Abstract
Recent studies have identified a series of estrogen receptor (ER)-interacting peptides that recognize sites that are distinct from the classic coregulator recruitment (AF2) region. Here, we report the structural and functional characterization of an ERα-specific peptide that binds to the liganded receptor in an AF2-independent manner. The 2-Å crystal structure of the ER/peptide complex reveals a binding site that is centered on a shallow depression on the β-hairpin face of the ligand-binding domain. The peptide binds in an unusual extended conformation and makes multiple contacts with the ligand-binding domain. The location and architecture of the binding site provides an insight into the peptide's ER subtype specificity and ligand interaction preferences. In vivo, an engineered coactivator containing the peptide motif is able to strongly enhance the transcriptional activity of liganded ERα, particularly in the presence of 4-hydroxytamoxifen. Furthermore, disruption of this binding surface alters ER's response to the coregulator TIF2. Together, these results indicate that this previously unknown interaction site represents a bona fide control surface involved in regulating receptor activity.
Keywords: coregulator, phage display, structure
The estrogen receptor (ER) functions as a ligand-activated transcription factor and is responsible for mediating the physiological effects of the steroid hormone 17β-estradiol (E2). Transcriptional activation is facilitated by two distinct activation functions (AF), the constitutively active AF1 located at the N terminus of the receptor and a ligand-dependent AF2 that resides in the C-terminal ligand-binding domain (LBD) (1), which recruit a range of coregulatory protein complexes to the DNA-bound receptor (2). ER agonists promote association with coactivators, whereas antagonists favor recruitment of corepressors.
The LBD serves as the major interaction point between nuclear receptors (NRs) and coregulatory proteins. Coactivator recruitment is mediated by short α-helical, leucine-rich LxxLL consensus motifs (NR-box) that bind to the AF2 region of the LBD (3–5). Structural analysis of NR LBDs has established that agonist binding stabilizes a receptor conformation in which elements of AF2 form a hydrophobic groove that can accommodate the LxxLL motifs found within coactivators (6–8). In contrast, ER antagonists affect the positioning of the AF2's mobile C-terminal helix (H12), thereby disrupting the LxxLL-binding site and preventing coactivator recruitment (7, 9). The resultant antagonist-induced conformation favors recruitment of corepressor complexes (10). Although the details of corepressor binding to ERLBD are not known, antagonist-induced displacement of H12 in other NRs promotes corepressor recruitment to an interaction surface that overlaps with the AF2 groove (11–13).
Although this conformational mechanism can be used to rationalize the differential recruitment of coactivators and corepressors to a common surface on ER, it is not immediately apparent how this alone can account for the diverse pharmacology of selective ER receptor modulators (SERMs). SERMs, such as raloxifene and 4-hydroxytamoxifen (OHT), are a class of synthetic ER ligands that exhibit tissue-dependent pharmacology, imitating the action of estrogens in certain tissues while opposing their action in others (14). These compounds appear to elicit their effects by inducing distinct conformational changes in ER that favor interaction with specific subsets of coactivators (15). The mechanisms of such coactivator binding are unclear but are likely to involve recognition surfaces, which are unrelated to AF2, that are revealed in response to a particular modulator.
The screening of random peptide libraries is a powerful method for isolating motifs that mimic protein–protein interaction surfaces. Phage display techniques using both focused and random peptide libraries have been used to investigate the interaction of coregulators with liganded ER (13, 16–19). These studies have isolated a number of short peptides with characteristic sequence motifs that specifically recognize different liganded states of ER. Such peptides, in addition to acting as conformational probes (16), have been shown to interfere with receptor activity both in vitro and in cell-based assays (18, 19). These observations have led to the suggestion that such motifs represent biologically relevant interaction modules or, alternatively, that they mimic receptor–cofactor interactions. Although some of the isolated motifs are directed to the region around ER's AF2 binding groove, several possess previously undescribed consensus sequences and appear to interact with distinct regions of the receptor.
To address whether ERLBD harbors additional protein–protein interaction sites, we have studied an ER interaction motif that binds specifically to ERα. The 11-residue peptide antagonist, referred to as αII, was initially isolated from a random peptide phage display library by using E2- or OHT-bound ERα and found to interact in the presence of a broad spectrum of receptor modulators (16). In this study, we have used a combination of techniques to both delineate the αII binding site on ERαLBD and demonstrate that this surface is involved in ER's response to certain ligands.
Materials and Methods
Protein Expression and Purification. The H12 truncated mutant (ERαΔH12) used for structural analysis was generated by inserting a stop codon after Val-533 in a pET15b-ERαLBD construct (20). ERαΔH12 was expressed in Escherichia coli strain C41 (DE3). ERαΔH12 was extracted from inclusion bodies with a buffer (150 mM NaCl/1 mM EDTA/2 mM DTT/10% glycerol/1 mM PMSF/50 mM Tris, pH 8.0) containing 1% (wt/vol) Zwittergent 3-12. The detergent was removed by dialysis, and the extract was applied to an E2–Sepharose column. The E2-affinity matrix was prepared as described (21). Bound ERαΔH12 was carboxymethylated overnight with 10 mM iodoacetic acid and eluted with 50 μM OHT in a buffer containing 1 mM DTT, 1 mM EDTA, 250 mM NaSCN, and 25 mM Tris (pH 8.5). ERαΔH12 was further purified by using ion exchange chromatography. Pooled fractions containing ERαΔH12 were combined with a 1.5-fold molar excess of αII peptide and concentrated to 10 mg/ml. Additional peptide was added to obtain a final peptide/LBD molar ratio of 3:1.
Crystallization and Structure Determination. Hanging drops composed of equal volumes of ERαΔH12-OHT-αII complex and reservoir solution [20% (wt/vol) polyethylene glycol (PEG) 2000 monomethyl ether (Mme)/620 mM sodium formate/3% (wt/vol) glucose in 100 mM Tris, pH 8.0] were equilibrated against reservoir solution at 19°C. Crystals were cryoprotected in mother liquor containing 32.5% (wt/vol) PEG 2000 Mme and vitrified in liquid nitrogen. Crystals belong to space group P212121 and have a single LBD homodimer per asymmetric unit. Diffraction data, collected to a resolution of 2 Å on beamline ID29 at the European Synchrotron Radiation Facility (Grenoble, France), were indexed, reduced, and scaled by using the hkl suite of programs (22). The structure was solved by molecular replacement using amore (23). Initial electron density maps clearly indicated the position of OHT and the αII peptide. The structure was refined with refmac5 (24) using all available data. All model building was carried out with quanta (Accelrys, San Diego). The final model has an Rcryst of 18.7% and a Rfree of 21.9%. Data collection and refinement statistics are shown in Table 1, which is published as supporting information on the PNAS web site.
Surface Plasmon Resonance (SPR). Measurements were performed by using a BIAcore X instrument and streptavidin-coated sensor chips. Experiments were carried out as described (8). Biotinylated αII peptide (SGSGLTSRDFGSWYA) was immobilized to variable responses (60–160 relative units). Qualitative binding experiments with liganded ERα/βLBD and LBD mutants were performed by flowing each complex (1 μM dimeric concentration) over the αII chip for 2 min at 5 μl·min-1. A sensor chip comprising an immobilized biotinylated SRC2–2 peptide (EKHKILHRLLQDS) (8) was used as a control binding surface to assess the structural integrity of the LBD mutants. Methods used for construction of the LBD mutants, the kinetic analysis and preparation of protein used for SPR studies are described in Supporting Text, which is published as supporting information on the PNAS web site.
Mammalian Two-Hybrid and Reporter Assays. Details of the ER and TIF2 expression plasmids used are described in Supporting Text. Mammalian two-hybrid experiments were carried out as described (19). For the transcriptional assays with the engineered TIF2 coactivator, Cos-7 cells were cotransfected with expression vectors (pSG5) for wild-type ERα and either TIF2-NR12 or TIF2αII together with the estrogen responsive 3× ERE-TATA-luc reporter. Experiments were performed in triplicate and contained 30 ng of ERα construct, 30 ng of β-gal construct, 150 ng of reporter construct, and 150 ng of TIF2 expression plasmid. After transfection, cells were treated with 10 nM E2, 100 nM OHT, or dimethylsulphoxide for 16 h before analyzing luciferase and β-gal activity. For the transcriptional assays comparing wild-type and mutant (ERα G442H) receptor activity, Cos-7 cells were cotransfected with expression vectors (pSG5 or Gal4DBD) for wild-type ERα, ERα G442H, Gal4-tagged ERα DEFΔH12, or Gal4-tagged ERα DEFG442HΔH12 and either empty expression vector (pSG5) or TIF2wt expression vector together with either the estrogen responsive 3× ERE-TATA-luc reporter or the Gal4 responsive reporter 5× UAS-TATA-luc. Each triplicate contained 30 ng of ERα construct, 30 ng of β-gal construct, 150 ng of reporter construct, and 30 ng of TIF2 expression plasmid. After transfection, cells were treated with 100 nM OHT or dimethylsulphoxide for 16 h before analyzing luciferase and β-gal activity.
Results
αII Peptide Motif Interacts Specifically with Liganded ERαLBD. Previous reports of αII binding to ERα showed that the interaction occurs within the C-terminal LBD region between residues 282 and 535 (18). To confirm that αII interacts with the alkylated LBD used for crystallization (9), we carried out real-time analysis of the binding between ERLBD and the αII interaction motif by using SPR. A biotinylated αII peptide immobilized onto a streptavidin-coated sensor chip can specifically recognise various liganded forms of ERαLBD (Fig. 5A, which is published as supporting information on the PNAS web site). The αII peptide interacts with ERαLBD bound to both agonists (E2, diethylstilbestrol, genistein) and an AF2 antagonist (OHT). The ligand interaction preference is similar to that observed for full-length ERα (16). As previously reported (18), αII binding is not dependent on AF2, because an LBD mutant lacking the C-terminal activation helix H12 (ERαΔH12) retains its ability to bind to the αII peptide (Fig. 5B). Kinetic analysis demonstrates that the αII peptide binds to ERαLBD with moderate affinity and has an apparent dissociation constant (Kd) of 34 μM (Fig. 5C).
Structure of the ERαLBD-αII Peptide Complex. To get a better understanding of the nature and location of the αII-binding site on ERα, we cocrystallized ERαLBD with an 11-residue peptide corresponding to the αII interaction motif (LTSRDFGSWYA). Initial crystallization trials with various ERαLBD-αII complexes did not yield crystals suitable for structural analysis. Based on our observations that αII binding was independent of H12, we also set up trials using the truncated ERαΔH12 mutant. The resultant complex, liganded with OHT, produced well diffracting crystals. The overall ERαΔ12 mutant structure is very similar to that seen previously for ERαLBD in complex with either E2 (9) or OHT (7) (rms deviation for α-carbons of 0.51 and 0.69 Å, respectively). Removal of H12 and the preceding loop has no discernable effect on the integrity of the LBD fold.
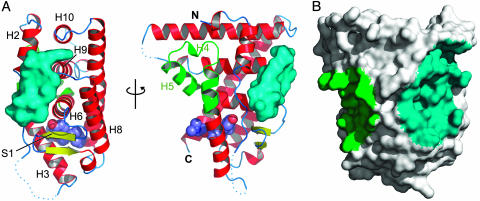
Peptide-Binding Site. The ERαΔH12-OHT-αII peptide complex crystallises with a single LBD homodimer in the asymmetric unit. Each monomer within the homodimer interacts with a peptide motif (Fig. 6A, which is published as supporting information on the PNAS web site). Difference Fourier maps revealed well defined electron density for the peptide, allowing us to model the entire 11-residue αII sequence (Fig. 6B). The αII motif binds across the β-hairpin face of the LBD (Fig. 1A). The interaction site is centered on a shallow depression between H2, H6, and H9 that surrounds the entrance of the solvent-filled channel that leads to the buried hormone-binding cavity. This surface lies diametrically opposite the receptor's AF2 coactivator recruitment site (Fig. 1B). Residues from H2, the H2–H3 loop, H6, the S1–S2 β-hairpin, H8–H9 loop, H9, and H10 contribute to an extensive binding surface (10 Å by 24 Å long).
Fig. 1.
Overall structure of ERαLBD in complex with the αII interaction motif. (A) Ribbon representation of the ERαΔH12-OHT-αII structure. Two perpendicular views of the LBD monomer are shown. The αII peptide is represented by a molecular surface (cyan). Dotted lines represent regions of the structure that are poorly defined in the experimental electron density. Secondary structure elements are labeled, and the AF2 region of the LBD (H3–H5) is colored green. (B) Composite surface representation of the ERαLBD monomer highlighting the location and extent of the known AF2 (green) and proposed αII (cyan) coregulator sites. The molecule is viewed in an identical orientation to the right ribbon representation in A.
The αII peptide adopts an extended conformation that closely matches the surface topology of the LBD (Fig. 2A). Residues Leu-1–Phe-6 run parallel to the S1 strand with residues Ser-3–Asp-5 adopting a 310 helical conformation. At Gly-7, the peptide backbone turns 90° and climbs toward H10. This bent conformation is stabilized by an intramolecular interaction between the backbone carbonyl of Phe-6 and the indole nitrogen of Trp-9. Gly-7 appears to fulfil a structural role because of the strained backbone dihedral angles required at this position.
Fig. 2.
Molecular detail of αII peptide interaction site. (A) View of the peptide interaction site. The αII peptide is shown in stick form with cyan carbons. LBD residues that make key contacts with the peptide are represented in ball-and-stick form with green carbons. (B) Schematic representation of the interactions made by the αII peptide. The αII peptide (purple bonds) and LBD residues (black bonds) are shown. Water molecules are depicted as cyan spheres. For clarity, only intermolecular hydrogen bonds are shown (dotted lines). Van der Waals contacts are represented by green spokes radiating between interacting residues. Figure produced with the program LIGPLOT (33). (C) Surface representation of αII site. The molecular surface of ERαΔH12 is shown and colored according to residue type (purple, hydrophobic; red, negative; blue, positive; yellow, polar; white, glycine). Binding site is viewed as in A.(D) Alignment of ERα and ERβ sequences in the vicinity of the αII binding site. Secondary structural elements and sequence numbering for ERα are shown along the top. Those residues that participate in hydrogen-bonded or nonpolar interactions with the αII motif are represented asterisks and circles respectively. Key sequence differences are boxed. (E) Binding of mutant E2-liganded ERαLBDs. Sensorgrams obtained from injection of 1 μM wild-type and mutant ERαLBDs over a surface immobilized with αII peptide. (Inset) Maximum binding response for each mutant to control SRC2–2 (LxxLL) peptide and αII peptide coated sensor chips. Responses shown are after 2-min continuous injection at a flow rate of 5 μl·min-1.
LBD recognition is mediated by an extensive network of hydrogen-bonded and nonpolar interactions. No single interaction appears to dominate, and the observed peptide conformation is reliant on multiple intra- and intermolecular contacts. Binding of the αII motif buries 650 Å2 of predominantly hydrophobic surface area on the LBD. Residues from the peptide make a total of eight direct hydrogen bonds with the LBD (Fig. 2B). The N terminus of the peptide is anchored to the loop between H2 and H3. Ser-3 is completely buried and plugs the entrance to the solvent channel. Contacts between the central portion of the motif and the LBD are mainly hydrophobic in nature (Fig. 2C). Phe-6 lies at the periphery of the site and packs against the aliphatic side chain of Arg-394, a residue that plays a critical role in recognition of the phenolic hydroxyl of ER ligands. Trp-9 is thrust toward the body of the LBD and packs against Gly-442 (H9) and Trp-393 (H6). Residues at the N-terminal end of H9 (Gln-441, Gly-442, Glu-443, and Glu-444) make multiple hydrogen-bonded interactions with C-terminal half of the αII motif (Fig. 2B). The C-terminal end of the motif is anchored by two separate interactions. The main chain amides of Tyr-11 and Ala-12 interact with carboxylate side chain of Glu-443. In addition, the side chain of Tyr-10 projects over the H8–H9 loop and binds in a narrow cleft between Gln-441 and Ala-493 (Fig. 2 A–C).
Comparison with all other ERαLBD ligand complexes demonstrates that the αII-binding site is structurally invariant. This observation underlies αII's ability to bind to any agonist-, AF2 antagonist-, or pure antagonist-bound ERα. The motif's specificity for ERα appears to derive from the fact that parts of the site are poorly conserved in ERβ and other NRs (Fig. 2D). In particular, sequence changes at the N-terminal end of H9 are likely to affect αII binding. In ERβ, Gly-442 and Glu-443 are replaced by a histidine and lysine, respectively.
Mutagenesis Studies of LBD Binding to αII. To validate the location of the interaction site delineated in the crystal structure and to investigate the ERα-specific nature of the interaction, we made a series of single site LBD mutants and evaluated αII-binding by using SPR. Choice of appropriate mutations was complicated by the fact that residues lining the binding site fulfil key roles in maintaining the structural integrity of the LBD. Consequently, only residues that anchored the termini of the αII motif were targeted (I326, Q441, G442, E443). These residues were either replaced with alanine (Q441, E443) or with the corresponding ERβ residue type (I326H, G442H, E443K).
Fig. 2E shows the binding response of each mutant (liganded to E2) to the αII sensor chip. Changes at all sites were poorly tolerated and severely impaired the interaction. In contrast, binding of the mutants to an immobilized LxxLL peptide, derived from the NRbox2 region of TIF2 (SRC2–2) (8), was similar to that observed for the wild-type LBD (Fig. 2E Inset). These control binding experiments suggest that none of the mutations impacted on the LBD's ability to bind E2 and adopt an activated conformation capable of recruiting coactivator. The mutant binding data indicates that the inability of the αII motif to interact with ERβ derives primarily from steric clashes with the bulkier residues found at positions 326 and 442 (279 and 394 in ERβ) in this isoform (Fig. 2D).
Binding of αII Peptide in Mammalian Cells. Previous studies have demonstrated αII binding to ER in a cellular context (18, 25). To confirm that the αII binding site observed in the crystal structure of the LBD complex is maintained in the full-length receptor in vivo, we investigated the interaction by using a well established mammalian two-hybrid (M2H) assay (19). As demonstrated in Fig. 3A, the αII peptide interacted with full-length ERα in the presence of both E2 and OHT, but not in the absence of ligand. Furthermore, whereas this interaction was independent of ER's AF2 domain (ΔH12), disruption of the αII site by the introduction of the G442H mutation abolished receptor–peptide association (Fig. 3 B and C). In contrast, an LxxLL peptide (19) bound equally well to both the wild-type and G442H-mutated full-length receptor in the presence of E2.
Fig. 3.
ERα-peptide interaction in mammalian cells. Interaction of GAL4-DBD-tagged αII with VP16-tagged ERs. Full-length ERα (A), ERαΔH12 (B), and ERαG442H (C). (D and E) Amino acid requirements for αII interaction. (D) Amino acid sequences of peptides used. (E) Mammalian two-hybrid receptor binding assay of GAL4-DBD-tagged wild type and mutant peptides to VP16-ERα wild type. Binding of the αII and LxxLL peptides were evaluated both in the absence of ligand (NH) and in the presence of E2 and OHT. Normalized luciferase activity is shown.
To elucidate which amino acid residues of the αII motif are essential for functional peptide–receptor interactions, we performed limited alanine scanning mutagenesis on the αII sequence and tested variants for ER interaction in the M2H system (Fig. 3 D and E). Replacement of Phe-6, Gly-7, Ser-8, Trp-9, or Tyr-10 completely abolished receptor interaction (Fig. 3E). In contrast, substitution of Ser-3 had a moderate effect on αII binding. In the crystal structure, the Oγ of Ser-3 is buried and makes a single hydrogen bond with the carboxylate of Glu-323. Although this interaction appears to be dispensable for binding, the motif is otherwise highly sensitive to sequence changes. Such a high degree of sequence dependence reflects the nature of αII binding, which relies on multiple interactions with the LBD.
Enhancement of OHT-Dependent Transcriptional Activity by an Engineered Coactivator. To investigate whether the αII interaction surface can be used as a functional docking site for coregulators, we attempted to modify the behavior of the p160 coregulator TIF2 (GRIP1, SRC2, NcoA-2). TIF2 enhances ER-mediated transcriptional activity in an AF2- and hormone-dependent manner (26). TIF2's receptor-interacting domain contains three NR boxes that are involved in ligand-dependent NR interaction. Of these, NR box 1 and 2 have been implicated in high-affinity binding to ER (8, 27). We inactivated NR box3 (TIF2–NR12) and replaced NR box 1 and 2 with the αII sequence by mutagenesis to create TIF2αII (Fig. 4A). When Cos-7 cells were cotransfected with expression vector for full-length ERα along with reporter plasmids, we found that coexpression of TIF2αII, but not TIF2–NR12, could markedly increase ER activity in the presence of OHT (Fig. 4B). Identical behavior was also observed with a truncated ER lacking the AF1 domain (ERα–DEF; data not shown). Similar results were observed in HuH7 and HeLa cells. As expected, when the G442H mutant receptor was analyzed under similar conditions, the modified TIF2αII failed to increase ER activity in the presence of OHT, whereas the observed coactivation effect of wild-type TIF2 was unaffected (data not shown).
Fig. 4.
Enhancement of tamoxifen-dependent transcriptional activity. (A) Schematic representation of NR-box modified coregulator TIF2 containing the αII peptide sequence. (B) Cell-based reporter assay of transcriptional activity of engineered TIF2 variants in the presence of full-length ERα. (C and D) Reporter activity of wild-type and mutant ER in the presence of 100 nM OHT. Data are presented as percent activation, where the activity of full-length ERα (C) and ERαDEFΔH12 (D) are set to 100%.
Mutation of αII Surface Alters ER's Sensitivity to Coregulators. Finally, we examined whether mutations of the αII-binding surface are able to affect ERα's transcriptional behavior. The transactivation characteristics of the G442H mutant receptor were studied by using either an ERE-luciferase (full-length ERα) or a Gal4-responsive (ERαDEF) reporter assay. The full-length wild-type and mutant receptors responded in an identical manner to E2. Surprisingly, the mutant receptor exhibited a substantial increase in transcriptional activity in response to OHT when TIF2 was coexpressed (Fig. 4C). Removal of AF1 resulted in similar behavior for the wild-type or mutant receptors regardless of whether TIF2 was coexpressed (data not shown). However, although the wild-type receptor requires AF2 for reporter activation, the G442H mutant responds to the addition of TIF2 even in the absence of H12 (Fig. 4D). These data suggest that the αII-binding surface mediates the observed OHT response via TIF2 itself or via unknown cellular coregulators that antagonize TIF2's effects.
Discussion
Comparison with Known Interaction Sites. The structure of ERαLBD in complex with the phage display-derived αII motif defines a unique interaction surface that is distinct from the AF2 recruitment site. The ability of the αII motif to recognize ERα regardless of the bound ligand derives from the fact that its binding site is located in a region of the LBD that is not affected by the distinct conformational effects induced by receptor agonists and antagonists. Previous studies have shown that the AF2 region of the LBD serves as the primary docking site for coactivators and corepressors (4–8, 12, 13). The αII site identified here is located on the opposite face of the LBD (Fig. 1B). Both surfaces are predominantly hydrophobic in character but differ in topology. The LxxLL binding groove is compact, encompasses a relatively small surface area (450 Å2), and is conserved in most NRs. In contrast, the αII site covers a larger area (650 Å2) and is specific to ERα. The concave αII binding surface incorporates conserved regions of the LBD that flank the entrance to a channel that leads to the buried hormone binding cavity. Binding of the αII peptide blocks the entrance to this channel and creates a 300-Å3 water-filled chamber. It is noted this chamber corresponds to the so-called “second-binding site” that has been evaluated as an alternative ligand-binding cavity (28). However, no experimentally validated function has been attributed to this region of ERαLBD, and its role in ligand-binding remains controversial. Consequently, it is highly unlikely that the αII site is involved in binding small molecule ligands.
Biological Significance of the αII Sequence. What is the likelihood that the αII sequence represents a previously unknown, biologically relevant recruitment motif? In the original phage display study, the αII motif was only observed once and, therefore, no consensus information is available (16). Our mutagenesis studies demonstrate that its interaction with ER is highly sequence-dependent. Database searches with either the αII sequence or a redundant motif based on the binding mode observed in the crystal structure do not yield any obvious homologies. Based on the apparent lack of key hotspots within the sequence, we currently favor a model in which the αII peptide acts as a structural, rather than sequence mimic of the true interaction partner. Consequently, the possibility that elements of the αII motif are incorporated into a nonlinear recognition module, formed by discontinuous regions of primary sequence, cannot be ruled out.
αII Interaction Surface: A Possible Coregulator Recruitment Site? The question remains as to whether the αII motif acts as a fortuitous conformational probe or an actual mimic of a bona fide ER interaction partner? Norris et al. (18) demonstrated that αII is able to inhibit the partial agonist activity of OHT while having a minimal effect on E2-mediated transcription. This observation strongly suggests that αII's cognate binding surface on ERα represents an important control site that is involved in regulating receptor activity under certain conditions. Short peptides that act as antagonists of protein-protein interactions have been isolated from naive libraries for a number of systems. Subsequent structural analysis has shown that the peptide binding site overlaps with that of a known binding partner (29). In the case of αII, the crystal structure of the complex indicates that peptide binding to the isolated LBD does not induce any significant conformational changes. This finding suggests that αII's antagonistic properties are manifested by a direct steric effect in which the peptide occludes a key binding site. At this stage, it is unclear whether the αII site is a docking surface for another region of ER (i.e., involved in intramolecular domain signaling) or for an as yet unidentified coregulator. The coactivator engineering experiments clearly demonstrate that the αII binding surface can serve as a valid receptor–coregulator interaction site. By replacing the NR-box/LxxLL regions of TIF2 with the αII sequence, we generated a coactivator (TIF2αII) that could be recruited to the αII surface by both agonist and antagonist-bound ER in an AF1/AF2-independent fashion.
Our preliminary functional analysis indicates that the αII surface may play a role in ER's sensitivity to coregulators in the presence of selective ER receptor modulators such as OHT (Fig. 4C). No differences in behavior were observed between wild-type and αII-mutated ER in the presence of E2. This ligand-dependency mirrors the previously reported antagonistic effects of the αII peptide on E2- and OHT-dependent receptor activity (18). The surprising result that the TIF2-mediated enhancement of OHT activity seen for the G442H mutant requires a functional AF2 H12/F domain only in the context of the full-length ER, but not in the context of the ER DEF, reinforces the idea that the functional interplay between different ER domains and coregulators determines the transcriptional properties of the wild type receptor (30). Furthermore, our demonstration that disruption of the αII binding site enhanced rather than decreased the response to TIF2 does not necessarily contradict the currently held view that TIF2 acts primarily as a coactivator. Although TIF2 coactivator function is usually associated with recruitment to a functional AF1/2 surface, targeting to other surfaces cannot be discounted. Recent studies by Yamamoto and coworkers (31, 32) discovered a context-dependent corepressor function in TIF2, which may indeed account for the effect seen in our study. Alternatively, TIF2's role could be indirect and mediated by either endogenous coregulators known to cooperate with TIF2 (e.g., CBP/p300, CARM-1) or unidentified corepressors that bind directly to the αII surface. Indeed, although our coregulator engineering experiment targeted a coactivator to the αII surface, similar experiments utilizing an engineered corepressor would probably have illustrated that the αII site may alternatively be involved in corepressor binding.
In summary, the site identified in this study appears to be involved in maintaining a reduced level of receptor activation in the presence of TIF2. Further studies are required to establish whether other recognized or unrecognized ER coactivators and corepressors also display sensitivity to the disruption of this unique control surface.
Supplementary Material
Acknowledgments
We thank Benita Katzenellenbogen for providing the ERαLBD expression construct, Geoffrey Greene for advice on the preparation of the E2 affinity resin, and Peter O'Brien for synthesis of the epoxypropyl-E2 used in the resin coupling reaction. We also thank staff on ID29 (European Synchrotron Radiation Facility, Grenoble, France) for technical support during data collection. A.C.W.P. is funded by a Wellcome Trust Career Development Fellowship, and E.H.K. is funded by the Biotechnology and Biological Sciences Research Council. J.-Å.G., E.T., and N.H. are supported by grants from the Swedish Research Council, the Swedish Cancer Society, and Karo Bio AB.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, estrogen receptor; E2 17β-estradiol; AF, activation function; LBD, ligand-binding domain; NR, nuclear receptor; OHT, 4-hydroxytamoxifen; SPR, surface plasmon resonance.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2BJ4).
References
- 1.Tora, L., White, J., Brou, C., Tasset, D., Webster, N., Scheer, E. & Chambon, P. (1989) Cell 59, 477-487. [DOI] [PubMed] [Google Scholar]
- 2.Smith, C. L. & O'Malley, B. W. (2004) Endocr. Rev. 25, 45-71. [DOI] [PubMed] [Google Scholar]
- 3.Heery, D. M., Kalkhoven, E., Hoare, S. & Parker, M. G. (1997) Nature 387, 733-736. [DOI] [PubMed] [Google Scholar]
- 4.Darimont, B. D., Wagner, R. L., Apriletti, J. W., Stallcup, M. R., Kushner, P. J., Baxter, J. D., Fletterick, R. J. & Yamamoto, K. R. (1998) Genes Dev. 12, 3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McInerney, E. M., Rose, D. W., Flynn, S. E., Westin, S., Mullen, T. M., Krones, A., Inostroza, J., Torchia, J., Nolte, R. T., Assa-Munt, N., et al. (1998) Genes Dev. 12, 3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolte, R. T., Wisely, G. B., Westin, S., Cobb, J. E., Lambert, M. H., Kurokawa, R., Rosenfeld, M. G., Willson, T. M., Glass, C. K. & Milburn, M. V. (1998) Nature 395, 137-143. [DOI] [PubMed] [Google Scholar]
- 7.Shiau, A. K., Barstad, D., Loria, P. M., Cheng, L., Kushner, P. J., Agard, D. A. & Greene, G. L. (1998) Cell 95, 927-937. [DOI] [PubMed] [Google Scholar]
- 8.Wärnmark, A., Treuter, E., Gustafsson, J.-Å., Hubbard, R. E., Brzozowski, A. M. & Pike, A. C. W. (2002) J. Biol. Chem. 277, 21862-21868. [DOI] [PubMed] [Google Scholar]
- 9.Brzozowski, A. M., Pike, A. C. W., Dauter, Z., Hubbard, R. E., Bonn, T., Engstrom, O., Ohman, L., Greene, G. L., Gustafsson, J.-Å. & Carlquist, M. (1997) Nature 389, 753-758. [DOI] [PubMed] [Google Scholar]
- 10.Shang, Y. F., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. (2000) Cell 103, 843-852. [DOI] [PubMed] [Google Scholar]
- 11.Hu, X. & Lazar, M. A. (1999) Nature 402, 93-96. [DOI] [PubMed] [Google Scholar]
- 12.Xu, H. E., Stanley, T. B., Montana, V. G., Lambert, M. H., Shearer, B. G., Cobb, J. E., McKee, D. D., Galardi, C. M., Plunket, K. D., Nolte, R. T., et al. (2002) Nature 415, 813-817. [DOI] [PubMed] [Google Scholar]
- 13.Huang, H. J., Norris, J. D. & McDonnell, D. P. (2002) Mol. Endocrinol. 16, 1778-1792. [DOI] [PubMed] [Google Scholar]
- 14.Katzenellenbogen, B. S. & Katzenellenbogen, J. A. (2002) Science 295, 2380-2381. [DOI] [PubMed] [Google Scholar]
- 15.Shang, Y. F. & Brown, M. (2002) Science 295, 2465-2468. [DOI] [PubMed] [Google Scholar]
- 16.Paige, L. A., Christensen, D. J., Gron, H., Norris, J. D., Gottlin, E. B., Padilla, K. M., Chang, C. Y., Ballas, L. M., Hamilton, P. T., McDonnell, D. P. & Fowlkes, D. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, C. Y., Norris, J. D., Gron, H., Paige, L. A., Hamilton, P. T., Kenan, D. J., Fowlkes, D. & McDonnell, D. P. (1999) Mol. Cell. Biol. 19, 8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norris, J. D., Paige, L. A., Christensen, D. J., Chang, C. Y., Huacani, M. R., Fan, D. J., Hamilton, P. T., Fowlkes, D. M. & McDonnell, D. P. (1999) Science 285, 744-746. [DOI] [PubMed] [Google Scholar]
- 19.Heldring, N., Nilsson, M., Buehrer, B., Treuter, E. & Gustafsson, J.-Å. (2004) Mol. Cell. Biol. 24, 3445-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson, K. E., Choi, I., Gee, A., Katzenellenbogen, B. S. & Katzenellenbogen, J. A. (1997) Biochemistry 36, 14897-14905. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein, S. W., Bordner, J., Hoth, L. R. & Geoghegan, K. F. (2001) Bioconjug. Chem. 12, 406-413. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 24.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 25.Bapat, A. R. & Frail, D. E. (2003) J. Steroid Biochem. Mol. Biol. 86, 143-149. [DOI] [PubMed] [Google Scholar]
- 26.Hong, H., Kohli, K., Garabedian, M. J. & Stallcup, M. R. (1997) Mol. Cell. Biol. 17, 2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wärnmark, A., Almlof, T., Leers, J., Gustafsson, J.-Å. & Treuter, E. (2001) J. Biol. Chem. 276, 23397-23404. [DOI] [PubMed] [Google Scholar]
- 28.van Hoorn, W. P. (2002) J. Med. Chem. 45, 584-589. [DOI] [PubMed] [Google Scholar]
- 29.Sidhu, S. S., Fairbrother, W. J. & Deshayes, K. (2003) ChemBioChem 4, 14-25. [DOI] [PubMed] [Google Scholar]
- 30.Metivier, R., Stark, A., Flouriot, G., Hubner, M. R., Brand, H., Penot, G., Manu, D., Denger, S., Reid, G., Kos, M., et al. (2002) Mol. Cell 10, 1019-1032. [DOI] [PubMed] [Google Scholar]
- 31.Rogatsky, I., Zarember, K. A. & Yamamoto, K. R. (2001) EMBO J. 20, 6071-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogatsky, I., Luecke, H. F., Leitman, D. C. & Yamamoto, K. R. (2002) Proc. Natl. Acad. Sci. USA 99, 16701-16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995) Protein Eng. 8, 127-134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.