Abstract
Nuclear receptors activate transcription by recruiting multiple coactivators to the promoters of specific target genes. The functional synergy of the p160 coactivators [steroid receptor coactivator-1, glucocorticoid receptor interacting protein (GRIP1), or the activator for thyroid hormone and retinoid receptors], the histone acetyltransferases cAMP response element binding protein binding protein (CBP) and p300 and the histone methyltransferase coactivator-associated arginine methyltransferase (CARM1) depends on the methyltransferase activity of CARM1. CARM1 methylates histone H3 and other factors including the N-terminal region of p300. Here, we report that CARM1 also methylates Arg-2142 within the C-terminal GRIP1 binding domain (GBD) of p300. In the GBD, both Arg-2088 and Arg-2142 are important for binding GRIP1. Methylation of Arg-2142 inhibits the bimolecular interaction of GRIP1 to p300 in vitro and in vivo. This methylation mark of p300 GBD is removed by peptidyl deiminase 4, thereby enhancing the p300–GRIP1 interaction. These methylation and demethylimination events also alter the conformation and activity of the coactivator complex and regulate estrogen receptor-mediated transcription, and they thus represent unique mechanisms for regulating coactivator complex assembly, conformation, and function.
Keywords: transcriptional regulation
Hormone-activated nuclear receptors (NRs) activate transcription by binding to specific DNA regulatory elements in target gene promoters; there they recruit coactivator proteins to remodel chromatin structure and assemble the transcription complex containing RNA polymerase II (1, 2). The p160 coactivators [e.g., steroid receptor coactivator-1, glucocorticoid receptor interacting protein (GRIP1), and the activator for thyroid hormone and retinoid receptors] bind directly to hormone-activated NR and recruit secondary coactivators such as p300/cAMP response element binding protein (CREB) binding protein (CBP) and coactivator-associated arginine methyltransferase (CARM1), which acetylate and methylate (respectively) histones and other proteins in the transcription complex (3–5). These and other covalent modifications of histones and cofactors play important roles in chromatin remodeling and transcriptional regulation (6–10).
The importance of histone acetylation by p300/CBP and p300/CBP associated factor (p/CAF) and histone methylation by CARM1 and protein arginine methyltransferase 1 (PRMT1) has been clearly demonstrated (8, 11–15), although the mechanisms by which most of these histone modifications contribute to the transcriptional activation process still remain to be elucidated. However, in addition to histones, other proteins in the transcription machinery are modified posttranslationally. For example, acetylation of the DNA binding transcription factor MyoD enhances its activity (16), whereas acetylation of p160 coactivators within their NR-interaction domain inhibits their binding to NRs (8).
Although protein acetylation and phosphorylation are reversible, and thus are used for dynamic regulation, the reversibility of Lys and Arg methylation of proteins has been unclear and hotly debated (17). Recently peptidyl deiminase 4 (PAD4) was shown to remove methylamine groups from methylated Arg residues in histones, thereby changing methylarginine to citrulline on free histones and at the promoter of an endogenous, estrogen-activated gene (18, 19). Furthermore, the overexpression of this enzyme inhibited transcriptional activation by estrogen receptor (ER).
CARM1, p300/CBP, and p160 coactivators function synergistically as NR coactivators (13). In addition to histone H3 (14, 20), p300 and CBP are methylation substrates for CARM1. Methylation by CARM1 in the KIX domain in the N-terminal region of p300 and CBP inhibits binding of the transcription factor CREB (9). In addition, methylation of three Arg residues near the KIX domain by CARM1 is apparently required for the ability of CBP to enhance transcriptional activation by a p160 coactivator fused to the Gal4 DNA binding domain (DBD), and CBP with mutations at these three Arg residues had a dominant negative effect on transcriptional activation by ER but not by another NR, retinoic acid receptor (10), suggesting that methylation of these residues might be involved in the functional interactions between p160, CBP, and CARM1 coactivators.
Here we report a site in the C-terminal region of p300 (Arg-2142) that is methylated by CARM1 in vitro and in vivo. Arg-2142 is in the p160-binding domain (which we will call the GRIP1 binding domain or GBD) of p300, and its methylation inhibits the bimolecular interactions between p300 and the p160 coactivator GRIP1. PAD4 efficiently removes this methylation mark from p300 GBD and thereby enhances the bimolecular association of GRIP1 and p300. Using mutations of Arg-2142 and two other conserved Arg residues in the p300 GBD, we also demonstrate distinct and different roles for Arg-2088, Arg-2142, and the methylation and demethylimination of Arg-2142 in regulating the assembly, conformation, and function of the p160–p300–CARM1 coactivator complex. Thus, p300 methylation by CARM1 and demethylimination by PAD4 may contribute to the functional synergy of this coactivator complex and represent unique mechanisms for dynamic regulation of the transcriptional activation process.
Materials and Methods
Plasmids. For a description of plasmids used see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Transient Transfection Assay. For a description of the transient transfection assay see Supporting Materials and Methods. Luciferase activities shown are the mean and range of variation for two transfected cell cultures and are representative of at least three independent experiments.
GST Pull-Down Assay and in Vitro Translation. For a description of the GST pull-down assay and in vitro translation see Supporting Materials and Methods.
Peptide Competition Assay. GST pull-down assays were performed in the presence or absence of the following competitor peptides: p300 R2 peptide, NPQLLAAFIKQRAAKYANS; p300 R3 peptide, QAGVQRAGLP; and p300 R3-dimethyl peptide, the same as R3 but with asymmetric dimethylarginine substituted for R.
In Vitro and in Vivo Protein Methylation and Demethylimination Assays. Full-length p300 protein was prepared in a baculovirus system as described (11). Protein methylation in vitro was performed with GST-CARM1 or GST-PRMT1 as described (20). To detect methylation of full-length p300 in mammalian cells, coimmunoprecipitation was performed with antiserum against p300 (Santa Cruz Biotechnology), using extracts from 293T cells in 100-mm dishes transfected with plasmids encoding p300 and CARM1 or PRMT1 (13). Immunoblot analysis was performed with an affinity-purified rabbit antiserum (Upstate Biotechnology) against a peptide representing the R3 region of p300 GBD but with asymmetric dimethylarginine substituted for Arg-2142. To detect methylation of p300 GBD fragment in vivo, 1-ml extracts (prepared as above) from 293T cells transfected with pSG5.Flag-GST-p300GBD (WT or R3K mutant) and pSG5.hemagglutinin(HA)-CARM1 or pSG5.HA-PRMT1 were incubated with glutathione-Sepharose beads as in standard GST pull-down assays. Bound proteins were analyzed by immunoblot, using antibodies against dimethyl-R3 peptide or Flag epitope (Sigma F3165).
PAD4 proteins were expressed and purified from pGEX-PAD4 or pGEX-PAD4(C645S) mutant (18). To achieve demethylimination of CARM1-methylated p300, 1 μg of GST-PAD4 or mutant protein was coincubated with the methylation reaction.
Results
CARM1 Methylates Arg-2142 of p300 GBD in Vitro and in Mammalian Cells. The coactivator synergy between CARM1 and p300 in transient transfection assays (13) led us to investigate whether this synergy might involve methylation of p300 by CARM1. In vitro methylation assays using radioactively labeled S-adenosylmethionine (AdoMet) demonstrated that full-length p300 was methylated by CARM1 but not PRMT1 (Fig. 1A). In control experiments CARM1 and PRMT1 methylated histone H3 and histone H4, respectively. A large N-terminal fragment of p300 (amino acids 1–596) containing only part of the KIX domain (Fig. 1B) was very weakly methylated, whereas large C-terminal fragments of p300 (amino acids 1572–2414) and CBP (amino acids 1594–2441) were strongly methylated by CARM1 (Fig. 1C). Thus, the methylation site is likely conserved between p300 and CBP. Within the p300 C-terminal region the cysteine- and histidine-rich CH3 region and the glutamine-rich Q region (Fig. 1B) were methylated weakly, but the GBD was strongly methylated by CARM1 (Fig. 1D). The CBP GBD (amino acids 2041–2240) was also methylated (data not shown). As shown previously (9, 10), a p300 KIX domain fragment (amino acids 568–828) was also strongly methylated by CARM1 in vitro (data not shown). Each of the three Arg residues in p300 GBD that are conserved between p300 and CBP was changed to Lys in mutant GBD fragments: R1K (Arg-2056–Lys), R2K (Arg-2088–Lys), and R3K (Arg-2142–Lys). R1K and R2K mutants of p300 GBD were methylated by CARM1 as efficiently as WT GBD, but the R3K mutant was not methylated at all (Fig. 1E Upper). Thus the major methylation site of p300 GBD is Arg-2142.
Fig. 1.
CARM1 methylates the GBD of p300 and CBP. (A) Full-length p300 (200 ng) or core histones (Roche Molecular Biochemicals) (500 ng) were incubated with GST-CARM1 or GST-PRMT1 (1 μg) and [3H]AdoMet. Methylation products were analyzed by SDS/PAGE and autoradiography. (B) p300 fragments were analyzed as methylation substrates. (C and D) The indicated p300 and CBP fragments fused to GST were analyzed as methylation substrates for CARM1 as in A. (E) GST-GBD WT or mutant fragments were tested as methylation substrates for CARM1 as in A.
A rabbit antiserum was generated against a peptide representing the R3 region of p300 GBD and containing asymmetric dimethylarginine at residue 2142 (dimethyl-R3 peptide). In immunoblots this anti-dimethyl-R3 peptide antiserum strongly interacted with the methylated p300 GBD fragment, but not with the unmethylated WT GBD, the unmethylatable R3K GBD mutant, or the CARM1-methylated KIX fragment (amino acids 568–828) of p300 (Fig. 2 A and B). The specificity of this antiserum for methylated p300 GBD further confirmed Arg-2142 as a CARM1 methylation site in vitro.
Fig. 2.
Detection of p300-GBD methylation in vivo. (A) GST-p300 GBD protein (1 μg) was incubated with or without GST-CARM1, GST-PRMT1, and 500 μM unlabeled AdoMet for 1 h at 30°C. Methylation products were detected by immunoblot, using antibody against dimethyl-R3 peptide. (B) GST-p300 GBD, GST-p300 GBD(R3K mutant), or GST-p300 KIX (1 μg) was incubated with GST-CARM1 and AdoMet and analyzed by immunoblot as in A. (C) 293T cells were transfected with 2–3 μg each of plasmids for p300, CARM1, and PRMT1. Cell extracts were immunoprecipitated (IP) with antiserum against p300, and precipitated proteins were analyzed by immunoblot (IB) with antibody against dimethyl-R3 peptide. (D) 293T cells were transfected with plasmids for Flag-GST-p300 GBD (WT or R3K mutant), CARM1, and PRMT1. The Flag-GST-GBD proteins were precipitated (P) from transfected cell extracts with glutathione-Sepharose beads and analyzed by immunoblot (IB) with antibody against dimethyl-R3 peptide (Upper), then stripped and reprobed with anti-Flag antibody (Lower).
To test for p300 methylation in mammalian cells, full-length p300 was overexpressed in 293T cells with or without CARM1. Immunoprecipitated p300 was specifically recognized in an immunoblot by the antibody against dimethyl-R3 peptide (Fig. 2C, lane 2), and the strength of this 300-kDa band was enhanced by overexpression of CARM1 (Fig. 2C, lane 3) but not by overexpression of PRMT1 (Fig. 2C, lane 4). Thus Arg-2142 of full-length p300 is methylated in vivo by CARM1.
Methylation of the p300 GBD fragment by CARM1 was also observed in mammalian cells. A Flag-tagged GST-GBD fusion protein was transiently expressed in 293T cells and isolated from the transfected cell extracts by binding to glutathione-Sepharose beads; the bead-bound proteins were analyzed by immunoblot. The anti-dimethyl-R3 peptide antiserum produced a weak band in some experiments when WT Flag-GST-GBD was overexpressed alone (Fig. 6, which is published as supporting information on the PNAS web site), and this signal was strongly enhanced when CARM1 was overexpressed with the WT GBD fusion protein (Fig. 2D, lane 3 and Fig. 6), but not when PRMT1 was overexpressed (Fig. 2D, lane 4). No signal was detected when the GBD(R3K) mutant was coexpressed with CARM1 (Fig. 2D, lane 5 and Fig. 6). The weak signal observed in some experiments when WT Flag-GST-GBD was expressed alone apparently was caused by methylation by endogenous CARM1, because small interfering RNA directed against CARM1 reduced this signal further (Fig. 6). When the blot was stripped and reprobed with anti-Flag antibody, the p300 GBD fragments were detected in all of the transfected cell extracts (Fig. 2D Lower, lanes 2–5). Thus Arg-2142 is the CARM1 methylation site of p300 GBD in mammalian cells and is methylated by endogenous as well as overexpressed CARM1.
Both Arg-2088 (R2) and Arg-2142 (R3) of p300 GBD Are Important for Physical and Functional Interaction with GRIP1 AD1. To analyze the functional role of p300 GBD methylation, we first tested the functional roles of the three Arg residues in p300 GBD that are conserved in CBP. The GBD of CBP and p300 binds to the AD1 region of p160 coactivators (21, 22). In a mammalian one-hybrid assay, increasing expression of GRIP1 AD1 (amino acids 730-1121) enhanced the ability of p300 GBD fused to Gal4 DBD to activate expression of a reporter gene (Fig. 7, which is published as supporting information on the PNAS web site). The R1K mutant of GBD exhibited WT activity in this same assay (data not shown), but the R2K mutant was almost inactive, and the R3K mutant had reduced activity (Fig. 7). Thus both the R2 and R3 residues of p300 GBD are important for the functional interaction between p300 GBD and GRIP AD1, suggesting that they may also be important for the physical interaction.
In direct GBD-GRIP1 AD1 binding studies, short synthetic peptides representing the R2 and R3 regions of p300 GBD were used as competitors in GST pull-down assays. GRIP1 AD1 synthesized in vitro (Fig. 3A, lane 1) bound strongly to p300 GBD fused to GST (Fig. 3A, lane 3) but not to GST (Fig. 3A, lane 2). Neither the R2 peptide nor the R3 peptide (20 μg) caused inhibition of the binding of GRIP1 AD1 to GST-GBD (Fig. 3A, lanes 4 and 5). However, a combination of the R2 and R3 peptides (10 μg of each) effectively reduced the binding of GRIP1 AD1 to GST-GBD (Fig. 3A, lane 9). This finding indicates that both R2 and R3 regions are important for binding to GRIP1 AD1 and are thus both presumably in contact with GRIP1 AD1.
Fig. 3.
Roles of R2, R3, and R3 methylation in binding of p300 GBD to GRIP1. (A) GRIP AD1 (amino acids 730-1121) was synthesized in vitro with [35S]methionine and incubated with glutathione-Sepharose beads containing 1 μgof bound GST-p300 GBD, with or without R2, R3, or dimethyl-R3 competitor peptides. Bound GRIP1 AD1 was eluted and analyzed by SDS/PAGE and autoradiography. (B) GST-p300 GBD protein (0.5 μg) was incubated under methylation conditions with or without GST-CARM1 (0.1 μg), then incubated with in vitro-translated HA-tagged GRIP1 AD1 fragment, HA-specific antibody, and protein A/G beads. After immunoprecipitation (IP), GRIP1-bound and unbound fractions were analyzed in parallel immunoblots (IB) with antibodies against dimethyl-R3 peptide (Upper) or against GST (Santa Cruz Biotechnology) (Lower).
R3 Methylation of p300 GBD Inhibits Its Binding to GRIP1 AD1. To test the effect of R3 methylation on the binding of p300 GBD to GRIP1.AD1, a dimethyl-R3 peptide (identical to the R3 peptide, but with asymmetric dimethylarginine at residue 2142) was compared with the unmethylated R3 peptide in the peptide competition assay described above. Neither the dimethyl-R3 peptide alone (data not shown) nor the dimethyl-R3 peptide combined with the R2 peptide (Fig. 3A, lane 8) reduced the binding of GRIP1 AD1 to GST-GBD, whereas the combination of the unmethylated R3 peptide with the R2 peptide competed effectively in this assay (Fig. 3A, lane 9). The concentrations of the R2, R3, and dimethyl-R3 peptide stock solutions were confirmed by HPLC (data not shown). These results suggest that methylation of R3 reduces the ability of p300 GBD to bind to GRIP1 AD1. Thus, the methylation of R3 could be used as a switch to regulate the assembly/disassembly of p300 with p160 coactivators.
The effect of methylation of p300 GBD on its binding to GRIP1 AD1 was tested directly in a coimmunoprecipitation assay. The p300 GBD fragment fused to GST was methylated by CARM1 in vitro and then incubated with HA-tagged GRIP1 AD1 fragments synthesized in vitro. These reactions typically result in the methylation of only a fraction of the total protein substrate. After immunoprecipitation with anti-HA antibody, the resulting immune complex and supernatant fractions were analyzed by immunoblot, using anti-GST antibodies to detect total GST-GBD (Fig. 3B Lower) and anti-dimethyl-R3 peptide antibodies to detect the methylated GST-GBD (Fig. 3B Upper). Because limiting amounts of GRIP1 AD1 fragment were used, GST-GBD was detected in both the pellet (i.e., bound to GRIP1 AD1) and the supernatant (not bound to GRIP1 AD1) (Fig. 3B Lower, lanes 3 and 4 and lanes 6 and 7). However, the methylated GST-GBD was found exclusively in the unbound fraction (Fig. 3B Upper, lane 7), not in the GRIP1-bound fraction (Fig. 3B Upper, lane 4). This finding demonstrates directly that protein methylation of p300 GBD inhibits its interaction with GRIP1 AD1.
Methylation of p300 GBD by CARM1 and Demethylimination by PAD4 Competitively Regulate p300–GRIP1 Binding. Methyl groups on some Arg residues of histones can be removed by demethylimination by the enzyme PAD4. PAD4 converts methylarginine in histones to citrulline by removing methylamine. We tested whether PAD4 can remove the methyl mark from p300 GBD. GST-GBD was methylated by CARM1 by using [3H]AdoMet (Fig. 4A, lane 1). Most of the methyl label was removed by incubation with purified PAD4 in the presence of calcium (Fig. 4A, lane 2), which was necessary for its enzymatic activity (Fig. 4A, lane 4), as previously shown (18, 19). A catalytically inactive mutant of PAD4 failed to reduce p300 GBD methylation (Fig. 4A, lane 3). Thus, PAD4 may play active roles in removing methylation marks from both histones and transcriptional cofactors.
Fig. 4.
PAD4 removes the methylation mark of p300 GBD and regulates p300–GRIP1 interaction. (A) WT or catalytically inactive mutant PAD4 enzyme was included in methylation reactions containing p300 GBD, CARM1, radioactively labeled AdoMet, and 4 mM calcium chloride. Labeled GBD was detected by SDS/PAGE and autoradiography. (B) Plasmids encoding Gal4 DBD fused to p300 GBD WT or R3K mutant (500 ng each) were transfected into CV-1 cells with 250 ng of GK1 reporter plasmid and plasmids for GRIP1 AD1 fused to VP16 AD, CARM1, PRMT1, and PAD4 WT or mutant (500 ng each). Luciferase activities of the transfected cell extracts are shown.
Next, we investigated the effects of methylation and demethylimination of p300 GBD on its interaction with the GRIP1 AD1 domain in vivo, using a mammalian two-hybrid assay. The strong interaction of p300 GBD (fused to Gal4 DBD) with GRIP1 AD1 (fused to VP16 AD) in CV-1 cells (Fig. 4B, lane 3) was reduced by overexpression of CARM1 (Fig. 4B, lane 4), but not by PRMT1 (Fig. 4B, lane 5). Furthermore, overexpression of PAD4 enhanced the interaction of p300 GBD with GRIP1 AD1 (Fig. 4B, lane 6), whereas the catalytically inactive PAD4 mutant had no effect (Fig. 4B, lane 7). As an additional control to demonstrate the importance of R3 methylation in these effects, similar experiments were conducted with the R3K mutant of p300 GBD (fused to Gal4 DBD). As shown (Fig. 7), the R3K mutant bound to GRIP1 AD1, but somewhat less efficiently than to WT GBD (Fig. 4B, lane 9). Neither CARM1 nor PAD 4 had any effect on the interaction of GBD(R3K) with GRIP1 AD1 (Fig. 4B, lanes 10–12). Essentially the same results were obtained in a mammalian one-hybrid assay, where CARM1 or PRMT1 were tested for their effects on reporter gene activation by Gal4-BD (WT or R3K mutant) and GRIP1 AD1 that was not fused to VP16 AD (Fig. 8, which is published as supporting information on the PNAS web site). In addition, adenosine dialdehyde, a general inhibitor of AdoMet-dependent methylation, enhanced the interaction of p300 GBD and GRIP1 AD1 in the mammalian two-hybrid assay (Fig. 9, which is published as supporting information on the PNAS web site). Thus, in cultured cells methylation by CARM1 inhibits and demethylimination by PAD4 enhances the interaction between p300 GBD and GRIP1 AD1.
The ability of PAD4 to enhance binding of p300 GBD to GRIP1 AD1 (Fig. 4B) is consistent with our conclusion from studies with the antibody against dimethyl-R3 peptide (Fig. 6) that p300 GBD is methylated at a basal level by endogenous CARM1. This conclusion is further supported by the ability of small interfering RNA against CARM1 to reduce the immunoblot signal of p300 GBD with the anti-dimethyl-R3 antibody (Fig. 6) and enhance p300 GBD-GRIP1 AD1 binding in a mammalian one-hybrid assay (Fig. 8).
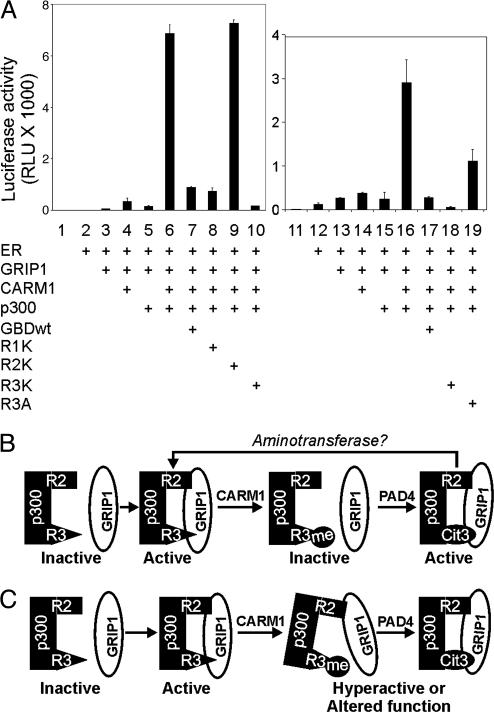
Different Roles of Arg Residues R2 and R3 of p300 GBD in Coactivator-Assisted Transcriptional Activation by ER. To test the effects of p300 GBD methylation on transcriptional activation, the p300 GBD WT and mutant fragments were used as dominant negative inhibitors for ER-mediated transcription. Because the interaction of GRIP1 and p300 is a critical step for coactivator assembly and transcriptional activation by NRs (12, 13, 23), the overexpressed p300 GBD fragment could act as a dominant negative inhibitor of NR-mediated transcription. As shown (13), coexpression of CARM1, p300, and GRIP1 synergistically enhanced the ability of ER to activate expression of a reporter gene controlled by an estrogen response element (Fig. 5A, lanes 1–6 and 11–16). Overexpression of the WT p300 GBD fragment dramatically reduced reporter gene expression (Fig. 5A, lanes 7 and 17), supporting the conclusion that the interaction between GRIP1 and p300 GBD is essential for ER-mediated transcription. The R1K mutant of p300 GBD, which bound GRIP1 as efficiently as the WT GBD (data not shown) had a dominant negative effect indistinguishable from that of WT GBD (Fig. 5A, lane 8). In contrast, the GBD(R2K) mutant exhibited no dominant negative activity (Fig. 5A, lane 9), consistent with its inability to bind GRIP1 (Fig. 7) and with the critical role of R2 in the interaction with GRIP1. The lost activity of the R2K mutant also demonstrates that the dominant negative effect of the WT GBD fragment is not caused by general squelching but is specifically caused by its ability to bind GRIP1 AD1 and thereby disrupt the interaction of GRIP1 with full-length p300.
Fig. 5.
Different roles of R2 and R3 of p300 GBD in coactivator assembly for ER-mediated transcription. (A) Plasmids for GRIP1 (250 ng), CARM1 (500 ng), p300 (500 ng), and ER (1 ng) were transfected into CV-1 cells with MMTV-ERELUC reporter plasmid (250 ng) and plasmids encoding p300 GBD WT or mutants (500 ng) as dominant negative competitors. Luciferase activity is shown. (B) The model shows the regulation of the p300–GRIP1 complex formation by CARM1 and PAD4. R2 and R3, Arg residues that contact GRIP1; me, methyl group; Cit3, R3 converted to citrulline by PAD4. (C) The model shows the regulation of the p300–GRIP1 complex conformation and function by CARM1 and PAD4.
Surprisingly, the GBD(R3K) mutant had a stronger dominant negative effect than WT GBD on ER-mediated transcription (Fig. 5A, lanes 10 and 18). To further investigate the role of R3 in ER-mediated transcription, another GBD mutant with R3 changed to Ala (R3A) was tested. Ala lacks the positive charge of Arg and Lys. The GBD(R3A) mutant did exhibit dominant negative activity, but less than the R3 WT or R3K mutant forms (Fig. 5A, lane 19). These results indicate that R3 plays an important but different role from R2 in the binding of GRIP1 and the synergistic coactivator function between GRIP1, p300, and CARM1.
Discussion
Importance of R2, R3, and R3 Methylation in the p300–GRIP1 Interaction. The binding interface between the GBD of p300/CBP and AD1 of p160 coactivators is critical for their synergistic cooperation as coactivators, presumably because p300 and CBP are anchored to the promoter by binding to p160 coactivators, which in turn bind directly to NRs (12, 23, 24). Here we demonstrate that two Arg residues in the GBD of p300 (Arg-2088 or R2; and Arg-2142 or R3) play critical but different roles in that binding interaction. Direct contact of the R2 residue with p160 AD1 was previously demonstrated in a solution structure of the activator for thyroid hormone and retinoid receptors AD1 fragment bound to a CBP fragment containing the R1 and R2 residues, but not the R3 residue (25). Our studies provide experimental confirmation of the critical importance of R2. Even a conservative substitution of Lys for R2 completely disrupted p300–GRIP1 binding (Figs. 5A and 7).
Whereas R2 is a major and critical contact site between p300 and GRIP1, our peptide competition data indicate that R3 also contributes to p300–GRIP1 binding (Fig. 3A) and acts as a regulatory contact site. R3 of p300 GBD was methylated by CARM1 (Figs. 1 and 2), and depending on the assay context, this methylation of R3 reduces or eliminates the binding of p300 to GRIP1, as indicated by peptide inhibition studies (Fig. 3A), in vitro binding studies (Fig. 3B), mammalian one-hybrid assay (Fig. 8), two-hybrid assay (Fig. 4B), reduction of endogenous CARM1 levels by small interfering RNA (Fig. 8), and inhibition of AdoMet-dependent methylation in cells by adenosine dialdehyde (Fig. 9). The fact that GBD-GRIP1 binding was eliminated by R3 methylation in direct bimolecular binding studies in vitro (Fig. 3), but was only reduced in assays where a larger functional complex of coactivators may be involved (Figs. 4B and 8), suggests that R3 methylation may not cause complete dissociation of the p300–GRIP1 interaction within the context of a larger coactivator complex.
In addition to p160 coactivators, the GBD region and sequences nearby interact with other transcription factors such as p53, Ets2, YY1, Smad proteins, KSHV IRF-1, IRF-3, and E1A (26–28). It will be interesting to determine whether methylation of the GBD also affects binding of these proteins to p300/CBP.
Removal of the p300 GBD Methylation Mark by PAD4: Reversal of CARM1's Inhibition of p300–GRIP1 Binding. PAD4 is recruited to the estrogen-activated pS2 promoter, where it demethyliminates methylated histones and plays a role in transcriptional repression of ER (18, 19). Here we show that the methylation of a nonhistone cofactor (p300) can be similarly reversed by PAD4 (Fig. 4A). PAD4 also enhanced the interaction of GRIP1 AD1 and p300 GBD, and this enhancement required Arg (not Lys) at the R3 position of the GBD (Fig. 4B). Thus methylation and demethylimination of the p300 GBD R3 residue is involved in modulating a key protein–protein interaction (p300–GRIP1) that affects the assembly or conformation of the p160 coactivator complex.
Because PAD4 can convert monomethylated or unmethylated Arg residues of histones to citrulline (18, 19), we cannot strictly distinguish whether the enhanced binding of p300 GBD to GRIP1 AD1 caused by PAD4 expression (Fig. 4B) is caused by PAD4 action on methylated or unmethylated GBD. Nevertheless, PAD4 can effectively remove labeled methyl groups from CARM1-methylated GBD (Fig. 4A). Thus replacing methylarginine with citrulline at R3 of p300 GBD restores the interaction with GRIP1. Unmodified Arg and citrulline at the R3 position apparently behaved similarly in terms of GRIP1 binding; however, considering the different side chains of arginine and citrulline, we cannot rule out the possibility that these two residues actually cause different coactivator complex conformations and function (Fig. 5 B and C).
Implications of p300 GBD Methylation and Demethylimination for Regulation of p160 Coactivator Complex Assembly/Disassembly, Conformation, and Function. Our data suggest two possible roles that methylation and demethylimination of p300 GBD by CARM1 and PAD4 may play in the transcriptional activation process. First, they could regulate the assembly and disassembly of the p160 coactivator complex. Second, these posttranslational modifications could alter the conformation and function of the p160 coactivator complex.
The first model would seem to predict that methylation of p300 GBD by CARM1 should have a negative effect on transcription. However, recent findings suggest that repetitive association and dissociation of steroid receptors and coactivators from their target promoters may be required to maintain an activated state of transcription. Hormone-activated steroid receptors and their coactivators dissociate from their target promoters after only a few seconds (29, 30), and the steady-state levels of these proteins on the promoters fluctuate up and down with a period of ≈45 min (31, 32). Several active mechanisms that may contribute to transcription complex dissociation include: NR chaperone proteins that associate with steroid receptors on the target gene promoter (33); inhibition of NR-p160 binding by acetylation of p160 coactivators by p300/CBP (8); and cyclical recruitment of proteasome components to the promoter (34). Thus, the inhibition of p300–GRIP1 binding by p300 GBD methylation may be important for the cyclical association and dissociation of coactivators at the promoter. In this model, the subsequent demethylimination of p300 GBD by PAD4 could be a way to restore p300-p160 binding, as our data suggest (Fig. 4B), or could even represent a recycling mechanism, which could conceivably be completed by a currently unknown aminotransferase reaction that converts citrulline back to arginine (Fig. 5B).
In our second model, methylation and demethylimination of the R3 residue of p300 GBD may not regulate coactivator complex assembly, but may cause conformational changes in the complex that facilitate subsequent steps in the transcriptional activation process (Fig. 5C). R3 methylation strongly inhibited bimolecular interactions of p300 and GRIP1 (Fig. 3), but in the context of a large coactivator complex, the remaining interaction between GRIP1 and the R2 region of p300 GBD might be sufficient for the integrity of the complex (Figs. 4B and 8). Furthermore, our data demonstrate that R3 is involved in p300–GRIP1 binding but plays a regulatory role in contrast to the critical role of R2. The R2K mutation completely abolished the ability of the p300 GBD fragment to interfere with ER-mediated transcription, but the R3K mutant of the GBD fragment was still effective as an inhibitor (Fig. 5A), indicating that it maintains the appropriate conformation for forming an active coactivator complex. The slightly stronger dominant negative effect of the R3K mutant, compared with WT GBD, may be caused by the different side-chain structure of Lys and Arg or the inability of the R3K fragment to be inactivated by methylation by endogenous CARM1. The R3K phenotype and the reduced dominant negative activity of the R3A mutant suggest significant roles for charge and modification status of R3 in ER-mediated transcription.
The strong coactivator synergy among CARM1, p300, and GRIP1 in NR-mediated transcription requires the methyltransferase activity of CARM1 (13) and cooperative histone modifications by CARM1 and p300 (15). In addition, it was previously proposed that the methylation of the KIX and KIX-adjacent regions of p300 and CBP might contribute to this synergy (9, 10). However, because there is no evidence of direct binding of the KIX region of p300/CBP to p160 coactivators, methylation of this region of p300/CBP might contribute to NR coactivator synergy by indirect mechanisms. Our data suggest that methylation and demethylimination of the R3 residue of p300 GBD may be a key mechanism in p300/CBP-p160-CARM1 coactivator synergy. Methylation of p300/CBP by CARM1 may promote a conformational change that loosens the p300–p160 interaction in the complex and facilitates additional steps in transcriptional activation (Fig. 5C). p300 methylation by CARM1 may be negatively regulated or functionally reversed by demethylimination by PAD4, or demethylimination by PAD4 may be a subsequent requisite step in coactivator complex maturation.
Supplementary Material
Acknowledgments
We thank Dr. T.-P. Yao (Duke University, Durham, NC) for providing plasmids encoding fragments of p300 and CBP and D. Gerke (University of Southern California) and R. Rice and W. Yuan (Upstate Biotechnology) for expert technical assistance. This work was supported by U.S. Public Health Service Grants DK55274 (to M.R.S.), HD38353 (to S.A.C.), and DK58110 (to W.L.K.) from the National Institutes of Health.
Author contributions: Y.-H.L., M.A.J., and M.R.S. designed research; Y.-H.L. performed research; S.A.C., W.L.K., and M.A.J. contributed new reagents/analytic tools; Y.-H.L., M.A.J., and M.R.S. analyzed data; and Y.-H.L. and M.R.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AdoMet, S-adenosylmethionine; CARM1, coactivator-associated arginine methyltransferase; CREB, cAMP response element binding protein; CBP, CREB binding protein; DBD, DNA binding domain; ER, estrogen receptor; GRIP1, glucocorticoid receptor interacting protein; GBD, GRIP1-binding domain; HA, hemagglutinin; NR, nuclear receptor; PAD4, peptidyl deiminase 4; PRMT1, protein arginine methyltransferase 1.
References
- 1.Mangelsdorf, D. J. & Evans, R. M. (1995) Cell 83, 841-850. [DOI] [PubMed] [Google Scholar]
- 2.Tsai, M.-J. & O'Malley, B. W. (1994) Annu. Rev. Biochem. 63, 451-486. [DOI] [PubMed] [Google Scholar]
- 3.Glass, C. K. & Rosenfeld, M. G. (2000) Genes Dev. 14, 121-141. [PubMed] [Google Scholar]
- 4.Hermanson, O., Glass, C. K. & Rosenfeld, M. G. (2002) Trends Endocrinol. Metab. 13, 55-60. [DOI] [PubMed] [Google Scholar]
- 5.McKenna, N. J. & O'Malley, B. W. (2002) Cell 108, 465-474. [DOI] [PubMed] [Google Scholar]
- 6.Rice, J. C. & Allis, C. D. (2001) Curr. Opin. Cell Biol. 13, 263-273. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Y. & Reinberg, D. (2001) Genes Dev. 15, 2343-2360. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., Lin, R. J., Xie, W., Wilpitz, D. & Evans, R. M. (1999) Cell 98, 675-686. [DOI] [PubMed] [Google Scholar]
- 9.Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B. M., Montminy, M. & Evans, R. M. (2001) Science 294, 2507-2511. [DOI] [PubMed] [Google Scholar]
- 10.Chevillard-Briet, M., Trouche, D. & Vandel, L. (2002) EMBO J. 21, 5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraus, W. L., Manning, E. T. & Kadonaga, J. T. (1999) Mol. Cell. Biol. 19, 8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, J., O'Malley, B. W. & Wong, J. (2000) Mol. Cell. Biol. 20, 2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, Y.-H., Koh, S. S., Zhang, X., Cheng, X. & Stallcup, M. R. (2002) Mol. Cell. Biol. 22, 3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, H., Baumann, C. T., Li, H., Strahl, B. D., Rice, R., Jelinek, M. A., Aswad, D. W., Allis, C. D., Hager, G. L. & Stallcup, M. R. (2001) Curr. Biol. 11, 1981-1985. [DOI] [PubMed] [Google Scholar]
- 15.An, W., Kim, J. & Roeder, R. G. (2004) Cell 117, 735-748. [DOI] [PubMed] [Google Scholar]
- 16.Sartorelli, V., Puri, P. L., Hamamori, Y., Ogryzko, V., Chung, G., Nakatani, Y., Wang, J. Y. & Kedes, L. (1999) Mol. Cell 4, 725-734. [DOI] [PubMed] [Google Scholar]
- 17.Bannister, A. J., Schneider, R. & Kouzarides, T. (2002) Cell 109, 801-806. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Y., Wysocka, J., Sayegh, J., Lee, Y. H., Perlin, J. R., Leonelli, L., Sonbuchner, L. S., McDonald, C. H., Cook, R. G., Dou, Y., et al. (2004) Science 306, 279-283. [DOI] [PubMed] [Google Scholar]
- 19.Cuthbert, G. L., Daujat, S., Snowden, A. W., Erdjument-Bromage, H., Hagiwara, T., Yamada, M., Schneider, R., Gregory, P. D., Tempst, P., Bannister, A. J., et al. (2004) Cell 118, 545-553. [DOI] [PubMed] [Google Scholar]
- 20.Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S.-M., Schurter, B. T., Aswad, D. W. & Stallcup, M. R. (1999) Science 284, 2174-2177. [DOI] [PubMed] [Google Scholar]
- 21.Kamei, Y., Xu, L., Heinzel, T., Torchia, J., Kurokawa, R., Gloss, B., Lin, S.-C., Heyman, R. A., Rose, D. W., Glass, C. K., et al. (1996) Cell 85, 403-414. [DOI] [PubMed] [Google Scholar]
- 22.Chen, H., Lin, R. J., Schiltz, R. L., Chakravarti, D., Nash, A., Nagy, L., Privalsky, M. L., Nakatani, Y. & Evans, R. M. (1997) Cell 90, 569-580. [DOI] [PubMed] [Google Scholar]
- 23.Chen, D., Huang, S.-M. & Stallcup, M. R. (2000) J. Biol. Chem. 275, 40810-40816. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M. Y., Hsiao, S. J. & Kraus, W. L. (2001) EMBO J. 20, 6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demarest, S. J., Martinez-Yamout, M., Chung, J., Chen, H., Xu, W., Dyson, H. J., Evans, R. M. & Wright, P. E. (2002) Nature 415, 549-553. [DOI] [PubMed] [Google Scholar]
- 26.Vo, N. & Goodman, R. H. (2001) J. Biol. Chem. 276, 13505-13508. [DOI] [PubMed] [Google Scholar]
- 27.Livengood, J. A., Scoggin, K. E., Van Orden, K., McBryant, S. J., Edayathumangalam, R. S., Laybourn, P. J. & Nyborg, J. K. (2002) J. Biol. Chem. 277, 9054-9061. [DOI] [PubMed] [Google Scholar]
- 28.Lin, C. H., Hare, B. J., Wagner, G., Harrison, S. C., Maniatis, T. & Fraenkel, E. (2001) Mol. Cell 8, 581-590. [DOI] [PubMed] [Google Scholar]
- 29.McNally, J. G., Muller, W. G., Walker, D., Wolford, R. & Hager, G. L. (2000) Science 287, 1262-1265. [DOI] [PubMed] [Google Scholar]
- 30.Becker, M., Baumann, C., John, S., Walker, D. A., Vigneron, M., McNally, J. G. & Hager, G. L. (2002) EMBO Rep. 3, 1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. (2000) Cell 103, 843-852. [DOI] [PubMed] [Google Scholar]
- 32.Metivier, R., Penot, G., Hubner, M. R., Reid, G., Brand, H., Kos, M. & Gannon, F. (2003) Cell 115, 751-763. [DOI] [PubMed] [Google Scholar]
- 33.Freeman, B. C. & Yamamoto, K. R. (2002) Science 296, 2232-2235. [DOI] [PubMed] [Google Scholar]
- 34.Reid, G., Hubner, M. R., Metivier, R., Brand, H., Denger, S., Manu, D., Beaudouin, J., Ellenberg, J. & Gannon, F. (2003) Mol. Cell 11, 695-707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.