adenovirus (8). In most cases, the rise in the percentage of CD8+CD4+ cells appears to be a direct result of cellular activation, with these cells displaying a phenotype of activated or previously activated T cells. We and others have previously shown that costimulation of human CD8+ T cells results in the de novo expression of CD4, consistent with the activated phenotype seen in vivo (9–13).
The CD4 molecule performs multiple functions on several cell types, including acting as an adhesion molecule, an inducer of cellular activation, and a chemotactic receptor (14, 15). We have previously determined that CD4 can function as a chemotactic receptor on activated human CD8+ T cells, allowing them to migrate in response to IL-16 (16). Furthermore, we have recently determined that CD4 can directly influence human CD8 T cell function in vitro (9). Ligation of CD4 on CD8 T cells sends a signal through the CD4-associated signaling molecule Lck, which results in up-regulation of Fas ligand (FasL) and IFN-γ, two molecules important in CD8 T cell function (9). Further, CD4 plays a direct role in CTL-mediated lysis of allogeneic B cell targets through interaction with MHC class II at the time of lytic activity. Infection of these CD8+CD4+ cells by HIV perturbs this activity by directly down-regulating cell-surface CD4 expression and consequently dysregulating FasL and IFN-γ production. Thus, CD4 plays a direct role in modulating CD8+ T cell responses in vitro, suggesting that CD4 has an important role in CD8 cell responses to antigen in vivo.
In the present study, we sought to investigate how CD4 expressed on the surface of CD8+ T cells contributes to the immune response in vivo. We used an adoptive transfer system in which purified CD8+ T cells obtained from normal wild-type (WT) mice or CD4 knockout (CD4-/-) mice were introduced into severe combined immunodeficient (SCID) mice. This system allowed us to examine the viral antigen- or alloantigen-specific responses of transferred CD8+ T cells, which either possessed or lack the ability to express CD4. We demonstrate that the CD4 molecule is required for optimal CD8+ T cell function in response to viral or alloantigens in vivo.
Methods
Cell Culture. CD8+ T cells from the spleens of lymphocytic choriomeningitis virus (LCMV) T cell receptor (TCR)-transgenic mice (B6 D2-TgNTCRLCMV 327sdz [P14] mice, The Jackson Laboratory) (17) were purified by magnetic cell sorting with CD8 microbeads (Miltenyi Biotec, Auburn, CA) and were stimulated in the presence of GP33–41 peptide (0.1 μg/ml) or plate-bound anti-mouse CD3 (5 μg/ml; Coulter) and soluble anti-mouse CD28 (5 μg/ml; Coulter). Cells were analyzed by flow cytometry at the specified time point after stimulation with CD8-PerCP and CD4-allophycocyanin mAbs (mouse) (Pharmingen); gating was used to highlight the CD8+ population. Dot-plot cursor settings were based on mouse isotype control staining and samples stained with single-color antibodies in the presence of isotype-control antibodies (12).
Adoptive Transfer. CD8+ T cells were purified by positive selection with magnetic activated cell sorting (MACS) (Miltenyi, Auburn, CA). Single-cell suspensions of splenocytes from WT C57BL/6 mice or CD4-/- C57BL/6-Cd4tm1Mak mice (The Jackson Laboratory) were stained with anti-CD8 microbeads and passed through the column/magnet apparatus. The positively selected fraction was restained with the same bead-conjugated antibodies and passed through the apparatus a second time. Typical purity was >97% CD8+ T cells, as assessed by flow cytometry. In some experiments, purified CD8+ cells were labeled with CFSE by incubating with 2.5 μM CFSE (Molecular Probes) for 7 min. Excess label was quenched with FCS, and cells were washed. CD8+ cells (1 × 107) were transferred into syngeneic C57BL/6-Prkdcscid (BL/6 SCID) mice (The Jackson Laboratory) or allogeneic C.B-17 scid/scid mice by i.v. tail injection in 0.5 ml of MEM. In some studies, purified CD4+ Th cells were isolated from WT mice by using MACS and anti-CD4 microbeads (typical purity >97%) and 1 × 107 cells introduced into SCID mice along with the CD8+ T cells.
LCMV Infection. BL/6 WT or BL/6 SCID mice were infected with 2 × 105 plaque-forming units (pfu) of LCMV-Armstrong through i.p. injection. Mice were killed at the indicated time points, and organs were harvested as described in refs. 18 and 19. In LCMV-infected mice in the adoptive transfer studies, the amount of CD8 cells recovered per spleen from the WT versus the CD4-/- groups was not statistically significant (P = 0.3356, Wilcoxon rank-sum test).
Assessment of Antiviral Immune Response. The antiviral response was assessed at the indicated times after cell transfer and/or LCMV infection as described in ref. 19. Phenotypic analysis of cell-surface marker and intracellular IFN-γ staining was performed as described by using CD4-FITC or -PE, CD8-allophycocyanin, and IFN-γ-FITC mAbs (Pharmingen). MHC class I tetramer staining was performed by using CD8-FITC, CD4-PE mAbs, and either DbNP396–404 (DbNP396) or DbGP33–41 (DbGP33)-allophycocyanin-conjugated tetramers (as indicated in the figures), and the numbers of CD8+, tetramer+ cells were determined by flow cytometry and gating (19). CTL activity was determined by a standard chromium release assay of LCMV-infected MC57 target cells (20). Virus titers were determined with infected mouse serum as described in ref. 18.
Assessment of Allogeneic Immune Response. Spleens from recipient C.B-17 scid/scid mice receiving allogeneic (CFSE-labeled) cells were assessed for transferred cells by flow cytometry for CFSE and staining with CD8-PE and CD4-allophycocyanin mAbs. Cytotoxicity assays were performed by using a standard chromium release assay on P815 (H2d) cells, which are a mastocytoma cell line derived from DBA/2 mice. IFN-γ enzyme-linked immunospots (ELISPOTs) were performed as described by using either irradiated (1,200 rad) P815 or WT BL/6 splenocytes as stimulator/feeder cells (19).
Results
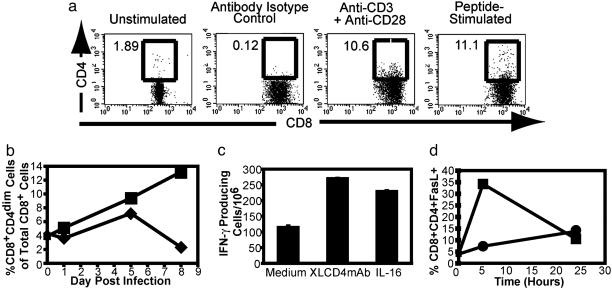
CD4 Expression on CD8+ T Cells. We have previously shown that antibody- or alloantigen-mediated costimulation results in de novo CD4 expression on human CD8+ T cells. In the current study, we first examined whether peptide-specific stimulation of murine CD8+ T cells results in de novo CD4 expression. CD8+ T cells from transgenic mice expressing an MHC class I-restricted TCR specific for LCMV glycoprotein residues 33–41 (GP33–41) (17) were stimulated with the GP33–41 peptide. After peptide stimulation, CD4 cell-surface expression was observed on a similar number of CD8+ T cells as that seen with antibody-mediated costimulation (Fig. 1a). These CD8+CD4+ cells also expressed high levels of the activation molecule CD25 and were observed to down-modulate cell-surface expression of the transgenic TCR (unpublished data), indicating that these cells were highly activated. We further analyzed the ability of LCMV to induce CD4 expression on CD8+ T cells after LCMV infection in WT C57BL/6 mice. Within 5 days of infection, we observed an increase in the percentages of CD8+CD4dim cells in the spleen and lymph nodes of infected mice (Fig. 1b). Increases in this population were modest in the spleen and appeared to resolve to preinfection levels by day 8. In the lymph nodes, however, we saw a dramatic increase in the percentages of CD8+CD4dim cells that continued through day 8 postinfection. No significant increases or decreases in the percentages of CD8+CD4dim cells were observed in the liver, lung, or intraepithelial lymphocytes, although increased total CD8 cell numbers were observed after antigen-driven expansion in all tissues tested (data not shown). The majority of the CD8+CD4dim cells also displayed a CD62Llow phenotype, indicating that they were activated effector cells, and 5–10% of these cells produced IFN-γ in response to stimulation with either the GP33–41 or NP396–404 peptide (data not shown). Although CD4 expression was less robust in mice ex vivo than we had previously seen on stimulated human CD8 cells ex vivo, in vitro-stimulated murine CD8+ T cells expressed IFN-γ and FasL after CD4 ligation (Fig. 1 c and d), consistent with the phenomena that were observed with human CD8+ T cells. Combined, these results allowed us to use the mouse system to examine the significance of CD4 expression on CD8+ T cells in vivo.
Fig. 1.
Expression of CD4 on murine CD8+ T cells. (a) CD4 expression on purified murine LCMV TCR-transgenic CD8+ splenocytes. Cells were analyzed by flow cytometry for CD4 and CD8 expression 2 days after stimulation; gating was used to highlight the CD8+ population. Numbers above each box in the dot plots represent the percentage of CD8+ cells that express CD4. (b) Percentages of CD8+CD4dim cells after LCMV infection of WT C57BL/6 mice. Cells from the spleen (diamonds) or lymph nodes (squares) were analyzed for expression of CD4 and CD8 at the indicated times postinfection with LCMV. Numbers represent the percentage of CD8+CD4dim cells of the total CD8+ T cell population and were determined by gating similar to that indicated above. (c) IFN-γ production after ligation of CD4 on in vitro-stimulated CD8 cells, as measured by ELISPOT. XL-CD4 mAb refers to treatment with cross-linked CD4 mAb. Data are expressed as the number of IFN-γ+ cells per million. (d) FasL expression on the surface of CD8+CD4+ T cells after treatment with medium (circles) or cross-linked CD4 mAb (XL-CD4; squares).
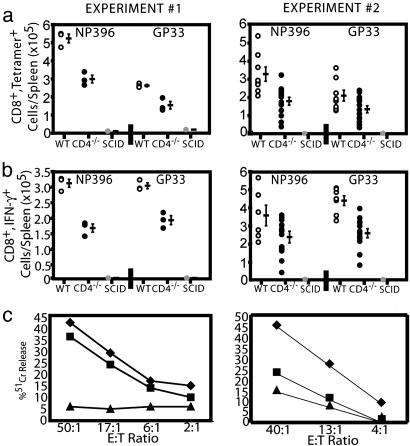
CD4 Expression and Antiviral Immunity. The CTL response to acute LCMV infection has previously been shown to not require CD4+ T cell help (21). To determine whether the induction of CD4 on CD8+ T cells that we observed after LCMV infection was important in the antiviral response in vivo, we used a murine adoptive-transfer system in which purified CD8+ T cells from WT or CD4-/- mice were transferred into syngeneic SCID mice, which themselves lack endogenous T cell responses but possess APCs. Mice were then immediately infected with LCMV, and the LCMV-specific CD8+ T cell response was assessed (Fig. 2). Although equivalent numbers of CD8 cells were transferred for all conditions and there was no difference in the number of CD8 cells residing in the spleens of each group 8 days after transfer and infection, the number of LCMV-specific cells in the spleens of mice transferred with CD8 cells from the WT mice was roughly twice that found in mice transferred with CD8 cells from CD4-/- mice (Fig. 2a). This phenomenon was also observed in the lymph nodes of these infected mice (unpublished data). CD8+ cells from infected CD4-/- mice generated significantly fewer IFN-γ-producing cells in response to stimulation with the two MHC class I-restricted immunodominant peptides GP33–41 and NP396–404 than did cells from infected WT mice (Fig. 2b), consistent with our earlier in vitro observations that CD4 ligation results in increases in IFN-γ production (Fig. 1) (9). The level of intracellular IFN-γ, however, was not different between the two groups as determined by mean florescence intensity of IFN-γ staining (unpublished data). No significant responses were observed to the MHC class II-restricted GP61–80 peptide, further demonstrating the purity of the transferred cells and the inability of the cells to react with a non-MHC class I-restricted peptide (data not shown). CD8+ cells from WT mice also displayed significantly (P ≤ 0.05) greater virus-specific CTL activity (Fig. 2c). When the number of CD8+ cells in the spleen was taken into account and the mice were assessed for CTL lytic units [one lytic unit (LU20) is the number of effector cells to give 20% lysis in a standard 51Cr release assay], mice transferred with CD8+ T cells from CD4-/- mice demonstrated reduced CTL activity (18 ± 0.5 LU20 versus 46 ± 2.7 LU20 for mice transferred with WT CD8+ T cells). These values were significantly different (P ≤ 0.05, Wilcoxon rank-sum test). Virus titers (18) in the peripheral blood of mice transferred with WT CD8+ T cells (2,837 ± 921 pfu/ml) were significantly lower than in those mice transferred with CD8+ T cells from CD4-/- mice (6,126 ± 1,578 pfu/ml) (P ≤ 0.05, Wilcoxon rank-sum test). Similar results were observed in five separate experiments, totaling 20 mice receiving WT CD8+ cells, 25 mice receiving CD4-/- cells, and 15 mice receiving no cells (control). Thus, those mice transferred with CD8+ T cells capable of expressing CD4 generated a significantly greater antiviral response after LCMV infection than those receiving CD8+ T cells lacking the ability to express CD4.
Fig. 2.
Role of CD4 in the CD8+ T cell-mediated antiviral response in vivo. This represents two experiments of six total in vivo adoptive transfer experiments. In experiment 1, 3 SCID mice received CD8+ T cells from WT mice (open circles), 3 SCID mice received CD8+ T cells from CD4-/- mice (black circles), and 2 SCID mice (denoted by “SCID”) received no CD8+ T cells (gray circles). In experiment 2, 8 SCID mice received CD8+ T cells from WT mice (open circles), 13 SCID mice received CD8+ T cells from CD4-/- mice (black circles), and 3 SCID mice received no CD8+ T cells (gray circles). (a) Quantitation of LCMV-specific CD8+ cell response by tetramer staining 8 days postinfection. Each circle represents an individual mouse, and the dashes represent the average values ± SD for each epitope indicated. Differences in CD8+, tetramer+ cell numbers between animals receiving WT and CD4-/-CD8+ T cells were statistically significant (P ≤ 0.05, Wilcoxon rank-sum test). These values are also significantly different from the negative (SCID) control group. (b) LCMV peptide-specific IFN-γ production 8 days postinfection. Each circle represents an individual mouse, and the dashes represent the average values for each epitope. Differences between groups receiving WT or CD4-/-CD8+ T cells are statistically significant (P ≤ 0.05, Wilcoxon rank-sum test). SCID controls showed undetectable staining. (c) CTL activity of splenocytes from SCID mice receiving no cells (triangles) and CD8 cells from WT (diamonds) or CD4-/- (squares) mice 8 days postinfection. Differences between groups receiving WT or CD4-/-CD8+ T cells were significant (P ≤ 0.05, Wilcoxon rank-sum test).
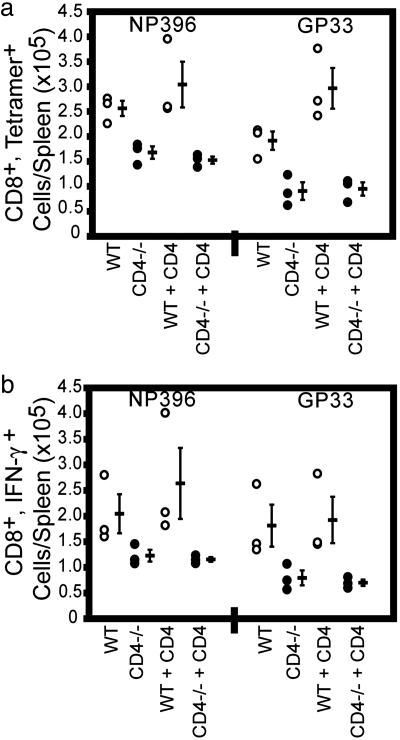
We next determined whether contamination with bona fide CD4+ Th cells could have had an effect on CD8+ T cells from WT mice during acute LCMV infection. SCID mice were transferred with purified CD8+ T cells as described above, and, in addition, some of these mice also received purified CD4+ cells from WT mice (Fig. 3). Reconstituted mice were infected with LCMV-Armstrong, and the LCMV-specific response generated by the transferred CD8+ cells was assessed. On day 8 postinfection, there were significant differences in the LCMV-specific CD8+ T cell responses between mice transferred with WT and CD4-/-CD8+ T cells, similar to that described above (P ≤ 0.05, Wilcoxon rank-sum test). The addition of Th cells to these mice did not result in increased numbers of LCMV-specific cells and did not alter the deficient response of mice transferred with CD8 cells from CD4-/- mice. In addition, there were significantly lower titers of virus in the sera of mice receiving WT CD8+ T cells plus Th cells (1,387 ± 371 pfu/ml) compared with mice receiving CD4-/-CD8+ T cells plus Th cells (2,613 ± 717 pfu/ml) (P ≤ 0.05, Wilcoxon rank-sum test). Thus, the decreased anti-LCMV response of CD8 cells from CD4-/- mice during acute infection is a result of the inability to express CD4 de novo rather than a lack of Th cell activity.
Fig. 3.
Antiviral response of CD8+ T cells from WT and CD4-/- mice in the presence and absence of Th cells. The data shown are derived from one of three similar experiments, with three mice in each group. The groups received cells as follows: CD8 cells from WT mice, CD8 cells from CD4-/- mice, CD8 cells from WT mice plus CD4 cells from WT mice, and CD8 cells from CD4-/- mice plus CD4 cells from WT mice. Each open circle represents an individual mouse transferred with CD8 cells from WT mice, each filled circle represents an individual mouse transferred with CD8 cells from CD4-/- mice, and dashes represent the average values ± SD of each population for each epitope. (a) Enumeration of LCMV-specific CD8+ T cells by tetramer staining 8 days postinfection. (b) Enumeration of IFN-γ-producing cells after ex vivo LCMV peptide stimulation 8 days postinfection.
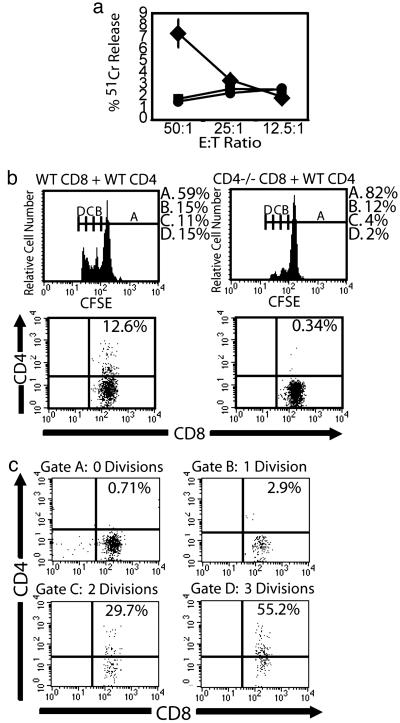
CD4 Expression on CD8+ T Cells After Allogeneic Stimulation in Vivo. We examined whether CD4 expression by CD8+ T cells influenced generation of an immune response in another model system, particularly one that requires bona fide CD4+ Th cells for the establishment of a CD8 cell response. We assessed CD8+ T cell reactivity against alloantigens by injecting C.B-17 scid/scid mice (H-2d) with purified, CFSE-labeled CD8+ cells from WT or CD4-/- CB57BL/6 (H-2b) mice. Some of these mice also received purified CD4+ cells from WT C57BL/6 mice. Five days after transfer, the spleens of the recipient mice were assessed for the adoptively transferred cells. Engraftment of CD8+ cells in the absence of CD4+ cells was relatively low, although CD8 cells from WT mice had a slightly higher level of engraftment than did CD8 cells from CD4-/- mice (data not shown). Because of this improved engraftment, we consequently performed detailed analysis of allogeneic responses only in animals receiving both CD4+ and CD8+ T cells (Fig. 4 and Table 1). In the presence of CD4+ cells, CD8+ cells displayed substantial levels of engraftment; however, mice receiving CD4-/-CD8+ (n = 6) cells had lower levels of engraftment than did mice receiving WT CD8+ (n = 5) cells (Table 1). The number of IFN-γ producing CD8+ cells was also highest in mice receiving WT CD8+ cells. These results were similar when the alloreactive cells were stimulated with either irradiated splenocytes or MHC class II negative P815 cells, thus reflecting IFN-γ production by MHC class I-restricted CD8+ T cells and not by CD4+ T cells or MHC class II-restricted CD8+ T cells. Ex vivo cytotoxic alloreactivity was found in mice receiving WT CD8+ plus CD4+ cells, whereas CTL activity was below the levels of detection in all other populations (Fig. 4a). These results are representative of three separate experiments involving similar numbers of mice. We further analyzed CD8+ cells in mice receiving WT CD8+ plus CD4+ cells (n = 9) or CD4-/-CD8+ cells plus CD4+ cells (n = 9) for cell-surface CD4 expression (Fig. 4 b and c). CD4 expression was detectable on CD8+ cells in mice receiving WT CD8+ cells plus CD4 cells, whereas no significant expression was detected in mice receiving CD4-/-CD8+ cells plus CD4 cells, indicating that the CD4 molecule observed on the CD8 cells was not the result of receptor shedding by the CD4+ helper cells. Those CD8+ cells that had undergone the most divisions displayed incrementally the highest levels of CD4 expression. WT CD8 cells also underwent more cell division in vivo than did CD8+ cells from CD4-/- mice. Thus, in the presence of CD4+ cell help, CD8+ T cells that are capable of expressing CD4 in vivo generate a much greater alloreactive response than do CD8+ T cells genetically incapable of expressing CD4.
Fig. 4.
Response of CD8+ T cells from WT and CD4-/- mice to alloantigen in vivo. C.B-17 scid/scid (H2d) mice received CD8+ T cells from either WT or CD4-/- BL/6 (H2b) mice plus or minus CD4+ T cells from WT BL/6 mice. (a) Alloantigen-specific CTL activity of splenocytes from C.B-17 scid/scid mice transferred with CD8 cells from WT mice plus CD4 cells from WT mice (diamonds; n = 5) and CD8 cells from CD4-/- mice plus CD4 cells from WT mice (squares; n = 6), and mice receiving no cells (circles; n = 2). The data shown are derived from one of three similar experiments and depict average values ± SD for mice in each group. (b) Flow cytometric analysis of C.B-17 scid/scid mice receiving WT CD8+ cells plus WT CD4 cells (Left) and CD4-/-CD8+ T cells plus WT CD4 cells (Right). (Upper) CFSE staining of transferred CD8+ cells, with the markers drawn to indicate cell populations after different numbers of cell divisions (indicated above each marker region). The legends at the right represent the percentage of cells within the marker region after the indicated number of cell divisions. (Lower) CD4 and CD8 analysis of transferred CFSE+ cells. (c) CD4 and CD8 staining profiles of populations within gates drawn at the indicated number of divisions of mice in b transferred with WT CD8+ cells plus WT CD4+ cells. Data represent one of a total of nine mice in each group from one of two similar experiments.
Table 1. Evaluation of CD8+ T cell responses in the spleens of C.B.-17 scid/scid mice.
| Criterion | WT CD8+ WT CD4 (n = 5) | CD4-/-CD8+ WT CD4 (n = 6) | SCID (n = 2) | P |
|---|---|---|---|---|
| No. of engrafted CD8+ cells per spleen | 227,213 (±61,428) | 80,409 (±23,681) | 0.00 | 0.041 |
| % CFSE+ cells ≥1 division | 41.6 (±6.65) | 21.7 (±3.75) | 0.00 | 0.014 |
| IFN-γ+ cells per 100,000 CD8+ cells (ELISPOT) | 3,114 (±1,438) | 214 (±194) | 0.00 | 0.017 |
The average values ± SD for the number of mice (n) indicated is provided.
Discussion
In this study, we have determined that the CD4 molecule expressed on CD8+ T cells has a functional role in the allogeneic and antiviral response in vivo. Our data suggest that the ligation of CD4, either by MHC class II on an APC or by secreted IL-16, modulates the CTL response. We have previously shown that ligation of CD4 on human CD8+ T cells results in increased expression of IFN-γ (9). IFN-γ may then have direct or indirect effects, including the up-regulation of Fas on APCs, rendering these cells more susceptible to killing by CD8+ T cells expressing FasL (22–25). IFN-γ has been determined to be important in controlling LCMV infection (26). Although the relatively low amount of CD8+ cells transferred into SCID mice in our studies was incapable of clearing LCMV-Armstrong within 8 days, significant differences were observed in the viral titers between mice transferred with WT and CD4-/-CD8+ T cells. This is consistent with the lower levels of IFN-γ produced by CD8+ cells incapable of expressing CD4 and further suggests that the IFN-γ produced after CD4 ligation has a role in the antiviral response. CD4 ligation is known to induce FasL expression on monocytes and primes Th cells to express FasL after TCR stimulation (23, 27). FasL expression by these cells has been demonstrated to subsequently induce apoptosis in cells expressing Fas. The similar effects seen after ligation of CD4 on CD8+ T cells suggest that reduced expression of FasL on CD8 cells from CD4-/- mice contributes to their decreased ability to clear virus in vivo.
CD4 is highly regulated during T cell development and expression is observed on most T cell progenitors at different times during thymopoiesis. The level of CD4 expression typically peaks at the same time as that of CD8 expression, at which time the CD4CD8 double-positive cell undergoes lineage commitment to become a CD4 or CD8 single-positive cell. These single-positive cells are then exported to the periphery where they travel to secondary lymphoid organs and tissues. Our data suggest that, after cellular activation, a subset of CD8+ T cells are induced to express CD4. We have previously determined that the newly expressed CD4 on human CD8+ T cell modulates CTL, FasL, and IFN-γ responses after ligation in vitro (9). Our current studies suggest that CD4 enhances the CD8+ T cell response against viral and alloantigens in vivo through a similar mechanism. This postthymic reexpression of CD4 represents another developmental step in CD8+ T cell differentiation.
The CD4 molecule could influence several aspects of CD8 cell function. Our previous studies (16) indicate that CD4 on the highly activated CD8+ T cells serves as a chemotactic receptor for IL-16. IL-16 is a proinflammatory cytokine produced by variety of cells including Th and CD8+ T cells (14). IL-16 production by these cells could attract activated CD8+ T cells to sites of inflammation and viral replication. Alternatively, our current results may reflect interaction of CD4 on CD8+ T cells with MHC class II on APCs. Our results suggest that, after ligation of CD4 on CD8+ T cells with its natural ligands MHC class II or IL-16, IFN-γ and FasL expression are up-regulated, inducing direct and indirect antiviral effects, including possible up-regulation of Fas antigen expression and induction of apoptosis of infected target cells. Together, these events could provide a mechanism through which CD4 directly modulates CD8+ CTL function. Interestingly, the CD4 molecule has been found, under various conditions, on a subset of hematopoietic progenitor cells (28, 29), B cells (30), NK cells (31), eosinophils (32), monocytes (33), neutrophils (34), mast cells/basophils (35), and microglia (36). Our current studies specifically examine the function of CD4 only on purified CD8+ T cells, and further studies are required to fully define the complex role that this molecule plays in the immune response.
CD8+ T cells can respond to antigen in the absence of CD4+ T cell help (21, 37–40). However, the absence of CD4+ T cells during the priming phase of the primary CD8+ T cell response does not allow the cell to differentiate into fully functional memory cells (38, 41, 42). Our data support the previous studies showing that CD8 cells can generate a strong response in the absence of CD4+ T cells, but we have determined that the CD4 molecule is required for this to occur optimally. We have not examined the effect(s) of the CD4 molecule on the generation of memory CD8 cells, which is the subject of future investigation. There is recent evidence from Tyznik et al. (43) and Pearce et al. (44) that a subset of CD8+ T cells in CD4-/- mice react with MHC class II-restricted epitopes. Our studies show a similar quantitative defect in CD8 cells from CD4-/- mice as that seen from these studies. Whereas the absence of CD4 expression during development in the thymus likely has the effect of selecting for these MHC class II-recognizing cells, our observations provide an additional mechanism for the defects observed in CD8+ T cells from CD4-/- mice. Our studies showed no increased activity to MHC class II-restricted LCMV peptides, decreased overall CD8+ T cell responses to allogeneic cells that would be disparate in MHC class II, and decreased responses to stimulating cells lacking MHC class II. This finding suggests that the observed defect in the CD8+ T cell response from CD4-/- mice is at the level of MHC class I-restricted CD8+ T cells. In addition, we show induced CD4 expression on truly MHC class I-restricted (TCR transgenic) CD8+ T cells in vitro, induction of CD4 expression on antigen-responsive CD8 cells in vivo, and that ligation of CD4 on these cells induces functional responses. Taken together, our data indicate that CD4 plays a direct role in the primary CD8 immune response and could have a role in “programming” effector and memory CD8 cell differentiation after response to antigen (45–48).
Acknowledgments
This work was supported by National Institutes of Health Grants AI057057, AI48392, and AI36059 and the University of California, Los Angeles, Center for AIDS Research (AI28697).
Author contributions: S.G.K., J.K.W., R.A., and J.A.Z. designed research; S.G.K., J.K.W., N.R.J., and Z.G. performed research; S.G.K., J.K.W., and C.M.R.K. analyzed data; and S.G.K. and J.A.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, antigen-presenting cell; CTL, cytotoxic T lymphocyte; LCMV, lymphocytic choriomeningitis virus; pfu, plaque-forming units; SCID, severe combined immunodeficient; TCR, T cell receptor; Th, T helper.
References
- 1.Seder, R. A. & Ahmed, R. (2003) Nat. Immunol. 4, 835-842. [DOI] [PubMed] [Google Scholar]
- 2.Zuckermann, F. A. (1999) Vet. Immunol. Immunopathol. 72, 55-66. [DOI] [PubMed] [Google Scholar]
- 3.Laux, I., Khoshnan, A., Tindell, C., Bae, D., Zhu, X., June, C. H., Effros, R. B. & Nel, A. (2000) Clin. Immunol. 96, 187-197. [DOI] [PubMed] [Google Scholar]
- 4.Imlach, S., McBreen, S., Shirafuji, T., Leen, C., Bell, J. E. & Simmonds, P. (2001) J. Virol. 75, 11555-11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macchi, B., Graziani, G., Zhang, J. & Mastino, A. (1993) Cell. Immunol. 149, 376-389. [DOI] [PubMed] [Google Scholar]
- 6.Lusso, P., De Maria, A., Malnati, M., Lori, F., DeRocco, S. E., Baseler, M. & Gallo, R. C. (1991) Nature 349, 533-535. [DOI] [PubMed] [Google Scholar]
- 7.Periwal, S. B. & Cebra, J. J. (1999) J. Virol. 73, 7633-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillemeyer, P., White, M. D. & Pascual, D. W. (2002) Cell. Immunol. 215, 173-185. [DOI] [PubMed] [Google Scholar]
- 9.Kitchen, S. G., Jones, N. R., LaForge, S., Whitmire, J. K., Vu, B. A., Galic, Z., Brooks, D. G., Brown, S. J., Kitchen, C. M. & Zack, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 8727-8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan, Y. B., Landay, A. L., Zack, J. A., Kitchen, S. G. & Al-Harthi, L. (2001) Immunology 103, 270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, L. P., Riley, J. L., Carroll, R. G., June, C. H., Hoxie, J., Patterson, B. K., Ohshima, Y., Hodes, R. J. & Delespesse, G. (1998) J. Exp. Med. 187, 1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitchen, S. G., Korin, Y., Roth, M. D., Landay, A. & Zack, J. A. (1998) J. Virol. 72, 9054-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flamand, L., Crowley, R. W., Lusso, P., Colombini-Hatch, S., Margolis, D. M. & Gallo, R. C. (1998) Proc. Natl. Acad. Sci. USA 95, 3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Center, D. M., Kornfeld, H., Ryan, T. C. & Cruikshank, W. W. (2000) Immunol. Today 21, 273-280. [DOI] [PubMed] [Google Scholar]
- 15.Ravichandran, K. S., Collins, T. L. & Burakoff, S. J. (1996) Curr. Top. Microbiol. Immunol. 205, 47-62. [DOI] [PubMed] [Google Scholar]
- 16.Kitchen, S. G., LaForge, S., Patel, V. P., Kitchen, C. M., Miceli, M. C. & Zack, J. A. (2002) Blood 99, 207-212. [DOI] [PubMed] [Google Scholar]
- 17.Pircher, H., Burki, K., Lang, R., Hengartner, H. & Zinkernagel, R. M. (1989) Nature 342, 559-561. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed, R., Salmi, A., Butler, L. D., Chiller, J. M. & Oldstone, M. B. (1984) J. Exp. Med. 160, 521-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murali-Krishna, K., Altman, J. D., Suresh, M., Sourdive, D. J., Zajac, A. J., Miller, J. D., Slansky, J. & Ahmed, R. (1998) Immunity 8, 177-187. [DOI] [PubMed] [Google Scholar]
- 20.van der Most, R. G., Sette, A., Oseroff, C., Alexander, J., Murali-Krishna, K., Lau, L. L., Southwood, S., Sidney, J., Chesnut, R. W., Matloubian, M. & Ahmed, R. (1996) J. Immunol. 157, 5543-5554. [PubMed] [Google Scholar]
- 21.Matloubian, M., Concepcion, R. J. & Ahmed, R. (1994) J. Virol. 68, 8056-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyaizu, N., McCloskey, T. W., Than, S., Hu, R., Kalyanaraman, V. S. & Pahwa, S. (1994) Blood 84, 2622-2631. [PubMed] [Google Scholar]
- 23.Tateyama, M., Oyaizu, N., McCloskey, T. W., Than, S. & Pahwa, S. (2000) Blood 96, 195-202. [PubMed] [Google Scholar]
- 24.Goodbourn, S., Didcock, L. & Randall, R. E. (2000) J. Gen. Virol. 81, 2341-2364. [DOI] [PubMed] [Google Scholar]
- 25.Siegel, R. M., Chan, F. K., Chun, H. J. & Lenardo, M. J. (2000) Nat. Immunol. 1, 469-474. [DOI] [PubMed] [Google Scholar]
- 26.Shtrichman, R. & Samuel, C. E. (2001) Curr. Opin. Microbiol. 4, 251-259. [DOI] [PubMed] [Google Scholar]
- 27.Oyaizu, N., Adachi, Y., Hashimoto, F., McCloskey, T. W., Hosaka, N., Kayagaki, N., Yagita, H. & Pahwa, S. (1997) J. Immunol. 158, 2456-2463. [PubMed] [Google Scholar]
- 28.Louache, F., Debili, N., Marandin, A., Coulombel, L. & Vainchenker, W. (1994) Blood 84, 3344-3355. [PubMed] [Google Scholar]
- 29.Muench, M. O., Roncarolo, M. G. & Namikawa, R. (1997) Blood 89, 1364-1375. [PubMed] [Google Scholar]
- 30.Moir, S., Lapointe, R., Malaspina, A., Ostrowski, M., Cole, C. E., Chun, T.-W., Adelsberger, J., Baseler, M., Hwu, P. & Fauci, A. S. (1999) J. Virol. 73, 7972-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentin, A., Rosati, M., Patenaude, D. J., Hatzakis, A., Kostrikis, L. G., Lazanas, M., Wyvill, K. M., Yarchoan, R. & Pavlakis, G. N. (2002) Proc. Natl. Acad. Sci. USA 99, 7015-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucey, D. R., Dorsky, D. I., Nicholson-Weller, A. & Weller, P. F. (1989) J. Exp. Med. 169, 327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filion, L. G., Izaguirre, C. A., Garber, G. E., Huebsh, L. & Aye, M. T. (1990) J. Immunol. Methods 135, 59-69. [DOI] [PubMed] [Google Scholar]
- 34.Biswas, P., Mantelli, B., Sica, A., Malnati, M., Panzeri, C., Saccani, A., Hasson, H., Vecchi, A., Saniabadi, A., Lusso, P., et al. (2003) Blood 101, 4452-4456. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., Li, L., Wadley, R., Reddel, S. W., Qi, J. C., Archis, C., Collins, A., Clark, E., Cooley, M., Kouts, S., et al. (2001) Blood 97, 3484-3490. [DOI] [PubMed] [Google Scholar]
- 36.Jordan, C. A., Watkins, B. A., Kufta, C. & Dubois-Dalcq, M. (1991) J. Virol. 65, 736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shedlock, D. J., Whitmire, J. K., Tan, J., MacDonald, A. S., Ahmed, R. & Shen, H. (2003) J. Immunol. 170, 2053-2063. [DOI] [PubMed] [Google Scholar]
- 38.Shedlock, D. J. & Shen, H. (2003) Science 300, 337-339. [DOI] [PubMed] [Google Scholar]
- 39.Rahemtulla, A., Fung-Leung, W. P., Schilham, M. W., Kundig, T. M., Sambhara, S. R., Narendran, A., Arabian, A., Wakeham, A., Paige, C. J., Zinkernagel, R. M., et al. (1991) Nature 353, 180-184. [DOI] [PubMed] [Google Scholar]
- 40.von Herrath, M. G., Yokoyama, M., Dockter, J., Oldstone, M. B. & Whitton, J. L. (1996) J. Virol. 70, 1072-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., von Herrath, M. G. & Schoenberger, S. P. (2003) Nature 421, 852-856. [DOI] [PubMed] [Google Scholar]
- 42.Sun, J. C. & Bevan, M. J. (2003) Science 300, 339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyznik, A. J., Sun, J. C. & Bevan, M. J. (2004) J. Exp. Med. 199, 559-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearce, E. L., Shedlock, D. J. & Shen, H. (2004) J. Immunol. 173, 2494-2499. [DOI] [PubMed] [Google Scholar]
- 45.Wong, P. & Pamer, E. G. (2001) J. Immunol. 166, 5864-5868. [DOI] [PubMed] [Google Scholar]
- 46.van Stipdonk, M. J., Hardenberg, G., Bijker, M. S., Lemmens, E. E., Droin, N. M., Green, D. R. & Schoenberger, S. P. (2003) Nat. Immunol. 4, 361-365. [DOI] [PubMed] [Google Scholar]
- 47.Mercado, R., Vijh, S., Allen, S. E., Kerksiek, K., Pilip, I. M. & Pamer, E. G. (2000) J. Immunol. 165, 6833-6839. [DOI] [PubMed] [Google Scholar]
- 48.Kaech, S. M. & Ahmed, R. (2001) Nat. Immunol. 2, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]