Abstract
Identification of T cell epitopes is a vital but often slow and difficult step in studying the immune response to infectious agents and autoantigens. We report a spatially addressable technique for screening large numbers of T cell epitopes for both specific antigen recognition and functional activity induced. This system uses microarrays of immobilized, recombinant MHC–peptide complexes, costimulatory molecules, and cytokine-capture antibodies. The array elements act as synthetic antigen-presenting cells and specifically elicit T cell responses, including adhesion, secretion of cytokines, and modulation of surface markers. The method allows facile identification of pertinent T cell epitopes in a large number of candidates and simultaneous determination of the functional outcome of the interaction. Using this method, we have characterized the activation of human CD4+ and CD8+ T cells responding to vaccinia, influenza, HIV-1, and Epstein–Barr viruses.
Keywords: MHC class I and class II, protein array, T cell epitopes
T cells recognize specific antigenic peptides bound to MHC proteins on the surface of antigen-presenting cells, triggering intracellular signaling and T cell effector functions. The particular functions expressed determine how successfully the infection can be eliminated, and insufficient or inappropriate responses can result in lack of clearance (1, 2), immunopathology (3), or autoimmunity (4). An important step in studying T cell responses is to identify the relevant epitopes, usually short peptides derived from pathogenic molecules that are recognized by clonotypic T cells present in the overall repertoire. The typically low frequency of specific T cells within the overall circulating T cell population and the large number of potential epitopes present in a pathogenic or autoimmune proteome present challenges to the facile and routine identification of T cell epitopes.
T cell epitopes currently are identified by testing overlapping peptides from pathogenic sequences in cellular assays such as T cell-proliferation (5), target-lysis (6), bulk or intracellularcytokine secretion (7), or ELISPOT (8) assays. Screening synthetic peptides that span the entire length of a pathogenic sequence in cellular assays is time- and reagent-intensive and, in particular, consumes large numbers of T cells, which are often a limiting factor when testing clinical samples. This normally tedious and expensive process can be simplified by using an array format that can both identify pathogen-derived epitopes from many candidates and give broad functional information about the nature of the T cell response induced.
Methods
Recombinant Production of Human MHC Proteins. Recombinant MHC proteins were prepared by adaptation of standard methods for multiple, parallel, small-scale preparations. Soluble, extracellular portions of HLA-DR1 (HLA-DRB1*0101 and HLA-DRA1*0101), including a uniquely reactive cysteine engineered at the C terminus of the alpha chain, were produced in S2 Schneider cells, chemically modified with PEO-maleimide–biotin (Pierce), and loaded with peptide as described in ref. 9. HLA-A2 (A*0201) extracellular heavy chain carrying a biotinylation-signal peptide (10) and β2 microglobulin light chain were produced in Escherichia coli as inclusion bodies, refolded in the presence of peptide, biotinylated, and purified by size-exclusion chromatography as described in ref. 11.
Peptides. Ha[306–318] (PKYVKQNTLKLAT) from influenza hemagglutinin, PP16 (PEVIPMFSALSEGATP) and PG13 (PEVIPMFSALSEG) from the HIV-1 p24 gag protein, QHY (QHYREVAAAKSSE) from the Epstein–Barr virus (EBV) protein BZLF-1, TT (QYIKANSKFIGITE) from tetanus toxin, MVA-74A (CLTEYILWV) and MVA-165 (KVDDTFYYV) from vaccinia, and null peptides A2[103–117] (VGSDWRFLRGYKQYA) from HLA-A2 and Tfr (RVEYHFLSPYVSPKESP) from transferrin receptor were chemically synthesized and verified by using mass spectrometry.
T Cell Clones and Lines, Growth, and Maintenance. The CD4+ HA1.7 human TH0 clone specific to the influenza Ha peptide bound to the class II MHC protein HLA-DR1 (12), CD4+ CTL cell clone AC25 specific to the HIV-1 gag peptide PP16 in complex with HLA-DR1 (13), the CD8+ T cell lines VA55 3.13 and VA49 3.12 specific to the vaccinia peptides MVA-74A and MVA-165, respectively, in complex with HLA-A2 (14), and a short-term polyclonal CD4+ T cell line specific to the EBV-derived peptide QHY in complex with HLA-DR1 (15) were maintained in RPMI medium 1640 (Invitrogen) and 10% FBS, with biweekly stimulation with irradiated allogeneic peripheral blood mononuclear cells, 12F6 α-CD3 antibody (from Johnson Wong, Massachusetts General Hospital, Boston), and IL-2 (BD Biosciences). A murine T cell hybridoma transfected with T cell receptor MHC binding domains derived from HA1.7 was maintained in Eagle's minimum essential medium, Spinner modification (Invitrogen) containing 10% FCS.
Tetramer Production and Staining. Fluorescently labeled streptavidin (SA) tetramers used for staining T cells were produced by the stepwise addition of SA-phycoerythrin (BioSource International, Camarillo, CA) or SA-allophycocyanin (BD Pharmingen) to purified, biotinylated MHC samples to a final molar ratio of 1:4, as described in refs. 9 and 10. Cells were stained at 37°C, fixed with 1% paraformaldehyde, and measured by using a FACSCalibur flow cytometer (BD Biosciences).
Production of Artificial Antigen-Presenting Arrays. Polystyrene, Permanox, and LabTek II CC2 slides were obtained from Nalge. MHC–peptide monomers (50 μg/ml) and unlabeled SA-linked tetramers (50 μg/ml bio-MHC with 14 μg/ml SA) were immobilized by spotting onto the surface in PBS (pH 7.4). We investigated several direct and indirect immobilization strategies and found simple adsorption to plastic or treated glass to be straightforward and reproducible. Costimulatory or adhesion antibodies [α-CD11a, α-CD2, and α-CD28 (Leinco Technologies, St. Louis)] were included at 5 μg/ml in the same solution. For cytokine-capture chips, the capture antibody [α-IFN-γ, α-TNF, α-IL-4, or α-granzyme B (BD Pharmingen)] was first spotted at 40 μg/ml and allowed to dry, and the MHC/adhesion antibody solution was spotted onto the dry spots. The arraying was accomplished by hand-spotting of 0.1–0.5 μl of solution, or by using a Cartesian Microsystems automatic noncontact array printer (Genomic Solutions, Ann Arbor, MI), or by using a manual microarrayer (Xenopore, Hawthorne, NJ). After airdrying, the chips can be stored for >3 months at 4°C without loss of activity. For more detailed descriptions of the manufacture and use of arrays, see Supporting Methods, which is published as supporting information on the PNAS web site.
Cytokine-Capture Detection by Using Artificial Antigen-Presenting Arrays. Spotted, dry chips were blocked by incubation with serum-containing T cell medium for 30 min at room temperature. Then, cells were added to the chip and incubated for 4–16 h at 37°C, contained within individual chamber slides (Nalge) or within a hydrophobic barrier (Ted Pella, Redding, CA). Typically, ≈106 cells per slide were used, although this amount was varied as noted. After removal of the supernatant, the chip was rinsed with fresh medium and examined by eye or microscope for cell adhesion. T cells were removed by washing twice with distilled water and three times with Tris-buffered saline solution containing 0.1% Tween 20 (TBST) (pH 7.6). Biotinylated detection antibody and fluorescently labeled SA [Alexa Fluor 555 or Alexa Fluor 647 (Molecular Probes)] were preincubated at high concentration (>1 mg/ml) and then diluted (1:250 final antibody dilution, 2 μg/ml final concentration SA) into TBST containing 0.3% BSA and 100 μM free biotin. Labeled MHC-detection antibodies were used at a final concentration of 1 μg/ml. Chips were stained for 1.5 h at room temperature. Then, the slide was washed three times with TBST, two times with ddH2O, and dried. The chips were scanned by using an Affymetrix 428 array scanner, and data were analyzed by using jaguar 2.0 software (Affymetrix, Santa Clara, CA). A single-pixel-wide median averaging was performed to remove speckle noise from the images (photoshop 7.0.1, Adobe Systems, San Jose, CA), and surface plots were prepared by using imagej 1.30v (National Institutes of Health, Bethesda). Spots exhibiting average fluorescence intensities significantly above no-MHC controls were considered positive. Typically, duplicate or triplicate spots were examined. For precipitating-substrate analysis, SA-horseradish peroxidase (Sigma) was used in place of fluorescently labeled SA, and the spots were developed by using a 3-amino-9-ethylcarbazole precipitating substrate (BD Pharmingen) and were observed by eye and under a dissection microscope.
Fluorescence Microscopy. After incubation, as described above, the chips were chilled to 4°C, and fluorescent antibodies (BD Pharmingen) and Hoechst stain (Molecular Probes) were added on ice for 40 min. The chip was then fixed with 1% paraformaldehyde for 5 min at room temperature, rinsed twice with PBS, and dehydrated once with 70% ethanol. A coverslip was applied with VECTASHIELD (Vector Laboratories), and the spots were observed by using a Nikon Eclipse E800 fluorescence microscope. Images were obtained by using a Spot RT Slider camera with spotbasic 4.0.2 software (Diagnostic Instruments, Sterling Heights, MI).
Results
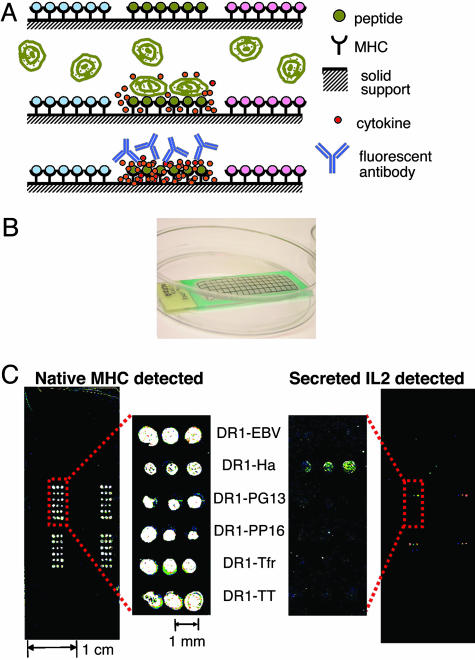
Artificial Antigen-Presenting-Element Arrays Can Be Used to Detect T Cell Activation. We prepared microarrays of soluble, recombinant MHC–peptide complexes immobilized together with cytokine-capture antibodies (see Methods). When T cells are incubated with such arrays, antigen-specific activation occurs in the particular array elements carrying appropriate MHC–peptide complexes, and this interaction leads to the secretion of cytokines and effector molecules. These molecules are captured locally by capture antibodies immobilized along with the MHC–peptide complexes and are detected in a position-specific manner by using fluorescent anti-cytokine detection antibodies (Fig. 1A). Shown is an example of such a microarray consisting of a polystyrene slide with a hydrophobic barrier surrounding the arrayed MHC–peptide complexes (Fig. 1B). Small volumes of cell suspension or staining antibody solutions (0.5–1.0 ml) are contained within the barrier. Up to ≈1,000 individual elements can be arrayed on a slide, with each element maintaining its ability to function as a complete antigen-presenting entity. Both class I and class II MHC complexes maintain their native conformation after immobilization as detected by conformationally specific antibodies (Fig. 1C Left; see also Fig. 6, which is published as supporting information on the PNAS web site). Incubation of these arrays with T cell samples leads to T cell activation and the expression of measurable responses. In this example, a T cell hybridoma specific for HLA-DR1 in complex with an influenza peptide (Ha) was incubated with an array carrying various DR1–peptide complexes. Whereas native DR1 conformation was detected in all of the array spots, antigen-induced secretion of IL-2 was detected on only the spots that contain the specific DR1–Ha complex (Fig. 1C Right). Devices such as these arrays allow parallel testing of many potential T cell epitopes, conservation of T cells and other reagents, mass production, and convenient use.
Fig. 1.
Artificial antigen-presentation chips. (A) Schematic representation of artificial antigen-presenting microarray technology. (Top) MHC–peptide complexes immobilized with different peptide antigens in distinct areas. Costimulatory and cytokine-capture antibodies can be coimmobilized (not shown). (Middle) T cells are incubated with the array. Only specific MHC–peptide complexes induce T cell responses. Cytokines are captured locally by immobilized anti-cytokine antibodies. (Bottom) Captured cytokines are detected by labeled antibodies in the locations where they were secreted, identifying the activating epitopes. (B) Photograph of a microarray with solution held in place by a hydrophobic barrier. (C) Microarray carrying various DR1–peptide complexes on a polystyrene slide, incubated with 106 murine hybridoma cells specific to DR1–Ha for 16 h and stained for native MHC with LB3.1-CY5 (Left) and for captured mouse IL-2 by using biotinylated α-mouse-IL-2 preincubated with SA-Alexa Fluor 555 (Right). The pattern was repeated in four areas.
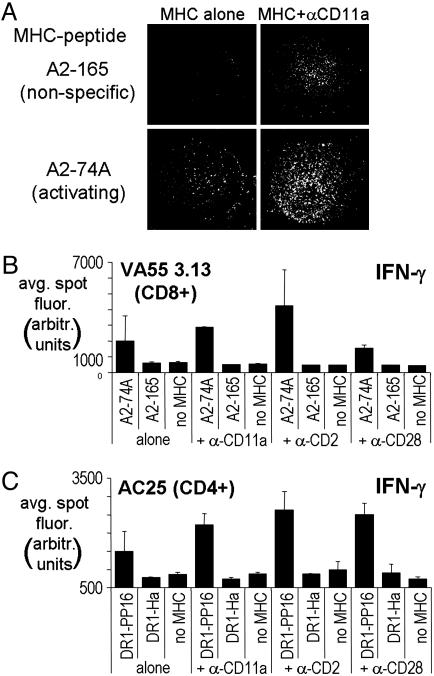
The Effect of Coimmobilized Costimulatory Molecules on MHC–Peptide Arrays. Most nontransformed and primary T cells require nonspecific costimulatory and/or adhesion signals in addition to a specific MHC–peptide stimulus for induction of full activation responses. The engagement of conventional costimulatory molecules (such as CD28) or integrins [such as CD2 (LFA-3) and CD11a (LFA-1)] on the T cell surface serves to amplify and/or stabilize signals that lead to cytokine secretion (16). In the absence of costimulatory signals, T cells adhere to the MHC–peptide-coated surfaces in a peptide-specific manner as has been described for murine T cells in ref. 17. VA55 3.13, a CD8+ human T cell line isolated from a vaccinia-vaccinated individual, specifically recognizes HLA-A2 bound to MVA-74A, a peptide derived from the vaccinia 189R gene product (14). VA55 T cells adhere to array elements containing the specific A2–74A complex but not the nonspecific A2–165 complex (Fig. 2A Left). When we included costimulatory signals in the form of antibodies to cell-surface molecules together with the MHC–peptide complexes, VA55 adhesion was more robust, and T cell spreading indicative of productive interaction was induced in the area containing the specific complex (shown for α-CD11a in Fig. 2A Lower Right). Although adhesion to nonspecific areas is observed in the presence of α-CD11a (Fig. 2A Upper Right), cytokine secretion is induced only in the specific areas (see below).
Fig. 2.
The effect of costimulation on T cell detection. (A) HLA-A2+ MVA-74A peptide-specific T cells incubated for5honan MHC–peptide array at ≈10 cells per mm2 (3 × 105 cells per chamber on a four-chamber slide) and stained with Hoechst nuclear stain. (Left) MHC complexes were arrayed alone at 50 μg/ml. (Right) α-CD11a adhesion antibodies were coimmobilized with MHCs. (Top) Nonactivating MHC–peptide array elements. (Bottom) Activating MHC–peptide spots. The single spot images shown are characteristic of such spots on replicate arrays. (B and C) Average spot intensities (in arbitrary fluorescence units) for IFN-γ secretion by 106 cells in response to 6-h incubation with array elements incorporating various types of costimulatory antibodies. (B) Response of VA55 3.13, a CD8+ line specific to A2–74A. (C) Response of AC25, a CD4+ clone specific to DR1–PP16.
Cytokine secretion is greatly enhanced by the inclusion of coimmobilized costimulatory molecules in the microarray elements. VA55 3.13 T cells were tested for IFN-γ secretion in response to activating and nonactivating MHC–peptide-array elements with and without various coimmobilized adhesion and costimulatory antibodies. IFN-γ secretion was observed in response to A2–74A immobilized on the array but not to nonspecific complexes or in the absence of MHCs (Fig. 2B). The IFN-γ detected was significantly enhanced in the presence of each of the costimulatory antibodies α-CD11a, α-CD2, and α-CD28, without the large increase in background as for adhesion (Fig. 2A). Similar results were obtained for AC25, a CD4+ T cell clone raised from an individual infected with HIV-1 that recognizes HLA-DR1 bound to PP16, a peptide from HIV-1 gag p24 (13). AC25 also secretes IFN-γ specifically in spots containing the appropriate MHC–peptide complex, and, again, the signal is enhanced in the presence of the costimulatory molecules tested (Fig. 2C). The combination of specific MHC–peptide complexes and costimulatory antibodies allows the array elements to act as artificial antigen-presenting cells with a single epitope localized to each area and enhances detection of relevant, specific T cell responses.
Specificity of the Response. Identical MHC–peptide chips were created with specific and nonspecific MHC–peptide complexes and were tested for IFN-γ induction. Incubations of both CD4+ AC25 (Fig. 3A) and CD8+ VA55 3.13 (Fig. 3B) on these chips result in clear, specific reactions with the correct complex with little or no detection on nonspecific spots. Tetramer-staining experiments performed for these same cells indicate the same specificity as observed by the microarrays (see Fig. 7, which is published as supporting information on the PNAS web site).
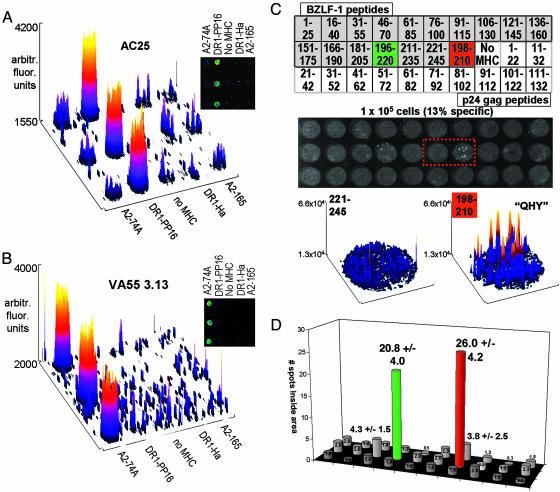
Fig. 3.
Arrayed class I and class II epitopes are specifically recognized by human CD8+ and CD4+ T cells. (A and B) MHC–peptide complexes and α-CD11a antibody were immobilized with α-IFN-γ capture antibody. The chips were incubated with 106 T cells for 6 h, and IFN-γ was detected. Fluorescence intensity on the chip is shown as a 3D surface plot of the array area, and a raw-fluorescence image is shown (Inset). (A) CD4+ clone AC25, specific to DR1–gag. (B) CD8+ line VA55 3.13, specific to A2–74A. (C) Response of a short-term polyclonal human T cell line against the EBV protein BZLF-1 (15) was analyzed by using overlapping peptides covering the sequence of the protein, each in complex with HLA-DR1. Additional spots contained the previously identified minimal epitope peptide QHY (BZLF-1 [198–210]), a series of HIV p24 gag overlapping peptides, and other control complexes for a total of 50 spots in an area of 192 mm2.(Top) A partial map of the array. The array was incubated with 100,000 T cells, of which ≈13% were specific to BZLF-1 by intracellular cytokine staining (≈13,000 specific cells or 60 per mm2). The secreted IFN-γ detected after a 6-h incubation is shown below the map. Small spots, corresponding to cytokines secreted by single responding cells, can be seen within the overall spot areas for two specific arrays; one is from the overlapping peptide series BZLF-1 [196–220] (green), and one is the previously identified minimal epitope BZLF-1 [198–210] (“QHY,” red). Surface plots for the boxed-array elements BZLF-1 [221–245] (nonactivating) and BZLF-1 [198–210] (activating) are shown below the array. Intensity spikes from individual secretion sources can be seen. (D) The average number of such spots observed for each element of the microarray in C is shown as a bar graph with the same layout (n = 4).
Often, to determine T cell epitopes, one must test the T cells with overlapping peptides covering the entire sequence of a known protein immunogen. We prepared microarrays carrying overlapping peptides covering the sequence of the lytic-stage antigen BZLF-1 from EBV and used the microarrays to screen for reactivity in a short-term T cell line raised from the peripheral blood mononuclear cells of an HLA-DR1+ patient presenting with acute infectious mononucleosis (15). In addition to the BZLF-1 overlapping 25-mer peptide series, the arrays also included the known minimal peptide epitope QHY (BZLF-1 [196–210]) and a second overlapping peptide series covering the sequence of HIV-1 gag p24.∥ Each peptide epitope was allowed to bind to peptide-receptive DR1, and the resulting complexes were spotted onto the array, resulting in 50 possible spots with which the cells could react. The spots were created by using 0.1 μl of MHC–peptide solution at 50 μg/ml per spot, with a resulting area of ≈0.44 mm2 for each spot. The short-term T cell line, which is ≈13% specific for BZLF-1 by intracellular cytokine staining (data not shown), was incubated on the chips at a density of 96,000 total cells per array (≈60 specific cells per mm2) (Fig. 3C). IFN-γ was specifically detected in the spots with previously identified EBV epitopes: peptide 196–220 in the overlapping peptide series [Fig. 3C (green)], and the minimal peptide epitope QHY [Fig. 3C (red)]. At these low cell densities, the inherent granularity of the assay is apparent, and the chips are more accurately analyzed by counting individual spots within the larger array spot areas, as in an ELISPOT analysis (8), instead of analyzing overall intensities. Surface plots for a nonactivating array element (BZLF-1 [221–245]) and the adjacent activating array element (QHY) clearly show spikes corresponding to individual secreting cells as opposed to higher-density cell incubations, where a more continuous peak is observed (compare surface plots in Fig. 3C with those in Fig. 3 A and B). The spots within each area were enumerated, and the average values for four experiments are shown in a bar graph with the same layout as the array (Fig. 3D). The positive elements are readily apparent, even at a low responding-cell density. An example of an identical array incubated with the EBV-specific T cell line at higher density can be seen in Fig. 8, which is published as supporting information on the PNAS web site.
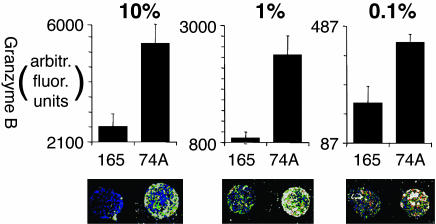
Sensitivity of Cytokine-Secretion Response. T cells responding to infection or vaccination often are present at low frequencies in the overall T cell population. As a test of the sensitivity of this technique, the VA55 3.13 T cell line was diluted into nonspecific (allogenic) peripheral blood mononuclear cells and tested on an array with specific and nonspecific HLA–A2 complexes. Secretion of granzyme B, a cytotoxic T cell marker, was detected. Fig. 4 shows VA55 3.13 tested at dilutions ranging from 10% to 0.1% of the total cells, and by using, in each case, 2.5 × 106 cells, a sample size corresponding to 1–3 ml of blood. A specific secretion signal was observed even after dilution to 0.1% of the total cells, a frequency of ≈1,000 specific cells per million (Fig. 4 Right) or ≈20–100 per spot, assuming an even distribution of cells around the array; however, at this level, interpretation is complicated by the relatively high nonspecific background approaching the level of sample-to-sample variability. This sensitivity is in the range of other current T cell-antigen-screening assays such as MHC tetramer staining (18) or ELISPOT assay (8); however, by using the microarray assay, many epitopes can be assayed in a single well. The sensitivity of epitope detection will depend on factors in addition to responder frequency, including the specifics of particular TCR/MHC–peptide interactions and the level and nature of effector activity induced by given epitopes.
Fig. 4.
Detection of low-frequency responses. VA55 3.13 CD8+ T cells (specific to HLA-A2 in complex with MVA-74A) were diluted into allogeneic peripheral blood mononuclear cells, and granzyme B secretion was detected on the chips after incubation. In each panel, the total number of cells per chamber was 2.5 × 106. The spots are ≈1 mm in diameter, for an average spot area of 0.75 mm2. Average intensities of spots (n = 2) and a raw-fluorescence image is shown after dilution of VA55 3.13 to 10% of the total cells (100,000 per million; Left), 1% of the total cells (10,000 per million; Center), and 0.1% of the total cells (1,000 per million; Right).
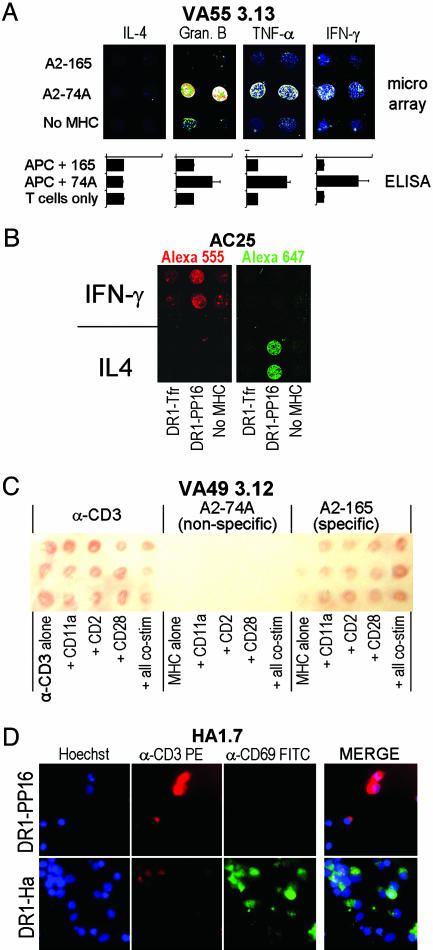
Multiple Cytokines Can Be Tested Simultaneously. The cytokine-capture chips can be designed for the simultaneous detection of multiple cytokines from a single responding T cell population. In Fig. 5A, VA55 3.13 was screened in a four-chambered slide, with different capture antibodies immobilized in each chamber. Specific secretion of IFN-γ, granzyme B, and TNF-α, but not IL-4, was observed. These results replicate those observed in conventional, bulk cytokine secretion ELISA experiments performed on the same cells (Fig. 5A). In another approach, multiple cytokines can be distinguished by using anti-cytokine antibodies labeled with different fluorescent probes. Fig. 5B shows an array carrying both IFN-γ- and IL-4-capture antibodies along with specific and nonspecific MHC–peptide complexes. By using two differently labeled cytokine-detection antibodies, both IL-4 and IFN-γ were detected in only the appropriate regions (Fig. 5B). The two strategies of spatial separation and multiple fluorescent labels for detection could be applied concurrently to obtain large amounts of information from a single experiment. Thus, very broad screens for functional responses could be carried out simultaneously with parallel screens of many different T cell epitopes.
Fig. 5.
Analysis of multiple T cell functions. (A) A multichamber LabTek II CC2 slide was prepared with a different capture antibody in each chamber but with identical MHC–peptide and α-CD11a patterns. 2.5 × 105 VA55 3.13 cells (specific to A2–74A) were incubated in each chamber for 6 h, and cytokine secretion was analyzed by using the appropriate detection antibody. Below each image is a bar graph showing the same cytokine detected by a bulk ELISA, where VA55 3.13 T cells are stimulated with peptide-pulsed, HLA-A2+ antigen-presenting cells. (B) A chip was spotted with α-IFN-γ- and α-IL-4-capture antibodies in different areas with the same MHC-peptide and α-CD11a stimuli coimmobilized. The chip was incubated for 6 h with 1 × 106 AC25 T cells (specific to DR1–PP16) and then stained with a mixture of biotinylated α-IFN-γ preincubated with SA-Alexa Fluor 555 and biotinylated α-IL-4 preincubated with SA-Alexa Fluor 647. The chip was scanned at both wavelengths. Red, Alexa Fluor 555; Green, Alexa Fluor 647. (C) Cytokine capture can be detected on the arrays by using precipitating substrate. The array shown was incubated for 24 h with 2 × 106 VA49 3.12 cells, a CD8+ T cell line specific to A2–165. IFN-γ secretion was detected in the regions containing α-CD3 and A2–165 but not A2–74A. (D) Cell-surface protein up-regulation in response to array elements can be detected by using fluorescently labeled antibodies. HA1.7, a CD4+ T cell clone specific to DR1–Ha, was incubated at 2.5 × 106 cells per chamber for 16 h on a microarray containing DR1–PP16 (null; Upper) and DR1–Ha (activating; Lower). Hoechst nuclear stain shows the presence of adhering cells, whereas α-CD3-phycoerythrin and α-CD69-FITC were used to show the relative levels of those activation-linked cell-surface markers. A three-color merge for each stimulus is shown at the right.
Nonfluorescent Detection of Cytokine Capture. In some cases, it may be advantageous to detect the cytokine secretion in a manner visible to the naked eye by using enzyme-linked antibody and precipitating substrate, as in a traditional ELISPOT assay, rather than fluorescent detection. Fig. 5C shows an array that was incubated with the CD8+ T cell line VA49 3.12, isolated from a vaccinia-immunized individual, which responds specifically to HLA–A2 in complex with the MVA-165 peptide (from the vaccinia gene product “018L”) but not the MVA-74A peptide (14). The array was analyzed by using biotinylated α-IFN-γ and peroxidase-coupled SA. Visually apparent development can be seen in areas where specific A2–165 peptide complex or positive control α-CD3 stimulus was immobilized but not in areas with nonactivating A2–74A complex (Fig. 5C).
Other Activation Markers. MHC–peptide microarrays may be used to detect other activation markers in T cells. For example, specific detection of transient calcium flux in cells on peptide–MHC tetramer arrays has been reported in murine T cells (17). We investigated whether T cell surface markers, conventionally assayed in flow-cytometry experiments, also could be evaluated in parallel on MHC–peptide and costimulatory microarrays. Fig. 5D shows a ×60 view of a single element of an MHC–peptide array with human CD4+ HA1.7 T cells specific for DR1–Ha (12) adhering. These cells have been stained with Hoechst nuclear stain, α-CD3-phycoetythrin, and α-CD69-FITC. CD69 is a common early activation marker that is up-regulated upon T cell activation (19), and CD3 is a component of the T cell receptor and is down-regulated upon activation (20). The HA1.7 T cells adhering to the specific DR1–Ha region are more numerous and clustered, as seen by the blue Hoechst stain, and exhibit down-regulated CD3 (red) and substantial CD69 up-regulation (green), as compared with cells in the nonspecific DR1–PP16 region, indicating an activation response to the specific complex. This technique could be applied to detect the expression of other cell-surface activation markers, or markers of intracellular signaling pathways.
Discussion
We have described a technique that can be used to screen small samples of T cells for specific peptide–MHC binding and for functional responses in a microarray format. Each array element consists of recombinant peptide–MHC complexes coimmobilized with costimulatory antibodies and anti-cytokine antibodies to locally detect T cell adhesion and activation responses; these elements act as individual, artificial antigen-presenting cells (21), with each element presenting a different T cell-epitope candidate. The arrays are convenient to use with small volumes of cell suspension or staining solutions, allowing for reagent conservation, and can be mass-produced with commercially available array spotters. The arrayed MHC–peptide complexes are largely native and remain spatially distinct after multiple washes, incubations, and stainings. The dry MHC–peptide arrays are stable for long periods, >3 months at 4°C. The functional responses induced when T cells recognize array elements, such as cytokine secretion or activation of cytolysis, are detected in a location dependent manner. In a previous study, microarrays of immobilized MHC–peptide complexes were used to observe adhesion and calcium flux in murine CD4+ T cells (17). In the work reported here, the combination of coimmobilized costimulatory antibodies with specific MHC–peptide complexes greatly enhances the detection level, and the use of cytokine-capture detection allows investigation of relevant functional responses.
The advantages of array technology that have revolutionized gene-expression research have already begun to be applied to important clinical problems of immunology and infection. Protein microarrays have been used to detect specific antibody responses in human serum (22), which may be applied conveniently to allergy testing, diagnosis of some diseases, and vaccine design for certain pathogens. However, many infections are not eliminated by antibody responses (23–25), and, in those cases, the ability to use array technology to analyze the cellular immune response would be beneficial. Using antigen-presentation microarrays with live immune cells allows broad, multidimensional information to be obtained quickly and easily. The ability to conveniently track T cells for specificity and functional response for many epitopes in parallel in small clinical samples has the potential to lend new levels to our understanding of the rise and fall of T cell populations during vaccination or infection.
Artificial antigen-presenting arrays show promise as convenient tools with which to discover T cell epitopes and characterize T cell responses. This technology is able to sensitively detect low-frequency responses and to give extensive information on the nature of the response to different T cell epitopes. This sort of data, conveniently and rapidly obtained, can be used to accelerate studies of the cellular immune response to novel pathogens and to promote the development of safe, effective vaccines or other innovative immunotherapies.
Supplementary Material
Acknowledgments
We thank Liying Lu, Iwona Strug, and Joyce Pepe (University of Massachusetts Medical School) for reagents; John Cruz, Masanori Terajima, Melissa Precopio (University of Massachusetts Medical School), Phillip Norris (Blood Systems Research Institute), Jonathan Lamb (University of Edinburgh Medical School), and Jerome Bill (National Jewish Medical and Research Center) for T cells; and Alan Rothman for helpful comments. This work was supported by National Institutes of Health Grants N01-AI95361 and U19-AI57319 and the University of Massachusetts Genomics Core and Fluorescent Microscopy facilities.
Author contributions: J.D.S. and L.J.S. designed research; J.D.S. performed research; J.D.S. and L.J.S. analyzed data; J.D.S. and L.J.S. wrote the paper; and W.E.D. grew T cell lines and clones.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SA, streptavidin; EBV, Epstein–Barr virus.
Footnotes
Note that for class I MHC proteins, the peptide termini typically bind within the MHC-binding site, and each potential epitope must be present as a separate peptide. For class II MHC proteins, the peptide termini can extend from the site, and a single long peptide can be used to evaluate multiple potential epitopes. Thus, many more individual peptides must be evaluated to screen a protein sequence for class I as compared with class II epitopes.
References
- 1.Collins, K. L. (2003) Curr. HIV Res. 1, 31-40. [DOI] [PubMed] [Google Scholar]
- 2.Levitsky, V. & Masucci, M. G. (2002) Virus Res. 88, 71-86. [DOI] [PubMed] [Google Scholar]
- 3.Terajima, M., Vapalahti, O., Van Epps, H. L., Vaheri, A. & Ennis, F. A. (2004) Microbes Infect. 6, 238-245. [DOI] [PubMed] [Google Scholar]
- 4.Christen, U. & von Herrath, M. G. (2004) Mol. Immunol. 40, 1113-1120. [DOI] [PubMed] [Google Scholar]
- 5.Yamada, A., Ziese, M. R., Young, J. F., Yamada, Y. K. & Ennis, F. A. (1985) J. Exp. Med. 162, 663-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engers, H. D., Thomas, K., Cerottini, J. C. & Brunner, K. T. (1975) J. Immunol. 115, 356-360. [PubMed] [Google Scholar]
- 7.Maecker, H. T., Dunn, H. S., Suni, M. A., Khatamzas, E., Pitcher, C. J., Bunde, T., Persaud, N., Trigona, W., Fu, T. M., Sinclair, E., et al. (2001) J. Immunol. Methods 255, 27-40. [DOI] [PubMed] [Google Scholar]
- 8.Czerkinsky, C., Andersson, G., Ekre, H. P., Nilsson, L. A., Klareskog, L. & Ouchterlony, O. (1988) J. Immunol. Methods 110, 29-36. [DOI] [PubMed] [Google Scholar]
- 9.Cameron, T. O., Norris, P. J., Patel, A., Moulon, C., Rosenberg, E. S., Mellins, E. D., Wedderburn, L. R. & Stern, L. J. (2002) J. Immunol. Methods 268, 51-69. [DOI] [PubMed] [Google Scholar]
- 10.Altman, J. D., Moss, P. A., Goulder, P. J., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996) Science 274, 94-96. [DOI] [PubMed] [Google Scholar]
- 11.Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992) Proc. Natl. Acad. Sci. USA 89, 3429-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb, J. R., Eckels, D. D., Lake, P., Woody, J. N. & Green, N. (1982) Nature 300, 66-69. [DOI] [PubMed] [Google Scholar]
- 13.Norris, P. J., Moffett, H. F., Brander, C., Allen, T. M., O'Sullivan, K. M., Cosimi, L. A., Kaufmann, D. E., Walker, B. D. & Rosenberg, E. S. (2004) AIDS Res. Hum. Retroviruses 20, 315-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terajima, M., Cruz, J., Raines, G., Kilpatrick, E. D., Kennedy, J. S., Rothman, A. L. & Ennis, F. A. (2003) J. Exp. Med. 197, 927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Precopio, M. L., Sullivan, J. L., Willard, C., Somasundaran, M. & Luzuriaga, K. (2003) J. Immunol. 170, 2590-2598. [DOI] [PubMed] [Google Scholar]
- 16.Davis, S. J., Ikemizu, S., Evans, E. J., Fugger, L., Bakker, T. R. & van der Merwe, P. A. (2003) Nat. Immunol. 4, 217-224. [DOI] [PubMed] [Google Scholar]
- 17.Soen, Y., Chen, D. S., Kraft, D. L., Davis, M. M. & Brown, P. O. (2003) PloS Biol. 1, E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar, P. R. & Ogg, G. S. (2002) J. Immunol. Methods 268, 3-7. [DOI] [PubMed] [Google Scholar]
- 19.Testi, R., D'Ambrosio, D., De Maria, R. & Santoni, A. (1994) Immunol. Today 15, 479-483. [DOI] [PubMed] [Google Scholar]
- 20.Valitutti, S., Muller, S., Salio, M. & Lanzavecchia, A. (1997) J. Exp. Med. 185, 1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oelke, M., Maus, M. V., Didiano, D., June, C. H., Mackensen, A. & Schneck, J. P. (2003) Nat. Med. 9, 619-624. [DOI] [PubMed] [Google Scholar]
- 22.Bacarese-Hamilton, T., Gray, J. & Crisanti, A. (2003) Curr. Opin. Mol. Ther. 5, 278-284. [PubMed] [Google Scholar]
- 23.Aasa-Chapman, M. M., Hayman, A., Newton, P., Cornforth, D., Williams, I., Borrow, P., Balfe, P. & McKnight, A. (2004) AIDS 18, 371-381. [DOI] [PubMed] [Google Scholar]
- 24.Khanna, R., Burrows, S. R. & Moss, D. J. (1995) Microbiol. Rev. 59, 387-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logvinoff, C., Major, M. E., Oldach, D., Heyward, S., Talal, A., Balfe, P., Feinstone, S. M., Alter, H., Rice, C. M. & McKeating, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.